Abstract
Recent studies have demonstrated distinctive motility and responses to extracellular cues of cells in isolation, cells collectively in groups, and cell fragments. Here we provide a protocol for generating cell sheets, isolated cells, and cell fragments of keratocytes from zebrafish scales. The protocol starts with a comprehensive fish preparation, followed by critical steps for scale processing and subsequent cell sheet generation, single cell isolation, and cell fragment induction, which can be accomplished in just 3 days including a 36–48 h incubation time. Compared to other approaches that usually produce single cells only or together with either fragments or cell groups, this facile and reliable methodology allows generation of all three motile forms simultaneously. With the powerful genetics in zebrafish our model system offers a useful tool for comparison of the mechanisms by which cell sheets, single cells, and cell fragments respond to extracellular stimuli.
Keywords: Cell migration, Collective cell migration, Cell fragment, Electric fields, Galvanotaxis, Electrotaxis, Zebrafish
1 Introduction
Cell migration is important in embryonic development, wound healing, and tumor metastasis [1–3], and is a critical process during immune response [4]. Cell motility may be roughly categorized in three forms scaled from cell fragments, as evidenced by cytokine-plasts penetrating interendothelial cell junctions in response to a chemoattractant [5], to single cells (cells in isolation), to cohesive cell groups and cell sheets.
The migration of both single cells and groups of cells has been extensively studied in tissue culture and are known to contribute to many physiological motility processes in vivo during embryogenesis and wound healing [6, 7]. Similar migratory behavior is also displayed by many invasive tumor types [8, 9].
Compared to single cells and cell sheets, migration of cell fragments has been under investigated. In vivo, viable cell fragments that pinch off directly from the cell body have been characterized and linked to various disease states [10, 11]. These anucleated fragments remain functional and migratory and can survive for hours or even days. For instance, fragments can function as inter-cellular “ferries” to transfer bioactive molecules [12], as marker “beacons” for tumor cells to navigate microenvironments [13] and as phagocytic “defenders” to ingest and kill bacteria [14].
Evaluating models of directional cell migration can provide more comprehensive insight into the complex mechanisms and signaling pathways involved in cellular motility [15]. Although studies of single cell migration, some in conjunction with either cell fragments or cell groups, have been reported [16, 17], a system containing all three motile units has not been reported before. A detailed comparison of the cells, and cell sheets or cell fragments, and their applications of spontaneous and directed migration are summarized in Table 1.
Table 1.
Comparison of cell types and their use in generating single cells, cell sheets, or cell fragment for migration study
| Study | Cell type | System of | Purpose | ||
|---|---|---|---|---|---|
| Cell sheets | Single cells | Cell fragments | |||
| Albrecht-Buehler [32] | Human skin fibroblast | No | No | Yes | Spontaneous migration |
| Malawista et al. [33] | Human PMN | No | No | Yes | Chemotaxis |
| Euteneuer et al. [16] | Fish keratocyte | No | Yes | Yes | Spontaneous migration |
| Cooper et al. [34] | Fish keratocyte | Yes | Yes | No | Electrotaxis |
| Verkhovsky et al. [27] | Fish keratocyte | No | Yes | Yes | Spontaneous and directed migration |
| Yount et al. [13] | Human glioblastoma | No | Yes | Yes | Spontaneous migration |
| Li et al. [35] | Many different kinds | Yes | Yes | No | Electrotaxis |
| Sun et al. [25] | Fish keratocyte | No | Yes | Yes | Electrotaxis |
| Cohen et al. [36] | HDCK | Yes | No | No | Electrotaxis |
Fish keratocytes provide an excellent locomotion model for better understanding polarization and migration [18–20]. An added advantage to using fish keratocytes from the zebrafish species [21, 22] is the availability of powerful molecular tools for genetic manipulation. Keratocyte motility is dependent on a synergistic actin treadmill of the self-organizing lamellipodia, which combines the protrusion of growing actin networks in the front and the retraction of the actomyosin contractile network in the rear. As a result, these cells move persistently with steady speeds, shape, and behavior and are thus ideal for in-depth analysis of directional response to extracellular cues, such as electric field [23, 24]. Interestingly, portions of keratocytes can spontaneously detach and form cytoplasmic fragments that lack major cell organelles and microtubules [16], yet still exhibit migrational movement similar to that of their whole cell counterparts. Thus, these keratocyte fragments represent an even simpler model of cell motility and are suitable for experimental cell migration studies [25]. In contrast to single cells or cell fragments, the collective response of cohorts of cells or cell sheets is much more complicated. Collective cell migration requires that all cells or otherwise selected cells sense a guidance cue and interpret it individually and/or cooperatively [26].
Here we describe an optimized protocol for preparing a unique system containing cell sheets, isolated cells, and cell fragments for use in spontaneous and directed migration studies. The protocol relies on the tissue culture of fish scales, and contains detailed step- by-step procedures for cell sheet generation, single cell isolation, and cell fragment induction. The quantitative analysis of these three motile units as a model system enables better insight into the cellular and molecular mechanisms that contribute to the intricate and coherent steps of polarization and directional migration as demonstrated in electrotaxis.
1.1 Overview of the Procedure
Essentially, the procedure described here consists of two parts: (1) isolating fish scales, (2) generating epidermal keratocyte sheets, isolating single keratocytes, and inducing cytoplasmic fragments.
An outline of our protocol is shown in Fig. 1. A detailed illustration of scale preparation and seeding to generate the three motile units of fish keratocytes is shown in Fig. 2. The morphology and characteristics of epithelial cell sheets, isolated cells, and cell fragments are provided in Fig. 3. Quantification of spontaneous migration of these three motile units and applications of these three motile units in electrotaxis are displayed in Figs. 4 and 5.
Fig. 1.
