Abstract
The physiological functions of phosphatidylinositol/phosphatidylcholine transfer proteins (PITPs) are poorly characterized, even though these proteins are conserved throughout the eukaryotic kingdom. Much of the progress in elucidating PITP functions has come from exploitation of genetically tractable model organisms, but the mechanisms for how PITPs execute their biological activities remain unclear. Structural and molecular dynamics approaches are filling in the details for how these proteins actually work as molecules. Herein, we discuss our recent work with Sec14-like PITPs, and describe how PITPs integrate diverse territories of the lipid metabolome with phosphoinositide signaling.
Keywords: lipid transfer proteins, Sec14, phosphatidylinositol, phosphoinositides
Introduction
Phosphoinositides are essential signaling molecules in eukaryotic cells, and these lipids serve dual signaling roles in that capacity (1,2). First, the phosphoinositide molecules themselves have intrinsic signaling capabilities. These activities function through the spatially and temporally regulated recruitment and activation of proteins that read the positional information encoded by the phosphorylation pattern of the inositol headgroup. Roles for phosphoinositides as co-factors in allosteric regulation of various enzymes and ion channels are other examples of such a function (3). Second, phosphoinositides not phosphorylated at the 3’-OH position of the inositol ring are substrates for phospholipases C and are therefore potential precursors for other second messengers – that is, soluble inositol phosphates and diacyglycerol. In this capacity, phosphoinositides are reservoirs for production of second messenger production. The prime example of this function involves phosphatidylinositol-4,5-bisphosphate (PtdIns-4,5-P2) . Hydrolysis of this phosphoinositide by phospholipase C produces the lipid second messenger diacylglycerol and the soluble second messenger inositol-1,4,5-trisphosphate (IP3) (4,5). Whereas the gating of intracellular calcium channels by this specific positional isomer of IP3 is well-established, elegant work from the York and Wente laboratories demonstrated that IP3 is the obligate precursor of the large cabal of other soluble inositol-polyphosphates and inositol-pyrophosphates. These soluble second messengers are involved in an astonishing variety of activities in cells ranging from nuclear functions to cell cycle regulation, to scaffolds for protein folding, allosteric regulation of protein function, etc (6,7). Given there are potentially ~70 chemically distinct soluble inositol phosphates in cells, their involvements in regulating a multitude of diverse cellular functions is not surprising.
What is surprising is the diversity of biological outcomes associated with phosphoinositide signaling given that the cohort of chemically distinct phosphoinositides (as defined by headgroup chemistry) is small. The most complex organisms produce six phosphoinositides (plus PtdIns), yet literally hundreds of cellular functions are subject to control by intact phosphoinositides molecules (i.e. independent of their hydrolytic products). How is such a large diversity in biological outcome generated? Current views see the solution to the problem through the lens of a combinatorial mechanism where phosphoinositide binding by some effector protein is accompanied by recognition of other binding partners. However, as discussed below, this concept fails to explain a more privileged biochemical circuitry that involves predetermined channeling of phosphoinositide directly to vicinal effector proteins. Such channeling mechanisms provide powerful new ideas for thinking about how biological outcomes for phosphoinositide signaling are diversified.
