Abstract
Adoptive cellular immunotherapy is a promising and powerful method for the treatment of a broad range of malignant and infectious diseases. Although the concept of cellular immunotherapy was originally proposed in the 1990s, it has not seen successful clinical application until recent years. Despite significant progress in creating engineered receptors against both malignant and viral epitopes, no efficient preclinical animal models exist for rapidly testing and directly comparing these engineered receptors. The use of matured human T cells in mice usually leads to graft-versus-host disease (GvHD), which severely limits the effectiveness of such studies. Alternatively, adult apheresis CD34+ cells engraft in neonatal non-obese diabetic (NOD)-severe combined immunodeficiency (SCID)-common γ chain–/– (NSG) mice and lead to the development of CD3+ T cells in peripheral circulation. We demonstrate that these in vivo murine-matured autologous CD3+ T cells from humans (MATCH) can be collected from the mice, engineered with lentiviral vectors, reinfused into the mice, and detected in multiple lymphoid compartments at stable levels over 50 days after injection. Unlike autologous CD3+ cells collected from human donors, these MATCH mice did not exhibit GvHD after T cell administration. This novel mouse model offers the opportunity to screen different immunotherapy-based treatments in a preclinical setting.
Keywords: Immunotherapy, chimeric antigen receptor, CAR, mouse model, adoptive cellular immunotherapy, ACI, NSG, T cell therapy, HIV, MATCH mice
Introduction
Advances in the understanding of the adaptive immune system and its signaling pathways have led to the development of gene-based therapies for a variety of diseases including cancer and chronic infections. The field of adoptive cellular immunotherapy (ACI) revolves around engineering specific cytotoxic immune responses against desired epitopes using synthetic receptor molecules.1, 2 This method supplants the normal pathways involved in T cell development where cells undergo sequential genetic rearrangements to create a functional T cell receptor (TCR). Instead of allowing the immune system to find a specific TCR against an antigen by happenchance, ACI is designed to “teach” the immune system to preferentially target desired antigens. This approach also circumvents the negative selection process during T cell maturation,3 enabling self-antigens to be targeted, an essential process for immunotherapy treatment of cancers. Typically, these engineered chimeric antigen receptors (CARs) do not utilize the standard major histocompatibility complex (MHC) signaling interactions and pathways,4 allowing them to recognize and bind intact molecules on cell surfaces with high affinity.5 Specifically engineering the immune system to eliminate cells based on desired criteria presents new possibilities for the treatment of a wide range of genetic or infectious diseases.
Currently, there are numerous clinical trials using ACI to treat cancers, including lymphomas, leukemias, and myelomas.6, 7 These trials use an array of engineered receptors containing different combinations of transmembrane domains, hinge regions, stimulators, and co-stimulatory molecules, and also differ in their ex vivo stimulation and expansion protocols. Additionally, recent CARs designed against conserved viral epitopes on the envelope glycoprotein of HIV have shown efficacy both in vitro and in vivo for eliminating virally infected cells.8, 9, 10 This strategy offers a new cellular-based treatment tool for HIV-infected individuals who remain infected despite effective anti-retroviral therapy (ART).11, 12 T cell-based immunotherapy approaches could augment current treatment regimens designed to reduce, and potentially eliminate, the latently infected reservoirs, while providing a long-term strategy to limit viral rebound.
Current T cell-based gene therapy approaches require the collection of a large number of white blood cells that are subsequently enriched for target cell populations. Cells are cultured and expanded through receptor stimulation methods such as CD3/CD28 beads13 or soluble antibodies,14 then exposed to gene-modifying constructs. After expansion and transduction, protocols differ in culture durations, ranging anywhere from 5 days to several weeks, after which the cells are collected and reinfused. A benefit of this process is the use of autologous cells, which limits potential graft-versus-host disease (GvHD) or rejection. While this therapy holds promise for treatment of diseases such as cancer and HIV, all new engineered receptors must be thoroughly tested in both in vitro cultures and relevant in vivo preclinical animal models to ensure their efficacy and safety.
After the first in vivo testing more than two decades ago,15 the only small-animal models being used to validate new engineered receptors are xenotransplantation experiments of modified human T cells into immunodeficient mice. Some models use immunocompetent mice by engineering the murine T cells,16, 17, 18 but these require adaptation of the receptors to function with mouse signaling pathways and prevent efficient comparison with the human immune system. The activity of the modified cells is then assessed by their ability to reduce or eliminate the target cell population. Several other mouse transplantation studies have demonstrated the potent effect of modified cells in this xenotransplantation setting.19, 20, 21 However, the infused T cells also frequently illicit a GvHD response in the mice,22, 23 resulting in death anywhere between 14 and 60 days, thus limiting the possibility for longer-term follow-up studies and potentially confounding results due to the concurrent immune responses against both mouse and the desired target epitopes. Potentially because of these factors, studies have often failed to find correlative data for the performance of engineered receptors during in vivo animal models and their clinical outcome in human patients.24, 25
Here, we present a novel small-animal model of autologous cell-based ACI using CD3+ T cells that eliminates GvHD symptoms observed in other models by using in vivo murine-matured autologous CD3+ T cells from humans (MATCH) after transplantation of adult human apheresis hematopoietic stem and progenitor cells (HSPCs) into immunodeficient mice. This model replicates current clinical methods for ACI and provides an essential platform for testing and comparing both the efficacy and the safety of newly engineered receptors in a relevant preclinical animal model.
Results
Adult Apheresis CD34+ HSPCs Engraft Neonatal NSG Mice and Develop into CD3+ T Cells
Mobilized adult human peripheral blood CD34+ HSPCs were collected from different donors (n = 4) and engrafted into neonatal non-obese diabetic (NOD)-severe combined immunodeficiency (SCID)-common γ chain–/– (NSG) mice. A summary of donor CD34+ cell information and the number of animals successfully engrafted in this study is included (Table S1). Cells were engrafted into five different cohorts of mice, with each mouse receiving 1 × 106 CD34+ cells. Peripheral blood was collected every other week starting at 8 weeks for tracking engraftment and lineage development (Figure S1). A total of 58 out of 61 injected mice (95%) successfully engrafted, which was defined as a minimum of 1% human CD45+ cells in the peripheral blood during the first three collections. Human cell engraftment consistently averaged 10%–20% in peripheral blood up to 34 weeks post-injection (Figure 1A). During early stages of engraftment (weeks 8–12), CD20+ B cells constitute the majority of circulating human CD45+ cells. At 12 weeks, CD3+ T cells begin to emerge and continue increasing in prevalence until 18 weeks, when they become the majority of peripheral human cells (Figure 1B). After the emergence of CD3+ T cells, we observe both CD4+ and CD8+ cells averaging 50%–70% and 30%–50%, respectively (Figure 1C). This CD4:CD8 ratio averaging 2.3 is similar to that observed in healthy humans. CD14+ monocytes are also consistently detected with decreasing frequency as the mice age. Several cohorts were sacrificed at either 18 or 22 weeks, resulting in fewer data points for later time points.
Figure 1.
Adult Apheresis CD34+ HSPCs Engraft Neonatal Mice and Develop into CD3+ T Cells
Compilation of five unique cohorts of mice engrafted with CD34+ cells from one of four human donors. (A) Peripheral human CD45+ engraftment levels observed in blood over time. Each dot represents an individual mouse with mean represented as black line. (B) Lineage development of human cells in peripheral blood. Different colors represent unique cell lineages with CD3+ T cells in blue, CD20+ B cells in red, and CD14+ monocytes in black. (C) T cell subset development over time within the CD3+ compartment with CD4+ cells in gray and CD8+ cells in orange. All x axes represent age of mice in weeks, which also corresponds to weeks post-transplant due to engraftment as neonates.
After observing appreciable levels of CD4+ T cells in peripheral blood of engrafted mice, we next wanted to determine whether such levels were capable of sustaining HIV infection, which is essential for any immunotherapy-based treatment strategies against HIV. At 18 weeks post-injection, a subset of mice (n = 8) was challenged with the CCR5 tropic laboratory strain HIV-1 BaL (Figure S2). Peripheral blood cell levels were monitored for CD4+ cell depletion as well as serum viral load. We observed a decrease in the percentage of CD4+ cells in the human population, and this was accompanied by a sustained average viral titer ranging between 105 and 107 over 16 weeks of infection. Mice were sacrificed at 34 weeks post-engraftment; blood and spleen were analyzed for CD4+ cell loss, with similar levels of depletion observed. These findings confirm that adult HSPCs are capable of engrafting NSG mice, undergoing multi-lineage hematopoietic development including CD3+ T cells, and sustaining active HIV replication.
