ABSTRACT
The evolution of natural modular proteins and domain swapping by protein engineers have shown the disruptive potential of non-homologous recombination to create proteins with novel functions or traits. Bacteriophage endolysins, cellulosomes and polyketide synthases are 3 examples of natural modular proteins with each module having a dedicated function. These modular architectures have been created by extensive duplication, shuffling of domains and insertion/deletion of new domains. Protein engineers mimic these natural processes in vitro to create chimeras with altered properties or novel functions by swapping modules between different parental genes. Most domain swapping efforts are realized with traditional restriction and ligation techniques, which become particularly restrictive when either a large number of variants, or variants of proteins with multiple domains have to be constructed. Recent advances in homology-independent shuffling techniques increasingly address this need, but to realize the full potential of the synthetic biology of modular proteins a complete homology-independent method for both rational and random shuffling of modules from an unlimited number of parental genes is still needed.
KEYWORDS: designer cellulosome, domain swapping, endolysin, horizontal transfer, modular protein, protein engineering, polyketide synthase
Protein engineers mimic natural evolution in vitro to create proteins that are better adapted to the application they have envisioned. Natural evolution is featured by selection of the fittest variant that arose either through mutation or exchange of genetic fragments. Whereas the accumulation of mutations represents a gradual, adaptive evolution process, genetic recombination leads to more disruptive evolution and even neofunctionalization. A range of mutagenesis and combinatorial engineering techniques has been developed to create in vitro libraries of variants used for directed evolution. Most commonly used combinatorial techniques such as DNA shuffling, in vivo homologous recombination, staggered extension process (StEP), random priming recombination, however, all rely on a high degree of homology (at least 70%). In contrast, evolutionary studies suggest that natural evolution is not limited to recombination of highly similar sequences only. Computational analyses and the prevalence of multidomain proteins demonstrate the success of non-homologous recombination to evolve completely novel protein functions in nature (reviewed by Lutz and Benkovic1). Nearly half of all proteomes consist of modular or multidomain proteins with each module or domain (both words are often used interchangeably, depending on the specific research field) having a dedicated function, ranging from substrate binding or interaction, transfer of intermediates, structural assembly to catalysis.2 Studies have shown that the number of known domains has been stagnating for almost a decade with only a few hundred new ones reported every year, while the number of new proteins with rearranged domains is still exponentially increasing.3 Natural evolutionary mechanisms that result in those new protein architectures include extensive duplication, shuffling of domains and insertion/deletion of a new domain, whereof insertion/deletion is the most frequent event for bacteria.4 The specific modular architecture and composition determines the function, specificity and performance of each modular enzyme.
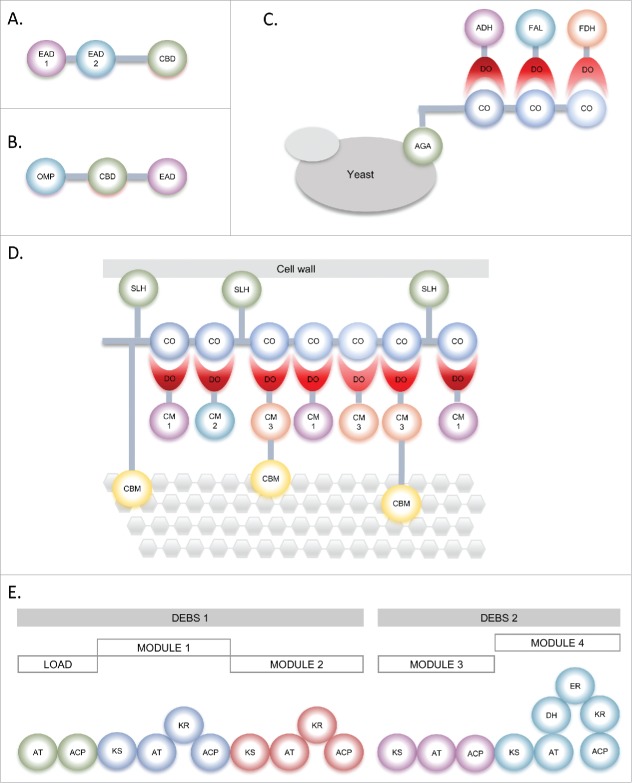