Schematic outline of production of cell sheets, isolating keratocytes, and cell fragments and examples of application. Fish scales produce keratocyte cell sheets, cells in isolation, and cell fragments. All maintain good motility, and respond to small applied electric fields by directional migration (electrotaxis/galvanotaxis). This provides a unique system to study molecular and genetic control of cell movement in collection and in isolation, and a powerful tool to study directional migration and the mechanisms
Fig. 2.
Critical steps to generate three motile units. (a) Autoclaved surgical tweezers, stainless steel nuts, 22 × 22 mm coverslips and a disposable 6-well tissue culture plate. (b ) Thorough washes with plenty distilled water are key to minimize bacterial or fungal contamination in subsequent steps. (c) An anesthetized zebrafish in an operation petri dish. (d) Pulling a scale from fish flank with sterilized surgical forceps. The exposed fish flank was sterilized with alcohol prep pads. (e) Scales are washed three times with complete culture medium containing antibiotics and antimycotics. (f) Evenly spread scales in a well of 6-well tissue culture plate. To prevent drying out of the scales, 20 μl of medium was pre-dropped in the center of the culture area. (g) A sterile 22 × 22 mm coverslip is laid over the wet scales. Care was taken to avoid bubble trapped between the coverslip and culture dish. (h) An autoclaved stainless steel nut is placed on the coverslip to hold scales in position. Culture medium (1.5 ml) is added to seal coverslip edges. (i) The whole plate is placed inside an incubator at room temperature. Sheets of keratocytes migrate off scales normally within 36 h
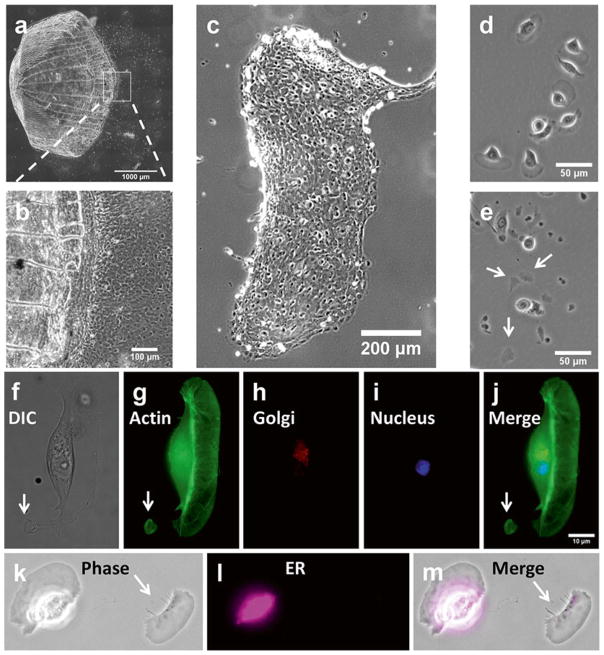
Fig. 3.
Micrographs of cell sheets, isolated keratocytes, and cell fragments. (a, b ) Sheets of epidermal keratocyte migrate out of fish scale. (c) A typical epidermal keratocyte sheet. (d) Isolated keratocytes. Most of the keratocytes have a typical canoe appearance. (e) Induced cell fragments (white arrows). (f–j) Cell fragment (white arrow ) lacked nucleus (blue ) as revealed by DAPI staining, and of Golgi body (red ), as revealed by antibody staining. Actin (green) as revealed by FITC-phalloidin presents in both keratocyte and fragment. (k–m ) Compared to the whole cell, a cell fragment (indicated by white arrow ) contains much less ER (cyan) as revealed by antibody staining (modified from [24])
Fig. 4.
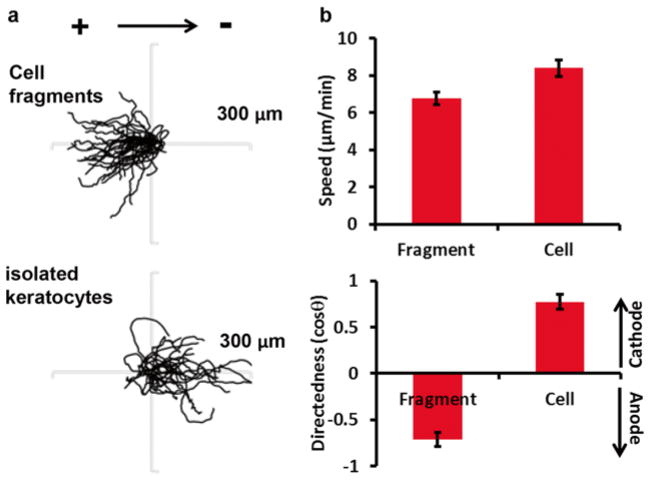
Migration of cell sheets, single cells, and cell fragments. Single cells and cell fragments migrate much fast, whereas cells in sheets showed very little movement. (a) Migration trajectories of cell fragments, cells in isolation, and cells in sheets. Duration: 30 min. Note the scales are different. (b) Quantification of migration
Fig. 5.
Contrasting difference in directional migration in response to electrical cues. (a) Migration trajectories of cell fragment s , single cells (cells in sheet are not shown) in the presence of an electric field with polarity as shown. Duration: 30 min. Note the scales are different. (b) Quantification of migration speed and directionality. Upon electric field application single cells migrate toward cathode. Note the cell fragments migrate in the opposite direction of their parental cells to anode. Cells in sheet are not shown
Fish preparation is critical for scale processing and isolation. Beakers and containers are pre-autoclaved. Fish net is sterilized by 70 % ethanol spray. Water needs to be at room temperature although autoclaving is not necessary. Tap water contains chlorine or chloramines, which can stress and possibly kill the fish. It is important to always use dechlorinated water, which can be achieved by exposing a container of water to air for at least 24 h or by running the water through a carbon filter. The fish must be washed thoroughly to avoid any bacterial or fungal contamination. Care should be taken when handling the fish to minimize stress and harm. Scales are taken from the fish flanks and laid in culture dish, covered with a 22 × 22 mm glass coverslip. A sterilized stainless steel hex nut is placed on top to hold the scales in place. Detailed illustration of these critical steps is shown in Fig. 2.
