Abstract
Arg-gingipain (Rgp) and Lys-gingipain (Kgp) are Porphyromonas gingivalis cysteine proteinases implicated as major virulence factors in pathologies of periodontitis. We purified a 660-kDa cell-associated gingipain complex existing as a homodimer of two catalytically active monomers which comprises their catalytic and adhesin domains. Electron microscopy revealed that the complex was composed of a globular particle with a 10-nm external diameter possessing one or two electron-dense hole-like structures. Two-dimensional gel electrophoresis and immunoblot analyses revealed the association of lipopolysaccharide (LPS) with the catalytic domains and a hemagglutinin domain, Hgp44, of Rgp and Kgp in the complex. The complex significantly degraded human type I collagen and elastin and strongly disrupted viability of human gingival fibroblasts and umbilical vein endotherial cells with an efficiency which was higher than that of the monomeric gingipains. The native complex produced only a small amount of nitrogen dioxide, tumor necrosis factor alpha, and interleukin-6 by macrophages, whereas the heat-denatured complex resulted in increased production. Inhibition of the proteolytic activities of the gingipain complex did not up-regulate the cytokine production, indicating that the functional domains in LPS are structurally masked by the complex proteins. These results indicate the importance of the complex in evasion of host defense mechanisms as well as in host tissue breakdown.
Gingipains are cysteine proteinases produced by Porphyromonas gingivalis, a gram-negative anaerobic bacterium associated with some types of periodontitis including chronic adult and progressive periodontitis (1, 14, 16, 18-20, 36, 38, 42). Gingipains are composed of Arg-specific (Arg-gingipains [Rgps]) and Lys-specific (Lys-gingipain [Kgp]) endopeptidases. Rgps are encoded by two rgp genes (rgpA and rgpB) (13, 24, 26, 27, 32, 35), whereas Kgp is encoded by a single kgp gene (33, 34). rgpA and rgpB are essentially identical, except that rgpB lacks most of the C-terminal adhesin domains of rgpA. Interestingly, the C-terminal adhesin domains of rgpA and kgp are highly homologous, although the propeptide and proteinase domains have no sequence similarity (21, 33, 36). Using various Rgp- and/or Kgp-deficient mutants as well as soluble gingipains purified from the culture supernatant of P. gingivalis strains, the virulence of the bacterium has been shown to be exclusively attributable to gingipains (1, 20, 26-28). These include extensive degradation of various host proteins including collagen, fibronectin, and fibrinogen (1, 20, 34), cytokines such as interleukin-6 (IL-6), IL-8, and tumor necrosis factor alpha (TNF-α) (7, 10, 31), complement factors C3 and C5 (47), and immunoglobulins (1, 20); disruption of the bactericidal activity of polymorphonuclear leukocytes (1, 20, 26); and strong induction of human fibroblast (3, 4) and human umbilical vein endothelial cell (HUVEC) (5) death. In addition, gingipains are also shown to be important for the bacterium to proliferate and survive in the periodontal pockets (26, 34, 39). The pathophysiological importance of gingipains has been further substantiated by newly developed gingipain inhibitors (22).
Gingipains are produced as secreted or membrane-associated forms on the cell surface (37, 41). The cell-associated gingipains comprise the majority (∼80%) of Rgp and Kgp activities (unpublished data) and are thus believed to be responsible for the virulence of the bacterium. Accordingly, the characterization and subsequent control of the cell-associated gingipain complex are thought to be the most promising therapeutic approaches for periodontitis and related systemic disorders including atherosclerosis and premature birth. Recently, Bhogal et al. (8) demonstrated a ∼300-kDa cell-associated gingipain complex composed of both catalytic domains (PrtR45 and PrtK48) and seven C-terminal hemagglutinin/adhesin domains (PrtR44, PrtR15, PrtR17, PrtR27, PrtK39, PrtK15, and PrtK44) encoded by two genes, prtR (rgpA) and prtK (kgp), in the cell sonicate of P. gingivalis. Whereas the structures of the gingipain complexes were well defined in the previous study, the functional significances of the complexes are incompletely understood.
Lipopolysaccharide (LPS), a component of the outer membrane of gram-negative bacteria, is a potent virulence factor causing toxic shock in the host (25). It has previously been shown that several proteins in P. gingivalis, including RgpB, RgpAA4 (Hgp27), and uncharacterized proteins P59 and P27, are modified with LPS (45). LPS is known to stimulate host cells mainly via the Toll-like receptor 4 (TLR4)/MD-2 pathway. However, previous studies have demonstrated that unlike enterobacterial LPS, P. gingivalis LPS uses TLR2 to induce innate immune responses in both human and mouse macrophages (15, 23). It has also previously been reported that P. gingivalis LPS is able to suppress the biological activity of TLR4 agonists (11, 48). Furthermore, the production of cytokines induced by P. gingivalis LPS has been shown to be negligible when compared with that of Escherichia coli LPS (15).
In the present study, we obtained a large cell-associated gingipain complex by detergent extraction. The purified gingipain complex was found to be modified by LPS. Nevertheless, LPS in the complex was barely able to stimulate human and mouse macrophages. Thus, the critical features of the gingipain complex, including high cytotoxicity, marked degradation of matrix proteins, and evasion of the host immune response, which are closely related to the virulence of the bacterium, were demonstrated.
MATERIALS AND METHODS
Chemicals.
Synthetic chromogenic substrates and protease inhibitors were purchased from the Peptide Institute (Osaka, Japan). HUVEC and gingival fibroblasts (Gin-1) were from Cell Applications Inc. and Dainippon Pharmaceutical Co. (Osaka, Japan). Human monocytic leukemia THP-1 cells were a kind gift from Hideaki Sakai, Nagasaki University, Nagasaki, Japan. Sucrose monolaurate and Detergent Starter Kit II were from DOJINDO (Kumamoto, Japan). A monoclonal antibody that recognizes lipid A of LPS was from HyCult Biotechnology B.V. (Uden, The Netherlands). Rgp- and Kgp-specific inhibitors, designated KYT-1 and KYT-36, respectively, were synthesized by procedures described previously elsewhere (22). All other chemicals were reagent grade or better.
Bacteria and culture conditions.
P. gingivalis ATCC 33277 was used as a wild-type strain. The Rgp-deficient (rgpA rgpB-deficient) mutant and the Kgp-deficient (kgp-deficient) mutant were described previously (26, 35, 39). The wild-type strain and these mutants were grown under anaerobic conditions (10% CO2, 10% H2, 80% N2) in enriched brain heart infusion broth at 37°C. Erythromycin (10 μg/ml) and tetracycline (1 μg/ml) were added to the medium if necessary.
Determinations.
Proteolytic activities of Rgp and Kgp were determined with benzyloxycarbonyl (Z)-Phe-Arg-4-methyl-7-coumarylamide (MCA) and Z-His-Glu-Lys-MCA as the respective substrates, as described previously (2, 20). One unit of each enzyme activity was defined as the amount of enzyme required for the release of 1 nmol of 7-amino-4-methylcoumarin/ml per min under the conditions used.
The collagenolytic activity assay was performed as previously described (6), with some modifications. Samples were added to a 96-well plate which was precoated with 500 μg of fluorescein isothiocyanate (FITC)-labeled type I collagen/ml and incubated for 6 h at 37°C. The inhibitor cocktail containing leupeptin (10 mM), EDTA (10 mM), and tosyl-l-phenylalanine chloromethyl ketone (1 mM) was then added to terminate the reaction, and the solubilized gelatin fragments were measured by using a Wallac 1420 ARVOsx (excitation and emission maxima at 490 and 535 nm, respectively; PerkinElmer). Collagenase from Clostridium histolyticum (type I; Sigma) was used as a positive control.
