Abstract
Interactions between Bacillus anthracis and host macrophages represent critical early events in anthrax pathogenesis, but their details are not clearly understood. Here we report the first genomewide characterization of the transcriptional changes within macrophages infected with B. anthracis and the identification of several hundred host genes that were differentially expressed during this intracellular stage of infection. These loci included both genes that are known to be regulated differentially in response to many other bacterial pathogens and those that appear to be differentially regulated in response to B. anthracis but not other bacterial species that have been tested. These data provide a transcriptional basis for a variety of physiological changes observed during infection, including the induction of apoptosis caused by the infecting bacteria. The expression patterns underlying B. anthracis-induced apoptosis led us to test further the importance of one very highly induced macrophage gene, that for ornithine decarboxylase. Our data show that this enzyme plays an important and previously unrecognized role in suppressing apoptosis in B. anthracis-infected cells. We have also characterized the transcriptional response to anthrax lethal toxin in activated macrophages and found that, following toxin treatment, many of the host inflammatory response pathways are dampened. These data provide insights into B. anthracis pathogenesis as well as potential leads for the development of new diagnostic and therapeutic options.
Bacillus anthracis is a gram-positive, endospore-forming bacterium that is the causative agent of anthrax. This disease is primarily zoonotic, affecting grazing herbivores, and naturally occurring human infections are rare in developed countries (13). Even so, the resilience of the B. anthracis endospore, the potentially lethal nature of the disease, and the efficiency with which it infects via an aerosol route make it an ideal biological weapon, and for these reasons the organism and the disease it causes have recently come under increased scrutiny (28, 29).
Infection by B. anthracis begins with entry of dormant endospores into the body through one of three possible routes: cutaneous exposure, ingestion, or inhalation. In systemic cases, spores are efficiently scavenged by resident phagocytes and transported to the regional lymph nodes (16, 17, 24, 47). The spores germinate en route, transforming into replicative vegetative cells. The nascent cells multiply within the phagocyte and begin to produce virulence factors such as the two binary exotoxins lethal toxin and edema toxin, eventually killing the host cell and escaping into the local environment (the bloodstream, in systemic anthrax). Up to this point, the symptoms of anthrax are relatively mild, but once in the blood the bacteria grow to a very high titer (sometimes approaching 108 organisms per ml of blood) (13, 47), and this combination of septicemia and toxemia quickly leads to shock and death of the host. Because anthrax is usually diagnosed at this late stage, the disease is very difficult to control. Understanding the events prior to this point—those occurring in association with the host phagocyte—is therefore critical for the development of new therapeutic options.
Although much of the B. anthracis-host interaction takes place within the phagocyte, the individual molecular events that make up this interaction are not well understood. Some of the factors that are critical for growth of B. anthracis inside the cell are beginning to be understood (23, 24, 30, 31), but the ways in which the macrophage responds to the growing infection remain unclear. In order to better define these responses and perhaps identify new points at which new therapeutic or diagnostic options might be possible, we sought to examine this response in detail on a global scale with DNA microarrays. This approach has been successful in several recent studies examining the response of a variety of mammalian cell types to a range of infecting bacteria, including both intra- and extracellular pathogens (3, 4, 6, 38, 46). These analyses have yielded insights into the common immune response, and they have also identified genes that are specifically involved in the response to a particular bacterium.
In this study DNA microarrays were used to examine the transcriptional response of the murine macrophage-like RAW 264.7 cell line during infection by B. anthracis and also in activated macrophages following treatment with B. anthracis lethal toxin. In comparing the expression patterns induced in each of these conditions, we anticipated being able to identify host genes specifically induced or repressed in response to toxin exposure as well as genes specifically regulated during infection. Within these response programs, we identified genes that are part of the common immune response induced by many or all bacterial pathogens as well as those that have not been reported previously in other systems and thus may be unique to the B. anthracis-macrophage interaction. Our findings confirm many of the biochemical and physiological phenomena previously observed during this interaction, including the importance of apoptosis in the pathogenic process and the significant modulation of cytokine synthesis by B. anthracis. In addition, we have identified a limited number of macrophage response genes that appear to be different from the common responses to previously characterized pathogenic bacteria and may serve as useful leads in the development of future diagnostic and/or therapeutic options targeting B. anthracis and its close relatives.
MATERIALS AND METHODS
Growth of bacteria and cell culture strains.
Bacillus anthracis Sterne 34F2 (containing plasmid pXO1 but not pXO2) was grown in brain-heart infusion broth or on sheep's blood agar plates, and spores were prepared as in Liu et al. (36). The murine macrophage-like RAW 264.7 cell line (ATCC TIB-71) was maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (Gibco-BRL) at 37°C with 5% CO2 in a humidified incubator. Cells were visualized by Diff-Quick staining (Baxter Scientific).
Infection and toxin exposure conditions.
Twenty-four h prior to infection, RAW 264.7 cells were counted and switched from DMEM-10% fetal bovine serum to minimal essential medium (MEM) with 10% horse serum, as this combination has been shown to inhibit outgrowth of extracellular spores. Cells were seeded at 3 × 107 cells per 30 ml of medium in 150-cm2 cell culture flasks and cultured overnight. Infections were begun by removing the medium and adding 15 ml of fresh MEM-10% horse serum containing B. anthracis Sterne 34F2 spores (a strain that carries pXO1 but lacks pXO2 and is thus able to produce both binary exotoxins but not capsule) at a multiplicity of infection of 10:1.
In experiments in which toxin exposure was being examined, the procedure was essentially the same except that the fresh medium contained B. anthracis protective antigen (500 ng/ml) and lethal factor (various concentrations for preliminary experiments, 8 ng/ml for treatments prior to RNA isolation). At these concentrations of protective antigen and lethal factor, previous studies have shown that essentially all of the lethal factor is bound by protective antigen, and thus the functional concentration of lethal toxin is the same as that of lethal factor (5). Note also that free protective antigen at these concentrations did not have an effect on cell viability, as seen in our own control experiments (data not shown) and previous studies (45). Salmonella lipopolysaccharide (0.5 μg/ml) was added simultaneously with lethal toxin when noted. For control experiments, fresh medium was added without spores or toxin.
RNA isolation.
Macrophage RNA was isolated at the desired time points by decanting the medium and extracting sequentially with Trizol (Invitrogen) and bromochloropropane according to the manufacturer's instructions. The RNA was then precipitated with isopropanol and washed with 75% ethanol and then dried. The pellets were resuspended in 200 μl of H2O. The RNA was further purified with RNeasy Midi columns (Qiagen) and again resuspended in H2O. RNA quality and purity were examined by measuring absorbance at 260 and 280 nm and by visualization on a denaturing agarose gel.
Generation of probes for microarray experiments.
DNA probes for microarray experiments were generated as described by Abulencia et al. (1). Briefly, 15 μg of total RNA was reversed transcribed in a mixture containing 6 μg of random hexamers (Invitrogen), 0.01 M dithiothreitol, an aminoallyl-deoxynucleoside triphosphate mixture containing 25 mM each dATP, dCTP, and dGTP, 15 mM dTTP, and 10 mM aminoallyl-dUTP (Sigma), reaction buffer, and 400 units of SuperScript II reverse transcriptase (Invitrogen) at 42°C overnight. The RNA template was then hydrolyzed by adding NaOH and EDTA to a final concentration of 0.2 and 0.1 M, respectively, and incubating at 70°C for 15 min. Unincorporated aminoallyl-dUTP was removed with a QIAquick column (Qiagen). The probe was eluted with a phosphate elution buffer (4 mM KPO4, pH 8.5, in ultrapure water), dried, and resuspended in 0.1 M sodium carbonate buffer (pH 9.0). To couple the aminoallyl with fluorescent labels, normal human serum-Cy3 or normal human serum-Cy5 (Amersham) was added in the dark at room temperature for 1 h. Uncoupled label was removed with the QIAquick column procedure.
Microarray hybridization, scanning, and image analysis.