Instructive regulation of phosphoinositide signaling
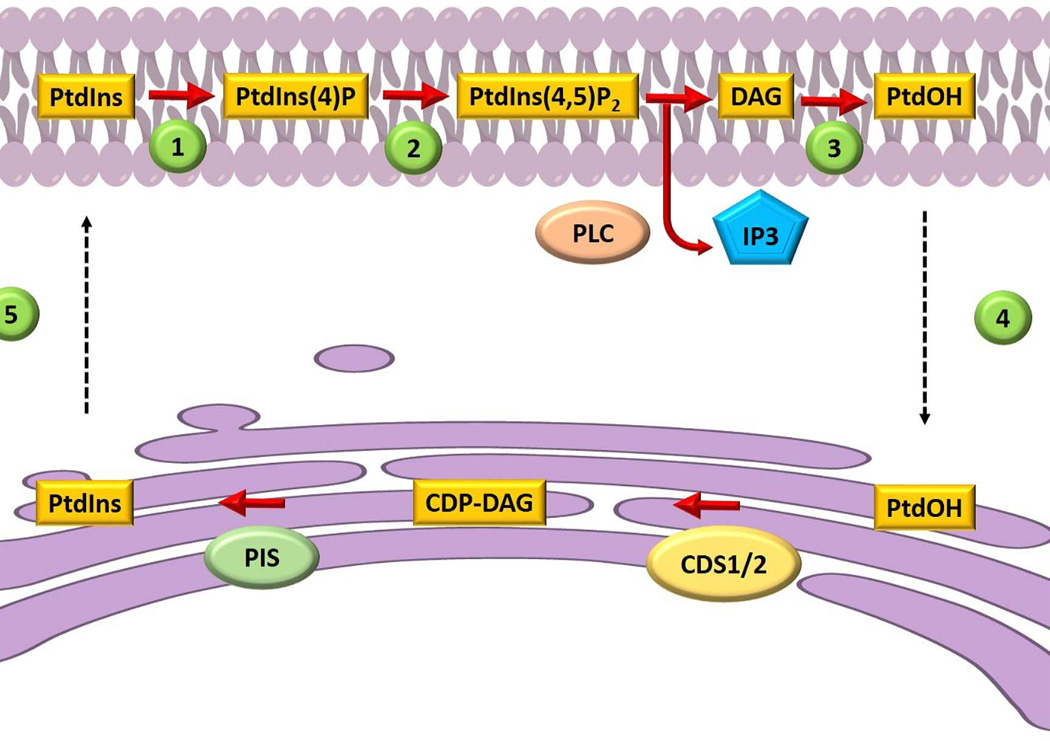
We believe the problem of diversifying phosphoinositide signaling outcomes is closely linked to initial ideas for how cells might solve topological problems associated with sustaining phosphoinositide signaling. The topological problem rests on the fact that the metabolic cycle for regenerating phosphoinositide molecules hydrolyzed during the course of agonist-stimulated signaling is confronted with the cell biology of PtdIns metabolism. PtdIns is a minor cellular phospholipid in many eukaryotic cells (including mammals) and is the metabolic precursor for phosphoinositides. PtdIns is produced de novo by PtdIns synthase which consumes the substrates inositol and cytidine-diphospho-diacylglycerol (CDP-DAG) to produce PtdIns and cytidine-monophosphate. CDP-DAG is generated by a synthase that consumes phosphatidic acid (PtdOH) and cytidine-trisphosphate. Both PtdIns- and CDP-DAG-synthases are integral membrane proteins of a compartment distinct from the major compartment of PtdIns-4,5-P2 signaling (i.e. the endoplasmic reticulum and plasma membrane, respectively). In the mid-1970s, Michell proposed the necessity of an ER-plasma membrane PtdIns/PtdOH transfer cycle to replenish the plasma membrane with PtdIns, and phosphoinositides, in the face of phospholipase C activity (8; Fig.1). In the first stage of the cycle, lipid carriers ferry DAG or PtdOH (produced by plasma membrane DAG kinases) from the plasma membrane back to the ER to recharge PtdIns synthesis. This newly synthesized PtdIns is subsequently mobilized from the ER to the plasma membrane by a second set of lipid carriers, the PtdIns transfer proteins (PITPs). This attractive, yet essentially untested, idea remains the prevailing concept for how PITPs are interpreted to execute their cellular functions (9).
Figure 1.
Schematic representation of a lipid exchange cycle between the plasma membrane and the endoplasmic reticulum. Phosphatidylinositol (PtdIns) is phosphorylated at the plasma membrane by a (1) PtdIns 4-OH kinase and a (2) 4-phosphate 5-OH kinase to produce PtdIns(4,5)P2. Phospholipase C (PLC) hydrolyzes PtdIns(4,5)P2 to produce inositol-trisphosphate (IP3) and diacylglycerol (DAG). Diacylglycerol is converted to phosphatidic acid (PtdOH) (3), and PtdOH is mobilized back to the ER (4) where it is converted to CDP-DAG. PtdIns synthase (PIS) consumes CDP-DAG to produce PtdIns molecules (5) that are subsequently trafficked by phosphatidylinositol transfer proteins to the plasma membrane. This figure is adapted from refs 8 and 30.
Our work with Sec14, the major yeast PITP, indicates at least PITPs of the Sec14-superfamily are unlikely to be bona fide lipid carriers. Rather, the evidence describes Sec14-like PITPs as regulated scaffolds that execute an interfacial presentation of PtdIns to PtdIns 4-OH kinases -- thereby making these PtdIns kinases better enzymes (10,11). That is, Sec14-like PITPs impose an important layer of control of PtdIns 4-OH kinase activities by boosting the activities of PtdIns-kinases -- a stimulation is made necessary by the biological inadequacy of PtdIns 4-OH kinase activities on membrane-incorporated PtdIns substrates. The presentation function is itself dependent on Sec14 binding to a second lipid species, in this case PtdCho, and that feature suggests new perspectives regarding the purpose for PITP-mediated lipid exchange (10,11). Herein, we discuss our views regarding how Sec14-like proteins stimulate PtdIns 4-OH kinases, and how those mechanisms are linked to diversification of biological outcomes for phosphoinositide signaling.
