Abstract
Post-traumatic stress disorder (PTSD) is a highly debilitating stress and anxiety-related disorder that occurs in response to specific trauma or abuse. Genetic risk factors may account for up to 30–40% of the heritability of PTSD. Understanding the gene pathways that are associated with PTSD, and how those genes interact with the fear and stress circuitry to mediate risk and resilience for PTSD will enable the development of targeted therapies to prevent the occurrence of or decrease the severity of this complex multi-gene disorder. This review will summarize recent research on genetic approaches to understanding PTSD risk and resilience in human populations, including candidate genes and their epigenetic modifications, genome-wide association studies and neural imaging genetics approaches. Despite challenges faced within this field of study such as inconsistent results and replications, genetic approaches still offer exciting opportunities for the identification and development of novel therapeutic targets and therapies in the future.
II. Introduction
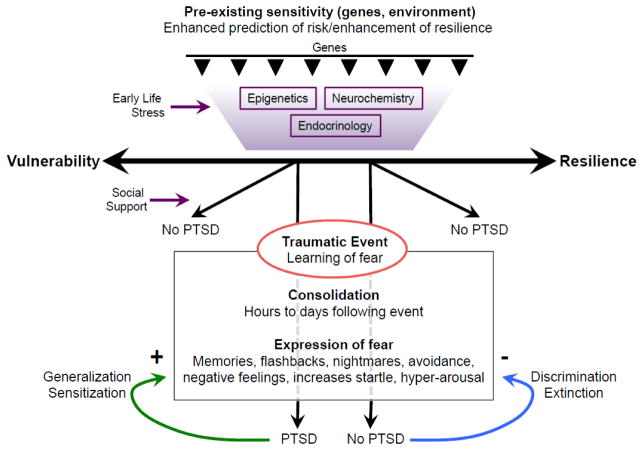
Post-traumatic Stress Disorder (PTSD) is a highly debilitating neuropsychiatric disorder in which fear-related memories of a traumatic event become overgeneralized and resistant to extinction (Figure 1). Symptoms include intrusive thoughts about the trauma accompanied with changes in physiology (increased heart rate and perspiration) as well as nightmares and flashbacks (1, 2).
Figure 1. Schematic of the development of gene by environment (G x E) interactions, and the development of post-traumatic stress disorder (PTSD).
Genetic heritability along with early life stress and childhood trauma comprise much of the risk for depression and anxiety related disorders such as PTSD. Following the traumatic event, the learned fear memory is consolidated to a more permanent state. The expression of the fear memory may manifest as the above described symptoms. Individuals that develop PTSD fail to discriminate and extinguish fear memories, and instead exhibit sensitization and generalization of the fear response. Enhancing the extinction and discrimination of learned fear memories are key behavioral responses that are targeted in the treatment of PTSD via psychotherapeutic approaches such as exposure based psychotherapy and pharmacological assisted psychotherapy.
Although estimates suggest that 90% of individuals will be exposed to a significant traumatic event in their lifetime (3), only about 5–10% of the general population suffers from PTSD (4, 5).
These studies suggest that individual vulnerability and resilience are key factors to consider in the pathology of PTSD. Furthermore, twin studies have demonstrated that genetic risk factors may account for up to 30–40% of the heritability of PTSD (6). The recent discovery of the Tac2 gene in mediating fear consolidation (7) is one example of how a gene discovery driven approach could shed light on the pathology and novel potential therapeutic targets in PTSD.
In addition to identifying genes that have been implicated in PTSD, we will discuss the epigenetic modifications of these risk genes. As such modifications are potentially reversible by drugs, behavioral therapy or environmental changes, this opens up new promising therapeutic avenues for PTSD. Beyond reviewing genetics studies, we also discuss the need for complementary approaches at multiple levels of biological complexity and emphasize the importance of combining and integrating findings across levels for a better understanding of biological pathways from gene to disease. These may include multi-modal imaging genetics studies, bioinformatic analyses, and functional analyses of cell and animal models.
III. Candidate gene studies
Candidate gene studies for PTSD involve the identification of genetic risk variants using prior knowledge and literature. In this section, we will discuss current findings in monoaminergic neurotransmitters, the hypothalamic-pituitary-adrenal (HPA) axis, neurotrophin, neuropeptide and receptor genes. While we will only discuss a small number of examples of candidate gene studies for PTSD, many excellent reviews have been published to date and the authors direct the reader to a number of such publications (6, 8–14). Such targeted studies have been at times criticized for not being unbiased, as compared with genome-wide association studies, or subject to file-drawer effects and other potential biases. Nonetheless, a number of pathways have been replicated multiple times and have survived meta-analytic analyses suggesting some possibly important genetic signals, as outlined below.