Aminosilane-coated slides printed with a set of 27,648 target sequences were prehybridized in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (Invitrogen), 0.1% sodium dodecyl sulfate, and 1% bovine serum albumin at 42°C for 60 min. The slides were then washed at room temperature with distilled water, dipped in isopropanol, and allowed to dry. Equal volumes of the appropriate Cy3- and Cy5-labeled probes were combined and dried and then resuspended in a solution of 50% formamide, 5× SSC, and 0.1% sodium dodecyl sulfate. The RNA was supplemented with 20 μg of mouse COT1 DNA and 20 μg of poly(A) DNA and then heated (95°C, 3 min) and quickly cooled on ice for 30 s. The probe mixture was then added to the microarray slide and allowed to hybridize overnight at 42°C.
Hybridized slides were washed sequentially in solutions of 1× SSC-0.2% SDS, 0.1× SSC-0.2% SDS, and 0.1× SSC at room temperature, then dried in air, and scanned with an Axon GenePix 4000 scanner. Individual TIFF images from each channel were analyzed with TIGR Spotfinder (available at http://www.tigr.org/software/tm4) (50). Spots whose shape deviated significantly from a true circle and those with signal-to-noise ratios of <3.0 were eliminated from further analysis.
Microarray normalization and data analysis.
Microarray data sets were normalized with a locally weighted linear regression (LOWESS) algorithm as implemented in the TIGR MIDAS software package (50). This program also allowed us to filter the data in order to remove spots for which flip dye replicates deviated considerably from the expected reciprocal relationship. The resulting data were then log2 transformed, and adjusted relative to the time zero point (uninfected) as described in Baldwin et al. (3). In brief, this was done by subtracting the log (2) of the time zero point from each of the other data sets, such that the reference RNA was effectively eliminated and changes in gene expression during the course of the 6-h experiment could be measured relative to the zero time point. Following this step, the data were averaged over duplicate genes within each array (when present) and over duplicate arrays done for each experiment.
Differentially expressed genes were determined at the 99% confidence level for each experimental data set by assuming that the log2 ratios for each data set approximate a normal distribution (initial characterizations of the data sets obtained in this study showed that the data did not show an appreciably intensity-dependent structure after LOWESS normalization; see supplemental Fig. S1 and all other supplemental figures and tables mentioned in this article at http://www.sitemaker.umich.edu/hanna_lab), and selecting genes with log2 ratio values that deviated by >2.58 standard deviations from the mean. In almost every case this level of confidence translated to a >2-fold change in expression levels, but in the rare instance that this was not true, our filtration was adjusted such that significant differential expression was defined as deviating by >2.58 standard deviations and changing >2-fold in expression level. Filtration for significant differential expression and further analyses were done with MS Excel.
Gene ontology (GO) annotation of genes identified as differentially expressed was carried out with the Onto-Express software package (available at http://vortex.cs.wayne.edu/Projects.html) and the UniGene cluster identifiers for each target sequence on the microarray. Probabilities of overrepresentation were estimated with a chi-square-based probability model, with Fisher's exact test substituted when necessary because of small sample size. When both a category and its subcategories showed statistically significant overrepresentation, the smallest (most specific) functional group that showed significant overrepresentation was reported.
RT-PCR assays.
Reverse transcription (RT) and PCR were used to validate the microarray data corresponding to the expression profiles of selected genes. Primers were synthesized by Invitrogen, Inc., and Integrated DNA Technologies. The sequences used were 5′-ACATGATGCTGTAAACTTTATTTGCGA and 5′-CCTCTTTTGTTCCGCTATCGATC (methylmalonate-semialdehyde dehydrogenase); 5′-CAGCAGGCTTCTCTTGGAAC and 5′-CATGCATTTCAGGCAGGTTA (ornithine decarboxylase); and 5′-CCAGAGCAAGAGAGGTATCC and 5′-CTGTGGTGGTGAAGCTGTAG (β-actin). RT-PCRs were performed with One Step RT-PCR kits (Invitrogen), and the number of cycles for each reaction template was optimized to ensure that all reactions being compared were within the linear portion of the amplification curve. Reaction products were analyzed on 1.25% agarose gels, stained with ethidium bromide, and visualized under UV light. Control reactions were done without reverse transcriptase (with the thermostable DNA polymerase alone) to ensure that there was no contaminating DNA in the RNA samples being assayed.
Cell death assays.
Measurement of necrosis and apoptosis within RAW 264.7 cells was done with the Cell Death Detection ELISA Plus (Roche) according to the manufacturer's instructions. Briefly, 104 cells per sample were cultured in a 96-well plate, and B. anthracis 34F2 spores, camptothecin, d,l-α-difluoromethylornithine (DFMO), or a combination of these were added in fresh medium. Untreated cells were also given fresh medium without any added constituents. At the appropriate time points, the presence of histone-associated DNA fragments was assayed in both the culture supernatant and the cytoplasm of infected cells. As prescribed by the manufacturer, we assigned a value of 1.0 to the levels of histone-associated DNA detected in untreated control cells, and the relative values of experimental samples were recorded as a ratio to the control. Experiments were performed at least three times each, and within each experiment measurements were taken from at least three replicate samples. Statistical significance was measured by Student's two-tailed t test. Although different experiments varied as to the overall magnitude of cell death recorded (perhaps reflecting variations in the age of RAW 264.7 cells), the results depicted in Fig. 3 are typical, and in every instance the differences between untreated cells and infected cells, camptothecin-treated cells, and infected cells treated with DFMO were statistically significant (P < 0.05).
FIG. 3.
Cell death (necrosis and apoptosis) in B. anthracis infection as measured by enzyme-linked immunosorbent assay. Measurements of necrosis and apoptosis have been normalized in each case to measurements obtained from untreated cells, such that levels of cell death are expressed as the ratio of experimental to untreated cells. Data shown are from representative experiments, and each column is the average of at least three individual measurements. Error bars indicate the standard deviation of each data set.
Thiazolyl blue assays for measurement of cell viability.
Killing of RAW264.7 cells by B. anthracis lethal toxin was assayed with the thiazolyl blue (Sigma M5655) assay. Before assay, the medium was changed to DMEM plus 10 mM HEPES, pH 7.4 (DMEM-HEPES) and the cell concentration adjusted to 4 × 105 cells/ml. Cells (6 × 104 per well) were plated in a 96-well plate and allowed to settle at room temperature for 30 min. Cells were incubated at 37°C and 5% CO2 for 16 h. Medium was aspirated and replaced with DMEM-HEPES containing test toxin concentrations in triplicate. Lethal toxin-containing samples ranged from 4 to 500 ng/ml of lethal factor (recombinant lethal factor and protective antigen; List Biological Laboratories) plus a constant concentration (0.5 μg/ml) of protective antigen, with or without a constant concentration (0.5 μg/ml) of lipopolysaccharide (LPS; from Salmonella enteritidis, Sigma L-4774). Plates were incubated for 4 h at 37°C, 5% CO2, and 50 μl of thiazolyl blue dissolved in DMEM-HEPES at 1.5 mg/ml was added to each well and incubated for 1 h at 37°C and 5% CO2. Aspirated medium was then replaced with 100 μl of 0.5% sodium dodecyl sulfate-25 mM HCl in 90% isopropanol and vortexed gently, and absorption was read at 570 nm to 650 nm on a Molecular Devices SpectraMAX 250 microplate reader. Lysis of RAW 264.7 cells was also measured with the same range of lethal toxin and lethal toxin plus LPS with the trypan blue visual assay. Briefly, cells were grown and treated exactly as above, but scraped after 4 h and mixed in a 1:1 mixture of 0.1% trypan blue to cell suspension, counted on a hemacytometer, and percent lysis was calculated by percentage of blue cells.
Data availability.
The expression data collected in this study are publicly available via the TIGR website (http://pga.tigr.org) and the ArrayExpress database (accession number E-TIGR-84). Complete lists of genes that were identified as differentially expressed in response to any of the experimental conditions described here are in the supplemental material at http://www.sitemaker.umich.edu/hanna_lab.
RESULTS AND DISCUSSION
Although many of the individual virulence factors employed by B. anthracis in causing disease are known, the effects of the infecting bacteria on host gene expression have yet to be clearly defined. It was therefore of interest to examine the transcriptional changes that occur within infected macrophages and identify genes that are induced or repressed in response to B. anthracis. We also sought to compare these trends to those observed in macrophages exposed to lethal toxin, as well as to cells that had been exposed to Salmonella lipopolysaccharide (LPS), a reagent that is known to induce a widespread activation program within macrophages (46).