Protein engineers have been inspired by this modularity principle and have been swapping modules between different parental genes to create chimeras with altered properties or even novel functions (neofunctionalization). Indeed, the plasticity of modular proteins offers a large potential to engineer customized proteins that perform optimal under specific conditions, have a tailor-made specificity or synthesize designed products. A prominent example is domain swapping of endolysins. These are peptidoglycan hydrolases encoded by bacterial viruses or bacteriophages and produced at the end of the lytic cycle. They enzymatically degrade the peptidoglycan layer from within the infected bacterial cell.5 Endolysins from Gram-positive phages mostly display a modular structure, comprising one or more enzymatically active domains (EADs) and one, mostly C-terminal, cell wall binding domain (CBD) (Fig. 1A). Bacteriophages of Gram-negative organisms on the other hand produce primarily globular endolysins with a single EAD.6 Endolysins have high in vitro and in vivo antimicrobial activity and show a low probability to provoke resistance development. Furthermore, they are specific at the genus, species or even serovar level, leaving beneficial flora unaffected.7 Horizontal gene transfer between bacteriophages and their host bacterial cells has been suggested based on the evolutionary relationships between bacteriophage endolysins and bacterial lytic enzymes (autolysins).8 Several natural chimeric endolysins have been described, such as the Listeria phage endolysin PlyPSA which consists of an EAD that is related to the amidase domains of Bacillus and Clostridium phage endolysins and a CBD that is highly similar to those of other Listeria phage endolysins.9 Another example is the lytic enzyme of the pneumococcal phage Dp-1 which comprises an EAD that is related to the murein hydrolase of a Lactococcus lactis phage endolysin and a CBD homologous to the pneumococcal lytic system.10 The discovery of natural chimeras has supported the construction of synthetic chimeric endolysins consisting of modules from different origin.11 As such, Dong et al.12 have extended the lytic spectrum of a staphylococcal lysin by fusing its EAD with the CBD from a lysin with lytic spectra across multiple genera. Other researchers demonstrated that switching the CBDs of 2 endolysins results in a swap of their binding and lysis specificity and that combining 2 CBDs results in a binding spectrum comprising the spectra of both original CBDs.13 Furthermore, an endolysin with an improved thermostability has been made by fusing the N-acetylmuramoyl-L-alanine amidase domain from the endolysin of a thermophilic bacteriophage to the CBD from the Clostridium perfringens-specific bacteriophage endolysin.14 An example of neofunctionalization in endolysin research is the creation of Artilysin®s (Fig. 1B) that fuse modules of unrelated origin to create endolysin-based antibacterials against Gram-negative bacteria. Specifically, Briers et al.5 have fused the sheep myeloid antimicrobial peptide of 29 amino acids to the N-terminus of the endolysin KZ144. As such, Artilysin®s can pass the protective outer membrane of Gram-negative pathogens from the outside and overcome this limitation of endolysins.
Figure 1.
Natural modular proteins and 2 neofunctionalized chimeric enzymes. (A) Endolysin (adapted from Roach and Donovan38); (B) Artilysin39; (C) Scaffolded enzyme cascade22 ; (D) Simplified natural cellulosome with a linear scaffoldin16; (E) Two parts of the 6-deoxyerythronolide B synthase (DEBS) polyketide synthase (adapted from Weissman24). EAD = Enzymatically active domain; CBD = Cell wall binding domain; OMP = Outer membrane permeabilizing peptide; CO = Cohesin; DO = Dockerin; ADH = Alcohol dehydrogenase; FAL = Formaldehyde dehydrogenase; FDH = Formate dehydrogenase; SLH = S-layer homology module; CM = Catalytic module (each number refers to a different module); CBM = Carbohydrate binding module; AT = Acyl transferase; ACP = Acyl carrier protein; KS = Ketosynthase; KR = Ketoreductase; DH = Dehydratase; ER = Enoyl reductase.