A. Monoaminergic neurotransmitters
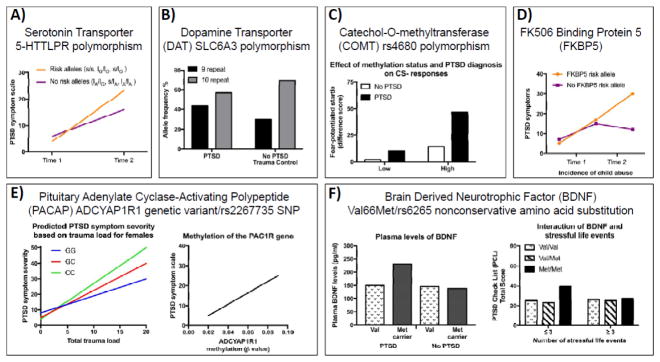
The serotonergic system has been studied extensively in the regulation of mood and the dysregulation of this system has been implicated in the pathophysiology of PTSD (15, 16). Several studies have found that a high risk of developing PTSD is associated with a specific polymorphism in the promoter region of the serotonin transporter gene SLC6A4. The 5-HTTLPR polymorphism contains two predominant alleles, L (long) and S (short) representing different lengths of a polymorphic promoter region, with the S-allele associated with reduced serotonin transporter gene expression, leading to reduced serotonin reuptake. Studies have indicated that risk for PTSD development is associated with genotype (mainly S-allele carriers, also associated with depression) and high levels of trauma/stress (Figure 2A) (17–20). However, several studies have reported no association (21, 22) and ultimately association studies of functional polymorphisms of the serotonin transporter gene have been inconclusive; in fact a meta-analysis of 12 studies did not find evidence of overall association, however homozygosity for the S allele was associated with PTSD in individuals with classified high trauma exposure (23). Other studies have demonstrated a correlation between methylation of SLC6A4 and PTSD symptoms following traumatic events. Individuals with more traumatic events were found to be at increased risk for PTSD, but only when they had lower methylation levels at the SLC6A4 locus. At higher methylation levels, individuals with more traumatic events were protected from developing PTSD (24, 25). Such findings are very interesting in their suggestion that methylation status of SLC6A4 interacts with the number of traumatic events to mediate the risk of PTSD. Importantly, this study did not observed an effect of SLC6A4 genotype on PTSD (26). Although these studies need replication in larger cohorts, they are consistent with a role for serotonergic regulation of emotional processing interacting with trauma and stress exposure to increase risk for (or resilience from) PTSD-related symptoms.
Figure 2. Examples of Genetic Variants associated with differential risk for Post traumatic Stress Symptoms.
A) The 5-HTTLPR multimarker genotype predicts posttraumatic stress disorder (PTSD) symptom scores and severity two to four weeks following trauma. The 5-HTTLPR risk alleles (orange line) result in increased PTSD symptoms compared to the non-risk alleles (purple line) (graph adapted from Mercer et al., 2012). B) The SLC6A3 polymorphism of the dopamine transporter 9 and 10 repeat (black and grey bars, respectively) allele and genotype frequencies in individuals with chronic PTSD and trauma control survivors without PTSD (Segman et al., 2002; Drury et al., 2009). C) Individuals with the Met/Met genotype in the COMT functional polymorphism at codon 158 (rs4680) demonstrate impaired fear inhibition as demonstrated by enhanced fear potentiated startle to the CS – (safety signal), which may be the result of icreased methylation of the COMT promoter region (graph adapted from Norrholm et al., 2013). D) The FKBP5 risk allele (orange line) results in increases in adult PTSD symptoms following exposure to childhood trauma compared to the protective allele (purple line) (graph adapted from Klengel et al., 2013). E) Females (but not males) with high levels of plasma PACAP38 display increased PTSD symptoms. Rs2267735 is a SNP spanning the PAC1R gene and there is an interaction between total trauma load and the risk CC genotype in females (left) (graph adapted from Almi et al., 2013). Methylation at the PAC1R locus is significantly positively correlated with PTSD symptoms (right) (graph adapted from Ressler et al., 2011). F) A nonconservative amino acid substitution (Val66Met, rs6265) in the BDNF gene has been identified. Met carriers with PTSD have significantly higher plasma levels of BDNF (left). Met/Met homozygote carriers who experienced fewer than four stressful life events had significantly higher PTSD check list (PCL) scores compared to Val/Val and Val/Met carriers (right) (graphs adapted from Zhang et al., 2013).