Injection of Autologous Mature CD3+ Cells into Engrafted Mice Leads to Rapid GvHD
We next wanted to determine whether engrafted and in-vivo-matured human cells were sufficient to tolerize animals to injection of autologous CD3+ cells and reduce the potential of GvHD. A schematic overview of the procedure is provided in the Supplemental Information (Figure S3). Autologous T cells were obtained in parallel with CD34+ HSPCs from human donors by enriching the CD34− cell product for CD3+ cells. These autologous CD3+ cells were then cryopreserved. Once mice engrafted with autologous HSPCs exhibited development of in-vivo-matured peripheral CD3+ cells at 14–16 weeks (Figures 2A and 2B), cryopreserved CD3+ cells were thawed and stimulated in vitro using CD3/CD28 beads. After bead removal, CD3+ cells were transduced with a lentiviral vector (LV) expressing the GFP for identification and in vivo tracking. After 10–12 days post-bead stimulation (8–10 days post-transduction), cells were collected, analyzed for GFP expression, and injected into mice previously humanized with the CD34+ HSPCs from the matched donor. Mice received various cell doses ranging from 1 × 105 to 1 × 107 cells at 18 weeks post-engraftment. At the time of injection, CD3+ cells were 75.8% positive for GFP expression (Figure S4).
Figure 2.
Injection of Autologous Mature CD3+ Cells into Engrafted Mice Leads to Rapid GvHD
Two cohorts of mice engrafted with adult apheresis CD34+ cells. (A) Mice were tracked over time for the level of human CD45+ cells in peripheral blood. (B) Mice were infused with autologous human CD3+ cells (n = 12) after in vitro transduction with a GFP-expressing lentiviral vector. Black circles represent percent of human CD45+ population in peripheral blood that is CD3+ over time and is plotted on the left y axis with the mean represented by solid black line. Green diamonds represent the percent of the total CD3+ cells that were GFP+ over time in each mouse and are plotted on the right y axis with the mean represented by a solid green line. Vertical green dashed line indicates time of autologous cell infusion for each cohort, and x axis represents age of mice (which also correlates with weeks post-engraftment). (C) Survival curves for the cohort receiving injection of autologous CD3+ cells with x axis representing days post-cell injection. (D and E) The second cohort of mice was engrafted with adult apheresis CD34+ cells (D) and infused with autologous CD4+-enriched cells (E) (n = 7) and graphically represented as previously described. (F) Survival curves for the cohort receiving injection of autologous CD4+ cells with x axis representing days post cell injection. Cross symbols (†) represent the last time point before sacrifice. Mock mice received an injection of PBS alone, without any cell products.
Engineered GFP+ T cells were detected in all mice and ranged from 5% to 46% of all CD3+ cells in peripheral circulation (Figure 2B). Despite the pre-existing human graft, we observed increasing severity of GvHD in a dose-dependent manner. GvHD was assessed through weekly weight measurements of engrafted animals, as well as visually through hunched posture, alopecia, and general systemic hair loss (Figure S5). All mice receiving the two highest cell doses (5 × 106 and 1 × 107) displayed GvHD within 2 weeks, while mice receiving lower doses had GvHD onset between 3 and 6 weeks (Figure 2C). This same trend was also observed in mice receiving an injection of autologous CD4+ T cells after depletion of CD8+ cells in vitro (Figures 2D–2F). The CD4+ cell population was 98.6% pure and 76.3% GFP+ at the time of infusion (Figure S4). These studies clearly indicated that prior engraftment of autologous HSPCs from matched human donors is not sufficient to prevent or mitigate the onset of GvHD in this mouse model.
In Vivo Murine-Matured Human CD3+ Cells as a Source for T Cell-Based Therapies
With the realization that autologous mature CD3+ cells would not serve as a good preclinical model, we hypothesized that human CD3+ cells that matured in vivo from engrafted HSCPs might serve this function (Figure S6). Because these MATCH cells matured in the background of mouse antigen presentation, they might not exhibit the same cytopathic activity that leads to GvHD onset. We first tested the ability of these cells to culture and expand ex vivo after collection. Spleens were harvested from engrafted mice and stimulated for 48 hr using CD3/CD28 beads. At time of collection, 58.2% of spleen cells were human CD45+, with 75% of those being CD3+ T cells. After bead stimulation, cells were transduced with a LV expressing GFP to compare transduction efficiency with mature CD3+ cells from human donors. After culture and transduction, CD3+ cells obtained from different mice (n = 3) that were infused with HSPCs from three different human donors averaged 65.6% GFP+ by flow cytometry as compared with 63.3% from mature human CD3+ cells isolated from apheresis products, indicating these cells are just as easily modified as their human-derived counterparts (Figure 3A). We next tested the expansion potential of stimulated cells collected from mice either in the presence or the absence of transduction. In both human- and murine-derived CD3+ cells following CD3/CD28 bead stimulation and removal, we saw an average 2-fold expansion over 4 days (Figure 3B). This observation is consistent independent of transduction, demonstrating that murine-derived CD3+ cells have a similar expansion potential compared with their human-derived counterpart during this short ex vivo expansion culture. After bead stimulation, upregulation of the early T cell activation marker CD69 was assessed. When compared with unstimulated murine-derived CD3+ cells from the same animal, stimulated cells exhibited a significant increase in cells expressing CD69 (Figure 3C), as well as an increase in mean florescence intensity (MFI) (Figure 3D).
Figure 3.
In Vivo Murine-Matured Human CD3+ Cells as Source for T Cell-Based Therapies
Cells isolated from the spleen of sacrificed mice were stimulated with CD3/CD28 beads and transduced with a GFP-expressing lentiviral vector. (A) Transduction efficiency of autologous human CD3+ (n = 3) cells isolated from apheresis product and cells derived from mice (n = 5) engrafted with CD34+ cells from the same human donors used for the autologous CD3+ cells. (B) Fold expansion of either mock (−) or transduced (+) cells 4 days after bead stimulation for both the autologous human CD3+ cells and the mouse-derived human CD3+ cells. (C and D) Percent (C) and mean fluorescence intensity (MFI) (D) of CD69+ on cells either unstimulated (Media) or stimulated (Stim) with CD3/CD28 beads. (E) Fold increase in production and secretion of human IFNγ from both autologous human CD3+ cells and murine-matured CD3+ cells resulting from bead stimulation (Stim) when compared with unstimulated control (media) (n = 2). (F) Fraction of either CD4+ or CD8+ cells that are positive for intracellular Ki67 expression resulting from bead-stimulating (Stim) compared with media-only controls (media) in either autologous human- (n = 4) or murine-matured CD3+ (n = 3) cells. (G) Human CD3+ cells isolated from two mice were analyzed by spectratyping for TCR rearrangements after CD3/CD28 bead stimulation. (H) Autologous CD3+ cells isolated from the same apheresis product used for mouse engraftment were also analyzed by spectratyping. Error bars represent SEM. **p < 0.05. n/s, not significant.
To determine whether MATCH cells were functionally capable of responding to stimulation and able to efficiently produce and secrete cytokines, we assayed for the presence of interferon γ (IFNγ) in the supernatant of cultured cells. CD3+ cells from autologous human donors or MATCH mouse splenocytes (n = 2) were split in half and put into culture either in media alone or in the presence of CD3/CD28 stimulation beads. Two days later, supernatant was collected from the cells and analyzed for IFNγ (Figure 3E). When compared with the unstimulated controls, both the human and MATCH CD3+ cells exhibited a 7- to 9-fold increase in secretion of IFNγ into the supernatant. This increase in IFNγ production and secretion trended slightly higher in the autologous human CD3+ cells, but not significantly over the MATCH cells.
In addition to IFNγ production, we also observed an increase in Ki67+ cells by flow cytometry in both the human and MATCH CD4+ and CD8+ cell populations, indicating similar levels of cell proliferation in response to stimulation (Figure 3F). The baseline level of Ki67+ cells without stimulation was slightly higher in the MATCH mice compared with their autologous human counterpart, which is probably due to the xenograft setting of the cells. Both cell sources were capable of responding to stimulation through increased proliferation without significantly altering their CD4:CD8 ratios (Figure S7), indicating these cells are functionally able to receive signal input and respond in a similar manner.
We also analyzed the TCR repertoire of cells collected from the mice by spectratyping analysis (Figure 3G). Spleens collected from two mice were stimulated with CD3/CD28 beads for 48 hr prior to TCR analysis. The autologous CD3+ cells from human donors were also analyzed both pre- and post-CD3/CD28 bead stimulation for comparison (Figure S8). A broad range of complementarity-determining region 3 (CDR3) lengths from mouse samples analyzed was observed and appeared qualitatively similar to human samples (Figure 3H), indicating that in vivo maturing CD3+ within the mice are capable of undergoing TCR rearrangement similar to human-derived CD3+ cells. Together, these data indicate that MATCH cells behave similarly to a human-derived counterpart, potentially serving as a genetically modifiable population for T cell immunotherapy studies.