A natural exponent of protein modularity that has been exploited by protein engineers are cellulosomes that degrade lignocellulose. Lignocellulosic biomass is the most abundant renewable resource and the most attractive substrate for biorefinery strategies to produce bio-based products such as biofuels, bioplastics or enzymes.15,16 Full enzymatic breakdown of lignocellulose requires the simultaneous action of several synergistic catalytic activities (e.g. cellulases, hemicellulases, lytic polysaccharide monooxygenases and pectinases). In nature, anaerobic bacteria have developed complex modular systems called cellulosomes (Fig. 1D) comprising a modular non-catalytic scaffoldin which can be linear as in most mesophilic Clostridia, or hierarchically branched as for Clostridium thermocellum. Each subunit of the scaffold can strongly interact with a complementary dockerin subunit, borne by enzymes with diverse catalytic activities. These multi-enzyme complexes give an advantage to aerobic cellulolytic bacteria that secrete free (hemi)cellulases because of the enhanced synergism among the catalytic units and close association between the cell-bound cellulosome and the substrate.17 The modular nature of the cellulosome complex and its beneficial properties led to the concept of designer cellulosomes (also called minicellulosomes) by incorporating recombinant (hemi)cellulose-degrading enzymes from diverse origin into the complex using the specific affinity of cognate cohesin-dockerin pairs.18 You et al.19 obtained an enhanced hydrolysis rate on Avicel and regenerated amorphous cellulose (RAC) using a four-component designer mini-cellulosome containing endoglucanases and a cellobiohydrolase from 3 different organisms compared to the non-complexed cellulose mixture. The addition of an expansin-like protein, i.e. a protein which disrupts hydrogen bonding between cellulose microfibrils and other cell wall polysaccharides without hydrolytic activity, to a bifunctional designer cellulosome of cellulases, enhanced the reducing sugar yield by 2-11 %.20 Other noncellulosomal enzymes have also been added to the designer cellulosome for synergistic effects. For example, Arfi et al.21 have developed a designer cellulosome consisting of a lytic polysaccharide monooxygenase of Thermobifida fusca and 2 cellulases resulting in a 63-68 % improvement in soluble sugars release. The cohesin-dockerin scaffold has also served to create other enzyme cascades. Liu et al.22 have created a protein scaffold with 3 enzymes docked to cohesins, namely 3 dehydrogenases responsible for sequential conversion of methanol to carbon dioxide (Fig. 1C), while You et al.23 used a similar assembled structure to convert glyceraldehyde-3-phosphate to fructose-6-phosphate.
Another striking example of modular enzymes subjected to swapping of non-homologous domains are the gigantic multi-enzyme type I polyketide synthases (PKSs). The biosynthesis of large numbers of structurally and stereochemically complex polyketides from a relatively small pool of simple precursors is achieved by cascades of individual enzymes present in the multi-enzyme complex.24 Structurally, type I PKSs consist of independently folded domains which are grouped into working units called modules for the introduction and functional tailoring of one building block into the growing chain (Fig. 1E). Two distinct groups of modular enzymes can be distinguished: the cis-acyl transferase (AT) PKS and the trans-AT PKS. The first one arose from repeated rounds of gene duplication coupled with domain diversification by homologous recombination, while the second one evolved through horizontal transfer of substrate-specific ketosynthase domains.25,26 These distinct modes of evolution resulted in structurally diverse classes of modular enzymes. The cis-AT PKS minimally contains one AT domain in each module, whereas AT is an isolated protein which acts iteratively in trans to deliver an extender unit to all of the modules in trans-AT PKS. Furthermore, extension modules of the cis-AT PKS contain a ketosynthase (KS), an acyl carrier protein (ACP), or some domains for optional processing activities, while the trans-AT PKS class consists of duplicated domains (e.g., doublets or triplets of ACPs), inactive domains with a cryptic function such as epimerase activity, and modules of nonribosomal peptide synthetase (NRPS) permitting the activation and incorporation of amino acids into the chain-extension intermediates.24,27 To date, several strategies have been applied to engineer PKSs with novel properties ranging from inactivation of specific reductive domains, over site-directed mutagenesis to domain and module swapping. Kushnir et al.28 have disabled ketoreductase, dehydratase and enoyl reductase domains to produce a library of 22 oxidized derivatives of premonensin, a shunt metabolite which structurally resembles an anti-cancer polyketide, while Kong et al.29 have introduced modifications within the polyene class of antifungal polyketides, leading to improved solubility and reduced hemolytic activity of the polyketide. Hagen et al.30 have produced adipic acid using a neofunctionalized, chimeric PKS consisting of heterologous reductive domains from various PKS clusters into the borrelidin PKS’ first extension module. However, most domain and module swapping approaches were less successful. Menzella et al.31 for example have interchanged 14 modules from 8 PKS clusters resulting in 154 bimodular combinations. Only half of the combinations gave the expected tri-ketide lactone product due to the introduction of unnatural intermodular junctions.
