ABSTRACT
Conjugated linolenic acid (CLNA) is a family of isomers of linolenic acid with a number of health-associated benefits, which has been attracting great interest. Microbial CLNA producers are potentially an alternative source of CLNA for human nutrition. In present study, 16 neonate feces were collected and used for Bifidobacteria isolation, from which 25 bifidobacteria isolates were obtained. The bifidobacteria isolates were identified using 16s rDNA sequencing as Bifidobacterium adolescentis, B. breve, B. longum and B. pseudocatenulatum. These isolates were further investigated for their ability to produce CLNA using linolenic acid as substrate via GC-MS. The results showed most of the isolates could convert free linolenic acid into c9,t11,c15-CLNA and t9,t11,c15-CLNA at different levels. B. pseudocatenulatum was the most effective CLNA producer, which converted 86.91% of linolenic acid to c9,t11,c15-CLNA and 3.59% of to t9,t11,c15-CLNA isomer and the isolate exhibited to accumulate CLNA during 72 h culturing in which most CLNA isomers were in the supernatant fluid. The results indicated that utilization of this isolate for CLNA production will eliminate the purification process.
KEYWORDS: bifidobacteria, bioconversion, conjugated linolenic acid, isolation
Introduction
Conjugated linolenic acid (CLNA) refers a general term for positional and geometric isomers of linolenic acid (Octadecatrienoic acid, C18:3) with conjugated double bonds. Nugteren and colleagues firstly demonstrated the effect of an oil extract on PGE2 biosynthesis, in which the effective content was confirmed as CLNA.1 More recently, CLNA has attracted increasing attention, especially focusing on its health-associated functions, such as anti-cancinogenesis, anti-obesity, anti-inflammation, anti-oxidant as well as positive effects on lipid metabolism.2,3
CLNA naturally occurs in dairy and meat products from ruminant as well as some plant seeds. In plant seeds, CLNA has different isomers such as punicic acid (c9,t11,c13-CLNA) from pomegranate seeds, α-eleostearic acid (c9,t11,t13-CLNA) from tung seeds and bitter seeds, α-calendic acid (t8,t10,c12-CLNA) from marigold seeds as well as jacaric acid (c8,t10,c12-CLNA) from jacaranda seeds. Although CLNA isomers are enriched in those plant seeds, the amount of CLNA is relatively small for further usage and those plant seeds could not be consumed by humans safely. Alkaline treatment of linolenic acid seems to be another effective approach for producing CLNA, whereas a number of harmful by-products will be brought which results in difficult on CLNA purification. At present CLNA has not been manufactured commercially. Therefore, alternative strategies are required increasing the supply of the functional fatty acid in the human diet.
Microbial conjugated linoleic acid (CLA) producers were widely reported,4 and researchers accidentally found some CLA producers could generate CLNA at an extremely high concentration.5-7 Compared to plant-derived CLNA, microbial CLNA has different conjugated double bond; c9,t11,c15-CLNA and t9,t11,c15-CLNA were generated with α-LNA as substrate, while c6,c9,t11-CLNA and c6,t9,t11 was produced from γ-LNA.5 Microbial CLNA has been identified with anti-cancer activities in vitro in line with those isomers from plant seeds extract.8 Additionally, most microbial CLNA was in the supernatant fluid which eliminates the process of further separation and purification.
Hitherto the most efficient microbial CLNA producers, Bifidobacteria, were isolated from human gastrointestinal tract especially infant-intestine. Bifidobacteria reach a high population in the infant gut microflora, and this early life prevalence supports their purported role as modulators of various metabolic and immune activities. Noteworthy, the colonization of the infant gastrointestinal tract with bifidobacteria provides us with the opportunity to determine whether or not CLNA production is a feature of the initial microbiota. The aim of the present study was to isolate the Bifidobacterium from breast-milk fed neonate feces and assess their ability for producing the beneficial conjugated linolenic acid from free linolenic acid.
Materials and methods
Subjects and fecal sampling
Faecal material was sampled from 16 neonates (full-term) aged from 1 d to 3 d. All samples were collected at the neonatal unit of the Wuxi Ninth People's Hospital, Wuxi, China. Fully informed consent was obtained from all the parents. All the neonates were breast fed and none had been on antibiotic treatment prior to sampling. Faecal samples were taken from the neonates and samples were stored at 4°C and delivered to the laboratory for processing in 5 h.