Epithelial cells are typically packed together with very little intercellular material between them. An extremely tight bond exists between adjacent cells such that dissociation of epithelium is a difficult process. Methods for isolating primary culture cells have been extensively developed. The culture systems presently used in most laboratories are based on mechanical disaggregation and/or enzymatic digestion of animal tissue into single cells. Currently, several commercialized products such as Gibco cell culture systems (Life Technologies), cell isolation optimizing systems (Worthington Biochemical Corporation), or PrimaCell (Chi Scientific) are available. However, these techniques are time-consuming, require optimization depending on tissue type and the procedures are varied in different laboratories. The technique we developed for generating epithelial cell sheets, isolating keratocytes, and inducing cytoplasmic fragments from the same fish scale samples requires neither mechanical tissue dissociation nor enzymatic digestion.
Traditional cell digestion with excessive volumes of trypsin–EDTA, followed by wash and centrifugation, typically in 15 ml Falcon tubes, may not be efficient for small amount of cells. In addition, inexperienced researchers may face technical difficulties when working with small cell pellets, thereby resulting in variable cell recovery. Thus, the use of small volume on-site trypsinization and subsequent wash and spin in Eppendorf tubes not only simplifies the cell digestion process, but also ensures maximal cell recovery.
The following procedures are based on our own experiences or are otherwise adapted from previous publications [27]. We integrated the procedures into a highly successful pipeline for easy and simultaneous generation of epithelial cell sheets, isolated kerato-cytes, and induced cytoplasmic fragments from the same sample of fish scales in just 1 h. Once the keratocyte sheets migrate off a scale and attach to the tissue culture treated plate surface (usually after 36–48 h incubation), the scale “sandwiches” are disassembled by carefully removing the weighing steel nuts and the coverslips. The cell sheets formed this way can be either directly used in experiment if they are generated over an appropriate carrier (an electrotaxis chamber in our case) or serve as sources for single cell isolation. Cells in six-well plates are washed and trypsinized by the small volume on-site digestion technique detailed in the procedures. In brief, 0.5 ml trypsin–EDTA is added in each well and cell detachment is constantly monitored under microscope. Once cells are completely dislodged, 0.5 ml complete culture medium containing bovine serum is added to halt and prevent over-digestion. The cells are then transferred into Eppendorf tubes for subsequent wash and spin at room temperature. Cell collection using Falcon tubes and centrifugation with cooling are not necessary as recommended. Concentrated single cells are then seeded in experiment carrier (electrotaxis chambers in our case) and cell density is appropriately adjusted by adding more complete culture medium. As soon as cells attach to the chamber surface, they can be directly assayed or further subject to fragment induction. Although cytoplasmic fragments pinch off spontaneously from keratocyte cells, the fastest and most efficient way to generate fragments is to treat with staurosporine, an alkaloid isolated from the bacterium Streptomyces staurosporeus [28]. Fragment production is facilitated by increasing the temperature to 35 °C without causing any cell death (see Supplementary movie S1). After complete removal of residual staurosporine, the resulting cytoplasmic fragments lack nuclei and other major organelles (Fig. 2), yet are still migratory and can be used for many downstream applications and side-by-side comparison with their parental cells.
In summary, there are many significant modifications and improvements compared to those previously reported in the literature. These are as follows: (1) use of zebrafish scales as keratocyte source; (2) wash fish multiple times (at least 5×) in beakers filled with clean room temperature distilled water; (3) use of stainless steel nuts to hold sandwiched scales in place; (4) use of a vacuum system and sterile pipettes to remove scales efficiently without dislodging attached cell sheets; (5) small volume on-site trypsinization and subsequent wash and centrifugation in Eppendorf tubes to minimize cell loss; (6) resuspending isolated cells in small volume and on-site dilution to achieve optimal cell density; (7) adaptation of treatment with staurosporine in culture media to induce fragment formation and subsequent incubation at 35 °C to facilitate fragment formation.
1.2 Advantages and Limitations of the Method
The present protocol has several limitations: (1) This protocol allows simultaneous generation of epithelial cell sheets and fragments from fish keratoctye model, limiting its use in mammalians cells. There are a number of publications that have demonstrated production of cell fragments in mammalian cells. Our attempt to induce cell fragment from neutrophils, human fibroblasts and gli-oma cells were unsuccessful. (2) Depending on species and age, not all fishes have equal ability to produce cell fragments, and their corresponding migratory behaviors may be different. (3) The use of staurosporine to induce fragments raises concerns that the treatment may cause cell stress or worse apoptosis, despite being the faster, more convenient and reliable method. It is recommended to monitor fragment formation under microscope, in addition to incubation at 35 °C to reduce drug exposure time. Nonetheless, thorough washes with excessive culture media to remove residual staurosporine results in negligible effect of apoptosis. (4) Our attempt to collect purified cell fragments alone has been unsuccessful. Hence, our electrotactic assay involving cell fragments are performed using a mixture of keratocyte cells and fragments, in some cases with the parent and fragment still tethered together. However, the latter may prove to be useful as an excellent model to study membrane tension in maintaining and regulating polarity and directional migration as demonstrated by Weiner’s group [29].