Another monoaminergic neurotransmitter that is likely involved in the pathophysiology of PTSD is dopamine. Attention, vigilance, arousal, and sleep are processes that are negatively impacted in PTSD, and dopamine is key for their regulation (27). A polymorphism in the gene encoding the dopamine transporter (SLC6A3 also known as DAT or DAT1) has a 40-base pair repeat that is polymorphic in the population. Genetic variants in dopamine transporters have been associated with PTSD (28–30). For example, 9-repeat (9R) allele of SLC6A3 is associated with PTSD symptoms. Interestingly, an increased risk for PTSD might be mediated by high methylation of the SLC6A3 promoter locus (CpG site cg13202751) in 9R allele carriers. (Figure 2B) (31). However, there has been at least one published study reporting no association of the 9-repeat allele of SLC6A3 with PTSD (32).
The dopamine receptor D2 (DRD2) contains rs1800497, a single nucleotide polymorphism (SNP) with T (A1) and C (A2) alleles. This allele is associated with PTSD in Caucasian war veterans (33) and excessive alcohol intake (34). The dopamine D3 receptor (DRD3) gene (involved in relevant processes such as executive functioning and emotional reactivity) has also been implicated in PTSD. In one study, 4 single nucleotide polymorphisms (SNPs), showed evidence of association with PTSD (35). However, two published studies in different cohorts have demonstrated no association between PTSD diagnosis or symptom severity with the A1 allele of DRD2 (32, 36), although one of these studies did find a significant interaction effect between DRD2 TaqIA and the BDNF Val66Met variants on score on the Davidson Trauma Scale (DTS)(36).
Two enzymes involved in monoamine metabolism are dopamine beta-hydroxylase (DBH) and catechol-O-methyltransferase (COMT). A polymorphism in DBH (-1021C/T; rs1611115) has been shown to account for 35–52% of the variation in plasma-DBH activity. War veterans with PTSD and a civilian sample of African Americans with PTSD who carried the CC genotype of the DBH–1021C/T variant had lower plasma DBH activity (37–39). COMT contains a functional polymorphism at codon 158 (rs4680) leading to a significant reduction in enzyme activity. The Met158 allele is also associated with a decreased ability to extinguish conditioned fear, a key feature of animal models of PTSD. Furthermore, a significant association between one or more copies of the Met158 allele and PTSD in addition to a gene–environment interaction between the Met158 allele and the number of traumatic event types in predicting PTSD has been described. Individuals with a Met/Met genotype display decreased fear inhibition, potentially as a result of higher methylation in the COMT promoter region (Figure 2C)(40–42). Thus, regulation of COMT and DBH may be important factors in the regulation of fear related processes in PTSD patients.
B. Hypothalamic-pituitary-adrenal axis candidate genes
A key feature of PTSD is the dysregulation of the hypothalamic-pituitary-adrenal (HPA) axis. A large body of work has shown that PTSD is often characterized by a dysregulation of the hypothalamic-pituitary-adrenal (HPA) axis, such that the system is altered and hyper-responsive to cortisol feedback (43–45). These effects may be observed in PTSD patients, of which a large number display consistently lower cortisol responses in dexamethasone suppression tests indicating enhanced negative feedback in the HPA axis (44, 46). The glucocorticoid receptor (GR) mediates negative feedback of the HPA axis, thus the HPA axis effects in individuals with PTSD are thought to occur, in part, via enhanced sensitivity of GR-mediated feedback mechanism, which acts to suppress stress-related cortisol release. Several polymorphisms of the glucocorticoid receptor gene have been observed in individuals with PTSD (47, 48). The SNP Bcl-1 is associated with hypersensitivity and enhanced feedback to glucocorticoid levels correlating with lower cortisol levels. Carriers who are homozygous for the Bcl-1 SNP (G allele) were found to have more traumatic memories than heterozygous or non-carriers of the SNP, 6 months after intensive care unit care (49). In addition, individuals with lifetime PTSD showed lower morning cortisol release, higher GR expression, and lower overall methylation levels in the examined GR promoter regions. Consistent with these findings, GR promoter methylation levels are lower in veterans with PTSD compared to those without PTSD (50). As with the other candidate gene studies described above, differing results have found no significant association between GR polymorphisms (N363S and BclI) and PTSD diagnosis in a group of Vietnam veterans (51).