T Cell Subset Composition and Activation of MATCH Mice Resembles Human Cells
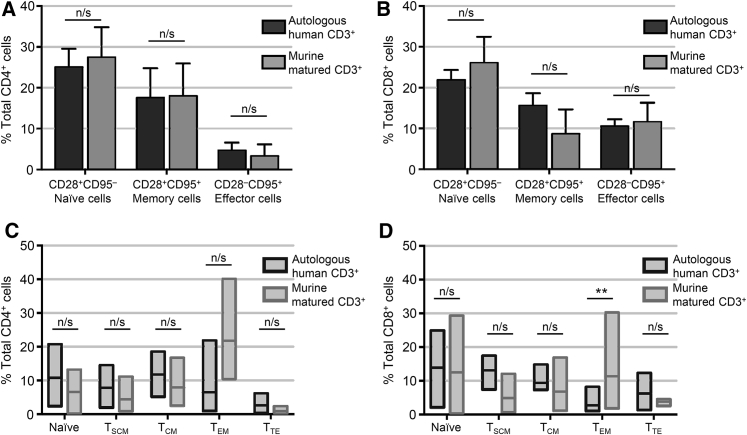
In order to further characterize the T cell repertoire that develops in our MATCH mouse model, we performed phenotypical analysis of T cell subsets in both the CD4+ and the CD8+ compartments as compared with the autologous human CD3+ products from the matched donors. Cells were stimulated for 48–72 hr using CD3/CD28 beads prior to analysis. The full staining panel and gating strategy for this analysis is provided in the Supplemental Information (Figure S9). The first layer of analysis determined the composition of naive T cells (CD28+CD95−), memory T cells (CD28+CD95+), and effector cells (CD28−CD95+) in both autologous human CD3+ cells and the MATCH mouse cells (Figures 4A and 4B). There was no statistically significant difference between human or MATCH cells in either CD4+ or CD8+ fractions for each of these subsets. We next further characterized the cells using additional markers for delineating memory and effector cell subsets including cell surface markers CD45RA and CCR7. Cells were characterized as naive (CD28+CD95−CD45RA+CCR7+), stem cell memory (TSCM; CD28+CD95+CD45RA+CCR7+), central memory (TCM; CD28+CD95+CD45RA−CCR7+), effector memory (TEM; CD28+CD95+CD45RA−CCR7−), and terminal effector cells (TTE; CD28−CD95+CD45RA+CCR7−) (Figures 4C and 4D). All five subsets of cells were present for both CD4+ and CD8+ cells in human-derived T cells (n = 4) and in MATCH T cells (n = 3) at comparable levels with no statistically significant differences except for TEM CD8+ cells (Figure 4D). These cells were slightly increased in prevalence for MATCH cells as compared with their autologous human cells counterparts.
Figure 4.
T Cell Subset Analysis of MATCH-Derived CD3+ Cells
T cells that matured in the mouse model were analyzed for the phenotypic composition as compared with their human-derived counterpart. (A and B) Percent contribution of naive, memory, or effector T cells in both the total (A) CD4+ or (B) CD8+ cell population for either autologous human CD3+ cells (dark bars, n = 4) or murine-matured CD3+ cells (light bars, n = 3) as defined by the combination of CD28 and CD95 expression. Error bars represent SEM. (C and D) Inclusion of additional surface markers CCR7 and CD45RA further delineate memory and effector cell subsets into stem cell memory (TSCM), central memory (TCM), effector memory (TEM), and terminal effector (TTE) T cells in either the total (C) CD4+ or (D) CD8+ cell populations in autologous human CD3+ cells (dark outlined boxes, n = 4) or murine-matured CD3+ cells (light outlined boxes, n = 3). The top and bottom bars of each box represent minimum and maximum percent observed with mean value represented by horizontal line within box. **p < 0.05. n/s, not significant.
Modified MATCH Cells Persist in Mice without Onset of GvHD
Two neonate litters of mice were injected with 1 × 106 adult human apheresis CD34+ cells from unique human donors. After 22 weeks post-injection, one mouse from each cohort was sacrificed and analyzed for peripheral and tissue engraftment levels. Total splenocytes were collected, placed in culture, and stimulated with CD3/CD28 beads, after which they were transduced with LV expressing GFP. Cells were allowed to expand in culture for an additional 3–5 days before being harvested, analyzed for GFP expression, and infused into remaining littermates humanized with the same CD34+ donor. Mice in both cohorts received a cell dose of either 1 × 106 or 2 × 106 total nucleated cells, of which 95.2%–99.6% was human CD45+. Within the human CD45+ fraction, 94.0%–96.3% was CD3+. At time of infusion, the GFP+ marking levels in CD3+ cells for the two cohorts were 66.3% and 73.0% (Figure S4). An administered cell dose of 1 × 106 or 2 × 106 in these mice corresponds to an average cell dose of 5.2 × 106 to 9.4 × 106 cells/kg, respectively. After injection of modified cells, we continued to monitor total human engraftment (Figure 5A), lineage frequency (Figures 5B and 5C), and modified cells in peripheral blood (Figure 5D). Average GFP+ cell levels ranged between 1.1% and 7.4% of all CD3+ cells observed. One animal was above this range, displaying 16% GFP+; however, this animal exhibited low CD45+ engraftment, resulting in low numbers of developed CD3+ cells, which inflated the proportion of modified cells.
Figure 5.
Modified Murine-Matured Human CD3+ Cells Persist in Mice without Onset of GvHD
(A) Neonatal mice were injected with adult apheresis-derived CD34+ cells, and peripheral CD45+ cell engraftment was tracked over time. (B) Lineage development of human cells in blood is shown with CD3+ cells in blue, CD20+ cells in red, and CD14+ cells in black. (C) T cell subsets were also tracked over time with CD4+ cells in gray and CD8+ cells in orange. (D) Total CD3+ content in the human population is shown in black circles plotted on the left y axis, and injected autologous GFP+ CD3+ cells are shown in green diamonds and plotted on the right y axis. Vertical green dashed line indicates time of autologous cell infusion, and all x axes represent age of mice (which also corresponds to weeks post-engraftment). (E–G) The total number of infused cells present in total peripheral blood of each mouse was calculated based on number of GFP+-modified cells observed by flow cytometry, the fraction of sample analyzed, and mouse weight for mice receiving (E) mature autologous CD3+ cells (n = 12), (F) mature autologous CD4+ cells (n = 7), or (G) murine-matured autologous CD3+ cells (n = 6). Each mouse is represented by one line, with different colors and patterns representing different cell doses administered. Number of mice (n) is shown for each group. All x axes represent weeks after modified cell infusion. Cross symbols (†) represent last time point of analysis before sacrifice.
To monitor for their persistence, we calculated the total number of modified cells circulating in peripheral blood of each mouse. Mice receiving an injection of human-matured autologous CD3+ or CD4+ cells exhibited an initial decrease of observed cells at the first time point post-injection (Figures 5E and 5F), most likely because of migration of cells into tissue compartments. The total number of modified cells detected at the second blood collection post-injection indicated significant cellular expansion, corresponding to the onset of GvHD previously described (Figure 2). In contrast, the MATCH cells maintained constant cell levels in peripheral blood over 7 weeks (52 days) after infusion, indicating maintained persistence without expansion (Figure 5G). These mice also did not exhibit the symptoms of GvHD such as weight loss or alopecia that were observed in the mice receiving human autologous T cells (Figure S5).
At 8 weeks post-CD3+ injection, mice were sacrificed and tissues were analyzed for the presence and frequency of modified GFP+ cells (Figure 6). Levels similar to those observed in blood (Figure 6A) were present in the bone marrow (Figure 6B), spleen (Figure 6C), and lung (Figure 6D) of each mouse. Interestingly, very few GFP+CD3+ cells were found in the thymus despite high levels of human engraftment (Figure 6E). The presence of modified cells in an array of tissues demonstrates their ability to circulate and perfuse into systemic tissues. In additional ongoing experiments using this model, animals have reached 20 weeks post-injection of MATCH cells without development of GvHD and with a 100% survival rate (data not shown).
Figure 6.