The mechanics of the Sec14 lipid exchange cycle
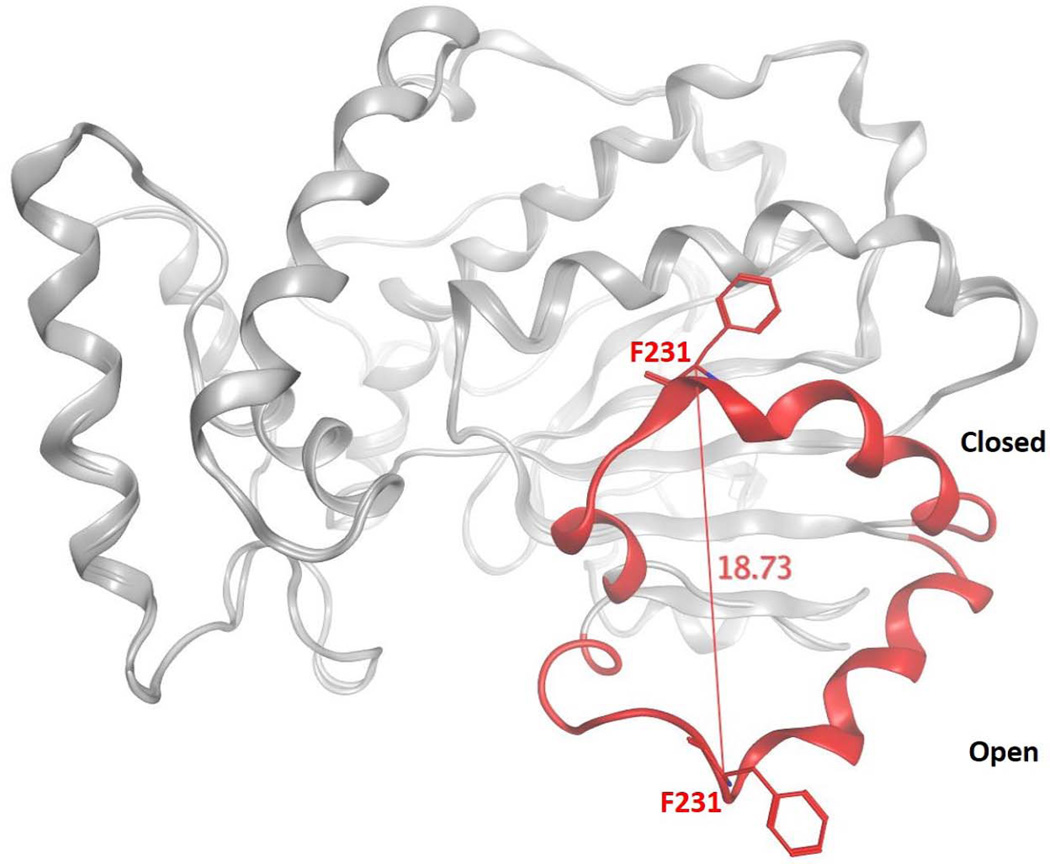
As the structural details of Sec14-like proteins continue to emerge, it becomes ever more clear that a detailed understanding of how PITPs execute lipid exchange is central to an understanding of how these proteins work in cells. Structural studies define two major Sec14 conformers. These represent an ‘open’ form which we interpret as the membrane-associated conformer engaged in lipid exchange, and the ‘closed’ form which is tightly bound to a single phospholipid molecule which we interpret as the cytoplasmic conformer of the protein (10,12). The conformational transitions between the ‘open’ and ‘closed’ conformers are governed by an 18Å displacement of a helical gate structure that governs access to the lipid binding pocket (Fig. 2). Reversible cysteine cross-linking experiments confirm that helical gate motions are essential for Sec14-mediated lipid exchange activity. These transitions involve large rigid body motions subject to control by an interesting conformational switch element, termed the G-module (13). This switch element is conserved in distant members of the Sec14-superfamily, and it serves an important function in these proteins as a number of human disease alleles involving these proteins are missense mutations that lie within the G-module itself (13).
Figure 2.