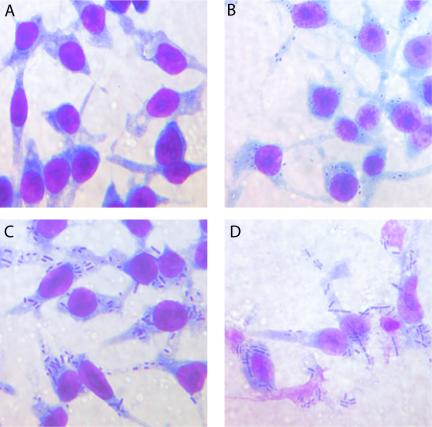
The RAW 264.7 murine macrophage-like cell line provided a convenient cell culture model system for these experiments, as it has been used widely in the past for studies of B. anthracis as well as other bacterial pathogens (12, 23, 26, 30, 56). Initial experiments matched previously published work (12, 48) and showed that 1 h after infection vegetative bacilli could be seen inside or closely associated with the macrophages (Fig. 1, panel B). After 3 h the bacteria were beginning to multiply (panel C), and 6 h after infection the majority of macrophages had lysed, and bacteria were growing in the extracellular medium (panel D).
FIG. 1.
Infection of RAW 264.7 cells by Bacillus anthracis. Murine macrophage-like RAW 264.7 cells are shown untreated (A) and 1 h (B), 3 h (C), and 6 h (D) after inoculation with B. anthracis 34F2 spores at a multiplicity of infection of 10:1.
Microarray analysis of macrophages infected with B. anthracis endospores.
In order to define the patterns of gene expression associated with these events, we isolated total RNA from RAW 264.7 cells following infection with B. anthracis Sterne 34F2 spores. Samples were harvested every hour for 6 h, after which relatively few intact murine cells were observed, and RNA yields decreased dramatically. RNA samples were used to make cDNA probe mixtures, which were then applied to DNA microarrays containing target sequences corresponding to 27,648 murine genes and expressed sequence tags. For comparison, mRNA samples from parallel mock infections, in which B. anthracis spores were omitted and only fresh medium was added to the cells, were also isolated and prepared in the same way. In each case data were collected and processed as described in Materials and Methods, and genes that were differentially expressed in a given sample were selected at a 99% confidence level.
Prior to analyzing gene expression in infected mouse macrophages, it was necessary to determine to what extent gene expression changed over time in the mock-infected cells, as these data provide an indication of the random fluctuation of gene expression levels within our experiment. Accordingly, we compared the expression profiles of cells at 0, 3, and 6 h after a mock infection in which the cells were given fresh medium (as in all other experiments) without challenge. Within this time interval, gene expression in RAW 264.7 macrophages changed very little: between 0 and 3 h after the mock infection, the median change in expression level was 1.0035-fold, with 95% changing by less than 1.26-fold and only six genes changing more than 2-fold (supplemental Fig. S1). Between 0 and 6 h, the overall level of change was even lower, with median change of 1.0025-fold, 95% changing by less than 1.21-fold, and only two genes changing in expression level by twofold or more. These measurements imply that changes observed in comparing different treatments, or cells treated the same way at different times, are likely to be due to the experimental difference and not due to stochastic changes in gene expression.
Previous studies with this experimental system and our own results have shown that roughly 3 h after inoculation, B. anthracis is able to escape from the phagolysosome into the host cell cytoplasm. This event thus separates the early phase of infection (those events occurring in the phagolysosome) from the late phase (the events occurring within the host cell cytoplasm, leading up to killing of the phagocyte and escape into the extracellular environment).
In our examination of macrophage gene expression during infection by B. anthracis, we analyzed these stages separately and began by assessing the gene expression patterns that occur early during infection, which encompasses phagocytosis of endospores, germination, and the beginning of bacterial replication. Within this time frame, 493 genes met our criteria for significant differential expression (see Materials and Methods for descriptions of these criteria), with 213 induced and 280 repressed in at least one of the three time points (see supplemental Table S1). These genes were sorted by functional family with the Onto-Express software package, which uses the gene ontology annotation system to sort genes by pathway and function (14). This allowed the identification of biological processes that were related to the genes within a given data set, and an assessment of the statistical significance of any overrepresentations that were found. Those biological processes with a statistically significant overrepresentation (P < 0.05) and at least three members present in the early (1 to 3 h postinoculation) data sets are shown in Table 1.
TABLE 1.
Functional families overrepresented in the macrophage transcriptional response to early stages of B. anthracis infection
| Induced
| ||||
|---|---|---|---|---|
| Biological process | P | |||
| Biogenic amine metabolism | 1.1 E-4 | |||
| Innate immune response (inflammatory response) | 1.2 E-4 | |||
| Response to stress (pathogen/DNA damage/wounding) | 1.8 E-4 | |||
| Cell migration | 2.2 E-4 | |||
| Regulation of apoptosis | 7.7 E-4 | |||
| Cytoskeleton organization and biogenesis | 0.0013 | |||
| Sterol metabolism | 0.0028 | |||
| Cell growth and/or maintenance | 0.0048 | |||
| DNA repair | 0.0088 | |||
| Regulation of cell cycle | 0.0151 | |||
| Receptor-linked signal transduction | 0.0164 | |||
| Amino acid metabolism | 0.0183 | |||
| Cell-cell adhesion | 0.0249 | |||
| GTPase-mediated signal transduction | 0.0264 | |||
| Morphogenesis | 0.0265 | |||
| Repressed
|
Biological process | P | ||
| Protein biosynthesis | 0.0351 | |||
| Chromosome organization and biogenesis | 0.0429 | |||
| Receptor-linked signal transduction | 1.3 E-4 | |||
| Protein folding | 6.5 E-4 | |||
| Protein biosynthesis | 0.0038 | |||
| Cell homeostasis | 0.0053 | |||
| Regulation of transcription | 0.0092 | |||
| Regulation of cell cycle | 0.0100 | |||
| Ion transport | 0.0145 | |||
| Protein kinase signaling cascades | 0.0158 | |||
| Proteolysis | 0.0253 | |||
| Defense response to pathogenic bacteria | 0.0347 | |||
| Protein phosphorylation and dephosphorylation | 0.0395 | |||
| Regulation of apoptosis | 0.0439 | |||
| Cell-cell adhesion | 0.0443 | |||
Gene expression during early infection by B. anthracis was marked by several distinct trends. Perhaps the most prominent of these is the induction of both innate immune response and stress-related genes. These loci include the CD14 gene, which encodes the receptor for the complex of bacterial lipopolysaccharide (LPS) and LPS-binding protein, and has been recently and previously shown to be essential for the innate immune response to bacterial infection (53). The product of this gene has also been recently shown to mediate the innate immune response to gram-positive bacteria by binding to cell wall-derived peptidoglycan-polysaccharide complexes, and this may explain its induction in response to B. anthracis (35). Also observed was an induction of genes associated with Toll-like receptor signaling (e.g., I-κB zeta, which has been seen in other studies to be rapidly induced in response to activation of the Myd88-dependent signaling pathway, commonly found downstream of different Toll-like receptors) (19, 34) as well as loci previously known to play a role in p38 mitogen-activated protein kinase activation (e.g., Mkk3, Mkk1ip1, phospholipase A2, and c-Myc), which is the pathway best characterized as the target for disruption by anthrax lethal toxin (7, 41).
It seems clear from the functional families represented in both the induced and the repressed data sets that the first 3 h following infection by B. anthracis spores are marked by major shifts in cellular signaling, as evidenced by the changes in the expression levels of genes involved in receptor-linked signal transduction, ion transport, protein phosphorylation and dephosphorylation, and GTP-linked signaling cascades. There were also significant changes in genes previously linked to control of cell cycle (e.g., c-Myc), regulation of apoptosis (e.g., Bcl-10), and cellular growth and homeostasis (e.g., Rab1). Collectively, these transcriptional patterns likely reflect the activation of the murine macrophages following phagocytosis of the B. anthracis spores. Concurrent with these changes, we noted expression level changes in loci involved in cell migration (e.g., somatostatin), cytoskeletal biogenesis and organization (e.g., γ-tubulin), and cell adhesion (e.g., protocadherin 10), which may reflect the fact that in each of the three forms of anthrax, phagocytosis and activation of the macrophage are closely followed by migration of the phagocyte to the regional lymph nodes.