A different gene, steroid receptor chaperone FK506 binding protein 5, (FKBP5) is thought to be involved in the pathogenesis of stress-related disorders as it regulates the stress system by altering GR sensitivity (52, 53). SNPs in FKBP5 result in enhanced glucocorticoid receptor sensitivity which lead to lower basal cortisol levels and increased risk for PTSD following a traumatic experience. Four SNPs in FKBP5 (rs9296158, rs3800373, 1360780, rs9470080, that are in high linkage disequilibrium with each other) have been shown to interact with child abuse severity to predict adult PTSD in a primarily African American civilian cohort (54, 55). More recently, one of these polymorphisms in FKBP5 (rs1360780), that increases the risk of developing stress-related psychiatric disorders in adulthood, was found to be dependent on changes in DNA methylation occurring as a consequence of childhood trauma–dependent stress (Figure 2D (56). Other studies indicate that genetic variation in FKBP5 influences the risk of anxiety and/or depressive disorders in early life and interact with childhood abuse to increase the risk of PTSD (57–59).
The neuropeptide, pituitary adenylate cyclase-activating polypeptide (PACAP), is involved in regulating corticotrophin-releasing hormone (CRH) a key mediator of the stress axis. Recently, a genetic variant in the PAC1 receptor (ADCYAP1R1; rs2267735) that disrupts a putative estrogen response element, has been found to be associated with PTSD in a primarily African-American cohort of women (Figure 2E, left) (60–62). In addition, genetic variation at the ADCYAP1R1 locus interacts with childhood maltreatment to influence the risk of developing PTSD later in life, specifically in women (63). Finally, PTSD patients display changes in peripheral blood DNA methylation and mRNA expression of the ADCYAP1R1 transcript (Figure 2E, right) (62). In contrast, in two separate, presumably less traumatized patient cohorts, the above association with the ADCYAP1R1 variant was not replicated (64). Together, these studies suggest that the PACAP/PAC1 pathway plays an important role in the abnormal stress responses associated with PTSD.
C. Additional candidate genes
Brain-derived neurotrophic factor (BDNF), a key regulator of neuronal plasticity, is known to be associated with depression as well as the extinction of fear, a key process disrupted in PTSD (65). A SNP in the gene encoding human BDNF gives rise to a functional variant at codon 66 where the amino acid valine (Val) is substituted by methionine (Met), also referred to as the Val66Met polymorphism, and has been suggested to be relevant in PTSD (for a review, please see Andero and Ressler, 2012). In humans, the frequency of the Met/Met in BDNF gene although low in certain populations (<5%), (66) was higher in individuals with PTSD compared to controls and individuals with the BDNF polymorphism Val66Met; the allelic frequencies of the Met/Met genotype and Met carriers was 33.3% in individuals with PTSD compared to 17.5% in non-PTSD controls (67, 68).
Met carriers have been shown to have significantly higher levels of startle responsiveness, an intermediate phenotype for PTSD, when compared to Val/Val carriers. Additionally, subjects with PTSD show higher levels of BDNF in their peripheral blood plasma compared to traumatized controls without PTSD (Figure 2F, left). Such investigations into BDNF levels in the periphery, while promising, cannot be equated with brain function. Further evidence has shown that Met/Met carriers who experienced fewer than four stressful life events had significantly higher PTSD symptoms compared to Val/Val and Val/Met carriers (Figure 2F, right). Finally, PTSD patients with the Met-66 allele of BDNF displayed a poorer response to exposure therapy compared with patients with the Val/Val allele (69). With regard to epigenetic modification of the BDNF gene and PTSD risk, there is a modest association between increased methylation of one CpG site within the BDNF gene and PTSD status (70, 71). Additionally, the Val66Met SNP has been shown to cause decreased hippocampal volume, deficits in declarative memory and impaired fear extinction (72–74). Despite the above positive findings, a number of studies have found no association (29, 75–77), (although a significant interaction has been reported between the above described DRD2 Taq1A (rs1800497) and Val66Met (rs6265) in predicting severity of PTSD (36).
GABAergic systems are known to play a role in the pathophysiology of anxiety and depression, both of which are common in individuals suffering from PTSD. Three polymorphisms in the GABAA receptor subunit alpha 2 (GABRA2) were found to have significant interactions with childhood trauma to predict PTSD (78).