Conformational transitions of Sec14 during lipid exchange. The ‘open’ and ‘closed’ conformers of Se14 are superimposed to highlight the motion of the gating helix that controls access to the Sec14 hydrophobic pocket. The Sec14 α-carbon backbone is represented in gray ribbon with the helical gate structural element rendered in red color. Distance monitoring of the Cα atom of residue F231 of the gate illustrates the 18 Å displacement of the helical gate.
Clearly, a major goal of future study is to determine how activity of the G-module is itself controlled. Membrane-binding must provide some sort of trigger, but what it may be is completely mysterious. But, there are indications that the triggering mechanism also involves events within the lipid binding pocket itself. Those insights came from directed evolution experiments where a naturally occurring Sec14-like protein lacking Sec14 biological activities (Sfh1) was functionally resurrected by gain-of-function mutations that restored Sec14-like activities to this highly related, but ‘dead’, Sec14-like protein. Remarkably, those resurrection substitutions altered amino acids buried deep within the protein hydrophobic pocket that were conserved between Sec14 and Sfh1, and the resurrection mechanism involved enhancement of helical gate dynamics and rates of lipid exchange (14). The available data suggest that water molecules residing within the hydrophobic pocket play important roles in facilitating the G-module conformational transitions required for gate opening and accessibility of the hydrophobic pocket for lipid exchange. Once the gate opens, biophysical studies indicate the phospholipid is presented with a choice of partitioning into one two chemically similar environments; i.e. from the membrane into the hydrophobic pocket and vice-versa (15). Simple partitioning accounts for the ATP-independence of the lipid exchange cycle, although it does not address how vectorial the lipid trajectories are, or how lipids are enticed to leave their resident environments in the first place.
How Sec14 binds its phospholipid ligands
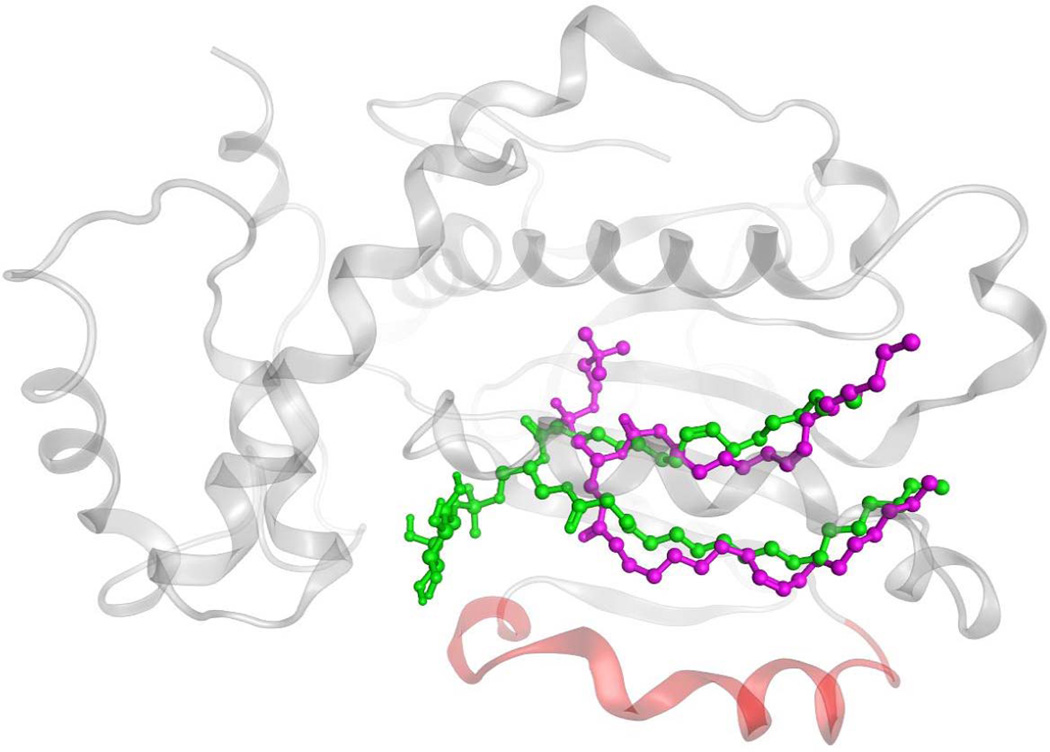
Simple partitioning models could be taken to imply that PtdIns and PtdCho enter and/or exit the hydrophobic pocket of Sec14-like proteins with similar trajectories, but structural studies indicate it unlikely to be so (10). Remarkably, Sec14 and Sfh1 bind the PtdCho and PtdIns headgroups at completely distinct sites. The PtdCho headgroup and glycerol backbone are buried deep within the Sec14 interior, while the corresponding regions of bound PtdIns are snorkel towards the protein surface (Fig. 3). This curious engineering guided rational mutagenesis studies which showed a Sec14 molecule must harbor both the ability to bind/exchange PtdCho and to bind/exchange PtdIns in order to stimulate production of PtdIns-4-phosphate (the product of PtdIns 4-OH kinase activity) in cells (10). Thus, homotypic PtdCho exchange reactions and homotypic PtdIns exchange reactions are biologically futile activities, and stimulation of PtdIns 4-OH kinase activity by Sec14 requires the PITP to undergo heterotypic exchange reactions (e.g. PtdIns for PtdCho or vice versa) to effect a suitable presentation of PtdIns to the PtdIns 4-OH kinase. Schaaf et al (10,11) propose a kinetic trap model to account for these results, and this model posits slow PtdCho exchange kinetics relative to those of PtdIns. Yet, the selectivity for PtdIns vs PtdCho is estimated to be small in energetic terms with H2O rearrangements facilitating negotiation of the energy barriers that confront the heterotypic phospholipid exchange cycle.