Modified CD3+ Cells Detected in Systemic Tissues
(A–E) Total human CD45+ engraftment as well as lineage contribution and detected frequency of GFP+ cells in tissues including (A) peripheral blood, (B) bone marrow, (C) spleen, (D) lung, and (E) thymus are shown. Black bars for CD3, CD20, and CD34 in marrow are represented as a proportion of total CD45+ cells, with CD4 and CD8 cells represented as a proportion of total CD3+ cells; all are plotted on the left y axis. Green bars represent the percent of total CD3+ cells detected that are GFP+, as well as for CD4+ and CD8+ cells, and are plotted on the right y axis. Error bars represent SEM. (F) Representative flow plots for GFP+ cell populations in the human CD45+ gate for each tissue, with numbers indicating % GFP+.
Retroviral IS Analysis Demonstrates Clonal Expansion Correlating with GvHD
Our laboratory has developed a method for clone tracking of lentiviral modified cells using the unique integration site (IS) of each transduced cell as a heritable mark.26 This type of analysis has primarily been used in CD34+ HSPC gene therapy studies to track hematopoietic development and differentiation of the engrafted cells, and to determine safety by monitoring for potential oncogenic transformations as evidenced by significant clonal expansion of a small subset of cells. We applied this technique to observe whether persistent cells in vivo represent a diverse subset of cells from the infusion product or result from antigenic stimulation of a small portion of cells. Necropsy tissues from several mice in each cohort were analyzed for their IS profile (Figure 7). Integrations were readily detected in each sample analyzed and ranged from 172 unique clones up to 6,882 clones in the animal receiving the highest cell dose. Animals receiving an injection of either mature autologous CD3+ cells (Figure 7A), mature autologous CD4+ cells (Figure 7B), or MATCH cells (Figure 7C) are shown.
Figure 7.
Integration Site Analysis Demonstrates Clonal Expansion of Autologous Cells, but Not Murine-Matured CD3+ Cells
(A–C) Retroviral integration site (RIS) analysis was used to measure the total number of integration sites detected in tissues of mice infused with (A) human-matured autologous CD3+ cells, (B) human-matured autologous CD4+ cells, or (C) murine-matured autologous human CD3+ cells. Individual mice, as well as cell dose received, are indicated on the x axis of each graph, with each column representing a unique mouse. All clones that represent >1% of sequences captured are shown as white or colored boxes, with the remainder represented by a single gray box. The total number of RIS clones detected in each sample is shown at the top of each column. Unique integration sites detected in multiple animals originating from the same pool of modified donor cells are color coded, and if the clone is not above 1% in the sample, it is represented by a colored number corresponding to its detected frequency. Chromosomal locations of the top five sites are listed below each column in order of most frequent. (D) Graph illustrating clonal expansion of all clones detected at >1% frequency. Individual mice grouped by cohort are listed on the x axis. The y axis represents the average number of unique fragments lengths (which approximately corresponds to number of unique cells) detected for clones contributing >1% to the total clonal population. A higher bar corresponds to greater cell division and expansion.
Although the total number of clones detected among these three conditions is relatively similar, the amount of clonal expansion was dramatically different (Figure 7D). Both cohorts receiving injections of human-matured CD3+ or CD4+ cells exhibited massive clonal expansion. Because of the random nature of acoustic shearing to fragment DNA, sequenced integrations with identical insertion points but different fragmented lengths of DNA must originate from different cells. By counting the number of unique fragment lengths observed, an estimate of clonal expansion is possible. Unique clonal integrations contributing to >1% of the total population had fragment counts on average ranging between 82.8 and 107.6. This was not observed in MATCH mice, for which the number of genomic fragment lengths ranged between 1.5 and 2.4. This clonal expansion most likely results from stimulation of infused cells as they react against mouse antigens, leading to onset of GvHD. This expansion also correlates with the increased frequency of total modified cells in peripheral blood (Figures 5E and 5F). The mouse receiving the highest cell dose of 1 × 107 cells (mouse 262; asterisk [*] in Figure 7D) also exhibited the greatest number of detected clones, resulting in no individual clone representing >1% of the total repertoire. This mouse was the first to succumb to GvHD in this cohort at 16 days post-cell infusion. One other mouse in this cohort (mouse 272; number sign [#] in Figure 7D) was an animal that survived cell infusion without the onset of GvHD. This mouse also did not exhibit the same clonal expansion seen in littermates that succumbed to GvHD, supporting the hypothesis that antigenic stimulation of infused cells resulted in disease onset. Importantly, all animals receiving the murine-matured CD3+ cells did not exhibit clonal expansion, consistent with their constant frequency of detection in the peripheral blood (Figure 5G).
Discussion
In this study, we describe for the first time to our knowledge an autologous T cell adaptive immunotherapy transplantation method using CD3+ cells derived from in-vivo-engrafted human adult apheresis CD34+ HSPCs. By using human CD3+ cells that derive from engrafted HSPCs, we mimic current clinical protocols for ACI. These murine-matured autologous T cells from humans (MATCH) are the ideal source for ACI, and we demonstrate the ability of these cells to persist at consistent levels in both peripheral blood and systemic tissues without the onset of GvHD often seen in T cell-based xenotransplant settings.22, 23 Similar to clinical protocols, this MATCH mouse model uses human T cells collected from the “patient,” expanded and modified ex vivo, and reinfused back into autologous engrafted recipients. Previous reports using adult apheresis HSPCs did not observe CD3+ cell maturation in their NSG mouse model.27 However, by using adult apheresis CD34+ HSPCs, we hereby demonstrate the ability to obtain peripheral human engraftment that averages 20% across multiple donors and mouse cohorts, the majority being CD3+ lymphocytes after 14–16 weeks. Within this CD3+ compartment, we observe ratios of CD4+ and CD8+ cells observed in normal healthy humans.28 Additionally, within both the CD4+ and CD8+ populations, we detect similar levels of different T cell subsets such as naive, memory, and effector cells between both human and MATCH-derived cells, indicating a diverse repertoire of cell products. These cells are functionally capable of both proliferating in response to stimulus and producing and secreting cytokines. We also demonstrate that the level of matured CD4+ cells in the mice are sufficient for sustaining HIV infection, allowing novel anti-HIV CAR T cell receptors targeted against the viral glycoprotein to be tested.29 Even though the mouse model presented here builds upon established models for HIV treatment therapies using the same mouse strain and engraftment procedures,30 additional characterization will be required to demonstrate functionality of this treatment for combination with other methods such as vaccine administration or genetic engineering.
Several previous reports have used mice as surrogates for testing ACI strategies.16, 17, 18, 19, 20, 21 However, most of these studies used either matured lymphocytes collected directly from human donors as the modified cell source or used murine T cell transplants in immunocompetent strains. When human T cells are engrafted into immunodeficient mice, they typically cause rapid GvHD,20, 31 limiting their usefulness for ACI strategies and potentially confounding results because of systemic immune activation. Alternatively, studies in immunocompetent mice such as C57B/6 mice show some promise;16, 17 however, these models transplant murine T cells that require different effector and signaling domains compared with human T cells. Additionally, in an immunocompetent setting, it is possible to illicit immune rejection of the infused cells. Another model that uses bone marrow, liver, thymus (BLT) mice has demonstrated potent T cell activity against HIV-infected cells.10, 32 However, BLT mice are laborious and expensive to generate because they require surgical implants of difficult to acquire fetal tissue samples, limiting the number of mice derived from one donor source, and these mice can also suffer from GvHD. The MATCH mice presented in this manuscript will circumvent this problem by using engrafted HSPCs as a source of CD3+ cells that have matured in vivo in the presence of mouse antigen. These cells do not recognize mouse antigen as foreign, thereby avoiding GvHD, and are capable of persisting at consistent levels within the mice. Mice receiving an injection of these cells do not exhibit the symptoms of GvHD, such as severe weight loss, hunched posture, and increasing hair loss. We observe peripheral and tissue resident levels of modified cells averaging 2% in each compartment analyzed, similar to what is currently reported in the clinic,33 with the exception of the thymus. Because cells present in thymic tissue are currently undergoing T cell maturation and differentiation, the majority should be either immature lymphocytes or lymphocytes migrating into peripheral circulation. Additionally, although we do observe infused modified T cells in the lungs of engrafted mice, we do not see high levels of tissue-specific retention, which enables greater systemic distribution.