Elastolytic activity was assessed by an elastin-fluorescein (200/400 mesh) degradation assay as previously described (30), with some modifications. The samples were incubated with 10 μl of elastin-fluorescein (5 mg/ml) in 10 mM phosphate buffer, pH 7.5, containing 0.5 mM CaCl2, 0.2 mg of bovine serum albumin/ml, 5 mM cysteine, and 0.025% sucrose monolaurate at 37°C for 2 h with rapid shaking. Ten microliters of 0.2 M EDTA and 10 mM tosyl-l-phenylalanine chloromethyl ketone was then added to the reaction mixtures and centrifuged at 8,000 × g rpm for 10 min. The fluorescence intensity in the supernatant was measured at an excitation wavelength of 490 nm and an emission wavelength of 535 nm.
Quantities of cytokines and nitrogen dioxide (NO2) were measured as follows. The cells were exposed to the freshly prepared or heat-denatured cell-associated gingipain complex (4 ng/well) in the medium at 37°C for 24 h. At the end of the incubation period, the supernatants were collected and analyzed. IL-6 and TNF-α were measured by enzyme-linked immunosorbent assay with specific reagent for each cytokine (BIOSOURCE, Camarillo, Calif.) according to the manufacturer's instructions. NO2, the end product of NO, was quantified using an NO2/NO3 Assay Kit-F II (DOJINDO).
Purification of the cell-associated gingipain complexes.
The purification steps are shown in Table 1. P. gingivalis cells were harvested by centrifugation at 10,000 × g for 20 min at 4°C and washed twice with phosphate-buffered saline (PBS). All steps were carried out at 4°C unless stated otherwise. The washed cells were resuspended in 0.5% sucrose monolaurate-PBS and allowed to stand for 3 h. After centrifugation at 105,000 × g for 30 min, the supernatant was filtered (0.22 μm) prior to gel filtration fast-performance liquid chromatography. The filtrate was applied to a column (1.8 by 70 cm) containing Sephacryl S-300 (Amersham Biosciences) that had been equilibrated with 20 mM sodium phosphate buffer containing 0.15 M NaCl and 0.5% sucrose monolaurate. The column was washed at a flow rate of 0.3 ml/min. The active enzyme fractions for Rgp and Kgp were pooled and concentrated. The enzyme solution containing both activities was passed through a column (2 by 6 cm) containing DEAE-Sepharose (Amersham Biosciences) equilibrated with the same buffer. After washing the column with the buffer, the adsorbed proteins were eluted by a stepwise gradient of NaCl (0, 0.1, 0.2, 0.3, and 0.5 M) in the buffer. The active enzyme fractions eluted at 0.2 M NaCl, which contained both Rgp and Kgp activities, were collected and concentrated by ultrafiltration (Centriprep YM30; Amicon). The enzyme solution was applied to a gel filtration column (2.6 by 60 cm) containing Superdex 200 (Amersham Pharmacia) equilibrated with 20 mM sodium phosphate buffer containing 0.15 M NaCl and 0.5% sucrose monolaurate. The column was washed at a flow rate of 1.1 ml/min. The active enzyme fractions were pooled and concentrated.
TABLE 1.
Purification of the gingipain complexes from P. gingivalisa
| Purification step | Rgp spec act (U/mg) | Kgp spec act (U/mg) |
|---|---|---|
| Soluble extract | 3,003 | 576 |
| Sephacryl S-300 | 8,688 | 1,747 |
| DEAE-sepharose | 5,893 | 3,488 |
| Superdex 200 | 9,976 | 7,880 |
Purification of monomeric Rgp and Kgp.
Monomeric Rgp and Kgp were purified from the supernatants of P. gingivalis KDP129 and KDP133, respectively, according to methods previously reported (1, 20, 26, 35).
Antibodies.
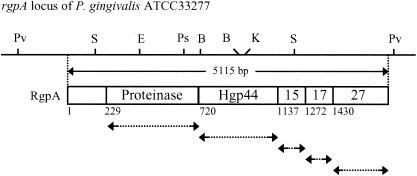
His6-tagged recombinant proteins of the proteinase domain, Hgp44, Hgp15, Hgp17, and Hgp27 of RgpA were overexpressed in E. coli BL21 carrying expression plasmids. The expression plasmids were constructed as follows: DNA for each domain of RgpA was amplified by PCR with various primers using P. gingivalis ATCC 33277 chromosomal DNA as a template (Fig. 1). The nucleotide sequences of the primers were as follows: 5′-ATCCATATGAAAAACTTGAACAAGTTTGTTTCG-3′ and 5′-AATGGATCCCGAAGAAGTTCGGGGGCATCGCTG-3′ for the proteinase domain of RgpA, 5′-GAATTCAGCGGTCAGGCCGAGATTGTTC-3′ and 5′-AAGCTTGCGCTTGCCGTTGGCCTTGATC-3′ for Hgp44, 5′-GAATTCGCAGACTTCACGGAAACGTTCG-3′ and 5′-AAGCTTTTTGGCGCCATTGGCTTCCGT-3′ for Hgp15, 5′-GAATTCCCTCAAAGTGTATGGATCGAGC-3′ and 5′-AAGCTTACGTACATCGTTTGCAGGTTCG-3′ for Hgp17, and 5′-GAATTCGCCAACGAAGCCAAGGTTGTGC-3′ and 5′-AAGCTTCTTTACAGCGAGTTTCTCTACG-3′ for Hgp27. The PCR products shown by arrows in Fig. 1 were inserted into the pGemT vector (Promega) and confirmed the sequences. The plasmids carrying hgp44, hgp15, hgp17, and hgp27 genes were digested with EcoRI and HindIII, and the plasmid of the proteinase domain gene was digested with NdeI and BamHI. The resulting DNA fragments were inserted into plasmid pET28a digested with the same restriction enzymes. His-tagged recombinant proteins were purified by using a HiTrap Chelating Sepharose column (5 ml; Amersham Pharmacia Biotech) loaded Ni2+ ions. Polyclonal antibodies to these recombinant proteins and the synthetic peptide NH2-DVYTDHGDLYNTPVRMC-COOH corresponding to the N-terminal 16-amino-acid sequence of the proteinase domain of Kgp were raised in rabbits. The immunoglobulin G fraction from each of these antisera was prepared by ammonium sulfate fractionation followed by protein A-Sepharose affinity chromatography.
FIG. 1.
Schematic representation of preparation of recombinant proteins of the proteinase and adhesin domains of RgpA. The DNA for each domain of RgpA was amplified by PCR with primers (arrows) using P. gingivalis ATCC 33277 chromosomal DNA as a template. The PCR products were inserted into plasmid pET28a. The numbers in the RgpA structure indicate the predicted amino acid residues assigned based on its nucleotide sequence.
Binding to lipids in liposomes.
Various synthetic phospholipids were solubilized in chloroform (10 mg/ml) and dried under nitrogen atmosphere and then suspended in 20 mM sodium phosphate buffer, pH 7.0, containing 0.15 M NaCl and 0.5% sucrose monolaurate. The suspension was dispersed by use of a bath sonicator and incubated for 30 min at room temperature. For preparation of lipid vesicles, the sonicated sample was diluted rapidly 100 times with 20 mM sodium phosphate buffer, pH 7.0, containing 0.15 M NaCl. Both the cell-associated gingipain complex and the monomeric enzymes were added to lipid vesicles with or without 1 mM Ca2+ and incubated for 1 h on ice. Lipid vesicles were then collected by centrifugation at 105,000 × g for 30 min at 4°C, and the amounts of the bound enzymes were determined.
Culture conditions of HUVEC, Gin-1, and THP-1.