Isolation of Bifidobacteria
One gram of each faecal sample was mixed with 9 ml phosphate buffered saline (PBS), pH 6.5. Serial dilutions and plating were performed in an anaerobic workstation (Whitley DG250 anaerobic workstation, Don Whitley Scientific, UK) that was continuously sparged with a mixture of 80% nitrogen, 10% carbon dioxide and 10% hydrogen. For selective growth of bifidobacteria, 100 μl of dilutions were spread-plated onto de Man, Rogosa, Sharpe agar supplemented with 0.05% (w/v) L-cysteine hydrochloride (mMRS) and 100 μg/ml mupirocin (Sangon Biotech, Shanghai, China). Agar plates were incubated in anaerobic workstation at 37°C for 72 h. Bacterial counts were recorded as colony forming units (CFU) per gram of faeces. Twelve colonies from each sample were randomly selected to analyze the dominant Bifidobacterium population and sub-cultured in mMRS agar and mMRS broth for 24 to 48 h. All the putative Bifidobacterium isolates were generated and maintained at −80°C in 30% glycerol.
F-6-PPK activity
The fructose-6-phosphate phosphoketolase (F-6-PPK) assay was carried out, which is an enzyme characteristic of bifidobacterial carbohydrate metabolism.9 The protocol of F-6-PPK was as follow. The washed cell pellets were incubated with 50 μl 6 mg/ml sodium fluoride, 50 μl 10 mg/ml sodium iodoacetate as well as 50 μl 80 mg/ml fructose-6-phospahte at 37°C for 60 min. Then 300 μl fresh 13.9 mg/ml hydroxylamine hydrochloride was added into the mixture and left at room temperature for 10 min. Lately 200 μl 15% trichloroacetic acid, 200 μl 4 M hydrochloric acid and 200 μl 5% iron chloride (with 0.1 M hydrochloric acid) were finally added into the mixture. If a red-violet color develops immediately this is taken as a positive for the presence of F-6-PPK. B. bifidum CCFM16 and L. plantarum ZS2058 were used as positive and negative controls, respectively.
16S rDNA sequencing
27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-TACGGCTACCTTGTTACGACT T-3′), the universal bacterial 16S rDNA (rDNA) primers, were used to generate an approximate 1500 bp 16S rDNA product under PCR conditions: 95°C 5 min; 35 cycles of 95°C 30 s, 55°C 30 s, 72°C 2 min; 72°C 10 min. This product was partially sequenced by 454 Roche sequencer (BGI, China). Analysis of the 16S rDNA sequences obtained via the BLAST program allowed the assignment of a strain to a particular species. Generally, when 16S rDNA similarity values exceed 98%, the strains are considered to be the same species.
Fat extraction from bacterial supernatant fluids and pellets
Prior to examination of the strains for CLNA production, each isolate was sub-cultured twice in mMRS broth, then cultured (2%) in broth spiked with 0.3 mg/ml free linolenic acid (99% purity; Nu-Chek). The linolenic acid was added as a 30 mg/ml stock solution containing 20% (v/v) Tween-80 and was previously filter-sterilized through a 0.45 μm Minisart filter (Sigma) and stored in the dark at −20°C. The strains were incubated anaerobically at 37°C.
Lipid extraction and FAME preparation and gas chromatography analysis
After 48 h incubation, the cultures were centrifuged at 5,000×g for 10 min at room temperature. The fat was extracted from the culture supernatant fluid and the bacterial pellet as previous described.10 C17:0 heptadecanoic acid (99% pure; Sigma) was used as the internal standard. Fatty acids were derived to corresponding methyl esters with (trimethylsilyl)-diazomethane (Sigma) as described previously.10,11 The FAMEs were extracted in hexane and loaded on an Rtx-wax column (30 m × 0.25 mm × 0.25 μm) using a gas chromatograph (GC2010 plus; Shimadzu, Japan) fitted with a QP2010 Ultra mass spectrometer. Injections of 1 μl were performed automatically at a split ratio of 10:1. Helium was used as the carrier gases. The temperature program was described previously.10 The injector and detector were operated at 240°C. Electron energy of 70 eV and ion source temperature of 220°C were used. The percentage conversion to CLNA isomers was calculated by dividing the amount of CLNA present in the broth after inoculation/incubation with the amount of linolenic acid present in the spiked broth before incubation.