Figure 3.
Differential binding strategies for PtdIns and PtdCho in the Sec14 hydrophobic pocket. Representation of Sec14 homology model in ‘closed’ conformation (as transparent gray Cα-carbon ribbon) with PtdIns (in green) and PtdCho (in magenta) in ball and stick mode. The Sec14 helical gate is rendered in red.
Biological function of Sec14
The Sec14 heterotypic exchange cycle is essential for support of optimal PtdIns 4-OH kinase activity in yeast cells – even though yeast membranes are PtdIns-rich. This lipid constitutes ~20 mol% of bulk glycerophospholipid in yeast. It is difficult to imagine an obligate need for a PITP-driven PtdIns supply mechanism in membranes so naturally rich in this phospholipid. Yet, Sec14 is essential for cell viability under these conditions, and genetic studies demonstrate it coordinates PtdCho and DAG metabolism with production of PtdIns-4-phosphate for the purpose of stimulating membrane trafficking in trans-Golgi network (TGN) and endosomal compartments (16–21). Thus, Sec14 can be viewed as a PtdCho sensor that links PtdCho metabolic information to production of PtdIns-4-phosphate. This is an example of what we term instructive regulation of PtdIns 4-OH kinase activity. That is, Sec14 primes PtdIns-4-phosphate production by the lipid kinase in response to PtdCho cues.
The idea that Sec14-like proteins diversify phosphoinositide signaling via such instructive mechanisms suggests interesting possibilities, and structural studies indicate just how broad this type of regulation might be. The structural signatures, or bar codes, involved in PtdIns headgroup coordinationare conserved throughout the Sec14 superfamily whereas the PtdCho-binding bar code is not (10,11,22). The clear implication is that Sec14-superfamily proteins have conserved inositol-lipid binding capacities, but are diversified in their binding capacities for alternate lipid ligands, and that Sec14-like proteins/domains serve as interfaces between the metabolism of diverse lipids/lipophiles (i.e. α-tocopherol, retinaldehyde, PtdCho, etc) with stimulated phosphoinositide synthesis.
This concept is supported by the observation that several naturally occurring human disease alleles, including the most common inherited alleles associated with retinal degeneration and acute vitamin E deficiency, directly alter residues of presumptive PtdIns binding bar-codes (10,11,22). That observation implies PtdIns, or some other inositol phospholipid, is normally a bona fide physiological ligand for the affected Sec14-like proteins (cellular retinadehyde binding protein and α-tocopherol transfer protein, respectively; 10,11,22, 23). Other Sec14-like proteins/domains associated with human disease (i.e. caytaxin and neurofibromin) also exhibit loss-of-function mutations that compromise their respective PtdIns-binding bar-code motifs (22).