HUVEC were grown in MCDB 151 medium (Sigma, St. Louis, Mo.) supplemented with 0.1% NaHCO3, 15% fetal bovine serum (FBS), 10 ng of recombinant acid fibroblast growth factor/ml, 10 μg of heparin/ml, and 60 μg of kanamycin/ml in humidified 5% CO2 at 37°C. Gin-1 was grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, 1% penicillin-streptomycin, and 1% l-glutamine in humidified 5% CO2 at 37°C. THP-1 cells were maintained in RPMI 1640 medium (GIBCO, Auckland, New Zealand) supplemented with 10% heat-inactivated FBS and 100 U of penicillin/ml and 100 μg of streptomycin/ml at 37°C and 5% CO2. THP-1 cells were then differentiated for 72 h in the presence of 10 ng of phorbol 12-myristate 13-acetate/ml, washed three times, and rested overnight.
Assessment of cell viability.
HUVEC monolayers and Gin-1 cultured overnight in each medium containing 15 and 10% FBS in 96-well microtiter plates were changed with each medium in the absence of FBS with or without the purified complex or the monomeric form of either Rgp or Kgp and then incubated up to 7 and 24 h, respectively. A total of 2.7 units of Rgp and 2.4 units of Kgp, based on units required for the release of synthetic substrate described above, were used for each cell. Viability of the cells was measured with Cell Counting Kit 8 (DOJINDO) as described previously (3-5).
Preparations of peritoneal macrophages.
Peritoneal cells were collected from inbred 8- to 12-week-old mice (C57BL/6 and LPS-nonresponsive C3H/HeJ) that had been intraperitoneally injected with 2 ml of 4% thioglycolate before 4 days by lavage of the peritoneal cavity with 10-ml portions of saline. The cell suspension was dispensed into plastic petri dishes. After incubation at 37°C for 1 h, the nonadherent cells were removed, and the remaining macrophage monolayers were rinsed three times with saline before fresh RPMI 1640 medium supplemented with 10% heat-inactivated FBS, 50 U of penicillin/ml, and 50 μg of streptomycin/ml was added. The macrophages were cultivated overnight at 37°C to give a uniform monolayer of well-spread cells composed of more than 95% macrophages.
Electron microscopy.
The purified membrane-associated enzyme complex absorbed to glow-discharged carbon-coated grids was washed twice with distilled water, blotted with filter paper, and then negatively stained with 0.5% uranyl formate for 30 to 60 s. The sample was analyzed with a JEM 2000EX electron microscope (JEOL Co., Ltd., Akishima, Japan) at 100 kV.
Two-dimensional electrophoresis.
IPG strips, pH 4 to 7 (Amersham Pharmacia Biotech), were rehydrated with gingipain complex for 12 h at room temperature. Isoelectric focusing was performed with a Multiphor II (Amersham Pharmacia Biotech) for a total of 8 kVh at 20°C. Strips were equilibrated for 15 min in 50 mM Tris-HCl, pH 8.8, 6 M urea, 30% glycerol, 1% sodium dodecyl sulfate (SDS), and 64 mM dithiothreitol and were then equilibrated for 15 min in the same buffer containing 135 mM iodoacetamide instead of dithiothreitol. Equilibrated IPG strips were transferred onto 10% polyacrylamide gels. SDS gels were electroblotted onto nitrocellulose membranes. Membranes were incubated with various primary antibodies (anti-Rgp, -Kgp, -Hgp44, or -Hgp27 or lipid A antibody) and visualized.
RESULTS
Characterization of a cell-associated gingipain complex obtained by detergent solubilization.
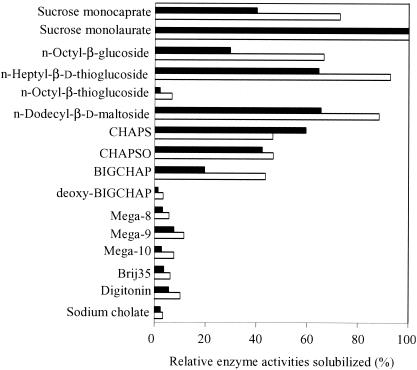
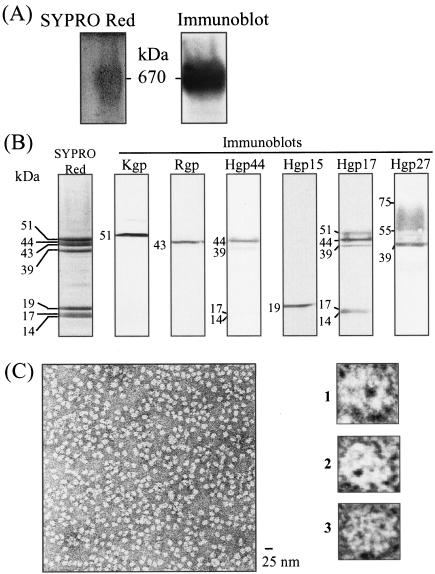
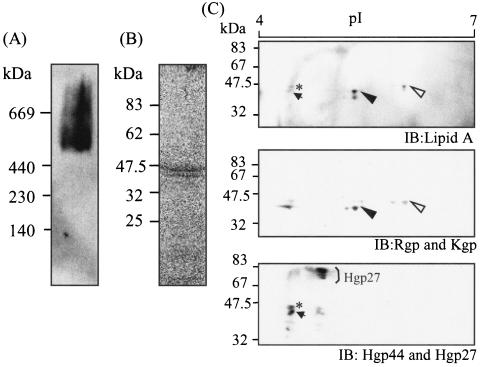
Gingipains exist mainly as cell-associated forms and partially as secreted forms. Several reports referred to the cell-associated gingipain complexes that were isolated by use of French press or sonication to release them from the bacterial cell surface. We adopted detergent solubilization to obtain the intact complexes comprising whole components. Various agents were tested for solubilization of the cell-associated gingipain activities. Both Rgp and Kgp activities were most effectively extracted from the bacterial cell surface with sucrose monolaurate, a nonionic detergent (Fig. 2). The extracted gingipain complexes were subsequently purified to homogeneity by conventional column chromatography (Table 1). The detergent-soluble fractions of P. gingivalis cells were subjected to gel filtration with Sephacryl S-300. The fraction of the major peak of proteolytic activity that contained 60 and 64% of the total cell-associated Rgp and Kgp, respectively, was applied to DEAE anion exchange chromatography. The fractions eluted by 0.2 M NaCl were applied to gel filtration columns containing Superdex-200. Both Rgp and Kgp activities were eluted at the same fraction of molecular mass of approximately 600 kDa, the fraction used as the cell-associated gingipain complex. During the purification steps, a major peak of Kgp activity was eluted, accompanied by Rgp activity. Coimmunoprecipitation assays using anti-Rgp and anti-Kgp antibodies revealed that both Rgp and Kgp were recovered in the same fraction (data not shown). As shown in Fig. 3A, the final preparation of the gingipain complex exhibited a broad but apparent single protein band with a molecular mass of 660 kDa with native polyacrylamide gel electrophoresis (PAGE), coincidently 400 to 700 kDa by gel filtration. Bhogal et al. (8) reported that the Rgp-Kgp complex purified from the soluble fraction extracted by sonication showed a molecular mass of 300 kDa, which was much smaller than that obtained in this study. When the gingipain complex was analyzed by SDS-PAGE, it contained seven apparent protein bands at 51, 44, 43, 39, 19, 17, and 14 kDa and faint diffuse band at 55 to 75 kDa (Fig. 3B). From immunoblot analyses with antibodies against the recombinant proteins including the catalytic domains of Rgp and Kgp and C-terminal adhesin domains of RgpA and Kgp, the eight protein bands were identified as Kgp (51 kDa), Rgp (43 kDa), Hgp44 (44 and 39 kDa), Hgp15 (19 kDa), Hgp17 (17 and 14 kDa), and Hgp27 (55- to 75-kDa smear band). This result indicates that components of the purified gingipain complex were similar to those of the complex previously described (8), except for the higher molecular mass of Hgp27. In spite of similar constituents, the molecular mass of the complex is twice that found by Bhogal et al. (8). The discrepancy may be due to differences in the extraction methods of the complex from the cells. In fact, the 660-kDa complex extracted by sucrose monolaurate was dissociated into a 200- to 300-kDa complex by sonication (data not shown). The 660-kDa complex is thus thought to be a dimer of the 300-kDa form.