Apolipoprotein E (ApoE) regulates the binding of lipoproteins to the low-density lipoprotein receptor and regulates neuronal and glial responses to stress. The gene APOE is polymorphic and a significant association between the ApoE2 allele and impaired memory, an increase in re-experiencing of traumatic memories and salivary cortisol levels, was reported in combat-exposed PTSD patients (78). Regulator of G-protein Signaling 2 (RGS2) has been implicated in learning and memory processes. An association with RGS2 (rs4606) and PTSD after traumatic hurricane experience was found under conditions of high stress and low social support (79). A different study found a significant association between RGS2 (rs4606) and post-traumatic growth thereby suggesting this gene may play a role in recovery from trauma (80).
Cholecystokinin (CCK) is a neuropeptide that has been implicated fear acquisition and extinction in rodents (81). A study in war veterans has demonstrated that a single nucleotide polymorphism in the promoter region of the CCK gene was associated with an increased prevalence of PTSD as well as with severity of PTSD symptoms (81).
Other studies have reported associations between PTSD and cannabinoid receptor (CNR1) gene variants. A polymorphism in the CNR1 gene (rs1049353) interacts with childhood physical abuse to predict posttraumatic threat symptoms, specifically to increase severity of threat or fear (82). A different study examined the association between genetic variation in the nicotinic receptor gene family and the PTSD. A novel association between rs12898919 in the cholinergic receptor nicotinic alpha-5 (CHRNA5) gene and PTSD was observed in non-hispanic white patients suggesting that CHRNA5 gene is associated with increased risk for PTSD (83).
Overall, candidate gene studies of PTSD, like those reviewed in the above section, have been underpowered, thus making many positive and negative results and findings difficult to interpret. In addition, mixed results have been observed across a subset of genes involved in serotonergic, dopaminergic, and neuroendocrine function. Furthermore, there are clearly limitations in study design and non-reproducibility of findings. Factors such as sex, race, trauma history are often not consistently addresses in study designs, thus causing difficulty in drawing conclusions across multiple different studies. Overall, although over-arching statements based on candidate gene approach studies are premature, genes identified through these avenues hold promise as potential biomarkers to undergo further investigation. For example, convincing evidence has been shown in the study of FKBP5, in which a gene by environment (G x E) effect between early adversity and variation in FKBP5 has been reported. Subsequent work has demonstrated a mechanistic account of the influence of this particular genetic variation on alteration of the stress response via epigenetic alterations of gene expression and downstream glucocorticoid function(56).
Genome-wide association studies (GWAS) large-scale, collaborative approaches, which will briefly be described below, offer beginning steps towards a goal of well-powered, hypothesis-neutral studies. Such approaches are needed to identify more robust candidate genes, which will be essential in identifying targets for research in the treatment of PTSD.
IV. Genome-wide association studies
Genome-wide association studies (GWAS) involve an unbiased approach to identify disease-associated genetic variants, across the vast genome of 3 billion base pairs of DNA and approximately 20,000 protein-coding genes that may predict disease. Most GWAS arrays test ~1 Million SNP variants, providing the most robust methods to perform unbiased genetic discovery. At least six successful GWAS studies have been reported for PTSD to date as outlined below.
The first GWAS of PTSD identified a genome-wide significant association between PTSD and a SNP (rs8042149) in the retinoid-related orphan receptor gene (RORA) (84). Furthermore, certain variations in this gene may predispose individuals with a history of child abuse to PTSD(85). Changes in RORA levels may decrease the ability of neurons to respond to oxidative stress, steroid hormone changes as well as inflammation often induced by trauma(84). The association between the RORA SNP rs8042129 and PTSD was also reported in a cohort of Florida hurricane survivors (86). In other replication cohorts, other SNPs within RORA have been nominally associated with PTSD (84), however, another study has found that associations between RORA SNPs were not detected in two independent replication samples (87).
In another very recent study using an unbiased gene-based approach, the Neuroligin or NLGN1 gene that encodes a synaptic adhesion molecule, was found to be associated with PTSD in two different civilian traumatized cohorts. In both cohorts, the NLGN1 SNP with the strongest association was also associated with NLGN1 expression in postmortem cerebellum or frontal cortex tissue, respectively. NLGN1 plays a role in synaptogenesis and synaptic maintenance, specifically in mediating the formation and maturation of synapses within the mammalian brain, and has been shown to be a potent trigger for the de novo formation of synaptic connections in addition to being implicated in other psychiatric disorders such as Autism (88–90). Furthermore, mouse model studies have found that depletion of neuroligin-1 in the lateral amygdala results in a deficit in storage of associative fear memory, suggesting an important role in fear learning and memory.