CLNA production by Bifidobacterium pseudocatenulatum PA3 in the presence of linolenic acid
B. pseudocatenulatum PA3 was inoculated from a fresh overnight culture and the medium incubated anaerobically at 37°C. Conversion of linolenic acid to CLNA was monitored in duplicate at regular time intervals over 72 h.
Results and discussion
Isolation of neonatal derived bifidobacteria
Bifidobacteria have been associated with several health and nutritional benefits especially for infant immune system development. Whereas the initial infant gastrointestinal tract is sterile and becomes colonized by different microorganism with Bifidobacterium being the dominant microbes after birth,12 which could be a veritable storehouse of Bifidobacterium. Several factors influence the diversity and content of the neonatal intestinal microflora including mode of delivery, feeding and antibiotic administration.13-16 In 5 of the 16 faecal samples, no isolates were obtained on the mupirocin containing mMRS agar; those samples might be meconium. For the remaining 11 faecal samples, 132 single colonies were picked for further analysis. Twenty five colonies from 7 samples were found to be F-6-PPK positive, which indicated their Bifidobacterium status (Table 1), whereas the colonies from sample A, sample B, sample D and sample G were non-F-6-PPK positive, those neonates were premature and/or delivered by caesarean birth, which had less access to obtain diverse microbes from the birth canal. Additionally, in sample E and sample F, there was a low isolation rate of bifidobacteria, which might indicate a more diverse microbiota, such as Clostridium and Bacteroides.12,17
Table 1.
Samples, description, and selection of bifidobacteria by F-6-PPK detection.
| Sample No. | Age | No. of colonies assessed | No. of F-6-PPK-positive colonies (%) |
|---|---|---|---|
| A | 1-day | 12 | 0 |
| B | 1-day | 12 | 0 |
| C | 2-day | 12 | 7 (58) |
| D | 1-day | 12 | 0 |
| E | 3-day | 12 | 2 (16) |
| F | 2-day | 12 | 2 (16) |
| G | 1-day | 12 | 0 |
| PA | 3-day | 12 | 4 (75) |
| GM | 1-day | 12 | 1 (8) |
| GN | 2-day | 12 | 5 (41) |
| NY | 2-day | 12 | 4 (32) |
With the 16S rDNA sequencing and BLAST analysis, those 25 F-6-PPK positive isolates were identified as different Bifidobacterium species, including B. longum, B. pseudocatenulatum, B. breve and B. adolescentis (Table 2). B. longum seems to be a major species which could be isolated from each F-6-PPK positive sample, for instance, C5, C9, C10, NY5, NY26, PA7A,PA34B as well as all the isolates from sample E, sample F, sample GN, sample GM were B. longum. In addition, among those samples, sample C shows the best Bifidobacterium diversity, in which 3 different Bifidobacterium species (B. longum, B. breve and B. pseudocatenulatum) were isolated.
Table 2.
16S rDNA and CLNA conversion of all the F-6-PPK positive isolates.