FIG. 2.
Solubilization of cell-associated gingipain complexes with various detergents. P. gingivalis bacterial cells were incubated with 20 mM phosphate buffer containing various detergents at 4°C for 3 h. After incubation, the solubilized materials were collected by centrifugation at 100,000 × g for 30 min. The Rgp (open column) and Kgp (closed column) activities in the supernatant fraction were determined. The values are expressed as the percentages of the maximum activities obtained with 0.5% sucrose monolaurate. CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate; CHAPSO, 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate; BIGCHAP, N,N-bis(3-d-gluconamidopropyl)cholamide.
FIG. 3.
Nondenatured polyacrylamide gel electrophoresis (A) and SDS-PAGE (B) followed by immunoblot analyses, and electron micrograph (C) of the final preparation of the cell-associated gingipain complex. (A) The final preparation of the cell-associated complex (27.5 μg) was examined by nondenatured PAGE containing 0.5% sucrose monolaurate, and the gel was then stained with SYPRO Red. For immunoblot analysis, the proteins separated by nondenatured PAGE were transferred onto a nitrocellulose membrane and immunostained with polyclonal antibodies that recognize the catalytic domains of both Rgp and Kgp. (B) The purified complex (3.5 μg) was boiled in an SDS-solubilizing buffer and separated by SDS-PAGE. Proteins were visualized with SYPRO Red staining (left panel). The proteins separated in the gel were transferred onto nitrocellulose membranes followed by immunostaining with polyclonal antibodies that recognize the catalytic domains of both Kgp and Rgp or various adhesin domains of RgpA. Numbers indicate molecular masses of the corresponding protein bands. (C) The purified complex was negatively stained with 0.5% uranyl formate and observed by electron micrography (magnification, ×280,000). The bar corresponds to 25 nm. Panels 1 to 3 were magnified images (magnification, ×4,480,000).
To further investigate the homogeneity and structural characteristics of the purified gingipain complex, the complex was imaged by electron microscopy after negative staining with uranyl formate (Fig. 3C). Homogeneous globular-shaped particles containing one or two electron-dense hole-like parts were observed. The external diameter of the complex was estimated to be approximately 10 nm.
Intense degradation of collagen and elastin by the complex.
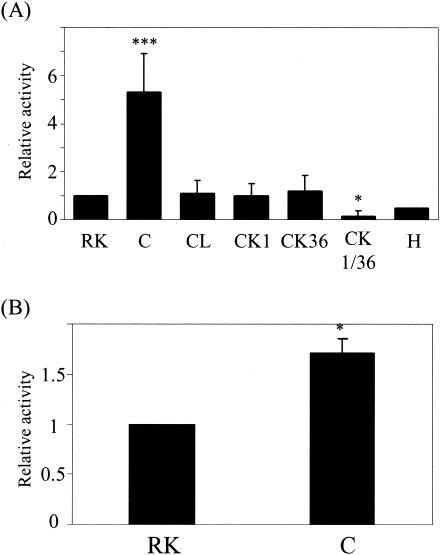
The gingipain complex contains C-terminal adhesin domains as well as Rgp and Kgp catalytic domains. Since the adhesin domain has been implicated as the “hemagglutination” and “hemoglobin-binding” domain, these domains are presumably important for a protease-substrate interaction in the complex (28, 36). When enzymatic properties were compared, no significant differences between the purified gingipain complex and the monomeric forms of Rgp and Kgp were found in substrate specificity toward synthetic substrates, optimal pH, thermal stabilities, and inhibitor profiles (data not shown). However, the complex exhibited several distinctive features. The rate of activation of Rgp by Ca2+ and Mg2+ was markedly greater for the complex form (314% by Ca2+ or Mg2+) than for the monomeric form (133% by Ca2+ and 139% by Mg2+). Although the degradation profiles of human soluble proteins such as fibronectin, fibrinogen, and gamma globulin were not different between the complex form and the combination of monomeric forms, the complex exhitied proteolytic activity against FITC-labeled human type I collagen which was more than fivefold more intense than that exhibited by the combined use of monomeric Rgp and Kgp (Fig. 4A). The degradation of collagen by the complex was most strongly inhibited by a combined action of Rgp- and Kgp-specific inhibitors (KYT-1 plus KYT-36), indicating that Rgp and Kgp are responsible for degradation of collagen. The collagenolytic activity of the gingipain complex exhibited 12.5-fold-higher specific activity than collagenase from C. histolyticum. The complex also showed a 1.75-fold-greater degradation against elastin-fluorescein than the combined use of monomeric enzymes (Fig. 4B). The results strongly suggested the assistance of adhesin domains for the catalytic domain to exhibit potent proteolytic activity and the importance of the gingipain complex in periodontal tissue breakdown.
FIG. 4.
Degradation of human type I collagen (A) and elastin (B) by the cell-associated complex and the monomeric forms of Rgp and Kgp. FITC-labeled collagen and elastin-fluorescein were incubated with the complex or the monomeric Rgp and/or Kgp in the presence or absence of Rgp and/or Kgp inhibitors as described in Materials and Methods. The collagenase from C. histolyticum was used as the positive control with the same protein amount as the complex. The values are expressed as a ratio to the activity obtained by treatment with the combination of monomeric Rgp and Kgp. RK, a combination of the monomeric forms of Rgp and Kgp; C, the complex; CL, the complex in the presence of leupeptin; CK1, the complex in the presence of KYT-1 (Rgp inhibitor); CK36, the complex in the presence of KYT-36 (Kgp inhibitor); CK1/36, the complex in the presence of KYT-1 and KYT-36; H, collagenase from C. histolyticum. *P < 0.05 and ***P < 0.005 compared with the value for the combination of monomeric Rgp and Kgp.
Enhanced cytotoxicity of gingipains by complex formation.