The lymph nodes are the site where the late stages of B. anthracis infection begin, and in cell culture this time frame is marked by the escape of the bacteria from the phagolysosome into the host cell cytoplasm, replication within the macrophage, and eventual killing of the host cell with subsequent release of the bacterium into the extracellular environment. These events were observed in previous work and in our own experiments to occur roughly 4 to 6 h postinoculation (Fig. 1). In order to identify the transcriptional patterns underlying the macrophage response to the later stages of infection, we analyzed total RNA samples taken 4, 5, and 6 h postinoculation in the same way the studies of the early stages of infection had been done. Within this time frame, 480 genes showed significant differential expression, with 292 genes induced and 188 genes repressed in at least one of the three time points examined (see supplemental Table S2).
With the methods described above, differentially expressed genes were sorted according to functional category, and biological processes whose components showed statistically significant (P < 0.05) overrepresentation are listed in Table 2. Salient findings included increases in the mRNA levels of many genes associated with the immune response and cytokine signaling (e.g., macrophage colony-stimulating factor, tumor necrosis factor alpha, and GADD45-β), reflecting the growing severity of the infection. Many of the immune response genes that were upregulated in early infection continued to increase in expression level into the later stages of the infection, and a variety of genes generally linked to the immune response were induced for the first time in this time frame. In general, the immune response genes differentially expressed during late infection by B. anthracis infection were the same genes identified as part of the stereotypical innate immune response transcriptional program. This set of transcriptional responses is triggered by a range of bacterial pathogens and has recently been characterized by several groups (4, 6, 38). One notable parallel is in the induction of NF-κB and a range of associated genes and regulatory targets (e.g., I-κB, I-κB kinase) which have been shown in several other studies to play pivotal roles in immune system activation (3, 4, 6, 38).
TABLE 2.
Functional families overrepresented in the macrophage transcriptional response to late stages of B. anthracis infection
| Induced
|
Repressed
|
|||
|---|---|---|---|---|
| Biological process | P | Biological process | P | |
| Regulation of apoptosis | 2.6 E-6 | Cell-matrix adhesion | 5.0 E-5 | |
| Regulation of transcription | 1.0 E-5 | G-protein coupled receptor protein signaling pathway | 3.1 E-4 | |
| Nitrogen metabolism | 2.0 E-5 | Angiogenesis | 0.0013 | |
| Immune response (inflammatory response) | 2.0 E-5 | Regulation of cell cycle | 0.0100 | |
| Regulation of cell cycle | 5.0 E-5 | Cell adhesion | 0.0109 | |
| Intracellular signaling cascade | 8.0 E-5 | Regulation of apoptosis | 0.0113 | |
| Protein phosphorylation and desphosphorylation | 9.0 E-5 | Immune response | 0.0175 | |
| Cytokine biosynthesis | 1.2 E-4 | Intracellular signaling cascade | 0.0212 | |
| Amine catabolism | 3.8 E-4 | |||
| Cytoskeleton organization and biogenesis | 0.0012 | |||
| Protein kinase cascade | 0.0025 | |||
| Cell differentiation | 0.0026 | |||
| Cell-matrix adhesion | 0.0034 | |||
| Cell adhesion | 0.0039 | |||
| Receptor-linked signal transduction | 0.0094 | |||
| GTPase-mediated signaling cascade | 0.0182 | |||
| Proteolysis | 0.0345 | |||
In addition to the “general immune response genes,” we continued to observe differential regulation of genes involved in regulation of cell cycle, intra- and extracellular signaling, and adhesion both to other cells and to the extracellular matrix. Many of these genes were identified early in infection, and it would appear that many of the processes begun soon after phagocytosis are still occurring late in B. anthracis infection. Intriguingly, the most significantly overrepresented functional family was the set of genes involved in regulating apoptosis. Several of these genes were observed to be differentially expressed during early infection, but the trend was much more pronounced 4 to 6 h after inoculation, and this particular functional family included several of the most highly induced genes identified (e.g., ornithine decarboxylase and the orphan nuclear receptor NURR1, both upregulated ≈14-fold late in infection). Significantly, the time frame in which the apoptosis-associated genes became most highly induced was essentially the same in which we first observed cell death occurring in infected macrophages (Fig. 1D), suggesting that the cell death caused by intracellular B. anthracis is apoptotic in nature. This is consistent with a recent study by Popov et al. (45), which reported membrane permeability changes commonly associated with early stages of apoptosis in B. anthracis-infected macrophages, as well as with several other studies that have shown that anthrax lethal toxin, a binary exotoxin which is required for full virulence in B. anthracis, is able to efficiently induce apoptosis and/or necrosis in a variety of cell types by inactivating a p38 mitogen-activated protein kinase signaling pathway (15, 33, 41, 44, 45).
Confirmation of specific gene expression trends by RT-PCR.
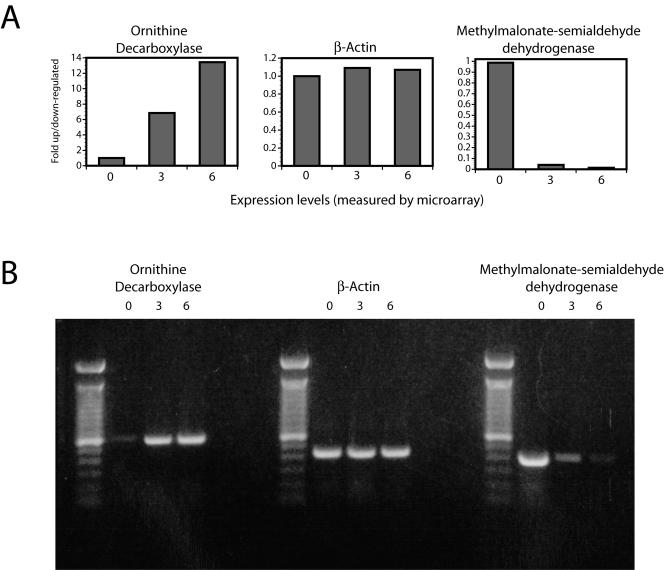
In general, our microarray analysis of B. anthracis infection suggested that many of the transcriptional patterns characteristic of a general macrophage response to bacterial pathogens are also found in the response to B. anthracis. In order to more completely assess the reliability of the arrays and the data derived from them, we sought to validate the RNA expression trends by an alternative method. To do this, we selected representative genes from each of three broad categories (induced following expression; repressed following infection; and unchanged during infection) and repeated our measurements of RNA expression with RT-PCR (Fig. 2). Total RNA samples were isolated as in the case of microarray analysis, and RT-PCRs were performed with equivalent amounts of starting RNA. Preliminary experiments were done to determine the optimal number of cycles for each PCR template such that all of the reactions being compared were within the linear portion of the amplification curve. The increase in the level of ornithine decarboxylase expression that we observed by microarray following infection was confirmed in our RT-PCR analysis, as was the decrease in the level of methylmalonate semialdehyde dehydrogenase expression. The gene encoding β-actin, which did not show an appreciable change in expression level in our microarray measurements and thus was an appropriate control, similarly remained at a relatively constant level of expression throughout the experimental time course when measured by RT-PCR.
FIG. 2.
RT-PCR measurement of expression levels for representative genes. A Expression levels for selected genes as measured by DNA microarray. Levels shown reflect induction or repression in cells 3 and 6 h postinfection relative to untreated cells. B RT-PCR analysis of gene expression. RT-PCRs were performed on equal amounts of starting RNA, and control reactions (not shown) demonstrated that there was no contaminating DNA present in the RNA samples.
Confirmation of B. anthracis-induced apoptosis and identification of a major antiapoptotic factor induced in infected macrophages.
