Abstract
Adult cardiac progenitor cells (CPCs) display a low capacity to differentiate into cardiomyocytes in injured hearts, strongly limiting the regenerative capacity of the mammalian myocardium. To identify new mechanisms regulating CPC differentiation, we used primary and clonally expanded Sca-1+ CPCs from murine adult hearts in homotypic culture or coculture with cardiomyocytes. Expression kinetics analysis during homotypic culture differentiation showed downregulation of Wnt target genes concomitant with increased expression of the Wnt antagonist, Wnt inhibitory factor 1 (Wif1), which is necessary to stimulate CPC differentiation. We show that the expression of the Wif1 gene is repressed by DNA methylation and regulated by the de novo DNA methyltransferase Dnmt3a. In addition, miR-29a is upregulated early during CPC differentiation and downregulates Dnmt3a expression, thereby decreasing Wif1 gene methylation and increasing the efficiency of differentiation of Sca-1+ CPCs in vitro. Extending these findings in vivo, transient silencing of Dnmt3a in CPCs subsequently injected in the border zone of infarcted mouse hearts improved CPC differentiation in situ and remote cardiac remodeling. In conclusion, miR-29a and Dnmt3a epigenetically regulate CPC differentiation through Wnt inhibition. Remote effects on cardiac remodeling support paracrine signaling beyond the local injection site, with potential therapeutic interest for cardiac repair.
Keywords: Cardiology, Stem cells
Dnmt3a and miR-29a coordinately regulate differentiation and paracrine signaling of cardiac progenitor cells to improve remodeling after infarction in mice.
Introduction
Healing of the infarcted myocardium proceeds through remodeling of the remaining tissue and, to a limited extent, regeneration of lost cardiac muscle. Both processes involve activation of endogenous cardiac progenitor cells (CPCs) in cardiac niches that, upon specific stimuli, undergo proliferation and differentiation toward vascular or muscle lineages and release paracrine factors acting on neighboring cells to modulate tissue repair (1). A number of progenitor cells have been identified in the adult mouse and/or human heart based on their expression of specific surface markers, e.g., c-kit (2) and Sca-1 (3), or transcription factors, such as homeobox gene islet1 (Isl1) (4), or their ability to efflux Hoechst dye 33342 (identified as side population cells, ref. 5). Although adult CPCs were shown to contribute to cell homeostasis during aging (6), their ability to regenerate cardiac muscle through durable engraftment, survival, and terminal differentiation seems marginal, at least in the adult mammalian heart (7, 8). An improved understanding of the mechanisms driving their proliferation, survival, and terminal differentiation, as well as paracrine signaling, would be highly desirable for their future therapeutic exploitation.
Just as sequential steps drive the specification of pluripotent stem cells to mesodermal and then cardiac lineages during embryogenesis, the terminal differentiation of adult CPCs follows similar signaling events that determine their fate, some of which recapitulate early specification during organogenesis. One of these signals is the bimodal activity (i.e., early activation followed by later extinction) of the canonical Wnt/β-catenin pathway that drives cardiac mesoderm formation in the embryo (9, 10). Recent studies have confirmed the importance of the same signaling sequence in human induced pluripotent stem cells (iPSCs) (11) and embryonic stem cells (ESCs) (12), in which early activation of BMP and Wnt, followed by Wnt inhibition, progressively removes repressive roadblocks in the path toward cardiac myocyte differentiation. We and others (13) have confirmed similar paradigms in adult CPCs (that have constitutive Wnt activity, ref. 14), in which secondary extinction of Wnt/β-catenin using genetic invalidation (15) or paracrine NO signaling (14) promotes the expression of late cardiac differentiation markers in CPCs in vitro and in vivo. Accordingly, small-molecule inhibitors of canonical Wnt similarly promote cardiac myocyte formation from mammalian (including human) ESCs or iPSCs in differentiation protocols in vitro (16).
However, transposition of the paradigm using pharmacologic Wnt blockade in vivo would be impractical due to expected adverse effects from indiscriminate Wnt inhibition in other cell types (17, 18). This calls for a better understanding of the dynamic regulation of Wnt signaling in the cardiac tissue, especially the mechanisms driving Wnt extinction in CPCs that promotes their progression toward cardiac myocyte differentiation.
We used a well-characterized model of adult mouse Sca-1+ CPCs (14, 19) to study the kinetics of expression of Wnt regulators and Wnt target genes in the course of in vitro differentiation into cardiomyocytes. We identified Wnt inhibitory factor 1 (Wif1) as a key modulator of their differentiation and found its expression to be epigenetically regulated by DNA methylation. Early expression of miR-29a downregulates the expression of the DNA methyltransferase Dnmt3a to demethylate the Wif1 gene, allowing its expression to promote CPC differentiation. Injection of CPCs with downregulated Dnmt3a in infarcted hearts attenuates adverse remodeling and preserves left ventricular function in vivo.
Results
Downregulation of Wnt/β-catenin commits adult mouse Sca-1+ CPCs to cardiac myocyte differentiation. After two rounds of MACS isolation, the positive fraction of isolated Sca-1+ population cells was purified to nearly 90% after exclusion of dead cells, as previously described (14, 19). Freshly isolated cells scored negative for myocyte (e.g., α-cardiac actinin, and cardiac troponin I and T), endothelial cell (e.g., CD31), and smooth muscle cell (e.g., α-smooth muscle actin) markers. However, transcription factors expressed early in the myocyte lineage (Nkx2.5, GATA-4) were each detected in the freshly isolated Sca-1+ CPCs. We next induced differentiation of CPCs toward the cardiac myocyte lineage using pretreatment with 5-azacytidine (AZA) for 3 days, followed by repetitive pulses of TGF-β, as previously described (20). Efficient early commitment to the myocyte lineage was assessed through the time-dependent expression of early cardiac-specific transcription factors, such as Nkx2.5, together with expression of cardiac structural genes, such as Tnnt2, for up to 26 days (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.91810DS1). At this time point, some primitive sarcomeric organization was also observed by immunohistochemistry (Supplemental Figure 1B).
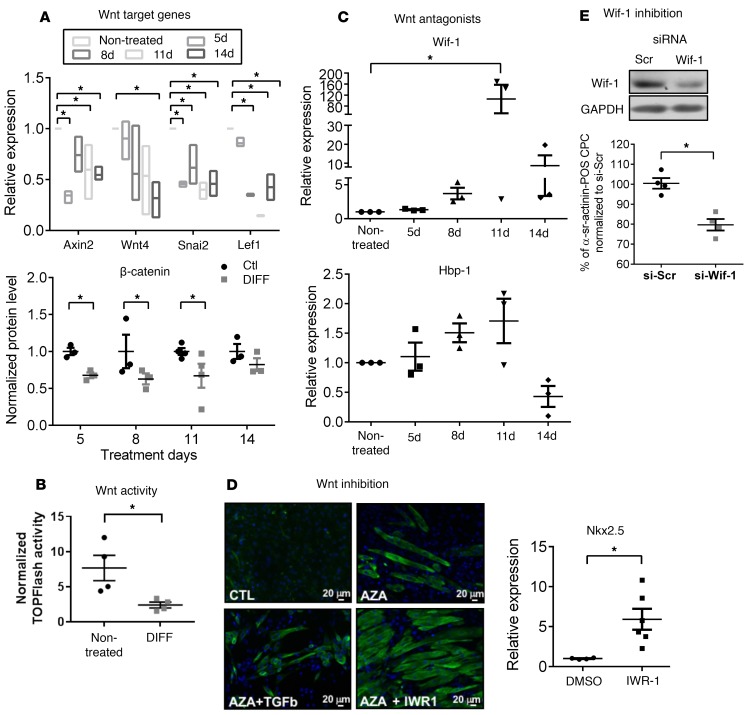
We previously demonstrated that adult mouse CPCs display a constitutive activity of the canonical Wnt/β-catenin pathway (14) and that their differentiation toward cardiomyocytes is enhanced upon genetic deletion of β-catenin in vitro and in vivo (14, 15). We therefore evaluated the dynamic activity of the endogenous Wnt/β-catenin pathway during in vitro differentiation of CPCs. We first monitored the time-dependent expression of Wnt target genes upon differentiation with AZA/TGF-β, as used above. Typical Wnt target genes, such as Axin2 and Wnt4, were downregulated in the course of differentiation, together with a decrease in β-catenin protein level. Moreover, this was paralleled with time-dependent downregulation of the Wnt activators Lef1 and Snai2 (Figure 1A); however, upregulation of Wnt repressors Hbp1 and Wif1 was evident (Figure 1C). The inhibition of Wnt signaling was further confirmed using the TOPflash reporter assay in HEK cells superfused with conditioned media from CPCs differentiated by AZA/TGF-β (Figure 1B). Altogether, this profile suggests repression of the constitutive Wnt/β-catenin activity in the course of CPC differentiation.
Figure 1. Cardiac progenitor cell differentiation is concomitant with a downregulation of Wnt signaling and upregulation of Wnt inhibitors and is potentiated by inhibition of Wnt/β-catenin.