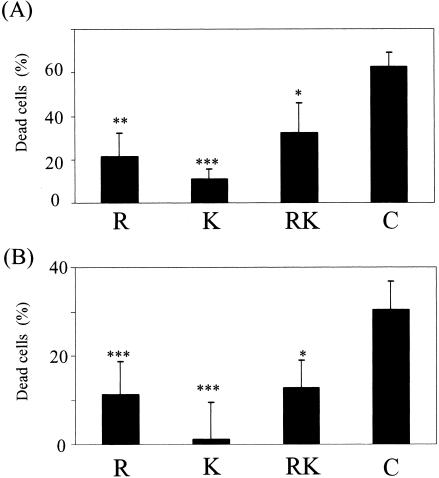
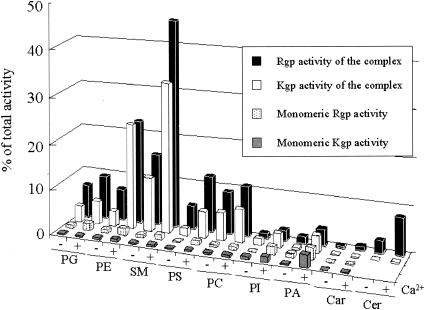
We previously reported that Rgp is responsible for the loss of adhesion activity and viability of human gingival fibroblasts (Gin-1) (3) and HUVEC (4, 5) induced by the culture supernatant of P. gingivalis. To investigate the effect of the cell-associated complex on viability of host cells, the purified gingipain complex, monomeric Rgp, monomeric Kgp, or the combination of monomeric Rgp and Kgp was added to cultures of Gin-1 and HUVEC. After 24 h of incubation, the complex induced a loss of viability of approximately 60% for Gin-1 (Fig. 5A), while the monomeric Rgp and Kgp induced only 20 and 10% cell death, respectively. With the combination of Rgp and Kgp, the extent of the viability loss was halted to 30%, which corresponded to the sum of effects of the respective enzymes. Obviously, the complex exhibited greater cytotoxicity than the monomeric enzymes. Similar results were obtained with HUVEC (Fig. 5B). P. gingivalis binds to a variety of host and bacterial cells through the C-terminal adhesin domains of RgpA and Kgp, resulting in bacterial coaggregation, colonization, and hemagglutination, which are important for invasion into host cells and bacterial survival. The high cytotoxicity is likely due to the existence of adhesin domains in the complex, and they seemed to accelerate the affinity of Rgp and Kgp catalytic domains for the host cells. To confirm this, we prepared liposomes consisting of various phospholipids and analyzed their preference for the complex or monomeric gingipains in the presence and absence of Ca2+. The monomeric Rgp and Kgp had little if any effect on binding to all types of liposomes regardless of the presence of Ca2+ (Fig. 6). In contrast, the complex showed significant binding to some phospholipids such as phosphatidylglycerol (PG), phosphatidylethanolamine (PE), sphingomyelin (SM), phosphatidylserine (PS), and phosphatidylcholine (PC) in the absence of Ca2+, as revealed by both Rgp and Kgp activities associated with these liposomes. The binding of the complex to PE-, SM-, and PS-containing liposomes was significantly increased in the presence of Ca2+. The Ca2+ ion is thought to be responsible for stabilizing the conformation of the complex. These results indicate that the complex binds to membranous lipids of host cells and induces cell death.
FIG. 5.
Cell viability of human gingival fibroblasts (A) and umbilical vein endothelial cells (B) after treatment with the complex or the monomeric forms of Rgp and Kgp. The cultures of Gin-1 and HUVEC were incubated with the purified complex (containing 2.7 units of Rgp activity and 2.4 units of Kgp activity) or the monomeric Rgp and Kgp equivalent to Rgp and Kgp activities of the complex, respectively. After incubation for 24 h (Gin-1) or 7 h (HUVEC), the cell viability was analyzed by using Cell Counting Kit 8. R, Rgp; K, Kgp; RK, the combined use of Rgp and Kgp; C, the purified complex. *P < 0.05, **P < 0.01, and ***P < 0.005 compared with the value for the complex.
FIG. 6.
Association of the complex with various phospholipids on liposomes. The purified complex and the monomeric forms of Rgp and Kgp were incubated with equal amounts of various liposomes at 4°C for 1 h in the presence or absence of 1 mM Ca2+. The liposomes were then collected by centrifugation at 105,000 × g for 30 min and resuspended in PBS. The Rgp and Kgp activities bound to liposomes were measured. The values were expressed as percentages of the total activities of both enzymes used. PG, phosphatidylglycerol; PE, phosphatidylethanolamine; SM, sphingomyelin; PS, phosphatidylserine; PC, phosphatidylcholine; PI, phosphatidylinositol; PA, phosphatidyc acid; Car, cardioliopin; Cer, cerebrosides.
Association of LPS to the complex.
Recent studies using the monoclonal antibody which recognizes sugar portions of LPS have suggested that RgpB and Hgp27 are covalently modified with LPS (12, 45). We thus examined whether the components in the purified complex are associated with LPS. Immunoblot analysis with the monoclonal antibody to lipid A after native PAGE revealed the apparent association of LPS with the complex (Fig. 7A). The band for the complex after native PAGE was also stained with Sudan III, which selectively reacts with lipid or lipid-containing molecule, suggesting the existence of LPS in the complex. To assess which components of the complex are modified with LPS, the components in the complex were separated by SDS-PAGE and then stained with Sudan III (Fig. 7B). Two protein bands with apparent molecular masses of 51 and 43 kDa were detected with Sudan III. Furthermore, the complex was subjected to two-dimensional PAGE followed by immunoblot analyses with antibodies to lipid A, Kgp, Rgp, Hgp44, and Hgp27 (Fig. 7C). Four protein spots were confirmed to show reactivity with anti-LPS antibody. Two of the spots clearly reacted with antibodies which recognized catalytic domains of Rgp (closed arrowhead) and Kgp (open arrowhead). The other two spots exhibited the reactivity with anti-Hgp44 and anti-Hgp27 antibodies. Considering the molecular masses of the bands, they correspond to Hgp44 derived from RgpA (asterisk) and from Kgp (arrow). In this study, we could not detect the obvious reactivity between Hgp27 and anti-lipid A antibody.
FIG. 7.
Modification of the cell-associated gingipain complex by attachment of LPS. (A) The purified complex was applied to native polyacrylamide gels. The proteins separated on the gel were then transferred to nitrocellulose membranes and immunostained with the monoclonal antibody to lipid A of LPS. (B) The complex was subjected to SDS-PAGE and stained with Sudan III. (C) The complex was subjected to two-dimensional PAGE followed by immunoblot (IB) analyses. Upper panel, anti-lipid A antibody; middle panel, antibodies recognizing the catalytic domains of both Kgp and Rgp; bottom panel, a combination of anti-Hgp44 antibodies and anti-Hgp27 antibodies, respectively. Immunoreacting spots to anti-lipid A antibody are coincident with the catalytic domains of Rgp (43 kDa) (closed arrowheads), Kgp (51 kDa) (open arrowheads), Hgp44, (asterisks), and Hgp39 (arrow).
Inactivation of LPS by gingipains and/or adhesins in the complex.
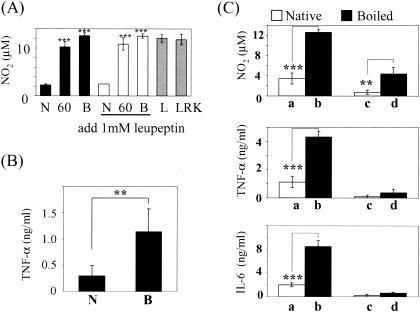
LPS stimulates host cells and induces the expression of cytokines such as TNF-α, IL-1, IL-6, and IL-12 that are important in the host response to infection. Since the purified complex included LPS, the effect of the complex on cytokine production from host cells was investigated. To represent the in vivo situations, the following experiments were performed in the presence of 10% fetal bovine serum. Treatment of mouse peritoneal macrophages with the gingipain complex induced only a small amount of NO2 production (∼2 μM) (Fig. 8A). Upon treatment with heat-inactivated complex, however, macrophages produced a high amount of NO2 (∼11 μM). Proteinase K treatment of the complex also up-regulated the production of NO2 by macrophages (data not shown). We then examined whether the proteolytic activities in the native complex participated in the down-regulation of LPS activity. Even in the presence of protease inhibitor leupeptin, the complex was confirmed to show little stimulatory activity to macrophages. In addition, the mixture of Rgp, Kgp, and LPS extracted from P. gingivalis by the hot phenol-water method (46) showed a strong NO2-inducing effect corresponding to LPS alone. Accordingly, it seemed that gingipains and/or adhesins in the complex could mask the LPS activity through nonproteolytic actions. To further confirm this, we used human phorbol 12-myristate 13-acetate-differentiated THP-1 macrophage (Fig. 8B). Similarly, TNF-α was markedly released from THP-1 cells upon treatment with the heat-denatured complex, whereas this cytokine was barely induced upon treatment with the native complex.
FIG. 8.