Intuitively, it seems that the ability to cause apoptosis in host macrophages would almost certainly be an advantage for B. anthracis, as apoptotic cells are much less likely than necrotic cells to burst and release cytokines and other proinflammatory signals into the extracellular environment. Since our data as well as those of other groups indicate that B. anthracis activates the host macrophage during infection, it may be that induction of apoptosis is a means by which the bacterium is able to kill macrophages silently and to a large extent delay or prevent a heightened immune response in the area of infection.
Because the microarray data seemed to point strongly toward apoptosis as the underlying mode of intracellular B. anthracis-induced cell death, we sought to confirm this possibility through other means. Accordingly, we infected RAW 264.7 cells with B. anthracis Sterne 34F2 spores and used a commercially available enzyme-linked immunosorbent assay kit (Cell Death Detection kit; Roche) to detect and differentiate between apoptosis and necrosis. Specifically, the kit detects histone-associated DNA fragments, which remain membrane bound following apoptosis but are released into the extracellular environment during necrosis. The two forms can thus be measured independently, and the precise mode of cell death that is occurring can be determined.
Microscopic observation of infected macrophages showed that 3 h after inoculation with B. anthracis Sterne 34F2 spores, bacteria were replicating within the host cells but had not escaped, while after 6 h there was appreciable cell death and bacteria could be seen in the extracellular medium (Fig. 1, panels C and D). Our cell death assays closely mirrored these observations and detected essentially no cell death 3 h postinoculation. We did, however, detect significant apoptosis 6 h after the beginning of infection (Fig. 3). The levels of apoptosis in B. anthracis-infected cells at the final time point were similar to the levels detected in cells treated with camptothecin, a well-characterized inducer of apoptosis (27).
As noted earlier, many of the most highly induced genes late in infection are related to the regulation of apoptosis. One particularly interesting example is the locus encoding the enzyme ornithine decarboxylase, which was induced by approximately 15-fold late in the infection. This enzyme catalyzes a key step in the synthesis of spermine and spermidine, key polyamines that are known to be essential for cell proliferation and differentiation. Significantly, these molecules also exert a powerful protective effect against cell death induced by a variety of cellular stresses (8, 32), and a recent study by Park et al. (40) showed that overexpression of ornithine decarboxylase is sufficient to protect cells from apoptosis induced by H2O2, γ-irradiation, and a variety of chemotherapeutic agents. Since it appeared that induction of apoptosis may be a key element in B. anthracis pathogenesis, and elevated levels of ornithine decarboxylase expression have not been reported in studies examining the macrophage response to a variety of other bacterial pathogens (3, 4, 6, 38), we sought to examine the role of ornithine decarboxylase in more detail and determine the extent of its importance in infection.
Ornithine decarboxylase expression has long been known to be a marker associated with a variety of cancers, and several chemotherapeutic agents are able to specifically inhibit ornithine decarboxylase. The most specific and best characterized of these is d,l-α-difluoromethylornithine (DFMO), which is an irreversible suicide inhibitor of ornithine decarboxylase. Conveniently, this compound has been used in previous studies examining ornithine decarboxylase activity in RAW 264.7 cells and has been shown to effectively inhibit the enzyme without affecting cell viability (21). We therefore took advantage of these properties and used DFMO to inhibit ornithine decarboxylase in B. anthracis-infected RAW cells in order to better understand the role of this enzyme in the response to infection.
Using the enzyme-linked immunosorbent assay system to assess cell death, we measured necrosis and apoptosis in macrophages that were infected with B. anthracis spores, treated with DFMO, or infected in the presence of DFMO, and compared them to untreated cells and cells treated with camptothecin (Fig. 3). Cells treated with inhibitor alone were essentially indistinguishable from untreated cells, indicating that, as previous studies had shown, DFMO did not have a discernible effect on RAW 264.7 cells. In comparing infected cells with those that had been infected in the presence of DFMO, we noted a significant difference in how rapidly apoptosis was induced. Infected cells treated with DFMO showed signs of apoptosis 3 h after infection began, while at this time point cells infected without the inhibitor did not show significant cell death. These data were consistent with microscopic observations, which showed infection progressing more rapidly in cells treated with inhibitor (data not shown), and suggest that ornithine decarboxylase is a key suppressor of apoptosis during infection by B. anthracis. Together, these findings further highlight the importance of apoptosis induction as an element of B. anthracis pathogenesis and are consistent with a recent study that suggested that small-molecule inhibitors of apoptosis may be therapeutically useful in treating anthrax and in preventing the disease in those exposed to spores (43).
Finally, it is of interest to point out that ornithine decarboxylase expression has been examined in response to a variety of bacterial pathogens, including enterohemorrhagic E. coli, Salmonella enterica serovar Typhi, Listeria monocytogenes, Mycobacterium tuberculosis, Bordetella pertussis, and Staphylococcus aureus, and in each case no discernible induction of expression was observed. It is tempting to speculate that a detectable increase in expression of this gene might be unique to B. anthracis and thus may serve as a useful marker for specific diagnosis of early-stage anthrax, though we note that the macrophage responses to close relatives like Bacillus cereus as well as other to biowarfare agents have not yet been characterized, and these data will be necessary to fully test this hypothesis.
Gene expression patterns induced by anthrax lethal toxin in macrophages.
Because anthrax lethal toxin is known to have profound effects on host macrophages, including disruption of a variety of signaling pathways and induction of apoptosis (20, 25), it was of interest to determine to what extent the gene expression patterns observed in infected cells could be attributed to the actions of lethal toxin alone. This toxin consists of the two proteins protective antigen, which facilitates entry into host cells, and lethal factor, which is a zinc metalloprotease that cleaves members of the MEK signaling family. It is known to disrupt a variety of signaling pathways within the host cell, and previous studies have shown that the effects of lethal toxin differ depending on the activation state of the macrophage (37, 41). We therefore examined gene expression in both activated and unactivated macrophages following treatment with anthrax lethal toxin and compared these expression patterns to those observed in untreated cells as well as those that were activated but not treated with toxin.
RAW 264.7 cells were treated with lethal toxin, Salmonella lipopolysaccharide (LPS), or a combination of the two, and total RNA was prepared as described for infected cells. LPS was used at a constant concentration of 500 ng/ml, and at this level it has been previously shown to induce a robust, general activation program within RAW 264.7 cells without significantly affecting cell viability (46). Lethal toxin was used at a concentration (8 ng/ml) that previous studies and our own preliminary experiments had shown was slightly below the threshold at which cell death began to occur (data not shown). RNA was collected 3 and 6 h after addition of toxin and/or LPS, and samples were analyzed and differentially expressed genes were selected at the 99% confidence level as described for infection.
As expected, the expression patterns observed in cells treated with Salmonella LPS were reflective of a profound change in cellular signaling, and in general these responses were somewhat more intense than those observed in any other experimental condition, in that the changes in both individual genes (see supplemental Table S3) and functional families (Table 3) showed higher levels of statistical significance. The genes identified in this set correspond well to those identified in previous studies examining expression trends in macrophages treated with a variety of bacterial pathogens (4, 6, 38, 46) and include many classical proteins involved in immune signaling (e.g., tumor necrosis factor alpha, interleukin-1α, macrophage colony-stimulating factor, and NF-κB).
TABLE 3.