Cultured cardiac progenitor cells (CPCs) were incubated or not (control cells [Ctl]) with 5-azacytidine (AZA) and TGF-β (DIFF) for 5, 8, 11, or 14 days. Gene expression (relative to respective time Ctl) was analyzed using RT-qPCR and normalized to GAPDH. (A) Relative expression of Wnt/β-catenin pathway target genes (Axin2, Wnt4) and Wnt activators (Lef1 and Snai2). Bounds of boxes represent minimum and maximum values, inner line represent means. *P < 0.05 vs. CTL; n = 3 different preparations; Kruskal-Wallis with Bonferroni correction. Expression of β-catenin protein levels in DIFF, relative to Ctl at each time point. *P < 0.05 vs. Ctl; n ≥ 3 different preparations; Mann-Whitney test. (B) CPCs were cultured in Ctl or DIFF medium for 8 days and their supernatant was incubated with HEK cells expressing the TOPflash reporter construct, indicative of Wnt morphogen production by and Wnt activity in donor CPCs. TOPflash signal was normalized for transfection efficiency (cotransfected TKRenilla). *P < 0.05 vs. Ctl; n = 4 different cultures; Mann-Whitney. (C) Relative (to respective time Ctl) expression of Wnt repressors Wif1 and Hbp1. *P < 0.05 vs. Ctl; n = 3 different preparations; Kruskal-Wallis with Bonferroni correction. (D) Representative images of CPCs treated or not with AZA and either TGF-β or the pharmacologic inhibitor of Wnt signaling response (IWR1, 10 μM) for 26 days. Immunocytochemistry was performed using an antibody against cardiac troponin T. Relative gene expression of Nkx2.5 in CPCs treated with IWR1 in absence of AZA for 11 days. *P < 0.05, n ≥ 4 experiments; Mann-Whitney test. Scale bar: 20 μm. (E) Wif1 expression modulates CPC differentiation in coculture. Expression of Wif1 in CPCs transfected with 50 nM siRNA targeting Wif1 (or siRNA scramble) for 48 hours. CPCs transfected with siRNA-Wif1 (si-Wif1) or scramble control (si-Scr) were cocultured with cardiomyocytes and their differentiation monitored by expression of α-sr-actinin. Results are reported as relative to differentiation in si-Scr–transfected CPCs (set at 100%). CPC differentiation is significantly decreased upon Wif1 inhibition. *P < 0.05; n = 4 different preparations; Mann-Whitney test.
To further assess the causality of Wnt repression in the differentiation process, we tested the effect of the pharmacologic inhibitor of Wnt/β-catenin, namely inhibitor of Wnt signaling response (IWR1) (21), on the differentiation of CPCs. Inhibitor treatment resulted in enhanced differentiation of CPCs, as reflected by the increased number of cardiac troponin T–expressing (cTnT-expressing) cells and by the increased expression of Nkx2.5 in CPCs treated with IWR1 with or without AZA (Figure 1D).
We next investigated the molecular mechanisms underlying Wnt signaling downregulation and particularly the functional significance of sustained upregulation of Wif1 during the early steps of CPC differentiation. To do this, we first silenced Wif1 expression in CPCs and tested the effect on their spontaneous (i.e., without pharmacologic treatment) differentiation in a coculture assay with neonatal rat cardiomyocytes (NNCMs), as previously described (14, 15). CPCs were first prelabeled with the permanent cell tracer CM-DiI, transfected with siRNA targeting Wif1 (or siRNA scramble) for 24 hours, and then mixed with NNCMs in 1:5 ratio and cocultured for 14 days. The efficiency of Wif1 inhibition on Wif1 expression was verified in parallel in homotypic CPC cultures (Figure 1E; see complete unedited blots in the supplemental material). Cells doubly stained cells for CM-DiI and α-sarcomeric actinin (α-sr-actinin) were considered to be CPCs undergoing cardiac differentiation; CPC differentiation was quantified as the ratio of cells doubly stained for CM-DiI and α-sr-actinin. We have previously excluded cell fusion using FISH and genetic techniques as confounders in the readout of this assay (14). Monocultures of CPCs were evaluated in parallel. No α-sr-actinin staining was observed in any monoculture (data not shown). As illustrated in Figure 1E, downregulation of Wif1 significantly decreased the number of differentiating CPCs cocultured with NNCMs by 21%, suggesting a role for this endogenous Wnt inhibitor in mediating CPC differentiation.
Wif1 gene expression is regulated by Dnmt3a-dependent methylation in CPCs, which regulates their differentiation to the myocyte lineage.
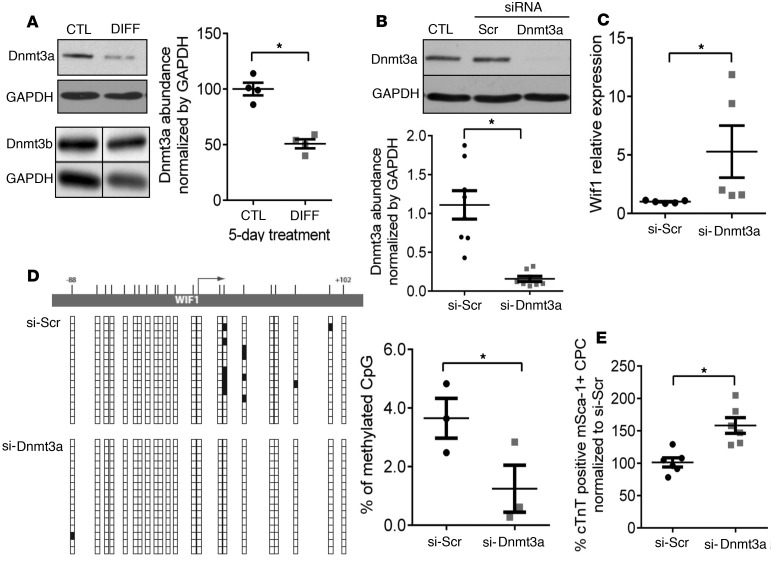
Wif1, a secreted antagonist of the Wnt/β-catenin pathway, was shown to be epigenetically regulated by DNA methylation in cancer cells (22–25). De novo DNA methylation is mediated mainly by the DNA methyltransferases Dnmt3a and Dnmt3b. To determine whether Dnmt3a and/or Dnmt3b could be putative regulators of Wif1 expression in CPCs, we first quantified protein abundance of both isoforms in CPCs during in vitro differentiation (Figure 2A; see complete unedited blots in the supplemental material). We found that the abundance of Dnmt3a, but not Dnmt3b, was strongly reduced early in the differentiation process (–48.77% ± 1.00% compared with the control). To verify the causality of Dnmt3a downregulation in Wif1 regulation, we treated naive CPCs with siRNA targeting Dnmt3a (Figure 2B; see complete unedited blots in the supplemental material) and measured the relative expression of Wif1 by quantitative real-time PCR (Figure 2C). Downregulation of Dnmt3a in CPCs triggered an upregulation of Wif1 expression (by 4-fold compared with the control siRNA scramble), supporting a negative regulatory role of Dnmt3a on Wif1 expression in CPCs. To verify that Dnmt3a affects Wif1 gene expression through its de novo methyltransferase activity, we analyzed the DNA methylation pattern of the Wif1 gene in untreated CPCs after siRNA downregulation of Dnmt3a for 72 hours. DNA methylation patterns were determined by bisulfite sequencing of the –88- to 102-bp region of Wif1, which contains a high number of CpG sites. Dnmt3a downregulation produced a significant decrease in the DNA methylation level within the Wif1 promoter region (–65.8% ± 13%, compared with control siRNA scramble), in particular at two CpG sites located immediately downstream of the transcription start site (Figure 2D). Therefore, downregulation of Dnmt3a, as observed during CPC differentiation, decreases DNA methylation and promotes Wif1 gene expression.
Figure 2. Dnmt3a downregulation upregulates Wif1 expression in cardiac progenitor cells and promotes their differentiation to the myocyte lineage.
(A) Cultured cardiac progenitor cells (CPCs) were treated with 5-azacytidine and TGF-β (DIFF medium) for 5 days, and Dnmt3a and Dnmt3b abundance was measured by immunoblotting (corrected for equal protein loading by GAPDH immunodetection). *P < 0.05 vs. CTL; n = 4 experiments; unpaired t test. (B and C) Untreated CPCs were transfected with siRNA targeting Dnmt3a for 72 hours, and (B) the effect on Dnmt3a protein level was measured by Western Blot and (C) the effect on Wif1 transcripts was measured by qRT-PCR. Results were normalized by GAPDH protein or mRNA abundance, respectively. *P < 0.05 vs. Si-Scr; n = 5–7 experiments; unpaired t test. (D) Analysis of the methylation pattern of the Wif1 gene in CPCs transfected with Si-Dnmt3a for 72 hours. CpG methylation in the Wif1 gene region extending from –88 to +102 bp was assessed by bisulfite sequencing. Representative images of (un)methylated CpG in corresponding Wif1 region. Vertical bars correspond to the location of CpG sites. Vertical lines of boxes below correspond to multiple clones of the corresponding sequences of Wif1 (white box: unmethylated; black box: methylated). Percentage of methylated CpGs in the Wif1 DNA fragments, calculated as a ratio of the number of methylated CpG to the total number of CpG sites from CPCs treated with Si-Dnmt3a (or Si-Scr). *P < 0.05 vs. Si-Scr; n = 3 experiments; unpaired t test. (E) CM-Dil–stained CPCs were first transfected with si-Dnmt3a (or si-Scramble) and then cocultured with neonatal cardiomyocytes (NNCM) for 14 days. Differentiation was assessed from the percentage of cardiac troponin T–positive (cTnT-positive) CPC. Dnmt3a inhibition increases the efficiency of CPC differentiation in coculture with NNCMs. *P < 0.05 vs. Si-Scr; n > 3 experiments; unpaired t test.
To further assess the causality of Dnmt3a downregulation in cardiac lineage specification, we analyzed the effect of Dnmt3a inhibition on the efficiency of differentiation of CPCs cocultured with NNCMs. Freshly isolated CPCs were prelabeled with the permanent cell tracer CM-Dil, transfected with siRNA targeting Dnmt3a or siRNA scramble constructs, and cocultured with NNCMs as described above (14). Cells doubly stained cells for CM-DiI and cTnT were considered to be CPCs undergoing cardiac differentiation; CPC differentiation was quantified as the ratio of cells doubly stained for CM-DiI and α-sr-actinin. Monocultures of CPCs were evaluated in parallel. No cTnT staining was observed in any monoculture (data not shown). By comparison, approximately 15%–20% of siRNA scramble–treated CPCs were doubly positive for cTnT and CM-DiI, indicative of cardiac differentiation, upon coculture with NNCMs. However, the number of differentiated CPCs was significantly higher when the cells were first transfected with siRNA-Dnmt3a (+61.6% ± 12 %, compared with siRNA scramble) (Figure 2E). This indicates that Dnmt3a regulates CPC differentiation to the cardiac lineage.
miR-29a downregulates Dnmt3a, with subsequent upregulation of Wif1 expression and CPC differentiation.