Production of NO2, TNF-α, and IL-6 from macrophages upon treatment with the complex. (A) NO2 production from macrophages treated with the complex, LPS, or a combination of Rgp and Kgp was measured. N, native complex; 60, the complex incubated at 60°C for 30 min; B, the boiled complex; L, 1 ng of LPS/ml; LRK, a mixture of LPS, Rgp, and Kgp. The cells were treated with the complex in the absence (black and gray columns) and presence (open columns) of 1 mM leupeptin. (B) TNF-α from THP-1 cells. THP-1 cells were incubated with the native (N) or boiled complex (B) for 24 h, and TNF-α in the medium was then measured. (C) The production of NO2, TNF-α, and IL-6 by macrophages of C57BL/6 mice (a, b) and C3H/HeJ mice (c, d) (n = 5) upon treatment with the native complex (open column) or heat-denatured complex (4 ng/well) (closed column) for 24 h in the presence (for NO2) and absence (for TNF-α and IL-6) of gamma interferon (100 U/ml) was analyzed.
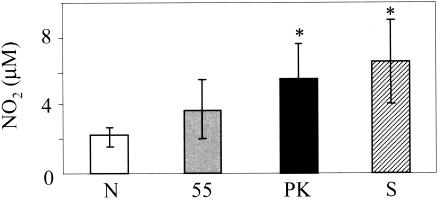
LPS derived from P. gingivalis is known to antagonize E. coli LPS-dependent activation of human endotherial cells through competition at the TLR4 signaling complex (11, 48). In contrast, the lipid A molecule of P. gingivalis is shown to activate cells through TLR4 and the MyD88-dependent signaling pathway (29). Next, we investigated whether LPS derived from P. gingivalis, like that from E. coli, was correctly recognized by TLR4. The peritoneal macrophages isolated from both C57BL/6 and LPS-hyporesponsive C3H/HeJ mice harboring a point mutation in the tlr4 gene (17) were incubated with the native or heat-denatured complex for 24 h. Macrophages from C57BL/6 mice incubated with the heat-denatured complex showed 3.5-fold-higher production of NO2 than that of the native complex (Fig. 8C). Similarly, the heat-denatured complex induced the enhanced production of both TNF-α and IL-6 by macrophages with much higher efficiency than the native protein. However, the production of NO2, TNF-α, and IL-6 by macrophages derived from C3H/HeJ mice was low or undetectable even upon treatment with the native or heat-denatured complex. This finding provides clear evidence that P. gingivalis LPS can be recognized by TLR4 but that the LPS domain responsible for interaction with TLR4 would be masked with the protein components of the complex. P. gingivalis might escape from recognition by inflammatory cells through this strategy. Recently, Calkins et al. (10) reported that TNF-α treated by gingipains was inactivated due to rapid degradation. Sugawara et al. (43) reported that CD14 on human monocytes was decreased by incubation with gingipains. We then examined whether the complex degradation produced TNF-α. Since the enzymatic activities of gingipains were not detectable under the experimental conditions in the presence of 10% serum, the decreased cytokine levels by the complex are unlikely induced by degradation of CD14 or cytokines. To exclude the possibility that the masked LPS is observed only in the purified complex, we examined the cytokine production by macrophages incubated with whole P. gingivalis cells or cell sonicate (Fig. 9). Upon stimulation of intact bacterial cells, the peritoneal macrophages produced a small amount of NO2, up to 2 μM. Mild heat treatment of the bacterium at 55°C, by which bacterial proteins were denatured but intracellular proteins were not leaked out, resulted in approximately 4 μM NO2 production. Degradation of bacterial surface proteins by proteinase K increased NO2 production by macrophages to 5 μM. LPS on the bacterial surface appears to be exposed by these treatments, thereby resulting in the stimulation of host cells. The bacterial cell sonicate induced the most potent NO2 production by the macrophages, probably due to the dissociation of protein components and LPS. Thus, the characteristic structure and features observed in the purified complex seem to represent the native gingipain complex in the bacterial cells.
FIG. 9.
Production of NO2 by mouse peritoneal macrophages incubated with P. gingivalis. Macrophages were incubated with various P. gingivalis cells in the presence of gamma interferon (100 U/ml). N, intact cells; 55, P. gingivalis treated at 55°C for 20 min; PK, P. gingivalis treated with proteinase K (0.5 mg/ml) at 37°C for 30 min; S, P. gingivalis sonicate. After 24 h of incubation, NO2 released into the medium was measured. *P < 0.05 compared with the value obtained with intact P. gingivalis (N).
DISCUSSION
Previously, Bhogal et al. (8) demonstrated the purification of a 300-kDa complex comprising eight protein subunits including catalytic and adhesin domains of RgpA and Kgp. The protein composition of the complex isolated by those authors appeared similar to that purified in this study except that the molecular mass of Hgp27 was much higher in the complex reported here than in the one reported previously. The predicted molecular mass of the present complex is estimated to be 294 to 323 kDa by assuming that the complex contains a single molecule for each subunit, which is in good agreement with that of the previous complex. The twofold-higher molecular mass of the complex reported here suggests dimerization of the 300-kDa complex. The discrepancy in the molecular mass may be due to the difference in the extraction methods. The sonication extract used in the previous study may dissociate a naturally occurring complex on the bacterial cells into basic units. Indeed, the 660-kDa complex extracted by sucrose monolaurate was dissociated into a 200- to 300-kDa complex by sonication. When the complex was treated with nonspecific phospholipase C from Bacillus cereus prior to native PAGE, the intensity of SYPRO Red staining was increased (unpublished data). Upon phase separation with Triton X-114 according to the method of Bordier (9), which separates amphiphilic integral membrane proteins and hydrophobic proteins into aqueous and detergent phases, respectively, more than 80% of proteolytic activities of both Rgp and Kgp in the complex were recovered in the detergent phase (unpublished data). In contrast, the monomeric forms of Rgp and Kgp were found in the aqueous phase under the same conditions. Taken together, these results indicate that the complex undergoes lipophilic modifications. This modification might contribute to dimerization of the 300-kDa complex.
We also demonstrated that the complex was modified with LPS through attachment to the protease and Hgp44 domains of Rgp and Kgp. Recent studies using the monoclonal antibody which recognizes sugar portions of LPS have suggested that membrane-associated forms of RgpA, RgpB, and Hgp27 are posttranslationally modified by LPS (12, 45). However, we could not detect the obvious reactivity between the monoclonal antibody to the backbone of lipid A and Hgp27. This result may be due to differences in the antibodies or in the samples used for immunoblot analysis. The crude cell sonicate or outer membrane fraction was used in the previous studies, whereas the purified complex was used in this study. Considering molecular size determined by SDS-PAGE, the protease domain of Rgp in the complex which reacted with anti-lipid A antibody was thought to be derived from RgpA; however, we cannot rule out the possibility that it came from rgpB, which is essentially identical to rgpA except for the lack of a 3′ region corresponding to C-terminal adhesin domains. Shoji et al. (40) reported that the porR mutant of P. gingivalis, which is defective in biosynthesis of sugar portions of cell surface polysaccharides, shows the preferential presence of Rgp in culture supernatant. Accordingly, the modification by LPS may contribute to the anchorage of the complex to the cell surface. RgpA and Kgp are synthesized as nascent precursor proteins containing an N-terminal prodomain, a protease domain, and C-terminal domains in the cytosol. They are likely transported across the inner membrane via an Sec translocon, because both RgpA and Kgp possess a signal sequence-like domain at their N termini and because P. gingivalis has homologs of E. coli secA, secD, secF, and secY. They are then transported through the outer membrane to the surface by an unidentified pathway. On the cell surface (or in periplasm), RgpA and Kgp are proteolytically processed, modified by LPS, and formed to the mature complex. As shown by electron microscopy, the complex is composed of a globular particle with an external diameter of 10 nm which contains one or two hole-like structures on the bacterial surface.