Another benefit of MATCH mice compared with others humanized using umbilical cord blood or fetal liver CD34+ cells is the quantity of HSPCs derived from one donor. It is possible to engraft numerous cohorts of mice from one donor, providing potentially dozens of matched humanized mice, allowing for the direct in vivo comparison of different components such as linker domains, co-stimulatory elements, or other factors. This model could potentially be used in parallel with strategies for testing augmentation methods for ACI such as vaccine administration34 or direct injection of immune-stimulating molecules,35, 36, 37 although these studies would require additional experiments to determine whether all the functional components for these responses are present. Other reports have suggested that using CAR T cell-mediated therapy in conjunction with antibody administration might result in synergistic approaches to treatment of diseases such as B cell lymphomas.38 Although this mouse model does result in a majority of circulating human CD45+ cells being CD3+, other cell populations of interest, such as CD20+ cells, are still present in lymphoid tissues, especially the spleen and bone marrow. The trend for a decreasing ratio of CD20+ cells compared with CD3+ cells occurs as T cells complete their long developmental process in the thymus and begin to migrate into the periphery beginning around week 14 post-engraftment. For testing ACI-mediated strategies against B cell epitopes, it is possible to inject bioluminescent tumor lines expressing these epitopes into these mice to assay for functionality of infused CAR T cells.39, 40
Another difference from current protocols for ACI is the duration of ex vivo culturing to which the cells were subjected. Currently in clinical settings, post-isolation and stimulation culture times range between 5 and 21 days. This extended duration allows for a significant expansion of cells ex vivo, so a large dose, sometimes as high as 1 × 109 cells/kg, can be infused into patients. We cultured cells for a total of 5 days after bead stimulation to obtain numbers of cells for infusion averaging 5–10 × 107 cells/kg, comparable with current doses administered in human patients. This shorter duration of culture might actually be beneficial compared with longer ex vivo culture times because reports have shown that over time, cells more frequently become exhausted and terminally differentiated into memory subsets not ideal for T cell therapy.41 Additionally, it remains unclear which cell populations within the greater CD3+ lymphocyte pool are the ideal effector cells for ACI.42, 43, 44 Some reports indicate that CD8+ cells alone are sufficient,45 whereas others indicate that specific subpopulations of CD8+ cells are specifically required for achieving desired cytotoxicity effects.46 Because both CD4+ and CD8+ cells mature in this model, these populations can be isolated to perform studies using purified cell products or specific ratios of CD4:CD8 cells.
An important consideration for ACI strategies is the evaluation of IS frequencies to determine the safety profile of gene therapy cassettes. Early gene therapy clinical trials using HSPCs demonstrated the propensity of some vectors to result in viral-mediated insertional mutagenesis that led to leukemic development in patients.47 Even though significant advancements have been made in vector design to prevent such oncogenic transformations, all integrating viral vectors should be assessed on a case-by-case basis. This is true not only for HSPC-based therapies, but for all cell populations undergoing vector-mediated gene insertion. Such studies are essential when the gene of interest being inserted contains singling domains such as CAR molecules. For these reasons, clone tracking methods will be essential in preclinical testing of all ACI vectors.7 We demonstrate the ability to perform these clone tracking studies to not only determine total genomic integration profile of engineered cells, but also to quantify clonal expansion of these cells. Ideally, an engineered cell population would only undergo clonal expansion when presented with the desired recognition antigen. Clonal expansion outside these conditions could indicate potential concerns for long-term vector safety or be a predictor of GvHD.
Another report documents that activity and persistence of transferred CD8+ T cells was enhanced by HSPC transplantation in mice.48 In this study, myeloablated and non-myeloablated transplants were performed in parallel with adoptive T cell therapy. They found that the presence of autologous HSCs in the bone marrow enhanced proliferation and survival of infused CD8+ cells and augmented tumor-directed cytotoxicity. This suggests an important consideration in testing and analyzing ACI strategies in mouse models in the presence of ongoing autologous hematopoietic development. The MATCH model is ideally situated to take advantage of this enhanced efficacy because T cell infusions take place in the presence of ongoing HSPC hematopoiesis.
The MATCH model we present here enables in vivo testing of a broad range of engineered immunotherapy or CAR molecules. By using human CD3+ T cells that mature from adult apheresis CD34+ HSPCs engrafted into NSG mice as the cell source for modification and transplantation, it is possible to model a clinically relevant ACI protocol that avoids the potential complications of GvHD or immune rejection of modified cells present in other models.
Materials and Methods
Ethics Statement
All animal studies were carried out in compliance with approved protocols (1864) by the Institutional Animal Care and Use Committee (IACUC) at the Fred Hutchinson Cancer Research Center.
Mice
NOD.Cg-PrkdcscidIl2rgtm1Wjl/Szj (NOD SCID gamma−/−, NSG) mice were purchased from The Jackson Laboratory or bred in-house under approved protocols and in pathogen-free housing conditions. Neonatal mice between 1 and 3 days postbirth received 150 cGy of radiation, followed 3–4 hr later by a single intrahepatic injection of 1 × 106 CD34+ cells resuspended in 30 μL of PBS containing 1% heparin (APP Pharmaceuticals). Blood samples were collected by retro-orbital puncture beginning at week 8 post-engraftment and continuing every other week until necropsy.
Human Cell Processing and Isolation
Human apheresis CD34+ HSPCs were obtained from two different sources. Some purified CD34+ cells were purchased from the Cell Processing Facility (CPF) at the Fred Hutchinson Cancer Research Center. Negative fractions from isolated CD34+ products were also purchased and used for sequential autologous CD3+ isolation using the positive selection kit from Miltenyi Biotec (Bergisch Gladbach) using the manufacturer’s suggested protocols. Isolated autologous CD3+ cells were cryopreserved for later use. Alternatively, some CD34+ cells were isolated from total apheresis products purchased from CPF using the CliniMACS Prodigy device (Miltenyi Biotec) using previously described protocols.49
Vector Production and Transduction
The vector used for transduction of CD3+ cells (pRSC-hPGK.eGFP) is an SIN LV produced with a third-generation split packaging system and pseudotyped by the vesicular stomatitis virus G protein (VSVG). Vectors for these studies were produced by our institutional Vector Production Core (Director: Hans-Peter Kiem) at the Fred Hutchinson Cancer Research Center. Infectious titer was determined by flow cytometry evaluating EGFP expression following titrated transduction of HT1080 human fibrosarcoma-derived cells with research grade LV vector preparations.
NSG Mouse Engraftment and Sample Processing
At each time point, a total of 200 μL of blood was collected by retro-orbital puncture. Blood was diluted 1:1 with PBS, and a portion was used for antibody staining and flow cytometry to determine engraftment levels and lineage contribution. At the time of necropsy, organ samples were harvested and filtered through a 70 μm filter (BD Biosciences) and washed with PBS. Blood and tissue samples were stained with appropriate fluorescence-activated cell sorting (FACS) antibodies for 15 min at room temperature. Red blood cells were removed by incubation in BD FACS Lysing Solution (BD Biosciences), which was then diluted out using PBS prior to analysis by flow cytometry. Stained cells were acquired on an FACS Canto II (BD Biosciences) and analyzed using FlowJo software v10.1 (Tree Star). Analysis was performed on up to 20,000 cells in the viable cell population for blood and 100,000 cells for tissues. Gates were established using full minus one (FMO)-stained controls. Samples were stained at a 1:20 dilution using human CD45-PerCP (Clone 2D1), mouse CD45.1/CD45.2-V500 (Clone 30-F11), CD3-FITC or allophycocyanin (APC) (Clone UCHT1), CD4-V450 (Clone RPA-T4), CD20-phycoerythrin (PE) (Clone 2H7), and CD14-APC or PE-Cy7 (Clone M5E2). Bone marrow was also stained with CD34-APC (Clone 581). All antibodies were acquired from BD Biosciences.
Autologous T Cell Injections
Autologous CD3+ cells were slow thawed into RPMI media (Life Technologies) containing 10% serum (Atlas Biologics), 1% penicillin-streptomycin (Pen/Strep) (Life Technologies), and 50 μg/mL interleukin (IL)-2 (Stem Cell Technologies). Cells were stimulated at 1:1 ratio with CD3/CD28 Beads (Life Technologies) for 48–72 hr at a concentration of 1.5–2.0 million cells/mL. After stimulation, beads were magnetically removed and cells were transduced with a third-generation LV at an MOI of 10. Cells were counted and adjusted to 1.5 million cells/mL with fresh media every 2 days until infusion. CD8+ cell depletion was performed using a CD8-positive selection kit (Stem Cell Technologies) following manufacturer’s protocol, keeping the wash instead of the cells attached to the magnet. Cells were suspended in 100 μL of PBS containing 1% heparin and infused by intravenous (i.v.) injection.