As noted from the examples above, natural protein modularity and domain swapping efforts demonstrate the disruptive potential of non-homologous domain shuffling. However, the majority of protein engineering studies has been based on a rational approach using classical restriction and ligation protocols, which becomes particularly cumbersome when a large collection of variants or variants of multidomain proteins (e.g. PKSs) have to be constructed. More diverse libraries of shuffled endolysins, designer cellulosomes and PKSs would allow to explore sequence space to a larger extent. Methods that allow full combinatorial, complete homology-independent shuffling from an unlimited number of parental genes would therefore significantly enhance the synthetic biology of modular proteins. Recent advances in the field such as the development of SHIPREC, ITCHY, SCRATCHY, SCOPE, SISDC and USERec32-37 increasingly respond to this unmet need with each method having its benefits and drawbacks. The strength of SHIPREC, ITCHY and SCRATCHY is the creation of non-biased and almost non-homologous recombination. Unfortunately, they are all limited to the shuffling of 2 parental genes. SISDC already allows for the facile recombination of 2 to 4 distantly related (or unrelated) proteins at multiple discrete sites. Moreover, this technique makes use of type IIb restriction-enzymes which cut outside the recognition site resulting in a scarless hybrid. The use of these restriction enzymes are actually also a disadvantage of the method as their introduction is laborious covering multiple PCRs. Furthermore, due to frameshifts a large number of non-functional sequences are obtained with the former methods.36,37 The most recently developed method, USERec, allows to shuffle several completely unrelated modules in a scarless manner, but a random combinatorial approach of unrelated fragments is complicated because specifically designed primers are required for each junction. It is expected that completely random shuffling of non-related fragments will – similar to random mutagenesis – generally yield many unexpectedly novel, improved chimeric proteins that combine fragments that could not be rationally envisioned beforehand. As such, unlimited, random shuffling of a high number of non-related gene fragments from many different sources remains the next big leap in the field of synthetic biology of modular proteins.
Funding Statement
This work was supported by research grant 1.5.171.15N of the Research Foundation – Flanders (FWO).