Our protocol has several unique advantages as follows: (1) This protocol is practical in that it allows for simultaneous generation of epithelial cell sheets, isolated keratocytes, and cytoplasmic fragments from the same fish scale samples within 2–3 days. (2) The method allows for the possibility of performing reliable integrative investigation of cell migratory behaviors using three different motile models from the same tissue samples (Figs. 4 and 5). (3) Researchers now take full advantage of the unique genetic manipulability of the zebrafish model organism since well-characterized mutant strains are readily available.
1.3 Applications of the Protocol
The protocol described here has been optimized for the generation of three motile units including epithelial cell sheets, single cells, and cell fragments derived from zebrafish scales. Downstream applications can be performed with this unique system by taking advantage of zebrafish as a powerful model organism and can be integrated into many studies such as basal motility, pharmacological perturbation and manipulation of cellular signaling networks, directional migration such as galvanotaxis/electrotaxis and mechanotaxis. We recently applied this approach to gain insights into the control of cell migration under a guidance cue of applied electric field [25]. In this current manuscript, electrotaxis experiment setup is not discussed, as it has been thoroughly described in detail elsewhere [30, 31]. We expect the new model system will facilitate studies on the mechanisms by which cells detect and respond to external signals.
2 Materials
2.1 Reagents
Zebrafish (Danio rerio), adult male and female, aged between 3 and 12 months. The line we used is wild type (AB), which is available from Zebrafish International Resource Center (ZIRC) (see Notes 1 and 2).
Distilled water.
70 % ethanol for sterilization.
Leibovitz’s L-15 medium (Life Technologies).
100× antibiotic–antimycotic (Life Technologies).
Fetal bovine serum (Life Technologies).
1 M HEPES pH 7.4 (Life Technologies).
0.25 % Trypsin–0.02 EDTA solution (Life Technologies).
Dulbecco’s phosphate-buffered saline (Life Technologies).
MS-222 (Sigma-Aldrich).
Sodium bicarbonate.
Dimethyl sulfoxide.
Staurosporine (Sigma-Aldrich).
Sodium chloride.
Potassium chloride.
Calcium chloride.
2.2 Equipment
Fish net.
Beakers, 4 l, plastic (Nalgene).
Solvent-resistant marker (Fisher).
Stainless steel hex nuts (Bolt Depot, size ¾).
No. 1 glass coverslip, size 22 × 22 mm (Corning Life Sciences).
Biohazard waste and sharps disposal container.
Forceps (Dumont FST, no. 5).
Sterile alcohol prep pads.
Pipettes for volumes 0.5–1000 μl.
Pipette filter tips for volumes 0.5–1000 μl.
Sterile plastic Pasteur pipettes (Fisherbrand).
Eppendorf tubes, 1.5 ml (Fisherbrand).
Falcon conical tubes, 15 ml (Corning Life Sciences).
Falcon conical tubes, 50 ml (Corning Life Sciences).
Six-well tissue culture treated plates (Corning Life Sciences).
Tissue culture petri dishes with 10 cm diameter (Corning Life Sciences).
Dissecting microscope.
Culture incubator (e.g., Quincy lab, model 12-140).
Benchtop centrifuges: non-refrigerated and refrigerated (e.g., Eppendorf 5415D and 5415R, respectively).
Autoclave.
Cell culture hood with laminar flow and UV light (The Baker Company, Bio-II-A).
Time-lapse imaging system, ideally with functions of X/Y/Z multiple position recording and multiple wavelength recording, as well as a CO2-supplied temperature control chamber incorporated onto the microscope. We currently use MetaMorph imaging software (Molecular Devices).
2.3 Reagent Setup
Complete culture medium: Leibovitz’s L-15 medium + 10 % FBS + 1× antibiotic–antimycotic + 14.2 mM HEPES pH 7.4. The complete medium is used as both cell culture and electro-taxis running buffer. For convenience, aliquot freshly made medium into 50 ml Falcon tubes and stored at 4 °C. Allow medium to reach room temperature before use.
MS-222 stock solution: Prepare a 0.4 % stock solution in ddH2O. Adjust pH to 7.2 using sodium bicarbonate as needed. Aliquots in 5 ml can be stored at −20 °C for a few months. To make euthanasia buffer thaw one vial of about 5 ml MS222 stock solution and add 15 ml of fish water (end concentration: 1 g/l).
Fish Ringer’s solution: 116 mM NaCl, 2.9 mM KCl, 1.8 mM CaCl2, 5 mM HEPES, pH 7.2. Fish Ringer’s solution can be prepared and sterilized by autoclaving in advance and stored at 4 °C for up to a month.
Staurosporine stock solution: For convenience it is recommended to make a 1 mM stock solution in DMSO and aliquot in tightly sealed vials at−20 °C. Prior to opening the vial allow it to equilibrate to room temperature for at least 20 min.
2.4 Equipment Setup
Preparation for fish sterilization: Clean work bench with 70 % ethanol. Fill an autoclaved plastic bucket with plenty of dechlorinated or distilled water that is enough to wash the fish and fish net as described in Subheading 3. Fill two autoclaved 5-l plastic beakers with distilled water. Make sure the water temperature is between 20 and 25 °C.
Preparation for scale processing: Thaw one vial of about 5 ml MS222 stock solution and mix into ~100 ml clean distilled water to make 200 mg/l buffer for anesthesia. Prepare two disposable tissue culture petri dishes and fill one with 100 ml freshly made 200 mg/l MS-222 in complete culture medium. Autoclave two surgical forceps, six stainless steel hex nuts, and a box of 22 × 22 mm coverslips. Prepare a stack of sterile alcohol prep pads. Make enough electrotaxis chambers using 10 cm tissue culture dishes if you plan to do experiment with cell sheets. Otherwise, label a six-well plate if you plan to isolate single cells and subsequently induce fragments. Drop 20 μl complete culture medium in the center of each chamber or well. Label three Eppendorf tubes on the side with a solvent-resistant marker and fill each tube with 1 ml complete medium.