Another gene that has been recently implicated in the development of PTSD risk is Tolloid-Like 1 or TLL-1, a zinc dependent metalloprotease which plays an important role in remodeling of the extracellular matrix (91). After GWAS analyses in a sample of European Americans and African Americans, TLL-1 was found to be a new susceptibility gene for PTSD(92).
In a different study, a GWAS design was used to identify genes associated with PTSD within a multi-racial sample largely composed of U.S. veterans. The authors identified SNPs within many candidate genes that were nominally significant. The most significant genes were UNC13C and DSCAM within the non-Hispanic black group whereas the most significant genes within the non-Hispanic white group were TBC1D2, SDC2 and PCDH7. Many of these genes have been previously implicated in both development and disorders of the central nervous system (93–95).
In a study performed in military personnel, whole transcriptome analysis was performed to compare gene-expression profiles in individuals with PTSD and matched controls without PTSD. Analysis of expression profiles led to the discovery of 203 differentially expressed genes in PTSD, of which 72% were upregulated, many of which were associated with the innate immune, neuroendocrine, and NF-κB systems (96).
The largest genome-wide association study of PTSD to date, carried out in a US military sample, found a genome-wide significant association with ANKRD55, a gene involved in autoimmune and inflammatory disorders(97). Furthermore, a multi-ethnic/racial GWAS of PTSD provided evidence for the phosphoribosyl transferase domain containing 1 gene or PRTFDC1, encoding an enzyme in the purine metabolic enzyme family, to be involved in PTSD pathology(98); PRTFDC1 has been shown to play a role as a possible tumor –suppressor gene (99), however, its role in the etiology of PTSD is unknown at present.
The recently formed Psychiatric Genomics Consortium-PTSD continues to encourage further discovery of genes involved in the pathology and susceptibility of or resilience to PTSD (100). A key challenge in GWAS approach is the replication of studies and findings from the different studies have not consistently implicated a primary set of PTSD risk loci. Several factors may play a role in non-replication, including false positives, heterogeneity across samples, as well as inflation of effect size estimates in moderately powered samples. Moving forward, progress in identifying the genetic factors underlying PTSD will hugely benefit from efforts such as the PGC’s PTSD Workgroup whose goal is to bring together GWAS data sets. Currently, genotyping has been completed or is underway for approximately 20,000 cases and 50,000 controls (100).
V. Neuroimaging genetics
Neuroimaging genetics provides another technique that has been used to identify genes relevant in PTSD and the neuroimaging intermediate phenotypes associated with PTSD. Individuals suffering from PTSD have been shown to exhibit specific patterns of information processing, such as attentional biases for trauma-related cues and impairments in sustained attention and memory that may be mediated by structural changes in the brain (101, 102).
Several genes appear to play a role in the pathophysiology of PTSD, while simultaneously exerting effects on specific brain structures and their corresponding functions. Studies of the serotonin transporter gene (SLC6A4) have focused on the SNP rs25531, present in a degenerate repeat in the promoter region known as 5-HTTLPR. This SNP is associated with heightened amygdala activation in S-allele carriers of this gene, in response to threat related cues(103). Other neuroimaging studies have found that allelic variants of FKBP5, COMT and NPY associate differentially with behavioral and neural responses to threat-relevant facial expressions. For example, there is increased activation in the amygdala and hippocampus in response to threat cues in carriers of risk alleles of FKBP5. Increased amygdala responses to angry and fearful faces have also been observed in risk allele carriers for COMT (Val-allele carriers for SNP rs4680) and NPY.
Recent work has shown that a variant in the opioid receptor-like 1 gene (OPRL1) was associated with PTSD symptoms and fear-potentiated startle, along with amygdala-insult connectivity by fMRI (Andero et al., 2013). Additionally, the variant in OPRL1 was also associated with PTSD symptoms and fear-potentiated startle. OPRL1 encoded the amygdala nociception (NOP)/orphanin FQ receptor (NOP-R), and mouse studies have shown that administration of NOP-R agonist into the central amygdala impairs fear consolidation (104), thus suggesting a potential “drug-able target” for future investigation. Such an approach is supported by the finding that administration of morphine, an opioid agonist, following trauma exposure, reduced the incidence of PTSD later on (105).