Functional families overrepresented in the macrophage transcriptional response to treatment with Salmonella lipopolysaccharide (500 ng/ml)
| Induced
|
Repressed
|
|||
|---|---|---|---|---|
| Biological process | P | Biological process | P | |
| Cell growth | 8.9 E-8 | Mitotic cell cycle, S phase | 4.5 E-8 | |
| Regulation of cell cycle | 5.2 E-7 | Regulation of cell cycle | 4.3 E-7 | |
| Lymph gland development | 1.0 E-5 | Cytokinesis | 1.3 E-5 | |
| Regulation of apoptosis | 2.0 E-5 | Lipid metabolism | 8.0 E-5 | |
| DNA repair | 9.2 E-5 | Cell motility | 3.2 E-4 | |
| Immune response | 1.9 E-4 | Protein phosphorylation | 2.1 E-4 | |
| Angiogenesis | 2.7 E-4 | Mitotic cell cycle, M phase | 7.5 E-4 | |
| Regulation of transcription | 4.5 E-4 | Endocytosis | 0.0032 | |
| Intracellular signaling cascade | 2.6 E-4 | Apoptosis | 0.0233 | |
| Cell differentiation | 0.0021 | Protein folding | 0.0074 | |
| Amine metabolism | 0.0026 | Regulation of transcription | 0.0031 | |
| Cell proliferation | 0.0055 | Intracellular signaling cascade | 0.0066 | |
| Stress response (to pathogen/DNA damage/wounding) | 0.0130 | |||
With the expression patterns from both untreated cells and LPS-activated cells as control baselines, we examined gene expression in both groups after treatment with lethal toxin. Surprisingly, the cells treated with lethal toxin alone (unactivated macrophages) showed essentially no significant changes in gene expression compared to untreated cells. The expression profiles from these cells and those from untreated control cells (described above) looked very similar, with only 17 genes changing by ≥2-fold throughout the 6-h time course and no coherent pattern with regard to function within this small group (supplemental Table S4). The finding that gene expression did not change appreciably throughout a toxin treatment that caused appreciable cell death (10 to 20%) was somewhat surprising, but it matched the observations in a similar study recently published by Tucker et al. (54). In this report, the authors assayed expression of roughly 1,200 genes in the same cell line after treatment with significantly higher concentrations of lethal toxin and noted that although widespread necrotic cell death was occurring within these cultures, essentially no changes in gene expression were detectable.
In contrast to the trend observed in unactivated macrophages, we identified 230 genes that changed significantly in expression level between macrophages that were activated with LPS and cells that were activated and treated with lethal toxin (supplemental Table S5). These genes were sorted by gene ontology annotation as described above in order to identify overrepresented functional families (Table 4). In general, most of the host genes that showed significantly changed expression levels following treatment with lethal toxin were associated with signaling and/or control of transcription, which is consistent with the known abilities of lethal toxin to disrupt and alter host cell signal transduction. Notably, apoptosis-related loci were not overrepresented in this set, though several key genes (e.g., ornithine decarboxylase) were present, albeit at lower levels than were observed during infection. We did observe some cell death in lethal toxin- or LPS-treated macrophages within the experimental time course, but it was not to the same levels that were observed in infected cells (as expected based on our use of a significant but sublethal dose). It is therefore difficult to assess whether the absence of induced apoptosis-related genes simply reflects lower levels of cell death or if other, as yet unidentified B. anthracis factors may play roles in inducing apoptosis, and further experiments will be necessary to answer these questions.
TABLE 4.
Functional families overrepresented in the transcriptional response to treatment with anthrax lethal toxin in activated macrophages
| Induced
|
Repressed
|
|||
|---|---|---|---|---|
| Biological process | P | Biological process | P | |
| Immune response | 5.0 E-5 | Innate immune response (inflammatory response) | 0.0033 | |
| Chromatin assembly/modification | 1.0 E-4 | Receptor-linked signal transduction | 0.0043 | |
| Regulation of cell cycle | 1.3 E-4 | Regulation of transcription | 0.0374 | |
| Cell matrix adhesion | 2.4 E-4 | |||
| Receptor-linked signal transduction | 5.2 E-4 | |||
| Regulation of transcription | 0.0149 | |||
| Intracellular signaling cascade | 0.0242 | |||
| Cytoskeleton organization | 0.0253 | |||
Perhaps the most striking finding observed in lethal toxin-treated cells was that many (142, or 62%) of the genes that changed significantly in activated macrophages treated with lethal toxin were also significantly changed when comparing LPS-treated cells and untreated cells (that is, they were also part of the LPS-triggered response). Given that treatment with LPS appears to trigger a widespread activation response, the significant overlap between these two data sets (genes present in both of these two sets are noted in supplemental Table S5) implies that one of the primary effects of lethal toxin within the macrophage is in modulating the stereotypical activation program induced by bacterial antigens (e.g., the patterns reported in reference 6). In general, many of the significant lethal toxin-induced changes in gene expression appear to be in the downward direction; that is, the induction of many of the classical immune response genes that was observed in LPS-treated cells is dampened in LPS- and lethal toxin-treated cells. Prominent examples of this phenomenon are shown in Fig. 4. Previous studies have shown that lethal toxin is able to suppress a variety of signals associated with activation of the immune system (2, 9, 10, 18, 42, 55). Our results are consistent with those reports, and they demonstrate this effect at the transcriptional level for the first time.
FIG. 4.
Dampening of immune response genes following treatment with anthrax lethal toxin. Bars indicate the level of induction relative to expression in untreated cells, as measured by DNA microarray. For each gene indicated, induction levels are shown for cells treated with LPS and cells treated with LPS and lethal toxin (LeTx).
Although many of the classical immune response genes were repressed in response to lethal toxin, a smaller subset of genes were upregulated in lethal toxin-treated activated macrophages. These genes, including the class I major histocompatibility complex locus, JAK-2, calcipressin, and macrophage colony-stimulating factor, are all associated with gamma interferon (IFN-γ)-mediated signaling and/or generation of TH-1 immunity (11, 39, 49). IFN-γ has been shown to play an important role in host responses to other intracellular bacterial pathogens, including Listeria monocytogenes and Mycobacterium tuberculosis (52), and a recent study by Gold et al. showed that exogenous IFN-γ is able to improve survival rates of human macrophages infected with B. anthracis (22). This report also showed that during infection by B. anthracis, IFN-α/β signaling is disrupted while IFN-γ is not, which is consistent with the implication from our data that although lethal toxin is able to dampen much of the stereotypical macrophage response, it appears to leave IFN-γ signaling intact.
It seems intuitive that IFN-γ and its associated signaling pathways would be important components in the macrophage response to B. anthracis, and it is somewhat interesting that lethal toxin has not evolved a means of IFN-γ signaling as it disrupts other pathways. Although IFN-γ has a protective effect on infected macrophages in cell culture and thus appears to work to the detriment of the infecting bacteria, it remains possible that there are more broad-reaching effects of IFN-γ signaling in vivo that may work to the advantage of B. anthracis at the organismal level and that these effects ensure that there is no selective pressure toward evolution of a means for IFN-γ disruption.
Conclusions.
This study represents the first characterization of the transcriptional changes within macrophages during infection by Bacillus anthracis and the first genomewide characterization of transcriptional changes in macrophages treated with anthrax lethal toxin. Although many of the genes identified as differentially regulated have been previously shown to play a role in the general innate immune response, we have also identified a limited set of macrophage genes that are induced or repressed in response to B. anthracis but have not yet been identified as such in other bacteria-host systems. These genes provide a transcriptional basis for many of the phenomena observed during anthrax, including the induction of apoptosis within host phagocytes, and it will be of considerable interest to see if future studies characterizing the macrophage response to other pathogens show the regulation of any of these genes to be truly specific to the B. anthracis-macrophage interaction.
We anticipate that this analysis of gene expression patterns during infection and toxin treatment will be valuable in ongoing efforts to develop new detection and therapeutic options for anthrax. It must be noted that these studies were all done with an attenuated strain of B. anthracis, and thus, although the data presented here are comprehensive relative to that strain, fully virulent B. anthracis (carrying pXO2) may cause other changes in host gene expression that were not identified here. It will therefore be of considerable interest to extend these studies to wild-type B. anthracis, and we are pursuing these studies.
We expect that these data will be of use to those in the B. anthracis field as well as to the general pathogenesis community, and we have made all microarray data collected in this study publicly available via the TIGR website (http://pga.tigr.org) and in the ArrayExpress database (http://www.ebi.ac.uk/arrayexpress/, accession number E-TIGR-84). We are continuing to examine these data for important insights, and we are also extending the techniques described in this work to the study of primary cells in culture and in vivo.
Acknowledgments
We thank members of the Hanna and Quackenbush groups for valuable discussions and Jeff McCarroll for help with database management.