Preparation for cell sheet generation: Set up a vacuum system. Set up a “sharps” container for properly disposing coverslips.
Preparation for single cell isolation: Set up a vacuum system. Arrange a benchtop centrifuge and a liquid aspiration system. It is recommended to label the tubes on the lid and on the side with a solvent-resistant marker.
Preparation for cell fragment induction: Make enough electrotaxis chambers using 10 cm tissue culture dishes. Turn on an incubator, set temperature to 35 °C, and allow to equilibrate. Set up a liquid aspiration system. Prepare 1 ml 100 nM staurosporine in complete culture medium. It is recommended to label the tubes on the lid and on the side with a solvent-resistant marker.
Liquid aspiration system setup: Connect the end of a 1-l vacuum flask to one end of a 0.45-μm membrane filter using a 20-cm rubber tube (in-house or water assisted). Connect the other end of the membrane filter to a vacuum unit using a rubber tube. Close the vacuum flask with a drilled rubber stopper that has a glass Pasteur pipette inserted in the hole. Connect the Pasteur pipette with a 30-cm rubber tube to another glass Pasteur pipette, which will be used to aspirate liquids using a 200-μl pipette tip.
2.5 Sample Handling Recommendations
Before starting to work with tissue culture, ensure that appropriate amounts of the required medium, buffers and enzymes have been pre-warmed to room temperature. It is recommended to aliquot complete culture medium into small quantities for single use only. Do not leave solutions open if they are not in use.
Like other primary cell cultures, tissue cultures from fish scales are very sensitive to contamination. Clean the area you will process the tissues including pipettes and centrifuges by spraying with 70 % (vol/vol) ethanol.
Always wear disposable gloves and replace them regularly.
Reusable stainless steel nuts, glassware and plasticware should be autoclaved to ensure sterility.
Use sterile instruments, aseptic techniques, and perform work in a laminar flow hoods to maintain sterility.
3 Methods
3.1 Fish Sterilization (Allow 25–30 min per Fish) (See Note 3)
Transfer fish into procedure room according to your institution’s regulations, and let it sit in a holding tank for at least 20 min.
Put a fish in an autoclaved 4-l plastic beaker filled with clean distilled or dechlorinated water and allow the fish to acclimate inside the fish net for 30 s. Water used for washing purposes must be kept at room temperature (20–25 °C) and dechlorinated to avoid stress caused by temperature fluctuations and chlorine.
Use the fish net to carefully transfer fish into another 5-l plastic beaker filled with clean distilled water. Repeat wash steps for total of five times. The fish net should be sanitized with 70 % ethanol and rinsed in clean distilled water before and after each use.
3.2 Scale Processing and Assembling (Allow up to 30 min per Fish)
Pour 100 ml anesthesia buffer containing 200 mg/l MS-222 into a clean petri dish. The 200 mg/l MS-222 buffer in distilled water must be freshly made for effective anesthesia.
Use a pipette to drop 20 μl complete culture medium in the center of each well.
Use a pipette to aliquot of 1 ml complete cell culture medium into three Eppendorf tubes.
Immerse the fish in anesthesia buffer containing 200 mg/l MS-222. During induction make sure the depth of anesthesia is appropriate by closely monitoring gill movement as an indicator.
Transfer anesthetized fish into a new clean sterile petri dish.
Gently hold the fish sideways by pressing its head and tail against the petri dish using your left thumb and middle finger.
Sterilize the fish flank where you plan to take scales by using alcohol prep pads.
With a pair of sterile surgical forceps, gently pull a scale off fish flank from where you just sterilized, and rinse it three times, 1 s each time by sequentially dipping the scale in the three Eppendorf tubes filled with complete cell culture medium (see Note 4).
Carefully lay individual scale into the medium at the center of the well. Use 10-cm tissue culture dishes if you plan to carry out an experiment with cell sheets right after they are formed. Otherwise use six-well tissue culture plates. A drop of complete culture medium helps to prevent the scale from drying out and to spread scales evenly.
Repeat steps 8 and 9 until there are 3–5 scales in each well.
Return fish into clean distilled water to facilitate recovery (see Note 5).
Use the same surgical forceps to spread scales evenly in the middle of each well (see Note 6).
Use new sterile surgical forceps to pick up a clean 22 × 22 mm glass coverslip. Tilt one side into the well such that it touches the medium, then slowly lower it to cover the evenly spread scales without trapping any bubbles in between. Care must be taken to avoid bubble as a gas–liquid interface interferes with keratocyte sheet formation.
Gently lay a sterile stainless steel hex nut on the top of the square coverslip. Make sure no scales float out from beneath the coverslip.
Add 1.5 ml complete culture medium surrounding the nut to immerse the coverslip. Don’t add too much medium. Excessive medium is easy to reach inner side of plate cover and to splash over during transportation, therefore increasing chance of contamination.
Repeat steps 12–15 until all the wells are processed.
Place covered culture plate in an incubator at room temperature and incubate for 36–48 h (see Note 7).
3.3 Cell Sheet Generation (Allow ~30 min)
Take the culture plate containing the scale assemblies from incubator.
Remove the steel nuts using sterile forceps and set them to the side.
Using sterile forceps with a sharp tip, carefully remove the coverslips that cover scales in each well. Some of the coverslips might be tightly attached to culture surface. In that case be patient and try to lift from one side (see Note 8).
Carefully aspirate culture medium, making sure not to disrupt the scales.
Use the vacuum aspiration system and sterile 200 μl pipette tips to remove scales from each well. Use just enough vacuum power to pick up a scale. Wipe off the suctioned scale carefully with alcohol prep pads (see Note 9).