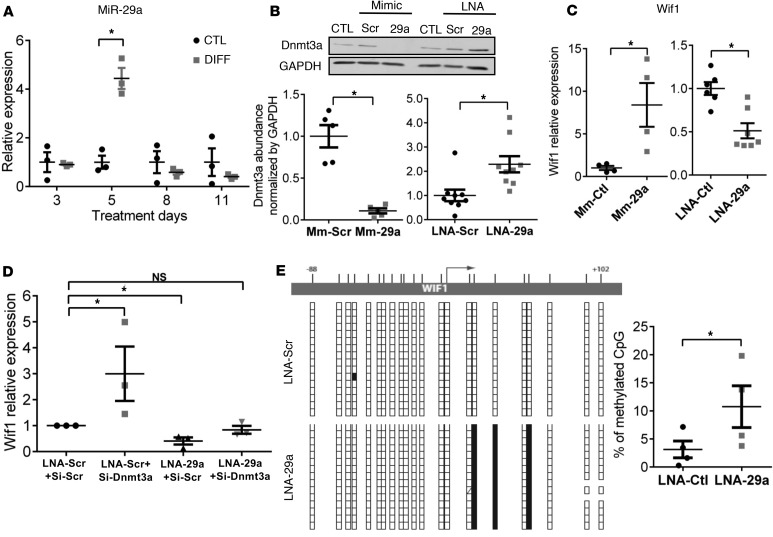
In search of epigenetic regulators of myocyte commitment that would coordinately regulate Dnmt3a downregulation and Wif1 upregulation, we performed an in silico bioinformatic search for microRNA putatively targeting Dnmt3a. Using 5 different algorithms, we identified the miR-29 family as strong potential candidates, as they were the only ones predicted by all 5 to target Dnmt3a 3′UTR (Supplemental Table 2). Therefore, we examined the expression level of miR-29a and miR-29b in the course of CPC differentiation and found an early (5 days) and transient upregulation of mature miR-29a in differentiation medium (+344% ± 50.8 %, compared with control, Figure 3A). The expression of miR-29b was also upregulated but with more variability among the independent replicates (Supplemental Figure 2). Of note, the upregulation of miR-29a at day 5 of the differentiation process coincided with the initial rise, followed by sustained upregulation of Wif1 (see Figure 1C). Using anti-miR (LNA-miR-29a) and a mimic construct of miR-29a in naive CPCs, we confirmed that miR-29a reciprocally regulates Dnmt3a protein level as well as Wif1 expression (Figure 3, B and C; see complete unedited blots in the supplemental material). To verify that miR-29a upregulates Wif1 expression through Dnmt3a repression, we treated CPCs with both anti-miR-29a and siRNA-Dnmt3a (and respective control constructs) and measured the relative expression level of Wif1 by qRT-PCR (Figure 3D). Combined inhibition of miR-29a and its target Dnmt3a abrogated the effect on Wif1 expression, showing that miR-29a indirectly regulates Wif1 expression through Dnmt3a downregulation. We further analyzed the methylation pattern of Wif1 by bisulfite sequencing in CPCs transfected with anti-miR-29a. miR-29a inhibition significantly increased the level of DNA methylation in the Wif1 gene (Figure 3E) and, in particular, at the two CpG sites that were found to be targeted by Dnmt3a (Figure 2D). These results demonstrate that miR-29a exerts epigenetic regulation of Wif1 via Dnmt3a.
Figure 3. miR-29a increases Wif1 through repression of Dnmt3a and its de novo methylation of Wif1 gene.
(A) Cultured cardiac progenitor cells (CPCs) were treated with 5-azacytidine/TGF-β (DIFF) or control medium (CTL) for 3 to 11 days. Expression of miR-29a was measured by qRT-PCR and normalized to 5S. *P < 0.05 vs. CTL at day 5; Mann-Whitney test. miR-29a in DIFF at day 5 is also different from DIFF at days 3, 8, and 11 by Kruskal-Wallis with Bonferroni correction; n = 3 experiments. (B) Untreated CPCs were transfected with either a miR-29a mimic construct (Mm-29a) or LNA-miR-29a (LNA-29a) or respective scramble constructs (Scr). Dnmt3a proteins (by Western Blot) and (C) Wif1 transcripts (by qRT-PCR), normalized by GAPDH protein or mRNA abundance, are reported as fold changes from CTL (set at 1). *P < 0.05 vs. mimic/LNA-scr; n = 4–8 experiments; unpaired t test. (D) Untreated CPCs were cotransfected with si-Dnmt3a and LNA-29a (or Scr controls). Relative expression of Wif1 mRNA (qRT-PCR, normalized to GAPDH) is expressed as fold change from CTL (transfected with both Scr constructs, set at 1). *P < 0.05 vs. LNA-Scr+si-Scr, 1-way ANOVA, Bonferroni’s test; n = 3 experiments. (E) Untreated CPCs were transfected with LNA-29a (or LNA-Scr) for 72 hours, and CpG methylation of the Wif1 gene region extending from –88 to +102 bp was assessed by bisulfite sequencing. Representative image of (un)methylated CpG in corresponding Wif1 region. Each vertical line of boxes below corresponds to multiple clones of the corresponding sequence of the Wif1 gene (white box: unmethylated; black box: methylated). Percentage of methylated CpG in the Wif1 DNA fragments. *P < 0.05 vs. LNA-Scr; n = 4 experiments; unpaired t test.
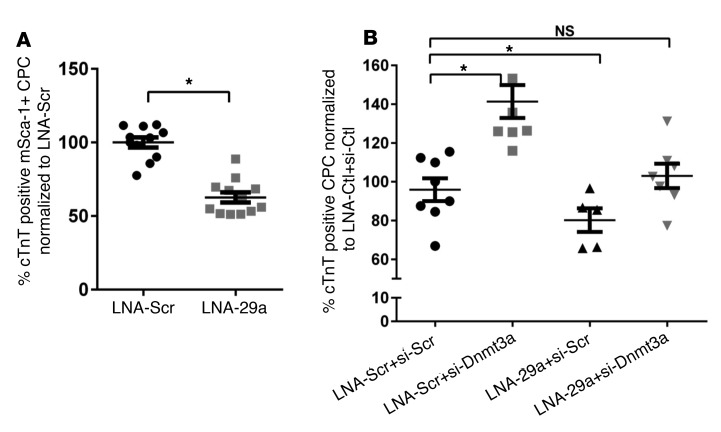
To confirm the involvement of miR-29a in cardiac differentiation of CPCs, we examined the effect of miR-29a inhibition on CPC differentiation upon coculture with NNCMs (as used above) (Figure 4A). Pretreatment of CPCs with LNA-miR-29a decreased their differentiation rate (63% ± 3.7 % of CPCs treated with the control LNA scramble). To test if miR-29a regulates differentiation through other putative targets (than Dnmt3a), we further cotransfected CPCs with LNA-miR-29a and siRNA-Dnmt3a (and respective control constructs) before coculture with NNCMs. Whereas miR-29a inhibition alone decreased CPC differentiation, as expected (Figure 4B, 3rd tick mark), combined miR-29a inhibition and repression of Dnmt3a abrogated the effect of miR-29a downregulation (Figure 4B, 4th tick mark). These data identified Dnmt3a as a functionally relevant target for miR-29a–mediated CPC differentiation.
Figure 4. miR-29a promotes cardiac progenitor cell differentiation to cardiomyocytes through Dnmt3a downregulation.
CM-Dil–stained cardiac progenitor cells (CPCs) were first transfected with locked nucleic acid targeting miR-29a (LNA-miR-29a) (or LNA-Scr) and/or siRNA-Dnmt3a (or si-Scr) and then cocultured with neonatal cardiomyocytes (NNCMs) for 14 days. The efficiency of CPC differentiation was determined by the percentage of doubly stained cells for CM-DiI and cardiac troponin T (cTnT). (A) CPCs transfected with LNA-miR-29a have a lower capacity to differentiate into cardiomyocytes, (B) whereas cotransfection of CPCs with both LNA-miR-29a and siRNA-Dnmt3a does not affect their efficiency of differentiation compared with cells cotransfected with respective controls (LNA-Scr+si-Scr). (A)*P < 0.05 vs. LNA-Scr; n > 3 experiments; unpaired t test. (B) *P < 0.05 vs. LNA-Scr+si-Scr; n > 3 experiments; 1-way ANOVA, with Bonferroni’s test.
Transient inhibition of Dnmt3a in CPCs promotes myocyte differentiation of CPCs in vivo.
We next examined the relevance of these epigenetic mechanisms on CPC differentiation in vivo in a mouse infarction model. CPCs were first transfected with siRNA-Dnmt3a (or scramble construct) ex vivo for 18 hours and then were directly injected in 3 distinct sites peripheral to the infarcted left ventricular (LV) area induced by permanent ligature of the left anterior descending (LAD) coronary artery. Efficient Dnmt3a downregulation was verified in parallel in a sample of each batch of cells. After injection, mice were phenotyped by cardiac MRI at 24 hours and then at 28 days after myocardial infarction (MI) before sacrifice. An additional group was sacrificed at day 15 after MI for histological analysis only.
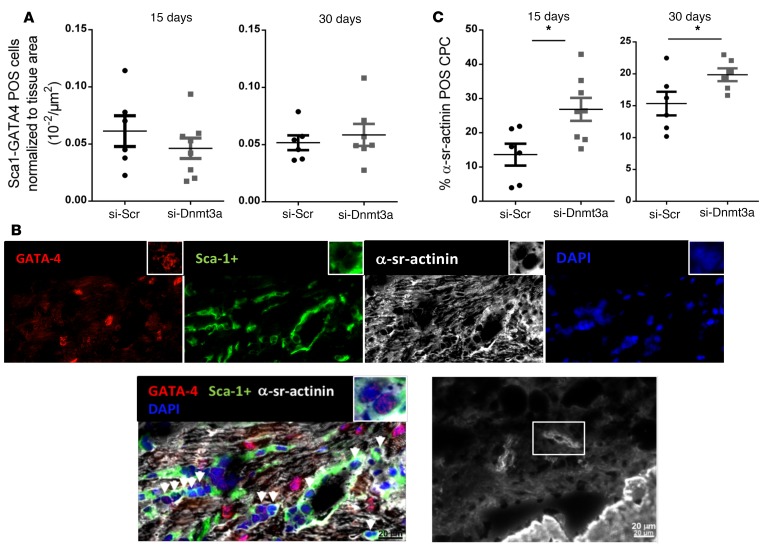
To measure the effect of the transient downregulation of Dnmt3a on CPC differentiation in vivo, we first identified cells doubly stained with Sca-1 and GATA4 markers as progenitor cells with cardiogenic potential (3, 20, 26). Histological analysis of LV sections (at 15 and 30 days) of the infarction border zones revealed the presence of small cells coexpressing GATA4 and Sca-1, albeit in very small proportion compared with the total number of GATA4-expressing cells (approximately 0.04% of GATA+ cells detected per μm2 tissue (Figure 5A). Among these doubly stained cells, we identified a small number of cells that coexpressed α-sr-actinin, indicating differentiation toward the cardiac lineage (Figure 5B), although the staining remained punctate, indicative of poor sarcomeric organization. No cell with adult-type differentiation was detected at either time point. Nevertheless, the number of Sca-1/GATA4/α-sr-actinin–expressing cells was almost double at 15 days after MI in hearts injected with siRNA-Dnmt3a–treated CPCs compared with controls (siRNA scramble CPCs) and increased by approximately 33% at 30 days after MI, revealing a positive effect of Dnmt3a downregulation on CPC differentiation in vivo (Figure 5C).