Murine-matured autologous T cells were isolated from the spleen. Following filtration through a 70 μm filter, red blood cells were removed by incubation in BD FACS Lysing Solution (BD Biosciences), which was diluted out using PBS prior to cell count. Cells were then stimulated with CD3/CD28 beads for 48–72 hr at a concentration of 1.5–2 million/mL in RPMI media containing 10% serum, 1% Pen/Strep, and 50 μg/mL IL-2. After stimulation, beads were magnetically removed and cells transduced with a third-generation LV at an MOI of 10. Cells were counted and adjusted to 1.5 million cells/mL with fresh media every 2 days until infusion. Cells were suspended in 100 μL of PBS containing 1% heparin and infused by i.v. injection.
The total number of modified cells in peripheral circulation was estimated for each mouse using counting beads (eBioscience) spiked into each blood sample at time of collection. By analyzing the number of beads observed during flow analysis and using an estimation of total blood volume of mice based on their weight (approximately 7% of volume by weight), it is possible to calculate the total number of modified cells circulating in peripheral blood of the mice.50
HIV Challenge of Humanized Mice
Mice were challenged by a single intraperitoneal injection of 200 μL of HIV-1 BaL containing 2.5 × 105 infectious units. HIV-1 BaL virus stock was obtained through the NIH AIDS Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases (NIAID), NIH, and deposited by Dr. Suzanne Garter, Dr. Mikulas Popovic, and Dr. Robert Gallo.51 Virus was propagated in PM1 cells obtained through NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, and deposited by Dr. Marbin Reitz.52 Viral supernatant was collected and filtered through 0.22 μm filters (EMD Millipore) and tittered on GHOST cells obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH, and deposited by Dr. Vineet N. KewalRamani and Dr. Dan R. Littman53 according to provided protocols. To determine viral load, we extracted RNA from mouse serum using the QIAamp Viral RNA Mini Kit (QIAGEN), which was then analyzed using the TaqMan RNA-to-Ct 1-Step Kit (Thermo Fisher) using primers and probes specific to the long terminal repeat (LTR) region (F: 5′-GCCTCAATAAAGCTTGCCTTGAG-3′, R: 5′-GGCGCCACTGCTAGAGATTTTC-3′, Probe: FAM 5′-AAGTAGTGTGTGCCCGTCTGTTRTKTGACT-3′ TAMRA). Plates were analyzed on an ABI TaqMan 7500 real-time PCR system (Thermo Fisher).
T Cell Subset Analysis and Intracellular Staining
A multicolor panel was used to assess cell surface and intracellular protein markers in this study. The T cell panel included CD3-PerCP clone SP34-2 (Thermo Fisher), CCR7-PE clone 3D12 (Thermo Fisher), CD45-BUV395 clone HI30 (BD), CCR5-APC clone 3A9 (BD), Ki67-ALX700 clone B56 (BD), CD95-PE-Cy7 clone DX2 (BD), CD45RA-V450 clone 5H9 (BD), CD8-BUV737 clone SK1 (BD), CD28-ECD clone CD28.2 (Beckman Coulter), and CD4-BV570 clone OKT4 (BioLegend). The panel also included a live/dead stain. Cell samples were stained with the above markers, as previously described.54 Acquisition was performed on LSR II and Fortessa instruments (BD). Flow cytometry data were analyzed using FlowJo (Tree Star). All gates were established based on FMO controls.
TCR Spectratyping
All TCR spectratyping was performed by the institutional Immune Monitoring Core (Director: Jianhong Cao) at the Fred Hutchinson Cancer Research Center. One million CD3+ cells were submitted as a frozen cell pellet for analysis.
Human IFNγ ELISA
Equal total cell numbers of either autologous human- or murine-matured CD3+ cells were placed into culture and stimulated using CD3/CD28 beads as previously described above. Supernatant was collected 48 hr after initiating bead stimulation and subsequently centrifuged to remove cellular debris. Supernatant was then assayed for human IFNγ content using a purchased ELISA kit (Thermo Fisher). Assay was performed according to manufacturer’s protocol.
IS Analysis
Lentiviral IS analysis was performed as previously described55, 56 with the following modifications. In brief, DNA was extracted from cell suspension of tissues using the DNeasy Blood and Tissue Kit (QIAGEN), and up to 3 μg was randomly sheared using an M220 focused ultrasonicator (Covaris). Fragmented DNA was purified, polished (End-It DNA End Repair Kit; Epicenter), and ligated to modified linker cassettes containing known primer binding sites. This product was amplified using sequential nested exponential PCRs. Product from the first PCR was purified, and eluted DNA was diluted prior to a second nested PCR, which added both barcodes and sequences required for compatibility with the next-generation sequencing MiSeq platform (Illumina). Sequencing was performed by the Genomics Core Facility at the Fred Hutchinson Cancer Research Center.
ISs were identified using a method similar to that described by Hocum et al.57 The forward and reverse reads were stitched using PEAR with the -q 30 option to trim sequence reads after two bases with a quality score below 30 were observed.58 FASTQ files were filtered using custom python scripts. Pairwise2 from the Bio module was used to confirm the presence of our primer sequence at the start of the sequence read using a gap open penalty of −2, a gap extension penalty of −1, and requiring a total mapping score of 25 or greater, equivalent to two mismatches or one insertion or deletion (indel). Presence or absence of the LTR region was determined using Pairwise2 to align a known 24-bp sequence from the LTR region to the sequence read using the same gap penalties described previously and requiring a total mapping score of 22 or greater, corresponding to two mismatches or one indel. To remove reads representing vector sequences (as opposed to genomic sequences), we aligned a known 24-bp sequence from the vector to the sequence read using Pairwise2 and the same settings as described for the primer alignment. The reads that contained the primer sequence and the LTR sequence, but not the vector sequence, were then trimmed and output in FASTQ format. Additionally, all reads were output to text files with relevant filtering information. FASTQ files were then converted to FASTA files using a custom python script. The Homo sapiens reference genome (GRCh38, GCA_000001305.2, December 2013) provided by the Genome Reference Consortium was downloaded from the University of California, Santa Cruz (UCSC) genome browser.59 The filtered and trimmed sequence reads were aligned to the reference genome using BLAT with options -out = blast8, -tileSize = 11, -stepSize = 5, and -ooc = hg11-2253.ooc.60 The hg11-2253.ooc file contains a list of 11-mers occurring at least 2,253 times in the genome to be masked by BLAT and was generated as recommended by UCSC using the following command: $blat hg38.2bit /dev/null /dev/null -tileSize = 11 -stepSize = 5 -makeOoc = hg11-2253.ooc -repMatch = 2253. The resulting blast8 files were parsed using a custom python script. The blast8 files contained multiple possible alignments for each sequence read, so any sequence read with a secondary alignment percent identity up to 95% of the best alignment percent identity was discarded. Sequence reads were then grouped based on their genomic alignment positions and orientation (sense versus antisense). Any alignments within 5 bp of one another and with identical integration orientations were considered to originate from the same IS; the genomic position with greatest number of contributing reads is reported as the IS. The total number of sequence reads contributing to a particular IS is reported as the number of genomically aligned reads for that IS.
Author Contributions
H.-P.K. is the principal investigator of the study, and designed and coordinated the overall execution of the project. K.G.H. conceived, designed, and coordinated the experiments. J.E.A. provided feedback and critical input. C.I., W.M.O., and Z.K.N. performed and analyzed experimental data. K.G.H. wrote the manuscript, which was critically reviewed by J.E.A. and H.-P.K.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Helen Crawford for help preparing and formatting this manuscript. We also thank Sarah Weitz, Melissa Comstock, and Don Parrilla for assistance with performing many of the mouse procedures and general colony maintenance, Sara Kubek and Shelly Heimfeld for processing and isolation of adult apheresis CD34+ cells, Jianhong Cao at the Immune Monitoring Core for TCR spectratyping analysis, Daniel Humphrys and Lauren Schefter for integration site analysis processing and analysis, and Martin Wohlfahrt and Donald Gisch for producing the lentiviral vectors used in this manuscript. This work was supported in part by grants from the National Institutes of Health (R21 AI110154 and R01 HL115128). H.-P.K. is a Markey Molecular Medicine Investigator and received support as the inaugural recipient of the Jose Carreras/E. Donnall Thomas Endowed Chair for Cancer Research and the Endowed Chair for Cell and Gene Therapy.
Footnotes
Supplemental Information includes nine figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.omtm.2017.05.004.