The PtdIns kinase activation mechanism
Each of the six Sec14-like PITPs of yeast specifies a unique biological outcome for PtdIns 4-OH kinase signaling, and most of these do not exhibit a PtdCho-binding bar-code (10,11). Indeed, the Sfh3 PITP acts in a biologically antagonistic way to Sec14, even though both PITPs activate the same PtdIns 4-OH kinase (24). In that case, Sfh3 directs PtdIns 4-OK kinase signaling towards lipid droplet metabolism while Sec14 directs it towards endosomal trafficking. These results indicate PITPs direct PtdIns 4-OH kinase activities towards distinct outcomes. This idea of instructive stimulation of PtdIns 4-OH kinases describes a new paradigm for how far-flung arms of lipid metabolism can commonly interface with phosphoinositide signaling but with distinct biological outcomes. This concept begs for identification of the various second ligands posited to prime PtdIns presentation to the lipid kinases. It also sets forth an emphasis on understanding the mechanics of PtdIns presentation to the lipid kinase, and the associated challenges now include detailed understanding of how Sec14 docks onto membrane surfaces, of the trajectories drawn by PtdIns and the priming ligands as these navigate entry and exit from the hydrophobic pocket, and of the relative kinetics of lipid entry and exit from the hydrophobic pocket.
How do Sec14-like PITPs interact with PtdIns 4-OH kinases? A direct physical interaction between enzyme and PITP offers the simplest configuration for ligand presentation after all. In some cases, such as in the collaboration between the yeast Sec14-like PITP Sfh4 and the Stt4 PtdIns 4-OH kinase in regulation of phosphatidylserine decarboxylation (25), an intimate physical interaction appears important. In other cases, such as stimulation of yeast Pik1 and Stt4 PtdIns 4-OH kinases by Sec14, a tight physical interaction does not seem essential. This point is highlighted by the fact that vertebrate PITPs, which have no sequence homology or structural similarity to Sec14, can serve as functional surrogates when expressed in yeast (26,27). Those data argue against a dedicated physical interaction between the PITP and the kinase. Perhaps the kinase registers the ‘presented’ headgroup only.
New opportunities for chemical interference with phosphoinositide signaling
The emerging evidence identifies PITPs as highly discriminating portals for interrogating phosphoinositide signaling. Recent advances in identifying specific small molecule inhibitors of Sec14 demonstrate that PITP-directed small molecule inhibitors offer new strategies for intervening with cellular phosphoinositide signaling pathways with much greater specificities than those possible with specific inhibition of individual PtdIns-kinases, or inducible depletion of compartment-specific pools of individual phosphoinositide species. The available data indicate Sec14 is susceptible to specific inhibition by compounds representing several different chemical scaffolds (28,29).
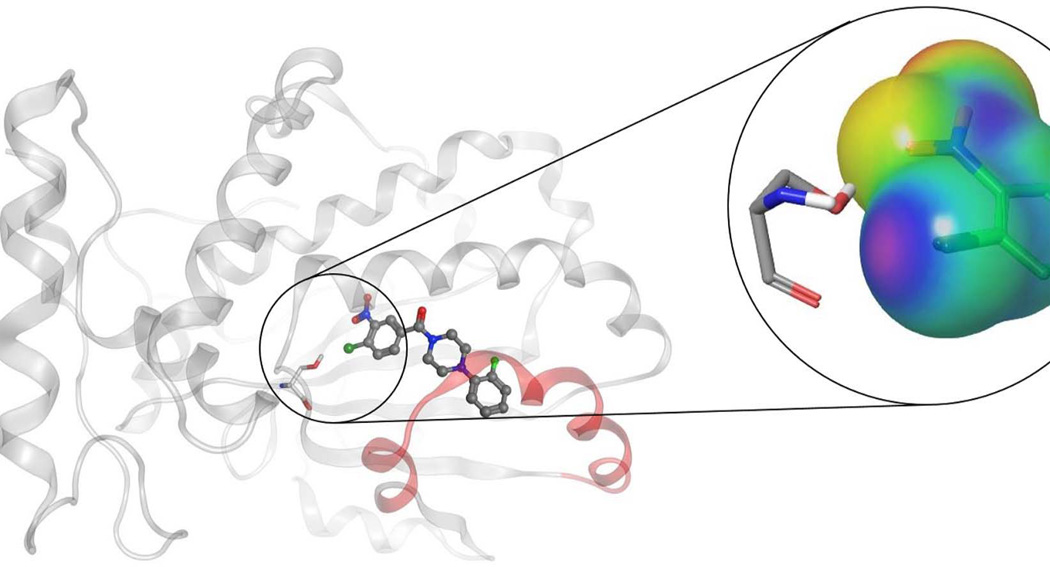
The best characterized of these Sec14-directed inhibitors are the nitrophenyl(4-(2-methoxyphenyl)piperazin-1-yl)methanones (NPPMs). Treatment of yeast with biologically active NPPMs recapitulates the effects seen by genetic ablation of Sec14 activity, and the evidence is strong that there are no off-target effects with these compounds (28). Indeed, their exquisite specificity is remarkable given that even very closely-related Sec14-like PITPs are not inhibited by Sec14-active compounds. The mechanism of inhibition involves entry of the active NPPM into the Sec14 hydrophobic pocket where it assumes a pose that occupies both PtdIns and PtdCho binding space. Several side-chain::NPPM interactions are important for NPPM binding, but the most critical is proposed to be a halogen bond interaction between the NPPM and a specific serine residue of Sec14 that also happens to play an important role in coordinating the headgroup phosphate of PtdCho (Fig 4; 28).