In each well, wash cells twice with 5 ml Fish Ringer’s Solution to remove any debris.
Add 3 ml complete culture medium to each well and locate the cell sheets under microscope. At this point the cell sheets can be kept in the dish and assayed later or used directly in the next step for single cell preparation.
3.4 Isolating Single Cells (Allow ~30 min)
-
1
Aspirate culture medium.
-
2
Wash cells once with PBS.
-
3
Add 0.5 ml 0.25 % trypsin–0.02 EDTA solution in each well. Continuously monitor cell detachment under microscope. Gently tilt well plate to ensure adequate coverage and to speed up digestion. Trypsinization is usually complete within 5 min at room temperature; confirm under microscope.
-
4
Add 0.5 ml complete culture medium to stop further digestion as soon as all cells are detached.
-
5
Transfer cells into 1.5 ml Eppendorf tubes, one well per tube.
-
6
Collect cells by centrifugation at 150 × g for 5 min in a bench-top centrifuge at room temperature.
-
7
Carefully aspirate supernatant using vacuum system without causing significant cell loss (see Note 10).
-
8
Combine cells from all wells into a new 1.5 ml Eppendorf tube, and add 1 ml complete culture medium.
-
9
Repeat steps 7 and 8 to wash and spin cells twice with complete culture medium.
-
10
Re-suspend cells in 0.2 ml complete culture medium. If needed single cell yield can be calculated by counting using a hemocytometer. The isolated cells can be assayed right away or proceed to next step for fragment induction.
3.5 Inducing Cell Fragments (Allow ~1 h 15 min)
-
11
Add 20 μl cell suspension in each experiment carrier (in our case an electrotaxis chamber).
-
12
Check under inverted microscope and add necessary amount of complete culture medium to achieve optimal cell density.
-
13
Let cells adhere to chamber surface at room temperature for 30 min.
-
14
Wash once with complete culture medium to remove unattached cells.
-
15
Add 0.5 ml complete culture medium containing 100 nM staurosporine.
-
16
Place the dish in an incubator with its temperature preset at 35 °C.
-
17
Incubate up to 30 min to induce fragments (see Note 11).
-
18
As soon as ideal fragments are formed, bring dish to room temperature and wash twice with complete culture medium.
-
19
Add enough complete culture medium and let cells and fragments recover for 10 min. Fragments made this way are ready for testing and can survive for a couple of hours (at least 3 h).
3.6 Troubleshooting
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting
| Methods section | Problem | Possible reason | Solution |
|---|---|---|---|
|
Fish sterilization Subheading 3.1, steps 2 and 3 |
Debris appears in the water during fish cleaning | Water temperature is too low or too high | Leave distill water in containers for longer time or measure water temperature and ensure it’s between 22 and 25 °C |
|
Scale processing Subheading 3.2, step 4 |
Fish moves and is difficult to hold | MS-222 is expired Not properly anesthetized |
Make new MS-222 stock solution. The 200 mg/l MS-222 in complete culture medium must be made freshly Return fish back to anesthetic buffer for further induction |
|
Scale processing Subheading 3.2, step 6 |
Scale is difficult to pull or multiple scales are pulled | Scales are too tiny | It fish is small and their scales are too tiny to see manipulation under a dissecting microscope is recommended |
|
Scale assembling Subheading 3.2, step 10 |
Scales are difficulty to spread | Scales are dried out | Add medium to pre-wet surface area of each culture well |
|
Scale assembling
Subheading 3.2, step 11 |
Scales float away when coverslip is laid | Too much medium around | Add just enough medium (20 μl) when pre-wetting the surface area |
|
Scale assembling Subheading 3.2, step 15 |
Bacterial and fungus in the culture dish | Contamination | The most common contamination lies on the shedding by fish. Therefore thorough wash with excessive clean water is the key to prevent bacterial and fungus contamination |
|
Cell sheet generation Subheading 3.3, step 1 |
Cell sheets disappear | Cell sheets adhere to coverslip | Use untreated clean coverslips to minimize undesired adherence. Otherwise, digest/recover cells from coverslips |
|
Isolating single cells Subheading 3.4, step 8 |
Low isolated cell yield | Room temperature is too low Scales are small Bubbles present during scale assembly Significant cell loss during trypsinization and subsequent wash steps |
Increase incubator temperature to 28 °C If this is turned out the case try to use more scales in each assembly. Up to 9 scales can be easily accommodated and covered by each 22 × 22 mm coverslip Make sure the coverslips used are clean and dust free Avoid dislodging cell pellet. Always leave a small amount of solution in the bottom when aspirating supernatant |
|
Inducing fragments Subheading 3.5, step 1 |
No or few cells attach | Bad tissue culture treatment | Change dishes or re-coat culture dishes with fibronectin |
|
Inducing fragments Subheading 3.5, step 15 |
Too many tiny fragments | Overtreatment of staurosporine | Either decrease staurosporine concentration to 50 nM or induce fragment at room temperature. In either case the induction process must be closely monitored under an inverted microscope |
3.7 Anticipated Results
This protocol provides detailed steps for the generation of epithelial cell sheets, single cells, and cell fragments derived from zebrafish keratocytes. The overall procedures, highlights of spontaneous migration and subsequent applications using electrotaxis as an example are summarized in Fig. 1. Figure 2 illustrates the key steps in processing fish scales. Supplementary movie S1 demonstrates formation of cytoplasmic fragment induced by staurosporine. The protocol allows for generation of a unique system of three motile units: a part of cell, a whole cell and a group of cells, from same tissue origin. The morphological characteristics of these three units are shown in Fig. 3. Also in Fig. 3, we confirm that the resulting cytoplasmic fragments lack nuclei and Golgi body, and contains little or no endoplasmic reticulum (ER) as we revealed previously [25].