Supplemental Information
References
- 1.Sadelain M., Brentjens R., Rivière I. The basic principles of chimeric antigen receptor design. Cancer Discov. 2013;3:388–398. doi: 10.1158/2159-8290.CD-12-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dotti G., Gottschalk S., Savoldo B., Brenner M.K. Design and development of therapies using chimeric antigen receptor-expressing T cells. Immunol. Rev. 2014;257:107–126. doi: 10.1111/imr.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Parijs L., Abbas A.K. Homeostasis and self-tolerance in the immune system: turning lymphocytes off. Science. 1998;280:243–248. doi: 10.1126/science.280.5361.243. [DOI] [PubMed] [Google Scholar]
- 4.Chmielewski M., Hombach A.A., Abken H. Antigen-specific T-cell activation independently of the MHC: Chimeric antigen receptor-redirected T cells. Front. Immunol. 2013;4:371. doi: 10.3389/fimmu.2013.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eshhar Z., Waks T., Bendavid A., Schindler D.G. Functional expression of chimeric receptor genes in human T cells. J. Immunol. Methods. 2001;248:67–76. doi: 10.1016/s0022-1759(00)00343-4. [DOI] [PubMed] [Google Scholar]
- 6.Geyer M.B., Brentjens R.J. Review: current clinical applications of chimeric antigen receptor (CAR) modified T cells. Cytotherapy. 2016;18:1393–1409. doi: 10.1016/j.jcyt.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.June C.H. Adoptive T cell therapy for cancer in the clinic. J. Clin. Invest. 2007;117:1466–1476. doi: 10.1172/JCI32446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B., Zou F., Lu L., Chen C., He D., Zhang X., Tang X., Liu C., Li L., Zhang H. Chimeric antigen receptor T cells guided by the single-chain Fv of a broadly neutralizing antibody specifically and effectively eradicate virus reactivated from latency in CD4+ T lymphocytes isolated from HIV-1-infected individuals receiving suppressive combined antiretroviral therapy. J. Virol. 2016;90:9712–9724. doi: 10.1128/JVI.00852-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ali A., Kitchen S.G., Chen I.S., Ng H.L., Zack J.A., Yang O.O. HIV-1-specific chimeric antigen receptors based on broadly neutralizing antibodies. J. Virol. 2016;90:6999–7006. doi: 10.1128/JVI.00805-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhen A., Kamata M., Rezek V., Rick J., Levin B., Kasparian S., Chen I.S., Yang O.O., Zack J.A., Kitchen S.G. HIV-specific immunity derived from chimeric antigen receptor-engineered stem cells. Mol. Ther. 2015;23:1358–1367. doi: 10.1038/mt.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer M., Hafner R., Schneider C., Trkola A., Joos B., Joller H., Hirschel B., Weber R., Günthard H.F., Swiss HIV Cohort Study HIV RNA in plasma rebounds within days during structured treatment interruptions. AIDS. 2003;17:195–199. doi: 10.1097/00002030-200301240-00009. [DOI] [PubMed] [Google Scholar]
- 12.Harrigan P.R., Whaley M., Montaner J.S. Rate of HIV-1 RNA rebound upon stopping antiretroviral therapy. AIDS. 1999;13:F59–F62. doi: 10.1097/00002030-199905280-00001. [DOI] [PubMed] [Google Scholar]
- 13.Trickett A., Kwan Y.L. T cell stimulation and expansion using anti-CD3/CD28 beads. J. Immunol. Methods. 2003;275:251–255. doi: 10.1016/s0022-1759(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 14.Riddell S.R., Greenberg P.D. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J. Immunol. Methods. 1990;128:189–201. doi: 10.1016/0022-1759(90)90210-m. [DOI] [PubMed] [Google Scholar]
- 15.Alexander R.B., Rosenberg S.A. Long-term survival of adoptively transferred tumor-infiltrating lymphocytes in mice. J. Immunol. 1990;145:1615–1620. [PubMed] [Google Scholar]
- 16.James S.E., Orgun N.N., Tedder T.F., Shlomchik M.J., Jensen M.C., Lin Y., Greenberg P.D., Press O.W. Antibody-mediated B-cell depletion before adoptive immunotherapy with T cells expressing CD20-specific chimeric T-cell receptors facilitates eradication of leukemia in immunocompetent mice. Blood. 2009;114:5454–5463. doi: 10.1182/blood-2009-08-232967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davila M.L., Kloss C.C., Gunset G., Sadelain M. CD19 CAR-targeted T cells induce long-term remission and B Cell Aplasia in an immunocompetent mouse model of B cell acute lymphoblastic leukemia. PLoS ONE. 2013;8:e61338. doi: 10.1371/journal.pone.0061338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacoby E., Yang Y., Qin H., Chien C.D., Kochenderfer J.N., Fry T.J. Murine allogeneic CD19 CAR T cells harbor potent antileukemic activity but have the potential to mediate lethal GVHD. Blood. 2016;127:1361–1370. doi: 10.1182/blood-2015-08-664250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milone M.C., Fish J.D., Carpenito C., Carroll R.G., Binder G.K., Teachey D., Samanta M., Lakhal M., Gloss B., Danet-Desnoyers G. Chimeric receptors containing CD137 signal transduction domains mediate enhanced survival of T cells and increased antileukemic efficacy in vivo. Mol. Ther. 2009;17:1453–1464. doi: 10.1038/mt.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Bonin M., Wermke M., Cosgun K.N., Thiede C., Bornhauser M., Wagemaker G., Waskow C. In vivo expansion of co-transplanted T cells impacts on tumor re-initiating activity of human acute myeloid leukemia in NSG mice. PLoS ONE. 2013;8:e60680. doi: 10.1371/journal.pone.0060680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velasquez M.P., Torres D., Iwahori K., Kakarla S., Arber C., Rodriguez-Cruz T., Szoor A., Bonifant C.L., Gerken C., Cooper L.J. T cells expressing CD19-specific engager molecules for the immunotherapy of CD19-positive malignancies. Sci. Rep. 2016;6:27130. doi: 10.1038/srep27130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bendle G.M., Linnemann C., Hooijkaas A.I., Bies L., de Witte M.A., Jorritsma A., Kaiser A.D., Pouw N., Debets R., Kieback E. Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat. Med. 2010;16:565–570. doi: 10.1038/nm.2128. [DOI] [PubMed] [Google Scholar]
- 23.Covassin L., Laning J., Abdi R., Langevin D.L., Phillips N.E., Shultz L.D., Brehm M.A. Human peripheral blood CD4 T cell-engrafted non-obese diabetic-scid IL2rγ(null) H2-Ab1 (tm1Gru) Tg (human leucocyte antigen D-related 4) mice: a mouse model of human allogeneic graft-versus-host disease. Clin. Exp. Immunol. 2011;166:269–280. doi: 10.1111/j.1365-2249.2011.04462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suntharalingam G., Perry M.R., Ward S., Brett S.J., Castello-Cortes A., Brunner M.D., Panoskaltsis N. Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N. Engl. J. Med. 2006;355:1018–1028. doi: 10.1056/NEJMoa063842. [DOI] [PubMed] [Google Scholar]
- 25.Takao K., Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2015;112:1167–1172. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beard B.C., Adair J.E., Trobridge G.D., Kiem H.P. High-throughput genomic mapping of vector integration sites in gene therapy studies. Methods Mol. Biol. 2014;1185:321–344. doi: 10.1007/978-1-4939-1133-2_22. [DOI] [PubMed] [Google Scholar]
- 27.Lepus C.M., Gibson T.F., Gerber S.A., Kawikova I., Szczepanik M., Hossain J., Ablamunits V., Kirkiles-Smith N., Herold K.C., Donis R.O. Comparison of human fetal liver, umbilical cord blood, and adult blood hematopoietic stem cell engraftment in NOD-scid/gammac-/-, Balb/c-Rag1-/-gammac-/-, and C.B-17-scid/bg immunodeficient mice. Hum. Immunol. 2009;70:790–802. doi: 10.1016/j.humimm.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amadori A., Zamarchi R., De Silvestro G., Forza G., Cavatton G., Danieli G.A., Clementi M., Chieco-Bianchi L. Genetic control of the CD4/CD8 T-cell ratio in humans. Nat. Med. 1995;1:1279–1283. doi: 10.1038/nm1295-1279. [DOI] [PubMed] [Google Scholar]