Figure 4.
Schematic model pose representation of an active NPPM in complex with Sec14. Figure shows NPPM-481 rendered in ball and stick mode, and inhibitor binding is stabilized via a network of halogen bond and hydrogen bond interaction with residue Ser173. Magnified figure shows the electrostatic potential (ESP) mapped over molecular surface of chloro-nitrophenyl moiety of NPPM-481, and illustrates the halogen-bonding interaction with Ser173 via an electro-positive a σ-hole on the chlorine moiety (see Nile et al., 2014).
The advent of PITP-directed inhibitors offers exciting new prospects for studying both PITP function and phosphoinositide signaling. We envision PITP-active inhibitors as potentially valuable tool compounds. First, PITP-directed inhibitors facilitate genome-scale functional interaction screens. Second, these inhibitors facilitate resolution of biochemical reconstitutions of lipid signaling from wild-type components. Finally, availability of well-characterized inhibitors empower study of PITPs and phosphoinositide signaling in systems for which genetic approaches are not available.
Acknowledgments
This work was supported by the Robert A. Welch Foundation and National Institutes of Health grant RO1 GM44530 to VAB.
References
- 1.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McLaughlin S, Murray D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438:605–611. doi: 10.1038/nature04398. [DOI] [PubMed] [Google Scholar]
- 4.Odom AR, Stahlberg A, Wente SR, York JD. A role for nuclear inositol 1,4,5-trisphosphate kinase in transcriptional control. Science. 2000;287:2026–2029. doi: 10.1126/science.287.5460.2026. [DOI] [PubMed] [Google Scholar]
- 5.Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr. Opin. Cell Biol. 2010;22:365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Berridge MJ, Irvine RF. Inositol trisphosphate, a novel second messenger in cellular signal transduction. Nature. 1984;312:315–321. doi: 10.1038/312315a0. [DOI] [PubMed] [Google Scholar]
- 7.Nishizuka Y. The role of protein kinase C in cell surface signal transduction and tumour promotion. Nature. 1984;308:693–698. doi: 10.1038/308693a0. [DOI] [PubMed] [Google Scholar]
- 8.Michell RH. Inositol phospholipids and cell surface receptor function. Biochim. Biophys. Acta. 1975;415:81–147. doi: 10.1016/0304-4157(75)90017-9. [DOI] [PubMed] [Google Scholar]
- 9.Cockcroft S, and NCarvou. Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta. 2007;1771:677–691. doi: 10.1016/j.bbalip.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Schaaf G, Ortlund E, Tyeryar K, Mousley C, Ile K, Woolls M, Garrett T, Raetz CRH, Redinbo M, Bankaitis VA. The functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the Sec14-superfamily. Molecular Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankaitis VA, Mousley CJ, Schaaf G. Sec14-superfamily proteins and the crosstalk between lipid signaling and membrane trafficking. Trends in Biochemical Sciences. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sha B, Phillips SE, Bankaitis VA, Luo M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein Sec14p. Nature. 1998;391:506–510. doi: 10.1038/35179. [DOI] [PubMed] [Google Scholar]
- 13.Ryan MM, Temple BRS, Phillips SE, Bankaitis VA. Conformational dynamics of the major yeast phosphatidylinositol transfer protein Sec14p: Insights into the mechanisms of phospholipid exchange and diseases of Sec14p-like protein deficiencies. Mol. Biol. Cell. 2007;18:1928–1942. doi: 10.1091/mbc.E06-11-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaaf G, Dynowski M, Mousley CJ, Shah SD, Yuan P, Winklbauer E, de Campos MKF, Trettin K, Quinones M-C, Smirnova T, Yanagisawa LL, Ortlund E, Bankaitis VA. Resurrection of a functional phosphatidylinositol transfer protein from a pseudo-Sec14 scaffold by directed evolution. Mol. Biol. Cell. 2011;22:892–905. doi: 10.1091/mbc.E10-11-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smirnova T, Chadwick TG, van Tol J, Ozarowski A, Poluektov O, Schaaf G, Ryan MM, Bankaitis VA. Local polarity and hydrogen bonding inside the Sec14p phospholipid-binding cavity: High-field multifrequency electron paramagnetic studies. Biophys. J. 2007;92:3686–3695. doi: 10.1529/biophysj.106.097899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleves AE, Novick PJ, Bankaitis VA. Mutations in the SAC1 gene suppress defects in yeast Golgi and yeast actin function. J. Cell Biol. 1989;109:2939–2950. doi: 10.1083/jcb.109.6.