The model system enabled us to yield some new insights that were not possible using previous model systems. First we compared the basal motility of the cell sheets, single cells, and cell fragments. Under no obvious directed cues cells in isolation migrate fast in a speed about 8 μm/min. While cell fragments migrate a little slower (6.76±0.49 μm/min) the cells in sheets are much less migratory (1.01±0.04 μm/min). Figure 4 shows the distinctive difference of spontaneous migration of the three motile units.
We also applied this protocol to investigate the migratory behaviors of cell fragments as well as cohesive cell sheets in comparison with isolated keratocytes under electrical stimulation. We found that the fragments, devoid of nuclei and major organelles, are still capable of sensing electrical field signals. The fragment, therefore, has the EF sensing mechanism and relevant signal transduction pathways leading to migration. Surprisingly, we observe that under exogenous EF guidance, fragments migrate in opposite directions compared to their intact mother cells (see Supplementary movies S2 and S3) [25].
The collective migration of keratocytes in cell sheets is also an interesting phenomenon. Compared to both isolated single cells and cell fragments, cell sheets are less migratory. However, upon electric field application, the cells in sheets immediately respond and migrate, resulting in a significant increase in migration speed (1.99±0.05 μm/min), nearly doubled compared to no EF control (1.01±0.04 μm/min), and in directionality (mean cosθ=0.98) that is much improved compared to that of single cells (mean cosθ=0.77) (see Supplementary movie S4). The quantification of the electrotactic migratory behaviors of the three motile units is summarized in Fig. 5.
There are many additional potential downstream applications. These include but not limited to pharmacological perturbation and manipulation of cellular signaling networks, especially when combined with the powerful zebrafish genetic tools, as well as other directed migration such as mechanotaxis. Our protocol provides a valuable system for investigating cell migratory behaviors and may help in dissecting mechanisms of cells in response to external signals.
Supplementary Material
Acknowledgments
We thank Y. Li for help with photography, B. Reid and T. Pfluger for critical reading of the manuscript, and K. Nakajima and Y. Shen for technical assistance. Y.H.S. was supported by NIH grant GM068952 to A.M. Work in the laboratory of M.Z. was supported by NIH grant 1R01EY019101 to M.Z., as well as by California Institute of Regenerative Medicine Research grant RB1-01417 and National Science Foundation Grant MCB-0951199 to M.Z. and P. Devreotes, whose support we gratefully acknowledge.
Footnotes
All experiments that use animal tissues should comply with all relevant institutional and governmental guidelines and regulations. Animal protocol must be developed and approved by relevant institute committees/authorities and be active when such experiments is implemented.
Animal tissues may contain human pathogens including viruses, bacterium and fungus, which may infect the researcher. Therefore, the use of protective equipment such as gloves, a lab coat, and goggles is recommended.
You must gently handle the fish throughout the procedure as detailed in sample handling recommendations in the instruction section to avoid stressing the animal.
Forceps are very sharp and there is a risk of laceration when using them, which is a potential source of infection by viruses.
After the recovery (usually about 10 min) fish will be returned back to standing water tank and can be reused for future experiment. In case of any sign of suffering immediately immerse fish in euthanasia buffer (distilled water containing 1 g/l MS222). Fish should be dead within minutes and must be disposed properly.
Although up to 9 scales can be easily accommodated in a well too many scales cause stack-over problem. Therefore, 3–5 scales in one well are recommended.
Occasionally, we see cell sheet formation after overnight incubation. However, incubation for 36–48 h gives us the most stable and maximal cell production. Increased incubation time does not produce more cells but increases chances of contamination.
Try to avoid scale shifting when lifting coverslips. Shifting scale may destroy a cell sheet where it migrated from. Dispose used coverslips in a sharps container to avoid potential injury.
Try to avoid scale shifting when picking up scales. Shifting scale may destroy a cell sheet where it migrated from.
Cell pellet could be very tiny. Try not to touch cell pellet when removing supernatant in this step and subsequent wash steps.
Fragmentation process can be monitored on a heated stage under inverted microscope. Do not leave the dish at 35 °C for extended periods of time. Overtreatment in staurosporine at high temperature produces many tiny fragments with decreased mobility and viability, compromising downstream applications such as electrotaxis and other motility assays.