Figure 5. Downregulation of Dnmt3a in cardiac progenitor cells injected in infarcted hearts improves cardiac progenitor cell differentiation in vivo.
Cardiac progenitor cells (CPCs) were first transfected with si-Dnmt3a and then injected in the border zone of infarcted mouse hearts after LAD ligature. (A) Mice were sacrificed at 15 or 30 days for immunohistochemical analysis of heart sections. Quantification of Sca-1/GATA4POS cells per μm2 tissue in the border zones of the myocardial infarct (MI). *P < 0.05 compared with respective si-Scr; n = 6–8 mice; Mann-Whitney test. (B) Representative section of mouse peri-infarct region stained for GATA4, Sca-1+, α-sr-actinin, and DAPI. Arrows indicate multiple small cells positive for Sca-1 and GATA4. Few of these doubly positive cells also express α-sr-actinin, although without visible sarcomeric organization. Representative cell with the beginning of a sarcomeric organization. Scale bar: 20 μm; original magnification, ×40 (inset). (C) Quantification of CPC differentiation in vivo 15 and 30 days after MI. Results are expressed as ratios of CPCs triple positive for GATA4/Sca-1/α-sr-actinin, as index of their differentiation, to the total number of GATA4/Sca-1+ CPCs. The percentage of CPCs expressing the cardiac sarcomeric protein is higher in hearts injected with CPCs with downregulated Dnmt3a at both time points. *P < 0.05 compared with respective si-Scr; n = 6–8 mice; Mann-Whitney test.
Injection of CPCs with downregulated Dnmt3a attenuates adverse cardiac remodeling in infarcted hearts.
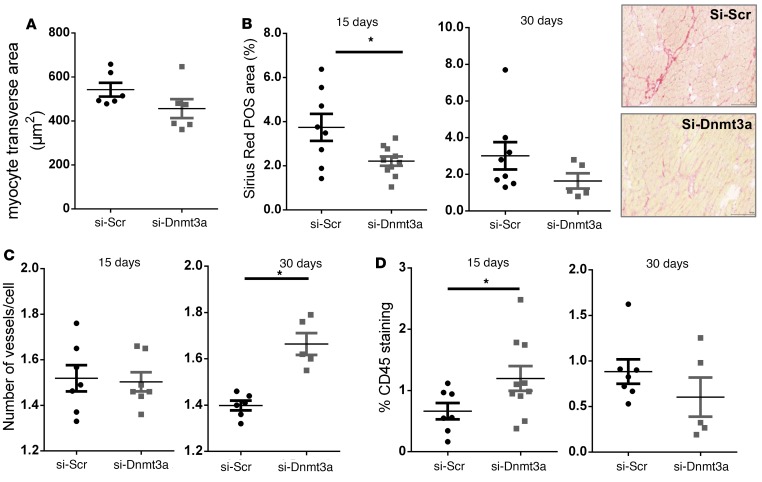
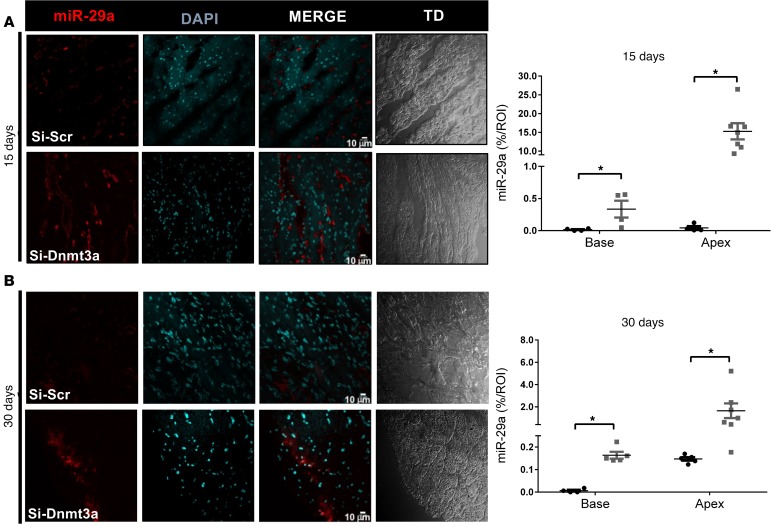
Interestingly, histological analysis of the remote myocardial areas revealed similar myocyte sizes (transverse areas) (Figure 6A), a transient increase in inflammatory infiltrates (CD45 staining) (Figure 6D), and decreased interstitial fibrosis (Sirius red staining) (Figure 6B) at 15 days as well as a significant increase in capillary densities (CD31 staining) at 30 days after MI (Figure 6C) in hearts injected with siRNA-Dnmt3a–treated CPCs compared with controls, suggesting potential paracrine effects of injected modified cells on cardiac tissue remodeling. We also examined the expression of miR-29a by in situ hybridization in the peri-infarct areas and found a significant increase in miR-29a abundance in hearts injected with siRNA-Dnmt3a–treated CPCs compared with controls both at 15 and 30 days after MI (Figure 7).
Figure 6. Injection of cardiac progenitor cells with downregulated Dnmt3a attenuates myocardial remodeling in areas remote from the infarcted region.
Cardiac progenitor cells (CPCs) were first transfected with si-Dnmt3a and then injected in the border zone of infarcted mouse hearts after LAD ligature. Mice were sacrificed at 15 or 30 days for immunohistochemical analysis of remote heart sections. (A) Myocyte transverse area (μm2) at 30 days. Remote myocardial transverse sections were stained with rhodamine wheat germ agglutinin (WGA) and quantified in 400 cells per heart in 6 hearts per group. (B) Interstitial fibrosis measured by Picrosirius red staining at 15 and 30 days. Areas of collagen fibers are identified in red (original magnification, ×20) were normalized to tissue area. Average of interstitial fibrosis was quantified in 3–6 sections per heart. n = 8–10 hearts at 15 days; n = 5–8 hearts at 30 days. *P < 0.05 compared with control; Mann-Whitney test. (C) Cardiac capillary density measured by isolectin staining and normalized to myocyte cell number at 15 and 30 days after surgery. Average capillary density was quantified in 3 sections /heart. n = 7 hearts at 15 days; n = 5–6 hearts at 30 days. *P < 0.05 compared with control; Mann-Whitney test. (D) Inflammatory cell infiltrate measured by immunostaining of CD45 15 and 30 days after surgery. n = 7–10 hearts at day 15; n = 5–7 hearts at day 30. *P < 0.05; Mann-Whitney test.
Figure 7. In situ hybridization of miR-29a in infarcted hearts with siRNA-Dnmt3a–injected cardiac progenitor cells.
Cardiac progenitor cells (CPCs) were first transfected with si-Dnmt3a and then injected in the border zone of infarcted mouse hearts after LAD ligature. Mice were sacrificed at 15 or 30 days for in situ hybridization analysis of remote heart sections. (A and B) Hybridization signals in remote myocardial areas at (A) 15 days or (B) 30 days after MI in hearts injected with si-Scr–treated CPCs or si-Dnmt3a–treated CPCs. Quantification of miR-29a is shown. Signals in hearts injected with Si-Scr CPCs are represented by black dots, and signals in hearts injected with siDnmt3a CPCs are represented by gray dots. *P < 0.05 compared with respective controls; n = 4–7 hearts; Mann-Whitney test.
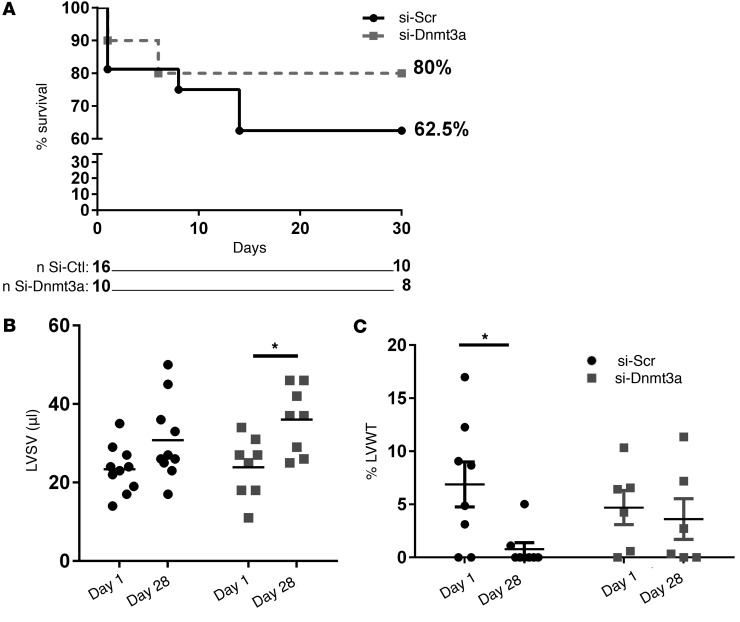
Finally, we examined the effect of the injection of epigenetically modified CPCs on the function of infarcted hearts. Infarct sizes (as determined from the number of akinetic/dyskinetic LV wall segments by MRI) were identical between treatment groups. Although the study was not powered for survival analysis, we observed a trend for better survival of mice injected with siRNA-Dnmt3a–treated cells compared with controls (Figure 8A). Likewise, mice in the active treatment group displayed a better recovery in LV functional parameters, such as LV stroke volume (Figure 8B) and a preserved LV wall thickening (Figure 8C), at 30 days after infarction.