This work was supported in part by the TIGR Program for Genomic Application Visiting Investigator Program (U01-HL66580, to J.Q.); the Great Lakes and Southeast Regional Centers of Excellence for Biodefense, HHS contract N266200400059C/N01-AI-40059, NIH grant AI08649, and ONR grants N00014-02-0061, N00014-00-1-0422, and N00014-01-1-1044 (to P.C.H.); a Pharmaceutical Research and Manufacturers Association Foundation Postdoctoral Research Award (to N.H.B.); a University of Michigan Cellular Biotechnology Training Program predoctoral fellowship (to K.D.C.); and a University of Michigan Medical School Molecular Mechanisms of Microbial Pathogenesis Postdoctoral Fellowship and a GLRCE Postdoctoral Training Award (to L.M.S.-R.).
Editor: D. L. Burns
REFERENCES
- 1.Abulencia, J. P., R. Gaspard, Z. R. Healy, W. A. Gaarde, J. Quackenbush, and K. Konstantopoulos. 2003. Shear-induced cyclooxygenase-2 via a JNK2/c-Jun-dependent pathway regulates prostaglandin receptor expression in chondrocytic cells. J. Biol. Chem. 278:28388-28394. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, A., J. Lingappa, S. H. Leppla, S. Agrawal, A. Jabbar, C. Quinn, and B. Pulendran. 2003. Impairment of dendritic cells and adaptive immunity by anthrax lethal toxin. Nature 424:329-334. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, D., V. Vanchinathan, P. Brown, and J. Theriot. 2003. A gene-expression program reflecting the innate immune response of cultured intestinal epithelial cells to infection by Listeria monocytogenes. Genome Biol. 4:R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belcher, C., J. Drenkow, B. Kehoe, T. Gingeras, N. McNamara, H. Lemjabbar, C. Basbaum, and D. Relman. 2000. The transcriptional responses of respiratory epithelial cells to Bordetella pertussis reveal host defensive and pathogen counter defensive strategies. Proc. Natl. Acad. Sci. USA 97:13847-13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhatnagar, R., and S. Batra. 2001. Anthrax toxin. Crit. Rev. Microbiol. 27:167-200. [DOI] [PubMed] [Google Scholar]
- 6.Boldrick, J., A. Alizadeh, M. Diehn, S. Dudoit, C. Liu, C. Belcher, D. Botstein, L. Staudt, P. Brown, and D. Relman. 2002. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc. Natl. Acad. Sci. USA 99:972-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brancho, D., N. Tanaka, A. Jaeschke, J. J. Ventura, N. Kelkar, Y. Tanaka, M. Kyuuma, T. Takeshita, R. A. Flavell, and R. J. Davis. 2003. Mechanism of p38 MAP kinase activation in vivo. Genes Dev. 17:1969-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brune, B., P. Hartzell, P. Nicotera, and S. Orrenius. 1991. Spermidine prevents endoclease activation and apoptosis in thymocytes. Exp. Cell Res. 195:323-329. [DOI] [PubMed] [Google Scholar]
- 9.Cordoba-Rodriguez, R., H. Fang, C. Lankford, and D. Frucht. 2004. Anthrax lethal toxin rapidly activates caspase-1/ICE and induces extracellular release of IL-1 beta and IL-18. J. Biol. Chem. 279:20563-20566. [DOI] [PubMed] [Google Scholar]
- 10.Dang, O., L. Navarro, K. Anderson, and M. David. 2004. Cutting edge: anthrax lethal toxin inhibits activation of IFN-regulatory factor 3 by lipopolysaccharide. J. Immunol. 172:747-751. [DOI] [PubMed] [Google Scholar]
- 11.Delneste, Y., P. Charbonnier, N. Herbault, G. Magistrelli, G. Caron, J. Bonnefoy, and P. Jeannin. 2003. Interferon-gamma switches monocyte differentiation from dentritic cells to macrophages. Blood 101:143-150. [DOI] [PubMed] [Google Scholar]
- 12.Dixon, T., A. Fahd, T. Koehler, J. Swanson, and P. Hanna. 2000. Early events in anthrax pathogenesis: Intracellular survival of B. anthracis and its escape from RAW264.7 macrophages. Cell. Microbiol. 2:453-463. [DOI] [PubMed] [Google Scholar]
- 13.Dixon, T., M. Meselson, J. Guillemin, and P. Hanna. 1999. Medical progress: anthrax. N. Engl. J. Med. 341:815-826. [DOI] [PubMed] [Google Scholar]
- 14.Draghici, S., P. Khatri, P. Bhavsar, A. Shah, S. A. Krawetz, and M. A. Tainsky. 2003. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 31:3775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duesbery, N., C. Webb, and S. Leppela, et al. 1998. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science 280:734-737. [DOI] [PubMed] [Google Scholar]
- 16.Dutz, W., and E. Kohout. 1971. Anthrax. Pathol. Annu. 6:209-248. [PubMed] [Google Scholar]
- 17.Dutz, W., and E. Kohout-Dutz. 1981. Anthrax. Int. J. Dermatol. 20:203-206. [DOI] [PubMed] [Google Scholar]
- 18.Erwin, J. L., L. M. DaSilva, S. Bavari, S. F. Little, A. M. Friedlander, and T. C. Chanh. 2001. Macrophage-derived cell lines do not express proinflammatory cytokines after exposure to Bacillus anthracis lethal toxin. Infect. Immun. 69:1175-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eto, A., T. Muta, S. Yamazaki, and K. Takeshige. 2003. Essential roles for NF-kappa B and a Toll/IL-1 receptor domain-specific signal(s) in the induction of I kappa B-zeta. Biochem. Biophys. Res. Commun. 301:495-501. [DOI] [PubMed] [Google Scholar]
- 20.Fukao, T. 2004. Immune system paralysis by anthrax lethal toxin: the roles of innate and adaptive immunity. Lancet Infect. Dis. 4:166-170. [DOI] [PubMed] [Google Scholar]
- 21.Gobert, A. P., Y. Cheng, J. Y. Wang, J. L. Boucher, R. K. Iyer, S. D. Cederbaum, R. A. Casero, Jr., J. C. Newton, and K. T. Wilson. 2002. Helicobacter pylori induces macrophage apoptosis by activation of arginase II. J. Immunol. 168:4692-4700. [DOI] [PubMed] [Google Scholar]
- 22.Gold, J., Y. Hoshino, S. Hoshino, M. Jones, A. Nolan, and M. Weiden. 2004. Exogenous gamma and alpha/beta interferon rescues human macrophages from cell death induced by Bacillus anthracis. Infect. Immun. 72:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guidi-Rontani, C., Y. Pereira, S. Ruffie, J. Sirard, M. Weber-Levy, and M. Mock. 1999. Identification and characterization of the germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Mol. Microbiol. 33:407-414. [DOI] [PubMed] [Google Scholar]
- 24.Guidi-Rontani, C., M. Weber-Levy, E. Labruyere, and M. Mock. 1999. Germination of Bacillus anthracis spores within alveolar macrophages. Mol. Microbiol. 31:9-17. [DOI] [PubMed] [Google Scholar]
- 25.Hanna, P. 1999. Lethal toxin actions and their consequences. J. Appl. Microbiol. 87:285-287. [DOI] [PubMed] [Google Scholar]
- 26.Hanna, P., B. Kruskal, R. Ezekowitz, B. Bloom, and R. Collier. 1994. Role of macrophages oxidative burst in the action of anthrax lethal toxin. Mol. Med. 1:1-12. [PMC free article] [PubMed] [Google Scholar]
- 27.Hortelano, S., M. Zeini, A. Castrillo, A. M. Alvarez, and L. Bosca. 2002. Induction of apoptosis by nitric oxide in macrophages is independent of apoptotic volume decrease. Cell Death Differ. 9:643-650. [DOI] [PubMed] [Google Scholar]
- 28.Inglesby, T., D. Henderson, Bartlett, J. G., and e. al. 1999. Anthrax as a biological weapon: medical and public health management. JAMA 281:1735-1745. [DOI] [PubMed] [Google Scholar]