| 16S rDNA |
CLNA production from 0.37 mg/ml linoleic acid |
|||||
|---|---|---|---|---|---|---|
| Isolate | Homology | Species | CLNA1 | % converted | CLNA2 | % converted |
| C5 | 99% | B. longum | 0.2545 ± 0.0022 | 67.69 | 0.0086 ± 0.0002 | 2.28 |
| C8 | 99% | B. pseudocatenulatum | 0.0911 ± 0.0022 | 24.24 | 0.0469 ± 0.0013 | 12.48 |
| C9 | 99% | B. longum | 0.2643 ± 0.0126 | 70.31 | 0.0175 ± 0.0009 | 4.66 |
| C10 | 99% | B. longum | 0.2637 ± 0.0015 | 70.16 | 0.0049 ± 0.0007 | 1.32 |
| C11 | 99% | B. breve | 0.3011 ± 0.0160 | 79.77 | 0.0190 ± 0.0024 | 5.06 |
| C21 | 99% | B. breve | 0.3201 ± 0.0005 | 85.16 | 0.0169 ± 0.0023 | 4.50 |
| C23 | 99% | B. longum | 0.2013 ± 0.0001 | 53.55 | 0.0055 ± 0.0001 | 1.46 |
| E38 | 99% | B. longum | 0.2322 ± 0.0050 | 61.78 | 0.0127 ± 0.0009 | 3.37 |
| E41 | 99% | B. longum | 0.2370 ± 0.0014 | 63.04 | 0.0116 ± 0.0004 | 3.08 |
| F45BB | 99% | B. longum | N.D. | — | N.D. | — |
| F47B | 99% | B. longum | N.D. | — | N.D. | — |
| NY5 | 99% | B. longum | 0.1439 ± 0.0056 | 38.9 | 0.0411 ± 0.0009 | 11.1 |
| NY6 | 99% | B. adolescentis | N.D. | — | N.D. | — |
| NY26 | 99% | B. longum | 0.1660 ± 0.078 | 44.86 | 0.0055 ± 0.018 | 1.48 |
| NY50 | 99% | B. adolescentis | N.D. | — | N.D. | — |
| PA3 | 99% | B. pseudocatenulatum | 0.3267 ± 0.0023 | 86.91 | 0.0135 ± 0.0009 | 3.59 |
| PA52 | 99% | B. pseudocatenulatum | 0.3196 ± 0.0071 | 85.02 | 0.0138 ± 0.0018 | 3.67 |
| PA7A | 99% | B. longum | 0.1805 ± 0.0099 | 48.02 | 0.0030 ± 0.0001 | 0.80 |
| PA34B | 99% | B. longum | 0.2759 ± 0.0052 | 73.39 | 0.0037 ± 0.0006 | 0.98 |
| GN22A2 | 99% | B. longum | 0.3184 ± 0.0226 | 84.70 | 0.0111 ± 0.0002 | 2.95 |
| GN32A | 99% | B. longum | 0.2720 ± 0.0065 | 72.36 | 0.0075 ± 0.0005 | 2.00 |
| GN40A | 99% | B. longum | 0.2674 ± 0.0023 | 71.14 | 0.0107 ± 0.0007 | 2.85 |
| GN45 | 99% | B. longum | 0.2622 ± 0.0002 | 69.75 | 0.0097 ± 0.0005 | 2.58 |
| GN57A | 99% | B. longum | 0.2039 ± 0.0030 | 54.24 | 0.0075 ± 0.0001 | 2.00 |
| GM59 | 99% | B. longum | 0.3027 ± 0.0036 | 80.53 | 0.0111 ± 0.0003 | 2.95 |
Microbial production of CLNA
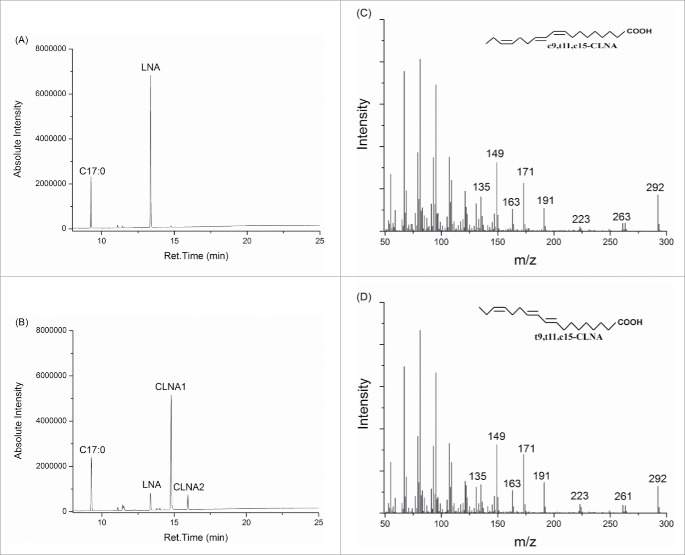
Bifidobacteria are able to produce different conjugated fatty acids including CLA, CLNA and conjugated stearidonic acid (CSA), when the relative substrates are freely present in the growth medium.5,8,18-20 All the F-6-PPK positive isolates were analyzed for CLNA production. After incubation for 72 h with 0.37 mg/ml of free linolenic acid, fatty acid profiles from all those isolates were analyzed via GC-MS. All of the isolates grew well in the mMRS broth plus linolenic acid, which means those isolates had better free fatty acid resistance, as in previous publications, some bifidobacteria strains could not grow in free fatty acid containing medium.6,19 As no standard CLNA was commercially available, the mass spectra of CLNA isomers was compared to previous results, which indicated that CLNA1 was c9,t11,c15-CLNA isomer and CLNA2 was t9,t11,c15-CLNA.5 In general, despite isolates from sample F and some isolates from sample NY, isolates from all the samples have different CLNA generation ability, which might indicate a feature of the initial microflora with CLNA production.