References
- 1.Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol. 2010;341:20–33. doi: 10.1016/j.ydbio.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 3.Calve S, Simon HG. Biochemical and mechanical environment cooperatively regulate skeletal muscle regeneration. FASEB J. 2012;26:2538–2545. doi: 10.1096/fj.11-200162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luster AD, Alon R, von Andrian UH. Immune cell migration in inflammation: present and future therapeutic targets. Nat Immunol. 2005;6:1182–1190. doi: 10.1038/ni1275. [DOI] [PubMed] [Google Scholar]
- 5.Malawista SE, Smith EO, Seibyl JP. Cryopreservable neutrophil surrogates: granule-poor, motile cytoplasts from polymor-phonuclear leukocytes home to inflammatory lesions in vivo. Cell Motil Cytoskeleton. 2006;63:254–257. doi: 10.1002/cm.20120. [DOI] [PubMed] [Google Scholar]
- 6.Locascio A, Nieto MA. Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration. Curr Opin Genet Dev. 2001;11:464–469. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- 7.Keller R. Cell migration during gastrulation. Curr Opin Cell Biol. 2005;17:533–541. doi: 10.1016/j.ceb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr Opin Cell Biol. 2005;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 9.Wang W, Goswami S, Sahai E, et al. Tumor cells caught in the act of invading: their strategy for enhanced cell motility. Trends Cell Biol. 2005;15:138–145. doi: 10.1016/j.tcb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Chironi GN, Boulanger CM, Simon A, et al. Endothelial microparticles in diseases. Cell Tissue Res. 2009;335:143–151. doi: 10.1007/s00441-008-0710-9. [DOI] [PubMed] [Google Scholar]
- 11.Markiewicz M, Richard E, Marks N, Ludwicka-Bradley A. Impact of endothelial microparticles on coagulation, inflammation, and angiogenesis in age-related vascular diseases. J Aging Res. 2013;2013:734509. doi: 10.1155/2013/734509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barry OP, Praticò D, Savani RC, FitzGerald GA. Modulationofmonocyte-endothelial cell interactions by platelet microparticles. J Clin Invest. 1998;102:136–144. doi: 10.1172/JCI2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yount G, Taft RJ, Luu T, et al. Independent motile microplast formation correlates with glioma cell invasiveness. J Neurooncol. 2007;81:113–121. doi: 10.1007/s11060-006-9211-4. [DOI] [PubMed] [Google Scholar]
- 14.Malawista SE, Van Blaricom G. Cytoplasts made from human blood polymor-phonuclear leukocytes with or without heat: preservation of both motile function and respiratory burst oxidase activity. Proc Natl Acad Sci U S A. 1987;84:454–458. doi: 10.1073/pnas.84.2.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mogilner A, Allard J, Wollman R. Cell polarity: quantitative modeling as a tool in cell biology. Science. 2012;336:175–179. doi: 10.1126/science.1216380. [DOI] [PubMed] [Google Scholar]
- 16.Euteneuer U, Schliwa M. Persistent, directional motility of cells and cytoplasmic fragments in the absence of microtubules. Nature. 1984;310:58–61. doi: 10.1038/310058a0. [DOI] [PubMed] [Google Scholar]
- 17.Zouani OF, Gocheva V, Durrieu MC. Membrane nanowaves in single and collective cell migration. PLoS One. 2014;9:e97855. doi: 10.1371/journal.pone.0097855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radice GP. Locomotion and cell-substratum contacts of Xenopus epidermal cells in vitro and in situ. J Cell Sci. 1980;44:201–223. doi: 10.1242/jcs.44.1.201. [DOI] [PubMed] [Google Scholar]
- 19.Keren K, Pincus Z, Allen GM, et al. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Ishihara A, Theriot JA, Jacobson K. Principles of locomotion for simple-shaped cells. Nature. 1993;362:167–171. doi: 10.1038/362167a0. [DOI] [PubMed] [Google Scholar]
- 21.Howe DG, Bradford YM, Conlin T, et al. ZFIN, the Zebrafish Model Organism Database: increased support for mutants and transgenics. Nucleic Acids Res. 2013;41:D854–D860. doi: 10.1093/nar/gks938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sprague J, Bayraktaroglu L, Bradford Y, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Cormie P, Messerli MA, Robinson KR. The involvement of Ca2+ and integrins in directional responses of zebrafish keratocytes to electric fields. J Cell Physiol. 2009;219:162–172. doi: 10.1002/jcp.21660. [DOI] [PubMed] [Google Scholar]
- 24.Allen GM, Mogilner A, Theriot JA. Electrophoresis of cellular membrane components creates the directional cue guiding keratocyte galvanotaxis. Curr Biol. 2013;23:560–568. doi: 10.1016/j.cub.2013.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y, Do H, Gao J, et al. Keratocyte fragments and cells utilize competing pathways to move in opposite directions in an electric field. Curr Biol. 2013;23:569–574. doi: 10.1016/j.cub.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weijer CJ. Collective cell migration in development. J Cell Sci. 2009;122:3215–3223. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- 27.Verkhovsky AB, Svitkina TM, Borisy GG. Self-polarization and directional motility of cytoplasm. Curr Biol. 1999;9:11–20. doi: 10.1016/s0960-9822(99)80042-6. [DOI] [PubMed] [Google Scholar]
- 28.Omura S, Iwai Y, Hirano A, et al. A new alkaloid AM-2282 of Streptomyces origin. Taxonomy, fermentation, isolation and preliminary characterization. J Antibiot (Tokyo) 1977;30:275–282. doi: 10.7164/antibiotics.30.275. [DOI] [PubMed] [Google Scholar]
- 29.Houk AR, Jilkine A, Mejean CO, et al. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song B, Gu Y, Pu J, et al. Application of direct current electric fields to cells and tissues in vitro and modulation of wound electric field in vivo. Nat Protoc. 2007;2:1479–1489. doi: 10.1038/nprot.2007.205. [DOI] [PubMed] [Google Scholar]
- 31.Zhao M, Song B, Pu J, et al. Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN. Nature. 2006;442:457–460. doi: 10.1038/nature04925. [DOI] [PubMed] [Google Scholar]
- 32.Albrecht-Buehler G. Autonomous movements of cytoplasmic fragments. Proc Natl Acad Sci U S A. 1980;77:6639–6643. doi: 10.1073/pnas.77.11.6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malawista SE, De Boisfleury Chevance A. The cytokineplast: purified, stable, and functional motile machinery from human blood polymorphonuclear leukocytes. J Cell Biol. 1982;95:960–973. doi: 10.1083/jcb.95.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cooper MS, Schliwa M. Motility of cultured fish epidermal cells in the presence and absence of direct current electric fields. J Cell Biol. 1986;102:1384–1399. doi: 10.1083/jcb.102.4.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L, Hartley R, Reiss B, et al. E-cadherin plays an essential role in collective directional migration of large epithelial sheets. Cell Mol Life Sci. 2012;69:2779–2789. doi: 10.1007/s00018-012-0951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cohen DJ, Nelson WJ, Maharbiz MM. Galvanotactic control of collective cell migration in epithelial monolayers. Nat Mater. 2014;13:409–417. doi: 10.1038/nmat3891. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.