Abbreviations
- ACP
acyl carrier protein
- AT
acyl transferase
- CBD
cell wall binding domain
- EAD
enzymatically active domain
- ITCHY
incremental truncation for the creation of hybrid enzymes
- KS
ketosynthase
- PKS
polyketide synthase
- SCOPE
structure-based combinatorial protein engineering
- SHIPREC
sequence homology-independent protein recombination
- SISDC
sequence-independent site-directed chim-eragenesis
- StEP
staggered extension process
- USERec
uSER friendly DNA recombination.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Lutz S, Benkovic SJ. Homology-independent protein engineering. Curr Opin Biotechnol 2000; 11:319-24; PMID:10975450; https://doi.org/ 10.1016/S0958-1669(00)00106-3 [DOI] [PubMed] [Google Scholar]
- [2].Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol 2007; 8:319-30; PMID:17356578; https://doi.org/ 10.1038/nrm2144 [DOI] [PubMed] [Google Scholar]
- [3].Bornberg-Bauer E, Albà MM. Dynamics and adaptive benefits of modular protein evolution. Curr Opin Struct Biol 2013; 23:459-66; PMID:23562500; https://doi.org/ 10.1016/j.sbi.2013.02.012 [DOI] [PubMed] [Google Scholar]
- [4].Pasek S, Risler J-L, Brézellec P. Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics 2006; 22:1418-23; PMID:16601004; https://doi.org/ 10.1093/bioinformatics/btl135 [DOI] [PubMed] [Google Scholar]
- [5].Briers Y, Walmagh M, Grymonprez B, Biebl M, Pirnay JP, Defraine V, Michiels J, Cenens W, Aertsen A, Miller S, et al.. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob Agents Chemother 2014; 58:3774-84; PMID:24752267; https://doi.org/ 10.1128/AAC.02668-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Van Tassell ML, Angela Daum M, Kim J-S, Miller MJ. Creative lysins: Listeria and the engineering of antimicrobial enzymes. Curr Opin Biotechnol 2016; 37:88-96; PMID:26710271; https://doi.org/ 10.1016/j.copbio.2015.10.006 [DOI] [PubMed] [Google Scholar]
- [7].Gerstmans H, Rodriguez-Rubio L, Lavigne R, Briers Y. From endolysins to Artilysins: novel enzyme-based approaches to kill drug-resistant bacteria. Biocheml Soc Trans 2016; 44:123-8; PMID:26862197; https://doi.org/ 10.1042/BST20150192 [DOI] [PubMed] [Google Scholar]
- [8].Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol 2005; 8:444-50; PMID:15979387; https://doi.org/ 10.1016/j.mib.2005.06.005 [DOI] [PubMed] [Google Scholar]
- [9].Zimmer M, Sattelberger E, Inman RB, Calendar R, Loessner MJ. Genome and proteome of Listeria monocytogenes phage PSA: an unusual case for programmed + 1 translational frameshifting in structural protein synthesis. Mol Microbiol 2003; 50:303-17; PMID:14507382; https://doi.org/ 10.1046/j.1365-2958.2003.03684.x [DOI] [PubMed] [Google Scholar]
- [10].Sheehan MM, Garcia JL, Lopez R, Garcia P. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergeneric origin. Mol Microbiol 1997; 25:717-25; PMID:9379901; https://doi.org/ 10.1046/j.1365-2958.1997.5101880.x [DOI] [PubMed] [Google Scholar]
- [11].Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Fut Microbiol 2012; 7:1147-71; PMID:23030422; https://doi.org/ 10.2217/fmb.12.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dong Q, Wang J, Yang H, Wei C, Yu J, Zhang Y, Huang Y, Zhang XE, Wei H. Construction of a chimeric lysin Ply187N‐V12C with extended lytic activity against staphylococci and streptococci. Microb Biotechnol 2015; 8:210-20; PMID:25219798; https://doi.org/ 10.1111/1751-7915.12166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Schmelcher M, Tchang VS, Loessner MJ. Domain shuffling and module engineering of Listeria phage endolysins for enhanced lytic activity and binding affinity. Microb Biotechnol 2011; 4:651-62; PMID:21535426; https://doi.org/ 10.1111/j.1751-7915.2011.00263.