The ability to produce CLNA from free linolenic acid differed notably, ranging from a non-detectable level to a conversion level of 90%. The most efficient producers were belonged to B. breve and B. pseudocatenulatum species, exhibiting between 79.77–85.26%, 24.24–86.91% conversion of free linolenic acid to c9,t11,c15-CLNA (Table 2), with all the strains also producing small amount of t9,t11,t15-CLNA isomer. B. pseudocatenulatum PA3 was the most effective CLNA producer in all the assessed isolates. There was a total 86.91% conversion of linolenic acid into c9,t11,c15-CLNA and 3.59% conversion to t9,t11,c15-CLNA; totally 90.5% linolenic acid was converted. In addition, B. breve C21 converted 89.66% linolenic acid to c9,t11,c15-CLNA (85.16%) and t9,t11,c15-CLNA (4.5%) while B. breve C11 converted 85.16% linolenic acid to c9,t11,c15-CLNA and 4.5% linolenic acid to t9,t11,c15-CLNA.
For B. longum isolates, the ability for CLNA generation was shown to be strongly strain-dependent; a number of B. longum isolates could convert over 60% linolenic acid to conjugated linolenic acid, for instance, B. longum GN22A2 and B. longum GM59 could convert over 80% of linolenic acid into CLNA, and approximately 40%–50% linolenic acid could be generated to CLNA by B. longum NY26 and B. longum PA7A. In contrast, B. longum F45BB and B. longum F47B could not generate any CLNA. Additionally, B. adolescentis isolates could not generate any CLNA from linolenic acid, which is in line with previous publication that non-CLNA producers were identified in the B. adolescentis species.6,19
CLNA production by Bifidobacterium pseudocatenulatum PA3 in the presence of linolenic acid
After culturing with linolenic acid at regular time intervals till 72 h, fatty acid profiles from both supernatant fluid and cell pellets of B. pseudocatenulatum PA3 were monitored. After culturing for 12 h, B. pseudocatenulatum PA3 showed to accumulate CLNA. During the cell growth (24h), the concentration of conjugated linolenic acid increased significantly. Up to 36 h culturing, the increase of conjugated linolenic acid got slower, with the maximum conversion at 72 h (Fig. 1). The total CLNA concentration reached 0.3402 mg/ml, with 90.5% conversion of linolenic acid. The conversion rate is currently the highest as we know.
Figure 1.
GC-MS total ion chromatograms of the products from fatty acid contents of B. pseudocatenulatum PA3 grown in mMRS plus 0.37 mg/ml linolenic acid. (A) 0h; (B) 72h; (C) Mass spectra for CLNA1; (D) Mass spectra for CLNA2.
To analyze the isomers produced from the strain, c9,t11,c15-CLNA and t9,t11,c15-CLNA were both generated. In the initial 36 h, c9,t11,c15-CLNA rapidly increased with a stable concentration till 36 h. Where t9,t11,c15-CLNA isomer was at a low concentration in the beginning and showed accumulation with the cell growth. In the CLNA isomers, c9,t11,c15-CLNA was the majority isomer, which occupied over 90% of the total CLNA.
The fatty acid contents from the supernatant fluid and cell pellets were both analyzed after 72 h culture (Fig. 2). The results indicated that this isolate has strong ability in linolenic acid absorption and conversion, in which no significant linolenic acid was determined in the supernatant fluid as well as no significant substrate remaining in the cells. For the conjugated linolenic acid isomers, either c9,t11,c15-CLNA or t9,t11,c15-CLNA was accumulated in the supernatant fluid nor the cell pellets which indicated that the products could be secreted by the lactic acid bacteria. This result was in line with previous CLA work by Coakley and colleagues in which CLA produced by B. breve NCFB 2258 was mostly accumulated in the supernatant fluid rather than cell pellets.19
Figure 2.