As with the approaches described in the prior sections related to candidate gene studies and GWAS, genetic neuroimaging studies have also not been consistent. Such inconsistencies highlight the need for replication with larger sample sizes, as well as a greater effort to examine specific genetic effects in different demographic groups. Additionally, the majority of neuroimaging genetics studies have focused on the amygdala and hippocampus; a broader investigation of other regions known to play a role in PTSD and stress related disorders such as the prefrontal cortical regions, the anterior cingulate, dorsolateral prefrontal cortex, etc. will be important targets for future studies. Despite these challenges, the studies discussed in the above section highlight the importance of studying structural differences in the brains of individuals that possess specific SNPs in relevant genes which may cause changes in behavior along with dysregulation in physiological processes such as learning, memory and HPA axis activity, all of which influence the development of PTSD (6).
VI. Conclusions
As detailed in the present review, both candidate gene approaches and GWAS studies have shed light on a number of novel genes which may play a role in the pathophysiology of PTSD. Despite these important steps forward, many candidate gene approaches and GWAS studies have been inconsistent with many non-replicated effects. As such, candidate gene study approaches will require greater sample sizes in addition to being combined with convergent molecular biological evidence and research using animal models, as has been shown so robustly in the case of FKBP5. Additionally, recent work investigating the fatty acid amide hydrolase (FAAH) gene has nicely used a cross-species approach by studying genetic knock-in mouse model in parallel with human variant allele carriers to investigate alterations in neural connectivity and circuitry, biochemistry and anxiety-like and fear learning behavior(106, 107). Such translational approaches are promising in their investigation of both phenotype and mechanisms across species.
With regards to GWAS studies, there is a great need for large-scale unbiased genome-wide approaches which have been proven successful in the study of other psychiatric disorders such as Autism Spectrum Disorders, Schizophrenia and Bipolar Disorder. The field as a whole will benefit tremendously from current efforts to bring together GWAS data sets, led by the PGC’s PTSD Workgroup. The next step in the discovery of novel gene targets will be to manipulate identified genetic targets in animal models in parallel to performing neuroimaging genetics in humans. Additionally, the identification of genetic targets with available agonists or antagonists, may be investigated in the context of animal models of fear learning and memory to validate and provide preliminary evidence (blockade of consolidation or enhancement of extinction, etc.) for potential therapeutics for the treatment of PTSD. Such an approach may allow for the discovery of previously unknown genetic targets and novel therapies that may eventually decrease the risk and enhance the resilience of individuals towards developing PTSD.
Acknowledgments
This work was supported in part by the following sources of funding: KJR (1R01MH096764), the Howard Hughes Medical Institute, and Ruth L. Kirstein NRSA predoctoral fellowship to FGM (F31 MH105237-01). This project was also funded by the Office of Research Infrastructure Programs/OD P51OD011132 (formerly NCRR P51RR000165).
Footnotes
Disclosures:
Drs. Banerjee and Morrison have nothing to disclose. Dr. Ressler is on the Scientific Advisory Boards for Resilience Therapeutics, Sheppard Pratt-Lieber Research Institute, Laureate Institute for Brain Research, The Army STARRS Project, and the Anxiety and Depression Association of America. He holds patents, for which he has received no equity or income within the last 3 years, for use of D-cycloserine and psychotherapy, targeting PAC1 receptor for extinction, targeting tachykinin 2 for prevention of fear, targeting angiotensin to improve extinction of fear. He receives or has received research funding from NIMH, HHMI, NARSAD, and the Burroughs Wellcome Foundation.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bowers ME, Ressler KJ. An Overview of Translationally Informed Treatments for Posttraumatic Stress Disorder: Animal Models of Pavlovian Fear Conditioning to Human Clinical Trials. Biological Psychiatry. 78(5):E15–E27. doi: 10.1016/j.biopsych.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morrison FG, Ressler KJ. FROM THE NEUROBIOLOGY OF EXTINCTION TO IMPROVED CLINICAL TREATMENTS. Depression and anxiety. 2014;31(4):279–290. doi: 10.1002/da.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kessler RC, et al. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Archives of general psychiatry. 2005;62(6):593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 4.Yehuda R, et al. Post-traumatic stress disorder. Nature Reviews Disease Primers. 2015;1:15057. doi: 10.1038/nrdp.2015.57. [DOI] [PubMed] [Google Scholar]