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- 18.Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MKY, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, Rivas MP, Fang M, Marchena J, Mehotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mousley C, Yuan P, Gaur NA, Trettin KD, Nile AH, Deminoff S, Dewar BJ, Wolpert M, Macdonald JM, Herman PK, Hinnebusch AG, Bankaitis VA. A sterol binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell. 2012;148:702–715. doi: 10.1016/j.cell.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nile AH, Bankaitis VA, Grabon A. Mammalian diseases of phosphatidylinositol transfer proteins and their homologs. Clinical Lipidology. 2010;5:867–897. doi: 10.2217/clp.10.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kono N, Ohto U, Hiramatsu T, Urabe M, Uchida Y, Satow Y, Arai H. Impaired a-TTP-PIPs interaction underlies familial vitamin E deficiency. Science. 2013;340:1106–1110. doi: 10.1126/science.1233508. [DOI] [PubMed] [Google Scholar]
- 24.Ren J, Lin CP-C, Pathak M, Temple BRS, Nile AH, Mousley CJ, Duncan MC, Eckert D, Leiker TJ, Ivanova PT, Milne DS, Murphy RS, Brown HA, Verdaasdonk J, Bloom KS, Ortlund EA, Neiman AM, Bankaitis VA. A phosphatidylinositol transfer protein integrates phosphoinositide signaling with lipid droplet metabolism to regulate a developmental program of nutrient stress-induced membrane biogenesis. Molecular Biology of the Cell. 2014;25:712–727. doi: 10.1091/mbc.E13-11-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riekhof WR, Wu WI, Jones JL, Nikrad M, Chan MM, Loewen CJR. An assembly of proteins and lipid domains regulates transport of phosphatidylserine to phosphatidylserine decarboxylase 2 in Saccharomyces cerevisiae. Journal of Biological Chemistry. 2014;289:5809–5819. doi: 10.1074/jbc.M113.518217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skinner HB, Alb JG, Whitters EA, Helmkamp GM, Jr, Bankaitis VA. Phospholipid transfer activity is relevant to but not sufficient for the essential function of the yeast SEC14 gene product. EMBO J. 1993;12:4775–4784. doi: 10.1002/j.1460-2075.1993.tb06166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ile KE, Kassen S, Cao C, Vihtehlic T, Shah SD, Huijbregts RPH, Alb JG, Jr, Stearns GW, Brockerhoff SE, Hyde DR, Bankaitis VA. The zebrafish class 1 phosphatidylinositol transfer protein family: PITP**isoforms and double cone cell outer segment integrity in retina. Traffic. 2010;11:1151–1167. doi: 10.1111/j.1600-0854.2010.01085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nile AH, Tripathi A, Yuan P, Mousley CJ, Suresh S, Wallace IM, Shah SD, Teiotico-Pohlhaus D, Temple B, Nislow C, Giaever G, Tropsha A, Davis RW, St.Onge RP, Bankaitis VA. PITPs as targets for selectively interfering with phosphoinositide signaling in cells. Nature Chemical Biology. 2014;10:76–84. doi: 10.1038/nchembio.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee AY, St.Onge RP, Proctor MJ, Wallace IM, Nile AH, Spagnuolo PA, Jitkova Y, Gronda M, Wu Y, Kim MK, Cheung-Ong K, Torres NP, Spear ED, Han MK, Schlecht U, Suresh S, Duby G, Heisler LE, Surendra A, Fung E, Urbanus ML, Gebbia M, Lissina E, Miranda M, Chiang JH, Aparicio AM, Zeghouf M, Davis RW, Cherfils J, Boutry M, Kaiser CA, Cummins CL, Trimble WS, Brown GW, Schimmer AD, Bankaitis VA, Nislow C, Bader GD, Giaever G. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science. 2014;344:208–211. doi: 10.1126/science.1250217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bankaitis VA, Grabon A. Phosphatidylinositol synthase and diacylglycerol platforms bust a move. Developmental Cell. 2011;21:810–812. doi: 10.1016/j.devcel.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]