- 29.Kitchen S.G., Zack J.A. Engineering HIV-specific immunity with chimeric antigen receptors. AIDS Patient Care STDS. 2016;30:556–561. doi: 10.1089/apc.2016.0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holt N., Wang J., Kim K., Friedman G., Wang X., Taupin V., Crooks G.M., Kohn D.B., Gregory P.D., Holmes M.C., Cannon P.M. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat. Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali N., Flutter B., Sanchez Rodriguez R., Sharif-Paghaleh E., Barber L.D., Lombardi G., Nestle F.O. Xenogeneic graft-versus-host-disease in NOD-scid IL-2Rγnull mice display a T-effector memory phenotype. PLoS ONE. 2012;7:e44219. doi: 10.1371/journal.pone.0044219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitchen S.G., Bennett M., Galić Z., Kim J., Xu Q., Young A., Lieberman A., Joseph A., Goldstein H., Ng H. Engineering antigen-specific T cells from genetically modified human hematopoietic stem cells in immunodeficient mice. PLoS ONE. 2009;4:e8208. doi: 10.1371/journal.pone.0008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackensen A., Meidenbauer N., Vogl S., Laumer M., Berger J., Andreesen R. Phase I study of adoptive T-cell therapy using antigen-specific CD8+ T cells for the treatment of patients with metastatic melanoma. J. Clin. Oncol. 2006;24:5060–5069. doi: 10.1200/JCO.2006.07.1100. [DOI] [PubMed] [Google Scholar]
- 34.Parviz M., Chin C.S., Graham L.J., Miller C., Lee C., George K., Bear H.D. Successful adoptive immunotherapy with vaccine-sensitized T cells, despite no effect with vaccination alone in a weakly immunogenic tumor model. Cancer Immunol. Immunother. 2003;52:739–750. doi: 10.1007/s00262-003-0405-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheever M.A., Greenberg P.D., Fefer A., Gillis S. Augmentation of the anti-tumor therapeutic efficacy of long-term cultured T lymphocytes by in vivo administration of purified interleukin 2. J. Exp. Med. 1982;155:968–980. doi: 10.1084/jem.155.4.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki E., Kapoor V., Cheung H.K., Ling L.E., DeLong P.A., Kaiser L.R., Albelda S.M. Soluble type II transforming growth factor-beta receptor inhibits established murine malignant mesothelioma tumor growth by augmenting host antitumor immunity. Clin. Cancer Res. 2004;10:5907–5918. doi: 10.1158/1078-0432.CCR-03-0611. [DOI] [PubMed] [Google Scholar]
- 37.Paulos C.M., Wrzesinski C., Kaiser A., Hinrichs C.S., Chieppa M., Cassard L., Palmer D.C., Boni A., Muranski P., Yu Z. Microbial translocation augments the function of adoptively transferred self/tumor-specific CD8+ T cells via TLR4 signaling. J. Clin. Invest. 2007;117:2197–2204. doi: 10.1172/JCI32205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.John L.B., Devaud C., Duong C.P., Yong C.S., Beavis P.A., Haynes N.M., Chow M.T., Smyth M.J., Kershaw M.H., Darcy P.K. Anti-PD-1 antibody therapy potently enhances the eradication of established tumors by gene-modified T cells. Clin. Cancer Res. 2013;19:5636–5646. doi: 10.1158/1078-0432.CCR-13-0458. [DOI] [PubMed] [Google Scholar]
- 39.Green D.J., Shadman M., Jones J.C., Frayo S.L., Kenoyer A.L., Hylarides M.D., Hamlin D.K., Wilbur D.S., Balkan E.R., Lin Y. Astatine-211 conjugated to an anti-CD20 monoclonal antibody eradicates disseminated B-cell lymphoma in a mouse model. Blood. 2015;125:2111–2119. doi: 10.1182/blood-2014-11-612770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Budde L.E., Berger C., Lin Y., Wang J., Lin X., Frayo S.E., Brouns S.A., Spencer D.M., Till B.G., Jensen M.C. Combining a CD20 chimeric antigen receptor and an inducible caspase 9 suicide switch to improve the efficacy and safety of T cell adoptive immunotherapy for lymphoma. PLoS ONE. 2013;8:e82742. doi: 10.1371/journal.pone.0082742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gattinoni L., Klebanoff C.A., Palmer D.C., Wrzesinski C., Kerstann K., Yu Z., Finkelstein S.E., Theoret M.R., Rosenberg S.A., Restifo N.P. Acquisition of full effector function in vitro paradoxically impairs the in vivo antitumor efficacy of adoptively transferred CD8+ T cells. J. Clin. Invest. 2005;115:1616–1626. doi: 10.1172/JCI24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu H.M., Winter H., Urba W.J., Fox B.A. Divergent roles for CD4+ T cells in the priming and effector/memory phases of adoptive immunotherapy. J. Immunol. 2000;165:4246–4253. doi: 10.4049/jimmunol.165.8.4246. [DOI] [PubMed] [Google Scholar]
- 43.Perret R., Ronchese F. Memory T cells in cancer immunotherapy: which CD8 T-cell population provides the best protection against tumours? Tissue Antigens. 2008;72:187–194. doi: 10.1111/j.1399-0039.2008.01088.x. [DOI] [PubMed] [Google Scholar]
- 44.Klebanoff C.A., Gattinoni L., Restifo N.P. Sorting through subsets: which T-cell populations mediate highly effective adoptive immunotherapy? J. Immunother. 2012;35:651–660. doi: 10.1097/CJI.0b013e31827806e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marzo A.L., Vezys V., Klonowski K.D., Lee S.J., Muralimohan G., Moore M., Tough D.F., Lefrançois L. Fully functional memory CD8 T cells in the absence of CD4 T cells. J. Immunol. 2004;173:969–975. doi: 10.4049/jimmunol.173.2.969. [DOI] [PubMed] [Google Scholar]
- 46.Klebanoff C.A., Gattinoni L., Palmer D.C., Muranski P., Ji Y., Hinrichs C.S., Borman Z.A., Kerkar S.P., Scott C.D., Finkelstein S.E. Determinants of successful CD8+ T-cell adoptive immunotherapy for large established tumors in mice. Clin. Cancer Res. 2011;17:5343–5352. doi: 10.1158/1078-0432.CCR-11-0503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 48.Wrzesinski C., Paulos C.M., Gattinoni L., Palmer D.C., Kaiser A., Yu Z., Rosenberg S.A., Restifo N.P. Hematopoietic stem cells promote the expansion and function of adoptively transferred antitumor CD8 T cells. J. Clin. Invest. 2007;117:492–501. doi: 10.1172/JCI30414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adair J.E., Waters T., Haworth K.G., Kubek S.P., Trobridge G.D., Hocum J.D., Heimfeld S., Kiem H.P. Semi-automated closed system manufacturing of lentivirus gene-modified haematopoietic stem cells for gene therapy. Nat. Commun. 2016;7:13173. doi: 10.1038/ncomms13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thaler B., Hohensinner P.J., Krychtiuk K.A., Matzneller P., Koller L., Brekalo M., Maurer G., Huber K., Zeitlinger M., Jilma B. Differential in vivo activation of monocyte subsets during low-grade inflammation through experimental endotoxemia in humans. Sci. Rep. 2016;6:30162. doi: 10.1038/srep30162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gartner S., Markovits P., Markovitz D.M., Kaplan M.H., Gallo R.C., Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 52.Lusso P., Cocchi F., Balotta C., Markham P.D., Louie A., Farci P., Pal R., Gallo R.C., Reitz M.S., Jr. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 1995;69:3712–3720. doi: 10.1128/jvi.69.6.3712-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mörner A., Björndal A., Albert J., Kewalramani V.N., Littman D.R., Inoue R., Thorstensson R., Fenyö E.M., Björling E. Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. J. Virol. 1999;73:2343–2349. doi: 10.1128/jvi.73.3.2343-2349.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peterson C.W., Haworth K.G., Polacino P., Huang M.L., Sykes C., Obenza W.M., Repetto A.C., Kashuba A., Bumgarner R., DeRosa S.C. Lack of viral control and development of combination antiretroviral therapy escape mutations in macaques after bone marrow transplantation. AIDS. 2015;29:1597–1606. doi: 10.1097/QAD.0000000000000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Adair J.E., Beard B.C., Trobridge G.D., Neff T., Rockhill J.K., Silbergeld D.L., Mrugala M.M., Kiem H.P. Extended survival of glioblastoma patients after chemoprotective HSC gene therapy. Sci. Transl. Med. 2012;4:133ra57. doi: 10.1126/scitranslmed.3003425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Adair J.E., Johnston S.K., Mrugala M.M., Beard B.C., Guyman L.A., Baldock A.L., Bridge C.A., Hawkins-Daarud A., Gori J.L., Born D.E. Gene therapy enhances chemotherapy tolerance and efficacy in glioblastoma patients. J. Clin. Invest. 2014;124:4082–4092. doi: 10.1172/JCI76739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hocum J.D., Battrell L.R., Maynard R., Adair J.E., Beard B.C., Rawlings D.J., Kiem H.P., Miller D.G., Trobridge G.D. VISA--Vector Integration Site Analysis server: a web-based server to rapidly identify retroviral integration sites from next-generation sequencing. BMC Bioinformatics. 2015;16:212. doi: 10.1186/s12859-015-0653-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.