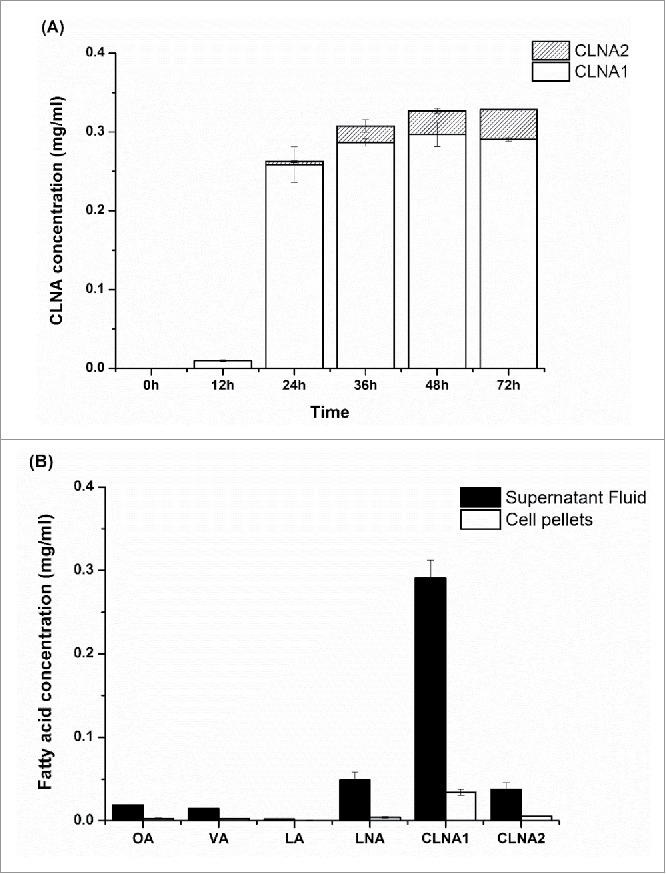
Conjugated linolenic acid production by B. pseudocatenulatum PA3. (A) CLNA concentration at different time; (B) Fatty acid profiles in the supernatant fluid and cell pellets.
Funding Statement
This research was supported by the National Natural Science Foundation of China (Nos. 31571810, 31530056), the Program for New Century Excellent Talents (NCET-13-0831), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1249), the National Natural Science Foundation of Jiangsu Province (No. BK20150141), and the Jiangsu Province “Collaborative Innovation Center for Food Safety and Quality Control.”
Disclosure of potential conflicts of interest
No conflicts of interest were disclosed.
References
- [1].Nugteren DH, Christ-Hazelhof E. Naturally occurring conjugated octadecatrienoic acids are strong inhibitors of prostaglandin biosynthesis. Prostaglandins 1987; 33:403-17; PMID:3107083; https://doi.org/ 10.1016/0090-6980(87)90022-0 [DOI] [PubMed] [Google Scholar]
- [2].Hennessy AA, Ross RP, Devery R, Stanton C. The health promoting properties of the conjugated isomers of alpha-linolenic acid. Lipids 2011; 46:105-19; PMID:21161605; https://doi.org/ 10.1007/s11745-010-3501-5 [DOI] [PubMed] [Google Scholar]
- [3].Yuan G, Chen X, Li D. Conjugated linolenic acids and their bioactivities: a review. Food Funct 2014; 5:1360-8; PMID:24760201; https://doi.org/ 10.1039/c4fo00037d [DOI] [PubMed] [Google Scholar]
- [4].O'Shea EF, Cotter PD, Stanton C, Ross RP, Hill C. Production of bioactive substances by intestinal bacteria as a basis for explaining probiotic mechanisms: bacteriocins and conjugated linoleic acid. Int J Food Microbiol 2012; 152:189-205; PMID:21742394; https://doi.org/ 10.1016/j.ijfoodmicro.2011.05.025 [DOI] [PubMed] [Google Scholar]
- [5].Hennessy A, Barrett E, Ross RP, Fitzgerald G, Devery R, Stanton C. The production of conjugated α-linolenic, γ-linolenic and stearidonic acids by strains of Bifidobacteria and Propionibacteria. Lipids 2012; 47:313-27; PMID:22160449; https://doi.org/ 10.1007/s11745-011-3636-z [DOI] [PubMed] [Google Scholar]
- [6].Gorissen L, Raes K, Weckx S, Dannenberger D, Leroy F, De Vuyst L, De Smet S. Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl Microbiol Biotechnol 2010; 87:2257-66; PMID:20556602; https://doi.org/ 10.1007/s00253-010-2713-1 [DOI] [PubMed] [Google Scholar]
- [7].Gorissen L, Vuyst LD, Raes K, De Smet S, Leroy F. Conjugated linoleic and linolenic acid production kinetics by bifidobacteria differ among strains. Int J Food Microbiol 2012; 155:234-40; PMID:22405353; https://doi.org/ 10.1016/j.ijfoodmicro.2012.02.012 [DOI] [PubMed] [Google Scholar]
- [8].Coakley M, Banni S, Johnson MC, Mills S, Devery R, Fitzgerald G, Paul Ross R, Stanton C. Inhibitory effect of conjugated alpha-linolenic acid from Bifidobacteria of intestinal origin on SW480 cancer cells. Lipids 2009; 44:249-56; PMID:19048324; https://doi.org/ 10.1007/s11745-008-3269-z [DOI] [PubMed] [Google Scholar]