Figure 8. Improved functional recovery of infarcted hearts after injection of cardiac progenitor cells with downregulated Dnmt3a.
Cardiac progenitor cells (CPCs) were first transfected with siRNA-targeting Dnmt3a (si-Dnmt3a) and then injected in the border zone of infarcted mouse hearts after LAD ligature. (A) Survival curves of mice injected with CPCs pretransfected with si-Dnmt3a (or scramble control). (B and C) Mice were phenotyped by cardiac magnetic resonance imaging (cMRI) at day 1 and 28 after surgery. (B) Left ventricular stroke volume (LVSV) at day 1 and 28 after myocardial infarction. Mice injected with CPCs after downregulation of Dnmt3a (si-Dnmt3a) show better recovery of LVSV compared with control CPCs (si-Scr) at day 28. (C) Increase in left ventricular wall thickening (LVWT) in systole (expressed as a percentage of diastolic thickness) in the same mice as in B. Mice injected with CPCs after downregulation of Dnmt3a (si-Dnmt3a) show better preservation of systolic LVWT compared with control CPCs (si-Scr) at day 28. *P < 0.05 compared with respective values at day 1; n = 8–10 mice; 2-way ANOVA, Sidak’s test.
Discussion
Using previously characterized adult CPCs and differentiation protocol, we show that the canonical Wnt/β-catenin pathway is downregulated in the early stages of differentiation and that its inhibition is sufficient to promote CPC differentiation to cardiomyocytes in vitro. This is consistent with ours and others’ previous data demonstrating the obligatory role of secondary Wnt/β-catenin inhibition for mesoderm specification toward the cardiac lineage as well as for the differentiation of adult Sca-1+ CPCs in vitro and in vivo (9, 12–15). Recent data in human ESCs have similarly identified the sequential role of synergistic activation of Wnt and BMP for initial mesoderm formation, followed by Wnt inhibition required for cardiac differentiation (12). Although this and other studies focused on downstream networks of transcriptional regulators, virtually nothing is known about the mechanism driving this secondary Wnt extinction.
Our expression kinetics and targeted RNA invalidation data point to the Wnt antagonist, Wif1, as a functionally relevant mediator of endogenous Wnt inhibition for CPC differentiation. Indeed, the progressive increase in Wif1 expression in AZA-treated CPCs coincides with the downregulation of Wnt target genes, and Wif1 siRNA targeting in naive (i.e., not treated with AZA) CPCs (that exhibit constitutive activity of Wnt signaling, ref. 14) attenuates their differentiation capacity in the coculture model, which is similar to the in vivo situation. The induction of Wif1 expression following demethylation treatment with AZA is in line with previous reports on the regulation (i.e., inhibition) of Wif1 gene expression by methylation in cancer cells (22–25). However, its progressive increase (peaking at day 11, i.e., 8 days after AZA was stopped) suggested intermediate epigenetic regulators. Indeed, we found that the de novo DNA methyltransferase Dnmt3a is strongly decreased early (at day 5, i.e., 2 days after AZA treatment arrest) during cardiac differentiation, concomitant with a strong increase in the expression of miR-29a. In addition, the functional importance of both Dnmt3a and miR-29a for Wif1 gene methylation and Wif1 expression as well as for CPC differentiation was verified in naive CPCs in monocultures as well as in the coculture model, respectively. Our identification of the miR-29 family based on bioinformatic analysis corroborates previous reports on Dnmt3a regulation by miR-29a in other cell types (27, 28) and is supported by our expression as well as miR-29a mimic and inhibition data. Altogether, we demonstrated that miR-29a indirectly modifies the methylation pattern of the Wif1 gene, promoting its expression through Dnmt3a downregulation, which is also key for the effect of miR-29a on differentiation. Wnt signaling regulators, such as Wif1, most probably belong to a subset of the methylation-dependent genes sensitive to Dnmt3a downregulation. Other genes coding cardiac-specific proteins, such as Myh6, Myh7, Tnni3, and Tnnt2, were reported to be similarly sensitive to DNA methylation (29–31). Likewise, besides its indirect regulation of methylation-sensitive genes through Dnmt3a targeting, miR-29a probably modulates the stability and/or translation of other transcripts regulating cardiogenesis that are not identified here. Notably, although the expression of miR-29a itself may also be methylation sensitive (32), its upregulation did not occur immediately after AZA treatment, suggesting that additional signaling events may support its temporary overexpression at day 5.
Our demonstration of similarly enhanced differentiation of CPCs with downregulated Dnmt3a in infarcted hearts extends our paradigm in vivo. We chose a model of transient decrease of Dnmt3a in injected CPCs because a prolonged downregulation could affect chromatin remodeling in an uncontrolled way and induce adverse effects (33). Likewise, we did not inject miR-29a mimics in the border zones of the injured tissue to avoid uncontrolled effects on multiple cell types and neither did we inject CPCs transfected with miR-29a mimics, as we questioned the concentration- and time-dependent efficiency of target(s) inhibition in vivo.
As observed in our in vitro experiments, transient Dnmt3a inhibition enhanced CPC differentiation in vivo, as revealed by costaining of Sca-1+ CPCs with GATA-4 and α-sr-actinin. Despite expression of some late markers of differentiation, most of the identified cells were still mononucleated and lacked organized sarcomeres even 4 weeks after injection, indicating incomplete maturation to the adult phenotype. Indeed, others have shown that only 50% of cells undergoing differentiation present a binucleation and an organized sarcomeric structure as late as 12 weeks after the injury (26), illustrating the long-term process of cardiac differentiation in vivo. We did not prelabel CPCs before injection, as it was not the purpose of this study to track cell survival/migration in the infarcted heart. Consistent with several previous studies (26, 34, 35), the number of cells differentiating to the cardiac myocyte lineage was minimal but nevertheless significantly increased by 30%–60% in hearts injected with Dnmt3a-targeted CPCs. As the cells were not labeled, part of the differentiating CPCs may include endogenous progenitors sensitive to paracrine signaling from the injected cells.
Such paracrine signals may also account for the preservation of cardiac function and prevention of wound expansion in mice injected with modified CPCs, with a significantly higher LV stroke volume and LV wall thickening at day 28 after acute MI (at similar initial MI sizes). Indeed, it is unlikely that the small number of differentiating CPCs, mostly at an immature state, were sufficient to restore contracting muscle. We also observed a trend toward higher survival rate in mice that received modified CPCs, suggestive of distal signaling to neighboring cells affecting broader aspects of cardiac function (36–39).
Our histomorphometric analysis of infarcted hearts suggests a modified secretome in siRNA-Dnmt3a–treated CPCs. Collagen measurements revealed a significant decrease in interstitial fibrosis as soon as 2 weeks after MI, possibly through differential expression of secreted antifibrotic factors. Interestingly, miR-29a is a well-known antifibrotic microRNA targeting various collagens and other extracellular matrix proteins as well as profibrotic Smad3 signaling (40, 41). Its expression was shown to be strongly decreased in the region of the fibrotic scar after MI, while its downregulation participates in the increased cardiac fibrosis in the injured heart (40). In lung cancer cells, Tan et al. have shown that miR-29a expression is methylation sensitive and that Dnmt3a/b downregulation regulates its expression in a positive feedback loop (32). An upregulation and increased secretion of miR-29a, as a secondary effect of Dnmt3a inhibition in modified CPCs, may participate to the beneficial antifibrotic effect we observed in vivo. Moreover, in human cardiac tissue, profibrotic changes induced by prolonged hypoxia during MI are associated with a global DNA hypermethylation and increased expression of Dnmt1 and Dnmt3b enzymes (42). Indeed, Dnmt3-induced hypermethylation of antifibrotic genes, such as THY-1 (43) or RASSF1A (44), was shown to contribute to fibroblast activation and fibrogenesis. Therefore, miR-29a overexpression, as observed in this study, could also affect cardiac fibrosis by targeting Dnmt3a/b, propagating Dnmt3 downregulation in surrounding cells.
Our histological analysis also revealed an increased capillary density in the remote myocardial area 4 weeks after MI, suggestive of increased neoangiogenesis in hearts injected with modified CPCs. Emerging evidence supports the role of epigenetic DNA methylation in endothelial cell differentiation and homeostasis. Knockdown of Dnmt1 and Dnmt3a promotes the angiogenesis of human mesenchymal stem cells, leading to arterial specific differentiation (45). In HUVECS and mouse carotid arteries endothelium, mechanosensitive genes, such as HoxA5, Klf3, and Klf4, are hypermethylated upon disturbed blood flow, as found in atherosclerotic vessels, while AZA treatment restores their methylation pattern (46–48). Again, the increased capillary density in hearts injected with modified CPCs may have been induced by modified paracrine factors and/or the overexpression of miR-29a in cardiac tissue, subsequently targeting Dnmt3a/b expression in neighboring endothelial cells. Altogether these data indicate propagation of differential cell signaling by modified CPCs beyond the injection site in the injured area.
The potential benefit of miR-29a overexpression in infarcted myocardium, as outlined above, could be used as a therapeutic opportunity, provided it is delivered at appropriate dose and targeted to specific cell types. However, circulating miR-29a levels were also shown to correlate with hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy (49). Other studies have also suggested adverse effects of miR-29a in the diabetic heart (50) or on vascular integrity in advanced age (51), raising some caution when considering systemic delivery of miR-29a as a therapeutic option. This also reinforces the advantage of our targeted approach with cell-specific regulation of endogenous expression rather than nonspecific supply of exogenous miR-29a mimics.
Methods
Cardiac progenitors and cardiomyocytes isolation.