The proteolytic activities of the purified complex toward collagen and elastin were very high, whereas those of the monomeric forms of Rgp and Kgp were relatively low (Fig. 4). The Km values for synthetic peptide substrates of the complex were not changed from those of monomeric forms (data not shown). The results thus suggest that the adhesin components raise the affinity of the catalytic domains to these proteins. The complex induced cell death in human gingival fibroblasts and HUVEC with higher efficiency than the combined use of monomeric Rgp and Kgp by a similar effect of adhesins (Fig. 5). We mimicked the adhesion of the complex to cells by using an in vitro liposome-binding experiment (Fig. 6). The monomeric Rgp and Kgp showed little or no binding to any types of liposomes; however, the complex showed significant binding to liposomes consisting of PG, PE, SM, PS, or PC. Eukaryotic plasma membranes preferentially include PC and SM on the outside. Bacterial cytoplasmic membranes are mainly composed of PE and PG. These results indicate that the complex binds directly to membranous lipids of both host and bacterial cells. This implies that the cell-associated complex may play a key role in colonization, coaggregation, hemagglutination, and infection of host cells by P. gingivalis.
In this study, we revealed the existence of LPS in the gingipain complex; however, the purified complex failed to induce the production of NO2, TNF-α, and IL-6 by murine and human macrophages (Fig. 8). The production of these cytokines was significantly increased when the complex was denatured by heat or proteinase K treatment. Inhibition of the proteolytic activities of gingipains in the complex did not stimulate macrophages. These results strongly suggest that LPS is conformationally concealed by protein subunits in the complex and thereby prevented from binding to receptors on the host cell surface. Upon heat treatment, the protein subunits were denatured and LPS was subsequently exposed. This hypothesis was also supported by the facts that TLR4 and TLR2 were not degraded by incubation with the complex or by P. gingivalis infection (unpublished data) and that the LPS purified by using hot phenol methods obviously activated the production of cytokines by the cells. It is generally accepted that TLR4 and its coreceptor MD-2 are the authentic LPS signaling transducers in many cell types. However, the cell activation by P. gingivalis LPS through the TLR4/MD-2-MyD88-dependent pathway is controversial (44). In the present study, P. gingivalis LPS and the denatured gingipain complex obviously increased the production of cytokines by macrophages from C57BL/6 but not from LPS-hyposensitive C3H/HeJ mice (17), indicating that P. gingivalis LPS, as well as classical enterobacterial LPS, has the ability to activate cells via the TLR4 pathway. It has been previously reported that gingipains degrade and inactivate cytokines such as IL-6, IL-8, and TNF-α (7, 10, 31); complement factors C3 and C5 (47); and immunoglobulins (21). We thus conclude that the cell-associated gingipain complex contributes to the suppression of the host immune response to recruit and localize neutrophils and macrophages to gingival inflammatory sites by bacterial infection through evasion of host LPS signaling as well as paralyses of cytokines and facilitates the progression of periodontitis.
Acknowledgments
We thank S. Yoshida and A. Takade, Department of Bacteriology, Kyushu University Graduate School of Medical Sciences, for help in electron microscopy.
This study was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, Technology, and Culture of Japan.
Editor: A. D. O'Brien
REFERENCES
- 1.Abe, N., T. Kadowaki, K. Okamoto, K. Nakayama, M. Ohishi, and K. Yamamoto. 1998. Biochemical and functional properties of lysine-specific cysteine proteinase (Lys-gingipain) as a virulence factor of Porphyromonas gingivalis in periodontal disease. J. Biochem. (Tokyo) 123:305-312. [DOI] [PubMed] [Google Scholar]
- 2.Abe, N., A. Baba, T. Kadowaki, K. Okamoto, S. Okazaki, T. Asao, and K. Yamamoto. 2000. Design and synthesis of sensitive fluorogenic substrates specific for Lys-gingipain. J. Biochem. (Tokyo) 128:877-881. [DOI] [PubMed] [Google Scholar]
- 3.Baba, A., N. Abe, T. Kadowaki, H. Nakanishi, M. Ohishi, T. Asao, and K. Yamamoto. 2001. Arg-gingipain is responsible for the degradation of cell adhesion molecules of human gingival fibroblasts and their death induced by Porphyromonas gingivalis. Biol. Chem. 382:817-824. [DOI] [PubMed] [Google Scholar]
- 4.Baba, A., T. Kadowaki, T. Asao, and K. Yamamoto. 2002. Cooperative action of Arg- and Lys-gingipains in disruption of fibronectin-integrin interactions in human gingival fibroblasts. Dent. Jpn. 38:40-43. [Google Scholar]
- 5.Baba, A., T. Kadowaki, T. Asao, and K. Yamamoto. 2002. Roles for Arg- and Lys-gingipains in the disruption of cytokine responses and loss of viability of human endothelial cells by Porphyromonas gingivalis infection. Biol. Chem. 383:1223-1230. [DOI] [PubMed] [Google Scholar]
- 6.Baici, A., G. Cohen, K. Fehr, and A. Boni. 1980. A handy assay for collagenase using reconstituted fluorescein-labeled collagen fibrils. Anal. Biochem. 108:230-232. [DOI] [PubMed] [Google Scholar]
- 7.Banbula, A., M. Bugno, A. Kuster, P. C. Heinrich, J. Travis, and J. Potempa. 1999. Rapid and efficient inactivation of IL-6 gingipains, lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Biochem. Biophys. Res. Commun. 261:598-602. [DOI] [PubMed] [Google Scholar]
- 8.Bhogal, P. S., N. Slakeski, and E. C. Reynolds. 1997. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesions. Microbiology 143:2485-2495. [DOI] [PubMed] [Google Scholar]
- 9.Bordier, C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 256:1604-1607. [PubMed] [Google Scholar]
- 10.Calkins, C. C., K. Platt, J. Potempa, and J. Travis. 1998. Inactivation of tumor necrosis factor-α by proteinases (gingipains) from the periodontal pathogen, Porphyromonas gingivalis. Implications of immune evasion. J. Biol. Chem. 273:6611-6614. [DOI] [PubMed] [Google Scholar]
- 11.Coats, S. R., R. A. Reife, B. W. Bainbridge, T.-T. T. Pham, and R. P. Darveau. 2003. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at Toll-like receptor 4 in human endothelial cells. Infect. Immun. 71:6799-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shephield, N. Paramonov, and E. F. Hounsel. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallagher, A., J. Aduse-Opoku, M. Rangarajan, J. M. Slaney, and M. A. Curtis. 2003. Glycosylation of the Arg-gingipain of Porphyromonas gingivalis and comparison with glycoconjugate structure and synthesis in other bacteria. Curr. Protein Pept. Sci. 4:427-441. [DOI] [PubMed] [Google Scholar]
- 14.Grenier, D. 1992. Inactivation of human serum bactericidal activity by a trypsin-like protease isolated from Porphyromonas gingivalis. Infect. Immun. 60:1854-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirschfeld, M., J. J. Weis, V. Toshchakov, C. A. Salkowski, M. J. Cody, D. C. Ward, N. Qureshi, S. M. Michalek, and S. N. Vogal. 2001. Signaling by Toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun. 69:1477-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt, S. C., J. Felton, M. Brunsvold, and K. S. Kornman. 1988. Implantation of Bacteroides gingivalis in non-human primate initiates progression of periodontitis. Science 239:55-57. [DOI] [PubMed] [Google Scholar]