- [9].Biavati B, Vescovo M, Torriani S, Bottazzi V. Bifidobacteria: history, ecology, physiology and applications. Ann Microbiol 2000; 50:117-31 [Google Scholar]
- [10].Yang B, Chen H, Gu Z, Tian F, Ross RP, Stanton C, Chen YQ, Chen W, Zhang H. Synthesis of conjugated linoleic acid by the linoleate isomerase complex in food-derived lactobacilli. J Appl Microbiol 2014; 117:430-9; PMID:24750362; https://doi.org/ 10.1111/jam.12524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yang B, Chen H, Song Y, Chen YQ, Zhang H, Chen W. Myosin-cross-reactive antigens from four different lactic acid bacteria are fatty acid hydratases. Biotechnol Lett 2013; 35:75-81; PMID:22955678; https://doi.org/ 10.1007/s10529-012-1044-y [DOI] [PubMed] [Google Scholar]
- [12].Mitsuoka T. Intestinal flora and human health. Asia Pac J Clin Nutr 1996; 5:2-9; PMID:24394457 [PubMed] [Google Scholar]
- [13].Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 2010; 107:11971-5; PMID:20566857; https://doi.org/ 10.1073/pnas.1002601107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 2011; 108:4578-85; PMID:20668239; https://doi.org/ 10.1073/pnas.1000081107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Andres A, Cleves MA, Bellando JB, Pivik RT, Casey PH, Badger TM. Developmental status of 1-year-old infants fed breast milk, cow's milk formula, or soy formula. Pediatrics 2012; 129:1134-40; PMID:22641754; https://doi.org/ 10.1542/peds.2011-3121 [DOI] [PubMed] [Google Scholar]
- [16].Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, Murphy B, Ross RP, Fitzgerald GF, Stanton C, et al.. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother 2012; 56:5811-20; PMID:22948872; https://doi.org/ 10.1128/AAC.00789-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Favier CF, Vaughan EE, De Vos WM, Akkermans ADL. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 2002; 68:219-26; PMID:11772630; https://doi.org/ 10.1128/AEM.68.1.219-226.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Oh DK, Hong GH, Lee Y, Min S, Sin HS, Cho SK. Production of conjugated linoleic acid by isolated Bifidobacterium strains. World J Microbiol Biotech 2003; 19:907-12; https://doi.org/ 10.1023/B:WIBI.0000007313.90368.0c [DOI] [Google Scholar]
- [19].Coakley M, Ross RP, Nordgren M, Fitzgerald G, Devery R, Stanton C. Conjugated linoleic acid biosynthesis by human-derived Bifidobacterium species. J Appl Microbiol 2003; 94:138-45; PMID:12492934; https://doi.org/ 10.1046/j.1365-2672.2003.01814.x [DOI] [PubMed] [Google Scholar]
- [20].Park HG, Cho SD, Kim JH, Lee H, Chung SH, Kim SB, Kim HS, Kim T, Choi NJ, Kim YJ. Characterization of conjugated linoleic acid production by Bifidobacterium breve LMC 520. J Agric Food Chem 2009; 57:7571-5; PMID:20349924; https://doi.org/ 10.1021/jf9014813 [DOI] [PubMed] [Google Scholar]