Isolation of Sca-1+ CPCs was performed using hearts of 8- to 10-week-old male C57Bl/6 mice based on the protocol described previously (3, 15) with some modifications, as in ref. 14. In brief, harvested hearts were enzymatically and mechanically digested and then the resulting suspension was filtered twice and red blood cells were removed by osmotic shock. Enrichment of Sca-1+ cells was achieved by sorting the cells using the anti-Sca-1 MicroBead kit (FITC) (Miltenyi Biotec; 130-092-529) and the Magnetic Cell Sorting system (MACS MS column; Miltenyi Biotec; 130-042-201), as described in ref. 14. The quality of the Sca-1+ enrichment fraction was verified by flow cytometry. Cells were seeded on precoated gelatin plates in Ham’s F-12K Medium (Thermo Fisher Scientific; 21127022) with 20% FBS (Sigma-Aldrich; F7524), rhFGF basic (10 ng/ml; R&D Systems; 233-FB), LIF (103 unit/ml; Merck Millipore; ESG1107), and 1% penicillin/streptomycin (Thermo Fisher Scientific; 10378016) for 3 to 4 days and then subsequently switched and amplified in Sca-1+ expansion medium, composed of DMEM/F12 GlutaMAX (Thermo Fisher Scientific; 31331028) supplemented with ITS (10 μg/ml insulin, 5 μg/ml transferrin, 5 ng/ml medium sodium selenite; Thermo Fisher Scientific; 41400045), rhFGF basic (10 ng/ml), epidermal growth factor (20 ng/ml; PeproTech; 315-09), LIF (103 unit/ml), B27 serum-free supplement (×50 diluted; Thermo Fisher Scientific; 17504044), HEPES (5 mM; Thermo Fisher Scientific; 15630080), and 1% penicillin/streptomycin. CPCs were further clonally expanded by dilution cloning, as reported previously (14, 19).
NNCMs were isolated from 0- to 3-day-old rat pups (Wistar) by consecutive digestions of heart pieces with collagenase type II (Thermo Fisher Scientific; 17101-015) and pancreatin (Sigma-Aldrich; P3292) according to a previously described protocol (20). Cells were separated by using two successive Percoll gradient centrifugations and selective attachment of fibroblasts. Neonatal myocytes were cultured in DMEM/M199 (4:1) (Thermo Fisher Scientific; 11965092 and 41150020) supplemented with 100 mM BrdU (Sigma-Aldrich; B5002) and 1% penicillin/streptomycin.
Differentiation assay of Sca-1+ CPCs in monoculture or in coculture with NNCMs.
Differentiation of Sca-1+ CPCs toward the cardiac myocyte lineage was induced with AZA (Sigma-Aldrich; A2385) pretreatment for 3 days, followed by TGF-β (PeproTech; 240-B-002), as previously described (20). In some experiments, cells were treated with the Wnt pharmacological inhibitor IWR1 (Sigma-Aldrich; I0161) alone or following AZA treatment. The molecule was renewed every day.
For the coculture experiments, isolated Sca-1+ CPCs were first labeled with the permanent cell tracer CM-Dil (Molecular Probes; Thermo Fisher Scientific; C-7000) and then transfected or not with siRNA/anti-miR or mimic for 24 hours. NNCMs were added on top of labeled CPCs in a ratio 1:5 (Sca-1+ CPCs/NNCMs), in DMEM (low glucose) supplemented with 10% FBS. Cocultures were maintained for 14 days before proceeding to immunohistochemical analysis.
Superfusion and TOP-Flash reporter assays.
Sca-1+ CPCs were differentiated in monocultures as described above or cultured in control medium. After refreshing media 48 hours before, conditioned media were harvested at day 8 of differentiation and centrifuged in Amicon Ultra-10K columns (Millipore, UFC901024) at 3,200 g for 15 minutes at 4°C to concentrate samples to a volume of 1 ml/P100. Concentrated media were used freshly for superfusion on HEK293T reporter cells.
One day before transfection, HEK293T cells were seeded at 320,000 cells/well in a 12-well-plate format in DMEM (Thermo Fisher Scientific, 61965-026) containing 10% fetal calf serum. Cells were transfected with 0.5 μg TOPFlash reporter construct (Addgene, 12456) and 0.05 μg TK-Renilla luciferase plasmid (as control for transfection efficiency, Promega, E2241) using Lipofectamine 2000 Transfection Reagent (Thermo Fisher Scientific; 11668027) in a DNA/Lipofectamine ratio of 1:3. Four hours after transfection, cell media were removed and transfected cells were superfused with conditioned media from differentiated or control Sca-1+ CPCs for 24 hours, using concentrated medium from 1 P100/well. The activity of TOPFlash and TK-Renilla were measured using the Dual-Luciferase Reporter Assay System (Promega; E1910).
Sca-1+ CPC transfection with siRNA, anti-miR, or mimic.
CPCs were seeded at 50% confluency the day before transfection. Cells were transfected with 25 nM siRNA targeting mouse Dnmt3a (Dharmacon; L-065433-01-0005), mouse Wif1 (Dharmacon; L-046832-00-0005), or siRNA-negative control (Dharmacon; D-001810-10-05) using jetPRIME transfection reagent according to the manufacturer’s recommendations (Polyplus transfection; 114-07). To mimic miR-29a activity, CPCs were transfected with 25 nM miR-29a mimic (Dharmacon; C-310521-07-0005) or mimic negative control (Dharmacon; CN-001000-01-05) using jetPRIME transfection reagent as well. To inhibit miR-29a activity, CPCs were transfected with 75 nM anti-miR–targeting miR-29a (LNA-29a, Exiqon; 427005-00) or anti-miR negative control (LNA-Scr, Exiqon; 199020-00) using Lipofectamine RNAiMAX Transfection Reagent (Thermo Fisher Scientific; 13778075). Four hours after transfection, medium was changed for complete Sca-1+ expansion medium. Depending on experiments, cells were harvested 24 or 72 hours after transfection. In coculture experiments, CM-Dil–stained CPCs were first transfected with siRNA/mimic/LNA for 4 hours and then washed twice with PBS; NNCMs were added on top of CPCs in a 1:5 ratio (Sca-1+ CPCs/NNCM) as previously described.
Western blot.
Protein levels of β-catenin, Wif1, Dnmt3a, and Dnmt3b were assessed in CPCs treated either with AZA and TGF-β or siRNA/mimic/anti-miR. For protein extraction, cells were solubilized in RIPA buffer containing proteinases inhibitors. Specific mouse monoclonal antibody against β-catenin (Thermo Fisher Scientific; 13-8400), rabbit polyclonal antibody against Wif1 (Abcam; ab33281), rabbit monoclonal antibody against Dnmt3a (Cell Signaling, 3598), and rabbit polyclonal antibody against Dnmt3b (Abcam; ab16049) were incubated at dilutions of 1:1,000, 1:500, and 1:1,000, respectively, for both Dnmt3a and Dnmt3b for 1 hour at room temperature. The blots were reprobed with a rabbit monoclonal anti-GAPDH antibody (Cell Signaling, 2118) as a loading control. After chemiluminescence revelation of proteins subjected to immunoblotting, signals were analyzed using digital image processing program (ImageJ, Wayne Rasband, NIH). The data are expressed as relative to GAPDH expression (in arbitrary units).
Immunohistochemical and histomorphometric analysis.
For immunocytochemical analysis of in vitro treatment, cells were fixed with 4% formaldehyde, permeabilized in 0.1% Triton/PBS buffer, and then incubated with primary antibody against cTnT (Abcam; ab10214) at 1:200 overnight and Alexa fluor 488 goat anti-mouse antibody (Thermo Fisher Scientific; A-11029) at 1:500 for 1 hour. To evaluate the number of differentiating cells in coculture assays, the ratio of doubly stained CPCs for CM-Dil and cTnT, normalized to the total number of CM-Dil+ CPCs, was used as an index of differentiation. A minimum of 200 cells per well were counted; each condition was performed in 4 replicates (4 wells), and a minimum of 3 independent experiments were performed. Therefore, approximately 800 cells/condition/experiment were counted.
For in vivo experiments, hearts were harvested at 15 or 30 days after surgery, cut in 3 pieces (base, median, and apex), and immediately cryopreserved. To proceed to histological analysis, myocardial samples were serially cut into 5-mm-thick sections and fixed in acetone.
To evaluate the amount of differentiating CPCs in the border zone of the MI, tissue was successively stained with rabbit anti-GATA4 antibody (Thermo Fisher Scientific; PA1-102) at 1:500 overnight, mouse anti-α-sr-actinin (Sigma-Aldrich; A7811) at 1:150 for 30 minutes, and rat FITC-anti-Sca-1 (Miltenyi Biotec; 130-102-297) at 1:10 for 1 hour. GATA-4 and α-sr-actinin stainings were revealed with Alexa Fluor 568 and Alexa Fluor 647 conjugates, respectively (Thermo Fisher Scientific; A11036 and A21247). Nuclei were counterstained with DAPI (Sigma-Aldrich; D9542). To reduce autofluorescence background and remove nonspecific signal from mouse Ig staining, tissues were preincubated with the Avidin/Biotin Blocking kit (Vector Laboratories; SP-2001) and the Mouse on Mouse kit (M.O.M., Vector Laboratories; BMK-2202). The number of CPCs localized in the border zone of the infarction was determined as the doubly stained cells for GATA-4 and Sca-1 per tissue area (in μm2). A minimum of 5 images (at ×40 magnification) per myocardial section were analyzed on 2 to 6 sections per heart. The number of differentiating cells was calculated as the ratio between triple-stained cells (GATA-4–, Sca-1–, and α-sr-actinin–positive cells) and doubly positive cells for GATA-4 and Sca-1.
To assess cardiac capillary density, tissue was costained with rhodamine-conjugated wheat germ agglutinin (plasma membrane staining, Vector Laboratories; RL-1022) and biotinylated isolectin B4 (GS-IB4, endothelial staining; Vector Laboratories; B-1205) and revealed with Fluorescein Avidin (Vector Laboratories; A-2001). Data from 200 to 400 cells were determined per slide using AxioVision software 4.8.2.0 (Zeiss).
Fluorescent stainings described above were imaged with an AxioImager.z1 microscope equipped with an ApoTome module for structured illumination and AxioVision 4.8.2.0 software (Zeiss).