- 17.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 18.Imamura, T., R. N. Pike, J. Potempa, and J. Travis. 1994. Pathogenesis of periodontitis: a major arginine-specific cysteine proteinase from Porphyromonas gingivalis induces vascular permeability enhancement through activation of the kallikrein/kinin pathway. J. Clin. Investig. 94:361-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamura, T., J. Potempa, S. Tanase, and J. Travis. 1997. Activation of blood coagulation factor X by arginine-specific cysteine proteinase (gingipain-Rs) from Porphyromonas gingivalis. J. Biol. Chem. 272:16062-16067. [DOI] [PubMed] [Google Scholar]
- 20.Kadowaki, T., M. Yoneda, K. Okamoto, K. Maeda, and K. Yamamoto. 1994. Purification and characterization of a novel arginine-specific cysteine proteinase (argingipain) involved in the pathogenesis of periodontal disease from the culture supernatant of Porphyromonas gingivalis. J. Biol. Chem. 269:21371-21378. [PubMed] [Google Scholar]
- 21.Kadowaki, T., K. Nakayama, K. Okamoto, N. Abe, A. Baba, Y. Shi, D. B. Ratnayake, and K. Yamamoto 2000. Porphyromonas gingivalis proteinases as virulence determinants in progression of periodontal diseases. J. Biochem. (Tokyo) 128:153-159. [DOI] [PubMed] [Google Scholar]
- 22.Kadowaki, T., A. Baba, N. Abe, R. Takii, M. Hashimoto, T. Tsukuba, S. Okazaki, Y. Suda, T. Asao, and K. Yamamoto. 2004. Suppression of pathogenicity of Porphyromonas gingivalis by newly developed gingipain inhibitors. Mol. Pharmacol. 66:1599-1606. [DOI] [PubMed] [Google Scholar]
- 23.Martin, M., J. Katz, S. N. Vogel, and S. M. Michalek. 2001. Differential induction of endotoxin tolerance by lipopolysaccharides derived from Porphyromonas gingivalis and Escherichia coli. J. Immunol. 167:5278-5285. [DOI] [PubMed] [Google Scholar]
- 24.Mikolajczyk-Pawlinska, J., T. Kordula, N. Pavloff, P. A. Pemberton, W.-C. A. Chen, J. Travis, and J. Potempa. 1998. Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol. Chem. 379:205-211. [DOI] [PubMed] [Google Scholar]
- 25.Morrison, D. C., and J. L. Ryan. 1987. Endotoxins and disease mechanisms. Annu. Rev. Med. 38:417-432. [DOI] [PubMed] [Google Scholar]
- 26.Nakayama, K., T. Kadowaki, K. Okamoto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 27.Nakayama, K. 1997. Domain-specific rearrangement between the two Arg-gingipain-encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol. Immunol. 41:185-196. [DOI] [PubMed] [Google Scholar]
- 28.Nakayama, K., D. B. Ratnayake, T. Tsukuba, T. Kadowaki, K. Yamamoto, and S. Fujimura. 1998. Haemoglobin receptor protein is intragenically encoded by the cysteine proteinase-encoding genes and the haemagglutinin-encoding gene of Porphyromonas gingivalis. Mol. Microbiol. 27:51-61. [DOI] [PubMed] [Google Scholar]
- 29.Ogawa, T., Y. Asai, M. Hashimoto, O. Takeuchi, T. Kurita, Y. Yoshikai, K. Miyake, and S. Akira. 2002. Cell activation by Porphyromonas gingivalis lipid A molecule through Toll-like receptor 4- and myeloid differentiation factor 88-dependent signaling pathway. Int. Immunol. 14:1325-1332. [DOI] [PubMed] [Google Scholar]
- 30.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oido-Mori, M., R. Rezzonico, P.-L. Wang, Y. Kowashi, J.-M. Dayer, P. C. Baehni, and C. Chizzolini. 2001. Porphyromonas gingivalis gingipain-R enhances interleukin-8 but decreases gamma interferon-inducible protein 10 production by human gingival fibroblasts in response to T-cell contact. Infect. Immun. 69:4493-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto, K., Y. Misumi, T. Kadowaki, M. Yoneda, K. Yamamoto, and Y. Ikehara. 1995. Structural characterization of arginine, a novel arginine-specific cysteine proteinase as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch. Biochem. Biophys. 316:917-925. [DOI] [PubMed] [Google Scholar]
- 33.Okamoto, K., T. Kadowaki, K. Nakayama, and K. Yamamoto. 1996. Cloning and sequencing of the gene encoding a novel lysine-specific cysteine proteinase (Lys-gingipain) in Porphyromonas gingivalis: structural relationship with the arginine-specific cysteine proteinase (Arg-gingipain). J. Biochem. (Tokyo) 120:398-406. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-212315. [DOI] [PubMed] [Google Scholar]
- 35.Pavloff, N., J. Potempa, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1995. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. J. Biol. Chem. 270:1007-1010. [DOI] [PubMed] [Google Scholar]
- 36.Pike, R., W. McGraw, J. Potempa, and J. Travis. 1994. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J. Biol. Chem. 269:406-411. [PubMed] [Google Scholar]
- 37.Rajapakse, P. S., N. M. O'Brien-Simpson, N. Slakeski, B. Hoffmann, and E. C. Reynolds. 2002. Immunization with the Rgp-Kgp proteinase-adhesin complexes of Porphyromonas gingivalis protects against periodontal bone loss in the rat periodontitis model. Infect. Immun. 70:2480-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott, C. F., E. J. Whitaker, B. J. Hammond, and R. C. Colman. 1993. Purification and characterization of a potent 70-kDa thiol lysyl-proteinase (Lys-gingivain) from Porphyromonas gingivalis that cleaves kininogens and fibrinogens. J. Biol. Chem. 268:7935-7942. [PubMed] [Google Scholar]
- 39.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 40.Shoji, M., D. B. Ratnayake, Y. Shi, T. Kadowaki, K. Yamamoto, F. Yoshimura, A. Akamine, M. A. Curtis, and K. Nakayama. 2002. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology 148:1183-1191. [DOI] [PubMed] [Google Scholar]
- 41.Slakeshi, N., P. S. Bhogal, N. M. O'Brien-Simpson, and E. C. Reynolds. 1998. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology 144:1583-1592. [DOI] [PubMed] [Google Scholar]
- 42.Slots, J., L. Bragd, M. Wikström, and G. Dahlen. 1986. The occurrence of Actinobacillus actinomycetemcomitans, Bacteroides gingivalis and Bacteroides intermedius in destructive periodontal disease in adults. J. Clin. Periodontol. 13:570-577. [DOI] [PubMed] [Google Scholar]
- 43.Sugawara, S., E. Nemoto, H. Tada, K. Miyake, T. Imamura, and H. Takada. 2000. Proteolysis of human monocyte CD14 by cysteine proteinases (gingipains) from Porphyromonas gingivalis leading to lipopolysaccharide hyporesponsiveness. J. Immunol. 165:411-418. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi, O., K. Takeda, K. Hoshino, O. Adachi, T. Ogawa, and S. Akira. 2000. Cellular responses to bacterial cell wall components are mediated through MyD88-dependent signaling cascades. Int. Immunol. 12:113-117. [DOI] [PubMed] [Google Scholar]
- 45.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westphal, O., and K. Jann. 1965. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure, p. 83-92. In R. L. Whistler (ed.), Methods in carbohydrate chemistry. Academic Press, Inc., New York, N.Y.
- 47.Wingrove, J. A., R. G. DiScipio, Z. Chen, J. Potempa, J. Travis, and T. E. Hugli. 1992. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 267:18902-18907. [PubMed] [Google Scholar]
- 48.Yoshimura, A., T. Kaneko, Y. Kato, D. T. Golenbock, and Y. Hara. 2002. Lipopolysaccharides from periodontopathic bacteria Porphyromonas gingivalis and Capnocytophaga ochracea are antagonists for human Toll-like receptor 4. Infect. Immun. 70:218-225. [DOI] [PMC free article] [PubMed] [Google Scholar]