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Swift SM, Seal BS, Garrish JK, Oakley BB, Hiett K, Yeh HY, Woolsey R, Schegg KM, Line JE, Donovan DM. A thermophilic phage endolysin fusion to a Clostridium perfringens-specific cell wall binding domain creates an anti-Clostridium antimicrobial with improved thermostability. Viruses-Basel 2015; 7:3019-34; PMID:26075507; https://doi.org/ 10.3390/v7062758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fan LH, Zhang ZJ, Yu XY, Xue YX, Tan TW. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc Natl Acad Sci U S A 2012; 109:13260-5; PMID:22853950; https://doi.org/ 10.1073/pnas.1209856109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mazzoli R, Lamberti C, Pessione E. Engineering new metabolic capabilities in bacteria: lessons from recombinant cellulolytic strategies. Trends Biotechnol 2012; 30:111-9; PMID:21930321; https://doi.org/ 10.1016/j.tibtech.2011.08.003 [DOI] [PubMed] [Google Scholar]
- [17].Vazana Y, Barak Y, Unger T, Peleg Y, Shamshoum M, Ben-Yehezkel T, Mazor Y, Shapiro E, Lamed R, Bayer EA. A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates. Biotechnology Biofuels 2013; 6:182; PMID:24341331; https://doi.org/ 10.1186/1754-6834-6-182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bayer EA, Morag E, Lamed R. The cellulosome - A treasure-trove for biotechnology. Trends Biotechnol 1994; 12:379-86; PMID:7765191; https://doi.org/ 10.1016/0167-7799(94)90039-6 [DOI] [PubMed] [Google Scholar]
- [19].You C, Zhang XZ, Zhang YHP. Mini-scaffoldin enhanced mini-cellulosome hydrolysis performance on low-accessibility cellulose (Avicel) more than on high-accessibility amorphous cellulose. Biochem Eng J 2012; 63:57-65; https://doi.org/ 10.1016/j.bej.2012.01.011 [DOI] [Google Scholar]
- [20].Chen C, Cui Z, Song X, Liu Y-J, Cui Q, Feng Y. Integration of bacterial expansin-like proteins into cellulosome promotes the cellulose degradation. Appl Microbiol Biotechnol 2016; 100:2203-12; PMID:26521249; https://doi.org/ 10.1007/s00253-015-7071-6 [DOI] [PubMed] [Google Scholar]
- [21].Arfi Y, Shamshoum M, Rogachev I, Peleg Y, Bayer EA. Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc Natl Acad Sci U S A 2014; 111:9109-14; PMID:24927597; https://doi.org/ 10.1073/pnas.1404148111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu F, Banta S, Chen W. Functional assembly of a multi-enzyme methanol oxidation cascade on a surface-displayed trifunctional scaffold for enhanced NADH production. Chem Commun 2013; 49:3766-8; PMID:23535691; https://doi.org/ 10.1039/c3cc40454d [DOI] [PubMed] [Google Scholar]
- [23].You C, Myung S, Zhang YHP. Facilitated substrate channeling in a self-assembled trifunctional enzyme complex. Angew Chem Int Ed Engl 2012; 51:8787-90; https://doi.org/ 10.1002/anie.201202441 [DOI] [PubMed] [Google Scholar]
- [24].Weissman KJ. Genetic engineering of modular PKSs: from combinatorial biosynthesis to synthetic biology. Nat Prod Rep 2016; 33:203-30; PMID:26555805; https://doi.org/ 10.1039/C5NP00109A [DOI] [PubMed] [Google Scholar]
- [25].Jenke-Kodama H, Sandmann A, Muller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol 2005; 22:2027-39; PMID:15958783; https://doi.org/ 10.1093/molbev/msi193 [DOI] [PubMed] [Google Scholar]
- [26].Nguyen T, Ishida K, Jenke-Kodama H, Dittmann E, Gurgui C, Hochmuth T, Taudien S, Platzer M, Hertweck C, Piel J. Exploiting the mosaic structure of trans-acyltransferase polyketide synthases for natural product discovery and pathway dissection. Nat Biotechnol 2008; 26:225-33; PMID:18223641; https://doi.org/ 10.1038/nbt1379 [DOI] [PubMed] [Google Scholar]