To evaluate interstitial myocardial fibrosis and inflammatory cell infiltrate, sections were stained either with collagen-specific Picrosirius red (Sirius red in aqueous picric acid; 365548; Sigma-Aldrich) or with rat anti-CD45 (BD Biosciences; 550539) and revealed with biotinylated anti-rat immunoglobulins (Vector Laboratories; BA4001) followed by peroxidase-coupled streptavidin (BD Biosciences; 554066) and DAKO liquid DAB (Agilent; K346811). Sirius red– and CD45-stained sections were then digitalized with a SCN400 slide scanner (Leica Biosystems). The fraction of myocardial tissue occupied by collagen fibers or by CD45 staining was determined as a percentage by quantitative morphometry using TissueIA software (Leica Biosystems).
All histological quantifications were performed in a blinded manner.
FISH.
Double-DIG labeled miRCURY LNA detection probes (Exiqon) were used for miR-29a staining, and scrambled probes were used as negative control. In brief, the cryosections were thawed, fixed using fresh 4% (wt/vol) paraformaldehyde, and permeabilized with 0.5% PBS Triton X-100 for 5 minutes on ice. Slides were then incubated overnight with 2 pmoles of probes in a moist chamber at the hybridization temperature suggested by manufacturer’s instructions. The next day slides were stringently washed with ×0.1 SSC buffer (3 M NaCl, 0.3 M sodium citrate) (3 times for 10 minutes) at 4°C above the hybridization temperature and washed once for 5 minutes in ×2 SSC at room temperature. Following an incubation with a blocking solution for 30 minutes at room temperature, slides were treated with 1:50 Anti-Digoxigenin-POD, Fab fragments from sheep (Roche; 11207733910) for 2 hours at room temperature. Following 2 washes with 0.1% Tween PBS (PBS-T) and 1 wash in PBS for 5 minutes each, miRNA signal was detected with the TSA Alexa fluor-647 (Thermo Fisher Scientific; T20951). After a final wash with PBS-T and PBS, nuclei were counterstained with DAPI for 5 minutes at room temperature. Fluorescent images were taken with a laser scanning confocal microscope (Olympus, FV1000/SIMS). Quantification of miR-29a was performed using Fiji Imagej software, and values are expressed as the percentage of positive signal relative to background signal.
RNA analysis.
Total RNA from cultured cells was isolated using TRIzol (Thermo Fisher Scientific; 15596026). Isolated RNA was subjected to DNase treatment to eliminate genomic DNA contamination. SYBR Green I real-time reverse transcription–PCR (RT–PCR) was carried out with an RT step at 50°C for 30 minutes and an enzyme activation step at 95°C for 15 minutes, followed by 40 cycles of denaturation (95°C for 2 seconds), annealing (59°C for 5 seconds), and extension (72°C for 15 seconds). Analyses were performed on the Bio-Rad iQ5 software 2.0. Results were expressed as relative gene expression using DDCt method.
To detect the level of miRNA, 40 ng total RNA was reverse transcribed using the miRCURY LNATM Universal RT microRNA PCR polyadenylation and cDNA synthesis kit (Exiqon; 203301). qRT-PCR for miR-29a and U6 detection was then performed with microRNA LNATM PCR primers (Exiqon) using the GoTaq qPCR Master Mix (Promega). Relative expression was determined using the DDCt method. See Supplemental Table 1 for oligonucleotide sequences of forward and reverse primers.
Bisulfite sequencing.
Analysis of Wif1 promoter methylation was performed in CPCs transfected with siRNA or anti-miR for 72 hours. Genomic DNA was extracted using the chloroform/isoamyl technique, and 5 μg genomic DNA was submitted to sodium bisulfite treatment as previously described (52) and resuspended in 100 μl of DNase-free water. Methylated CpG content determination of Wif1 promoter was performed by PCR amplification and sequencing of bisulfite-treated DNA. For PCR amplification of bisulfite-converted Wif1 sequences, the following nested primers were used: BiWif1 S 5′-GGGTGGGTTGGTTATGTGTTTTT-3′; BiWif1 AS 5′-AGAATTTTATAAGTAGTATAGGTTGGG-3′; nBiWif1 S 5′-GTAGGAGGTTTGAGTGATGATTTAGAAGT-3′; nBiWif1 AS 5′-TGTTGGTATTGTTAGTTTTGTTAGTATCGTGTTTTTG-3′. Nested qPCR product was cloned in PCR4-TOPO plasmid (Invitrogen; P/N 46-0080) and transformed bacterial clones were used for sequencing (Macrogen Inc.).
Surgical procedures.
In vivo experiments were performed on 12-week-old C57Bl6/J male mice. Sca-1+ CPCs were transfected ex vivo with 25 nM siRNA targeting Dnmt3a (or siRNA-Scr) for 18 hours and then harvested, counted, and resuspended in 1,000,000 cells/75 μl PBS. Cells were directly injected in 3 distinct sites peripheral to the infarcted LV area (1,000,000 cells/heart) induced by permanent ligature of the LAD. Permanent ligation of the LAD was induced upon anesthesia (2,2,2-TriBromoEthanol, 310 mg/kg, i.p.) with a 8–0 prolene suture (Ethicon). After surgery, a pain relief drug was administered (buprenorphine, 0.1 mg/kg) for 24 hours. Mice were killed 15 or 30 days after surgery, and the hearts were removed for further examination.
Cardiac MRI measurements and image analysis.
29 animals were included in the experiment. Three mice died during follow-up. Mice were scanned on day 1 and day 28 after MI induction on an 11.7 T MRI scanner dedicated to small-animal applications (Biospec, Bruker). A quadrature 1H resonator was used for radiofrequency transmission (inner diameter = 72 mm, length = 6.6 cm) in conjunction with a surface receive-only coil array (length = 10.7 cm). Anesthesia was induced with 3% isoflurane in oxygen and then maintained with 0.5 to 2% isoflurane during the entire procedure. Mice were placed in prone position and monitored for electrocardiogram and respiration with neonatal electrodes wrapped around the paws and a pneumatic sensor placed under the animal. Body temperature was followed by using a rectal probe and regulated with a dedicated heating blanket. Cardiac scout images were obtained in the conventional planes with a tripilot sequence. Then, an Intragate 2D cine Fast Low Angle Shot (FLASH) sequence was applied to acquire a stack of seven to eight 1-mm-thick contiguous short-axis images covering the entire ventricles, perpendicular to the LV long axis. Imaging parameters were as follows: repetition time: 5.83 ms; echo time: 1.45 ms; flip angle: 25°; field of view: 30.0 × 30.0 mm; and matrix size: 256 × 256, resulting in an in-plane resolution of 0.12 × 0.12 mm².
All analyses were performed on a blinded basis using Segment imaging software (Medviso v1.8). The LV systolic function was assessed in the stack of short-axis images by tracing epicardial and endocardial borders. End-diastolic, end- systolic, and stroke volume were determined (μl). The number of dyskinetic segments was visually evaluated on short-axis cine sections, according to previously published standardized segmentation distinguishing 6 basal segments, 6 midcavity segments, and 4 apical segments (53).The 17th segment, corresponding to the apex, was not evaluated, as no cine image was acquired in 2- or 4-chamber view. A semiautomated technique was used for the determination of the ventricular thickness at the level of the MI. The “report per slice” tool from Segment Software (Medviso v1.8) was manually placed on a region covering the entire MI zone, and mean end-diastolic and end-systolic wall thickness were then reported.
Statistics.
Cell culture experiments were conducted on at least 3 independent cell culture isolates obtained on different days. Statistical analysis was performed using unpaired 2-tailed Student’s t test, nonparametric tests, or ANOVA followed by Bonferroni post-hoc test, when appropriate after verifying normal values distribution. Statistical tests were performed using GraphPad Prism (GraphPad Software Inc.). Results are reported as mean ± SEM, and P values of less than 0.05 were considered significant.
Animal care.
All animal procedures conformed to the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the Institutional Animal Care and Research Advisory Committee of the Université Catholique de Louvain.
Author contributions
ADP, EA, BS, CB, HE, NB, AL, LV, BG, and VDM acquired and analyzed data; CDS, SM, DC, OF, and DHK analyzed data and provided critical scientific input; ADP and JLB planned the experiments, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by grants from the Fonds National de la Recherche Scientifique (FNRS, PDR 1.B089.14 and 3.4643.10), the European Commission (UE LSHM-CT-05-018833), and the Federation Wallonie-Bruxelles (ARC 11/16-035) to JLB. ADP was Chargée de Recherche of the FNRS. We thank Delphine Demulder, Adrien Strapart, and Roxane Verdoy for excellent technical work.
Footnotes
BS’s present address is: Federal Agency for Medicines and Health Products, Brussels, Belgium
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:JCI Insight. 2017;2(12):e91810. https://doi.org/10.1172/jci.insight.91810.
Contributor Information
Aurelia De Pauw, Email: aurelia.depauw@gmail.com.
Emilie Andre, Email: e.andre@uclouvain.be.
Caroline Bouzin, Email: caroline.bouzin@uclouvain.be.
Hrag Esfahani, Email: hrag.esfahani@uclouvain.be.
Nicolas Barbier, Email: nicolas.barbier@student.uclouvain.be.
Axelle Loriot, Email: axelle.loriot@uclouvain.be.
Laetitia Vanhoutte, Email: laetitia.vanhoutte@uclouvain.be.
Bernhard Gerber, Email: bernhard.gerber@uclouvain.be.
Vittoria di Mauro, Email: Vittoria.Di_Mauro@humanitasresearch.it.
Daniele Catalucci, Email: daniele.catalucci@cnr.it.
Olivier Feron, Email: olivier.feron@uclouvain.be.
Denise Hilfiker-Kleiner, Email: hilfiker.denise@mh-hannover.de.
Jean-Luc Balligand, Email: Jl.Balligand@uclouvain.be.