- 29.Inglesby, T., T. O'Toole, D. Henderson, B. J. G., M. Ascher, E. Eitzen, A. Friedlander, J. Gerberding, J. Hauer, J. Hughes, J. McDade, M. Osterholm, G. Parker, T. Perl, P. Russell, and K. Tonat. 2002. Anthrax as a biological weapon-updated recommendations for management. JAMA 287:2236-2252. [DOI] [PubMed] [Google Scholar]
- 30.Ireland, J., and P. Hanna. 2002. Macrophage-enhanced germination of Bacillus anthracis endospores requires gerS. Infect. Immun. 70:5870-5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ireland, J. A., and P. C. Hanna. 2002. Amino acid- and purine ribonucleoside-induced germination of Bacillus anthracis DeltaSterne endospores: gerS mediates responses to aromatic ring structures. J. Bacteriol. 184:1296-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan, A., P. DiMascio, M. Medeiros, and T. Wilson. 1992. Spermine and spermidine protection of plasmid DNA against single-strand breaks induced by singlet oxygen. Proc. Natl. Acad. Sci. USA 89:11428-11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kirby, J. 2004. Anthrax lethal toxin induces human endothelial cell apoptosis. Infect. Immun. 72:430-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitamura, H., K. Kanehira, T. Shiina, M. Morimatsu, B. Jung, S. Akashi, and M. Saito. 2002. Bacterial lipopolysaccharide induces mRNA expression of an I-kappaB MAIL through Toll-like receptor 4. J. Vet. Med. Sci. 64:419-422. [DOI] [PubMed] [Google Scholar]
- 35.Li, X., B. U. Bradford, F. Dalldorf, S. M. Goyert, S. A. Stimpson, R. G. Thurman, and S. S. Makarov. 2004. CD14 mediates the innate immune responses to arthritopathogenic peptidoglycan-polysaccharide complexes of Gram-positive bacterial cell walls. Arthritis Res. Ther. 6:R273-R281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, H., N. H. Bergman, B. Thomason, S. Shallom, A. Hazen, J. Crossno, D. A. Rasko, J. Ravel, T. D. Read, S. N. Peterson, J. Yates 3rd, and P. C. Hanna. 2004. Formation and composition of the Bacillus anthracis endospore. J Bacteriol 186:164-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moayeri, M., and S. Leppela. 2004. The roles of anthrax toxin in pathogenesis. Curr. Opin. Microbiol. 7:19-24. [DOI] [PubMed] [Google Scholar]
- 38.Nau, G., J. Richmond, A. Schlesinger, E. Jennings, E. Lander, and R. Young. 2002. Human macrophage activation programs induced by bacterial pathogens. Proc. Natl. Acad. Sci. USA 99:1503-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parganas, E., D. Wang, D. Stravopodis, D. Topham, J. Marine, S. Teglund, E. Vanin, S. Bodner, O. Colamonici, J. van Deursen, G. Grosveld, and J. Ihle. 1998. Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93:385-395. [DOI] [PubMed] [Google Scholar]
- 40.Park, J., Y. Chung, S. Kang, J. Kim, Y. Kim, H. Kim, Y. Kim, J. Kim, and Y. Yoo. 2002. c-Myc exerts a protective function through ornithine decarboxylase against cellular insults. Mol. Pharmacol. 62:1400-1408. [DOI] [PubMed] [Google Scholar]
- 41.Park, J. M., F. R. Greten, Z. Li, and M. Karin. 2002. Macrophage apoptosis by anthrax lethal factor through p38 map kinase inhibition. Science 297:2048-2051. [DOI] [PubMed] [Google Scholar]
- 42.Pellizzari, R., C. Guidi-Rontani, G. Vitale, M. Mock, and C. Montecucco. 1999. Anthrax lethal factor cleaves MKK3 in macrophages and inhibits the LPS-IFN gamma-induced release of NO and TNFalpha. FEBS Lett. 462:199-204. [DOI] [PubMed] [Google Scholar]
- 43.Popov, S., T. Popova, E. Grene, F. Klotz, J. Cardwell, C. Bradburne, Y. JAMA, M. Maland, J. Wells, A. Nalca, T. Voss, C. Bailey, and K. Alibek. 2004. Systemic cytokine response in murine anthrax. Cell. Microbiol. 6:225-233. [DOI] [PubMed] [Google Scholar]
- 44.Popov, S. G., R. Villasmil, J. Bernardi, E. Grene, J. Cardwell, T. Popova, A. Wu, D. Alibek, C. Bailey, and K. Alibek. 2002. Effect of Bacillus anthracis lethal toxin on human peripheral blood mononuclear cells. FEBS Lett. 527:211-215. [DOI] [PubMed] [Google Scholar]
- 45.Popov, S. G., R. Villasmil, J. Bernardi, E. Grene, J. Cardwell, A. Wu, D. Alibek, C. Bailey, and K. Alibek. 2002. Lethal toxin of Bacillus anthracis causes apoptosis of macrophages. Biochem. Biophys. Res. Commun. 293:349-355. [DOI] [PubMed] [Google Scholar]
- 46.Rosenberger, C. M., M. G. Scott, M. R. Gold, R. E. Hancock, and B. B. Finlay. 2000. Salmonella typhimurium infection and lipopolysaccharide stimulation induce similar changes in macrophage gene expression. J. Immunol. 164:5894-5904. [DOI] [PubMed] [Google Scholar]
- 47.Ross, J. 1957. The pathogenesis of anthrax following the administration of spores by the respiratory route. J. Pathol. Bacteriol. 73:485-494. [Google Scholar]
- 48.Ruthel, G., W. J. Ribot, S. Bavari, and T. A. Hoover. 2004. Time-lapse confocal imaging of development of Bacillus anthracis in macrophages. J. Infect. Dis. 189:1313-1316. [DOI] [PubMed] [Google Scholar]
- 49.Ryeom, S., R. Greenwald, A. Sharpe, and F. McKeon. 2003. The threshold pattern of calcineurin-dependent gene expression is altered by loss of the endogenous inhibitor calcipressin. Nat. Immunol. 4:874-881. [DOI] [PubMed] [Google Scholar]
- 50.Saeed, A. I., V. Sharov, J. White, J. Li, W. Liang, N. Bhagabati, J. Braisted, M. Klapa, T. Currier, M. Thiagarajan, A. Sturn, M. Snuffin, A. Rezantsev, D. Popov, A. Ryltsov, E. Kostukovich, I. Borisovsky, Z. Liu, A. Vinsavich, V. Trush, and J. Quackenbush. 2003. TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374-378. [DOI] [PubMed] [Google Scholar]
- 51.Schipper, R., J. Romijn, V. Cuijpers, and A. Verhofstad. 2003. Polyamines and prostatic cancer. Biochem. Soc. Trans. 31:375-380. [DOI] [PubMed] [Google Scholar]
- 52.Stark, G., I. Kerr, B. Williams, R. Silverman, and R. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 53.Tasaka, S., A. Ishizaka, W. Yamada, M. Shimizu, H. Koh, N. Hasegawa, Y. Adachi, and K. Yamaguchi. 2003. Effect of CD14 blockade on endotoxin-induced acute lung injury in mice. Am. J. Respir. Cell. Mol. Biol. 29:252-258. [DOI] [PubMed] [Google Scholar]
- 54.Tucker, A., I. Salles, D. Voth, W. Ortiz-Leduc, H. Wang, I. Dozmorov, M. Centola, and J. Ballard. 2003. Decreased glycogen synthase kinase 3-beta levels and related physiological changes in Bacillus anthracis lethal toxin-treated macrophages. Cell. Microbiol. 5:523-532. [DOI] [PubMed] [Google Scholar]
- 55.Webster, J. I., L. H. Tonelli, M. Moayeri, S. S. Simons, Jr., S. H. Leppla, and E. M. Sternberg. 2003. Anthrax lethal factor represses glucocorticoid and progesterone receptor activity. Proc. Natl. Acad. Sci. USA 100:5706-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiner, M., and P. Hanna. 2003. Macrophage-mediated germination of Bacillus anthracis endospores requires the gerH operon. Infect. Immun. 7:3954-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The expression data collected in this study are publicly available via the TIGR website (http://pga.tigr.org) and the ArrayExpress database (accession number E-TIGR-84). Complete lists of genes that were identified as differentially expressed in response to any of the experimental conditions described here are in the supplemental material at http://www.sitemaker.umich.edu/hanna_lab.