- [27].Garg A, Xie X, Keatinge-Clay A, Khosla C, Cane DE. Elucidation of the cryptic epimerase activity of redox-inactive ketoreductase domains from modular polyketide synthases by tandem equilibrium isotope exchange. J Am Chem Soc 2014; 136:10190-3; PMID:25004372; https://doi.org/ 10.1021/ja5056998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kushnir S, Sundermann U, Yahiaoui S, Brockmeyer A, Janning P, Schulz F. Minimally invasive mutagenesis gives rise to a biosynthetic polyketide library. Angew Chem Int Ed Engl 2012; 51:10664-9; PMID:22996590; https://doi.org/ 10.1002/anie.201202438 [DOI] [PubMed] [Google Scholar]
- [29].Kong D, Lee M-J, Lin S, Kim E-S. Biosynthesis and pathway engineering of antifungal polyene macrolides in actinomycetes. J Ind Microbiol Biotechnol 2013; 40:529-43; PMID:23515854; https://doi.org/ 10.1007/s10295-013-1258-6 [DOI] [PubMed] [Google Scholar]
- [30].Hagen A, Poust S, de Rond T, Fortman JL, Katz L, Petzold CJ, Keasling JD. Engineering a polyketide synthase for in vitro production of adipic acid. ACS Synth Biol 2016; 5:21-7; PMID:26501439; https://doi.org/ 10.1021/acssynbio.5b00153 [DOI] [PubMed] [Google Scholar]
- [31].Menzella HG, Reid R, Carney JR, Chandran SS, Reisinger SJ, Patel KG, Hopwood DA, Santi DV. Combinatorial polyketide biosynthesis by de novo design and rearrangement of modular polyketide synthase genes. Nat Biotech 2005; 23:1171-6; https://doi.org/ 10.1038/nbt1128 [DOI] [PubMed] [Google Scholar]
- [32].Ostermeier M, Shim JH, Benkovic SJ. A combinatorial approach to hybrid enzymes independent of DNA homology. Nat Biotech 1999; 17:1205-9; PMID:10585719; https://doi.org/ 10.1038/10850 [DOI] [PubMed] [Google Scholar]
- [33].Sieber V, Martinez CA, Arnold FH. Libraries of hybrid proteins from distantly related sequences. Nat Biotech 2001; 19:456-60; PMID:11329016; https://doi.org/ 10.1038/88129 [DOI] [PubMed] [Google Scholar]
- [34].Kawarasaki Y, Griswold KE, Stevenson JD, Selzer T, Benkovic SJ, Iverson BL, Georgiou G. Enhanced crossover SCRATCHY: construction and high-throughput screening of a combinatorial library containing multiple non-homologous crossovers. Nucleic Acids Res 2003; 31:e126-e; PMID:14576326; https://doi.org/ 10.1093/nar/gng126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].O'Maille PE, Bakhtina M, Tsai MD. Structure-based combinatorial protein engineering (SCOPE). J Mol Biol 2002; 321:677-91; PMID:12206782; https://doi.org/ 10.1016/S0022-2836(02)00675-7 [DOI] [PubMed] [Google Scholar]
- [36].Villiers BRM, Stein V, Hollfelder F. USER friendly DNA recombination (USERec): a simple and flexible near homology-independent method for gene library construction. Protein Eng Des Sel 2010; 23:1-8; PMID:19897542; https://doi.org/ 10.1093/protein/gzp063 [DOI] [PubMed] [Google Scholar]
- [37].Hiraga K, Arnold FH. General method for sequence-independent site-directed chimeragenesis. J Mol Biol 2003; 330:287-96; PMID:12823968; https://doi.org/ 10.1016/S0022-2836(03)00590-4 [DOI] [PubMed] [Google Scholar]
- [38].Roach DR, Donovan DM. Antimicrobial bacteriophage-derived proteins and therapeutic applications. Bacteriophage 2015; 5:e1062590; PMID:26442196; https://doi.org/ 10.1080/21597081.2015.1062590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yang H, Yu J, Wei H. Engineered bacteriophage lysins as novel anti-infectives. Front Microbiol 2014; 5:542; PMID:25360133 [DOI] [PMC free article] [PubMed] [Google Scholar]