References
- 1.Sanganalmath SK, Bolli R. Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ Res. 2013;113(6):810–834. doi: 10.1161/CIRCRESAHA.113.300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith AJ, et al. Isolation and characterization of resident endogenous c-Kit+ cardiac stem cells from the adult mouse and rat heart. Nat Protoc. 2014;9(7):1662–1681. doi: 10.1038/nprot.2014.113. [DOI] [PubMed] [Google Scholar]
- 3.Oh H, et al. Cardiac progenitor cells from adult myocardium: homing, differentiation, and fusion after infarction. Proc Natl Acad Sci U S A. 2003;100(21):12313–12318. doi: 10.1073/pnas.2132126100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laugwitz KL, et al. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature. 2005;433(7026):647–653. doi: 10.1038/nature03215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh H, et al. Cardiac muscle plasticity in adult and embryo by heart-derived progenitor cells. Ann N Y Acad Sci. 2004;1015:182–189. doi: 10.1196/annals.1302.015. [DOI] [PubMed] [Google Scholar]
- 6.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473(7347):326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509(7500):337–341. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaruba MM, Soonpaa M, Reuter S, Field LJ. Cardiomyogenic potential of C-kit(+)-expressing cells derived from neonatal and adult mouse hearts. Circulation. 2010;121(18):1992–2000. doi: 10.1161/CIRCULATIONAHA.109.909093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naito AT, et al. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci U S A. 2006;103(52):19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clevers H, Loh KM, Nusse R. Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science. 2014;346(6205):1248012. doi: 10.1126/science.1248012. [DOI] [PubMed] [Google Scholar]
- 11.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8(2):228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 12.Rao J, et al. Stepwise clearance of repressive roadblocks drives cardiac induction in human ESCs. Cell Stem Cell. 2016;18(3):341–353. doi: 10.1016/j.stem.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 13.Qyang Y, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1(2):165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 14.De Pauw A, et al. Paracrine nitric oxide induces expression of cardiac sarcomeric proteins in adult progenitor cells through soluble guanylyl cyclase/cyclic-guanosine monophosphate and Wnt/β-catenin inhibition. Cardiovasc Res. 2016;112(1):478–490. doi: 10.1093/cvr/cvw196. [DOI] [PubMed] [Google Scholar]
- 15.Zelarayán LC, et al. Beta-Catenin downregulation attenuates ischemic cardiac remodeling through enhanced resident precursor cell differentiation. Proc Natl Acad Sci U S A. 2008;105(50):19762–19767. doi: 10.1073/pnas.0808393105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willems E, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell-derived mesoderm. Circ Res. 2011;109(4):360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duan J, et al. Wnt1/βcatenin injury response activates the epicardium and cardiac fibroblasts to promote cardiac repair. EMBO J. 2012;31(2):429–442. doi: 10.1038/emboj.2011.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oerlemans MI, Goumans MJ, van Middelaar B, Clevers H, Doevendans PA, Sluijter JP. Active Wnt signaling in response to cardiac injury. Basic Res Cardiol. 2010;105(5):631–641. doi: 10.1007/s00395-010-0100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoch M, et al. Erythropoietin preserves the endothelial differentiation capacity of cardiac progenitor cells and reduces heart failure during anticancer therapies. Cell Stem Cell. 2011;9(2):131–143. doi: 10.1016/j.stem.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Smits AM, et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc. 2009;4(2):232–243. doi: 10.1038/nprot.2008.229. [DOI] [PubMed] [Google Scholar]
- 21.Chen B, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5(2):100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urakami S, et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin Cancer Res. 2006;12(2):383–391. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- 23.Fendri A, et al. Epigenetic alteration of the Wnt inhibitory factor-1 promoter is common and occurs in advanced stage of Tunisian nasopharyngeal carcinoma. Cancer Invest. 2010;28(9):896–903. doi: 10.3109/07357907.2010.494324. [DOI] [PubMed] [Google Scholar]
- 24.Liu YL, Yang HP, Zhou XD, Gong L, Tang CL, Wang HJ. The hypomethylation agent bisdemethoxycurcumin acts on the WIF-1 promoter, inhibits the canonical Wnt pathway and induces apoptosis in human non-small-cell lung cancer. Curr Cancer Drug Targets. 2011;11(9):1098–1110. doi: 10.2174/156800911798073041. [DOI] [PubMed] [Google Scholar]
- 25.Hou HA, et al. Distinct association between aberrant methylation of Wnt inhibitors and genetic alterations in acute myeloid leukaemia. Br J Cancer. 2011;105(12):1927–1933. doi: 10.1038/bjc.2011.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noseda M, et al. PDGFRα demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat Commun. 2015;6:6930. doi: 10.1038/ncomms7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabbri M, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104(40):15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu W, et al. miR-29a maintains mouse hematopoietic stem cell self-renewal by regulating Dnmt3a. Blood. 2015;125(14):2206–2216. doi: 10.1182/blood-2014-06-585273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abbey D, Seshagiri PB. Aza-induced cardiomyocyte differentiation of P19 EC-cells by epigenetic co-regulation and ERK signaling. Gene. 2013;526(2):364–373. doi: 10.1016/j.gene.2013.05.044. [DOI] [PubMed] [Google Scholar]
- 30.Tompkins JD, et al. Mapping human pluripotent-to-cardiomyocyte differentiation: methylomes, transcriptomes, and exon DNA methylation “memories”. EBioMedicine. 2016;4:74–85. doi: 10.1016/j.ebiom.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fang X, et al. Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes. FASEB J. 2016;30(9):3238–3255. doi: 10.1096/fj.201600346R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan M, Wu J, Cai Y. Suppression of Wnt signaling by the miR-29 family is mediated by demethylation of WIF-1 in non-small-cell lung cancer. Biochem Biophys Res Commun. 2013;438(4):673–679. doi: 10.1016/j.bbrc.2013.07.123. [DOI] [PubMed] [Google Scholar]
- 33.Klutstein M, Nejman D, Greenfield R, Cedar H. DNA methylation in cancer and aging. Cancer Res. 2016;76(12):3446–3450. doi: 10.1158/0008-5472.CAN-15-3278. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh PC, et al. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med. 2007;13(8):970–974. doi: 10.1038/nm1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uchida S, et al. Sca1-derived cells are a source of myocardial renewal in the murine adult heart. Stem Cell Reports. 2013;1(5):397–410. doi: 10.1016/j.stemcr.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C, Gu H, Yu Q, Manukyan MC, Poynter JA, Wang M. Sca-1+ cardiac stem cells mediate acute cardioprotection via paracrine factor SDF-1 following myocardial ischemia/reperfusion. PLoS One. 2011;6(12):e29246. doi: 10.1371/journal.pone.0029246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li TS, et al. Direct comparison of different stem cell types and subpopulations reveals superior paracrine potency and myocardial repair efficacy with cardiosphere-derived cells. J Am Coll Cardiol. 2012;59(10):942–953. doi: 10.1016/j.jacc.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hong KU, et al. c-kit+ Cardiac stem cells alleviate post-myocardial infarction left ventricular dysfunction despite poor engraftment and negligible retention in the recipient heart. PLoS One. 2014;9(5):e96725. doi: 10.1371/journal.pone.0096725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu ML, et al. Anti-inflammatory peptides from cardiac progenitors ameliorate dysfunction after myocardial infarction. J Am Heart Assoc. 2014;3(6):e001101. doi: 10.1161/JAHA.114.001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Rooij E, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105(35):13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao J, et al. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther. 2012;20(6):1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Watson CJ, et al. Hypoxia-induced epigenetic modifications are associated with cardiac tissue fibrosis and the development of a myofibroblast-like phenotype. Hum Mol Genet. 2014;23(8):2176–2188. doi: 10.1093/hmg/ddt614. [DOI] [PubMed] [Google Scholar]
- 43.Neveu WA, Mills ST, Staitieh BS, Sueblinvong V. TGF-β1 epigenetically modifies Thy-1 expression in primary lung fibroblasts. Am J Physiol, Cell Physiol. 2015;309(9):C616–C626. doi: 10.1152/ajpcell.00086.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tao H, et al. DNMT3A silencing RASSF1A promotes cardiac fibrosis through upregulation of ERK1/2. Toxicology. 2014;323:42–50. doi: 10.1016/j.tox.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Zhang R, et al. Knockdown of DNMT1 and DNMT3a promotes the angiogenesis of human mesenchymal stem cells leading to arterial specific differentiation. Stem Cells. 2016;34(5):1273–1283. doi: 10.1002/stem.2288. [DOI] [PubMed] [Google Scholar]
- 46.Jiang YZ, Jiménez JM, Ou K, McCormick ME, Zhang LD, Davies PF. Hemodynamic disturbed flow induces differential DNA methylation of endothelial Kruppel-Like Factor 4 promoter in vitro and in vivo. Circ Res. 2014;115(1):32–43. doi: 10.1161/CIRCRESAHA.115.303883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dunn J, et al. Flow-dependent epigenetic DNA methylation regulates endothelial gene expression and atherosclerosis. J Clin Invest. 2014;124(7):3187–3199. doi: 10.1172/JCI74792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunn J, Thabet S, Jo H. Flow-dependent epigenetic DNA methylation in endothelial gene expression and atherosclerosis. Arterioscler Thromb Vasc Biol. 2015;35(7):1562–1569. doi: 10.1161/ATVBAHA.115.305042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roncarati R, et al. Circulating miR-29a, among other up-regulated microRNAs, is the only biomarker for both hypertrophy and fibrosis in patients with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2014;63(9):920–927. doi: 10.1016/j.jacc.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 50.Arnold N, Koppula PR, Gul R, Luck C, Pulakat L. Regulation of cardiac expression of the diabetic marker microRNA miR-29. PLoS One. 2014;9(7):e103284. doi: 10.1371/journal.pone.0103284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boon RA, et al. MicroRNA-29 in aortic dilation: implications for aneurysm formation. Circ Res. 2011;109(10):1115–1119. doi: 10.1161/CIRCRESAHA.111.255737. [DOI] [PubMed] [Google Scholar]
- 52.Loriot A, De Plaen E, Boon T, De Smet C. Transient down-regulation of DNMT1 methyltransferase leads to activation and stable hypomethylation of MAGE-A1 in melanoma cells. J Biol Chem. 2006;281(15):10118–10126. doi: 10.1074/jbc.M510469200. [DOI] [PubMed] [Google Scholar]
- 53.Cerqueira MD, et al. Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac Imaging Committee of the Council on Clinical Cardiology of the American Heart Association. Int J Cardiovasc Imaging. 2002;18(1):539–542. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.