- 5.Breslau N. The epidemiology of posttraumatic stress disorder: what is the extent of the problem? The Journal of clinical psychiatry. 2001 [PubMed] [Google Scholar]
- 6.Almli LM, Fani N, Smith AK, Ressler KJ. Genetic approaches to understanding post-traumatic stress disorder. The international journal of neuropsychopharmacology/official scientific journal of the Collegium Internationale Neuropsychopharmacologicum (CINP) 2014;17(2):355–370. doi: 10.1017/S1461145713001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andero R, Dias Brian G, Ressler Kerry J. A Role for Tac2, NkB, and Nk3 Receptor in Normal and Dysregulated Fear Memory Consolidation. Neuron. 2014;83(2):444–454. doi: 10.1016/j.neuron.2014.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Broekman BFP, Olff M, Boer F. The genetic background to PTSD. Neuroscience & Biobehavioral Reviews. 2007;31(3):348–362. doi: 10.1016/j.neubiorev.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 9.Amstadter AB, Nugent NR, Koenen KC. Genetics of PTSD: Fear Conditioning as a Model for Future Research. Psychiatric annals. 2009;39(6):358–367. doi: 10.3928/00485713-20090526-01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norrholm SD, Ressler KJ. Genetics of Anxiety and Trauma-Related Disorders. Neuroscience. 2009;164(1):272–287. doi: 10.1016/j.neuroscience.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta D, Binder EB. Gene × environment vulnerability factors for PTSD: The HPA-axis. Neuropharmacology. 2012;62(2):654–662. doi: 10.1016/j.neuropharm.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 12.Skelton K, Ressler KJ, Norrholm SD, Jovanovic T, Bradley-Davino B. PTSD and Gene Variants: New Pathways and New Thinking. Neuropharmacology. 2012;62(2):628–637. doi: 10.1016/j.neuropharm.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma S, Powers A, Bradley B, Ressler K. Gene × Environment Determinants of Stress- and Anxiety-Related Disorders. Annual review of psychology. 2016;67:239–261. doi: 10.1146/annurev-psych-122414-033408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cornelis MC, Nugent NR, Amstadter AB, Koenen KC. Genetics of Post-Traumatic Stress Disorder: Review and Recommendations for Genome-Wide Association Studies. Current psychiatry reports. 2010;12(4):313–326. doi: 10.1007/s11920-010-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernardy NC, Friedman MJ. Psychopharmacological Strategies in the Management of Posttraumatic Stress Disorder (PTSD): What Have We Learned? Current Psychiatry Reports. 2015;17(4):1–10. doi: 10.1007/s11920-015-0564-2. [DOI] [PubMed] [Google Scholar]
- 16.Hendriksen H, Olivier B, Oosting RS. From non-pharmacological treatments for post-traumatic stress disorder to novel therapeutic targets. Eur J Pharmacol. 2014;732:139–158. doi: 10.1016/j.ejphar.2014.03.031. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z, et al. The relationship between combat-related posttraumatic stress disorder and the 5-HTTLPR/rs25531 polymorphism. Depression and Anxiety. 2011;28(12):1067–1073. doi: 10.1002/da.20872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hans Jörgen Grabe MD, et al. Serotonin Transporter Gene (SLC6A4) Promoter Polymorphisms and the Susceptibility to Posttraumatic Stress Disorder in the General Population. American Journal of Psychiatry. 2009;166(8):926–933. doi: 10.1176/appi.ajp.2009.08101542. [DOI] [PubMed] [Google Scholar]
- 19.Telch MJ, et al. 5-HTTLPR genotype potentiates the effects of war zone stressors on the emergence of PTSD, depressive and anxiety symptoms in soldiers deployed to iraq. World Psychiatry. 2015;14(2):198–206. doi: 10.1002/wps.20215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, et al. An Examination of the Association between 5-HTTLPR, Combat Exposure, and PTSD Diagnosis among U.S. Veterans. PLoS ONE. 2015;10(3):e0119998. doi: 10.1371/journal.pone.0119998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellman TA, et al. Serotonin polymorphisms and posttraumatic stress disorder in a trauma exposed African American population. Depression and anxiety. 2009;26(11):993–997. doi: 10.1002/da.20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sayin A, et al. A prospective study of serotonin transporter gene promoter (5-HTT gene linked polymorphic region) and intron 2 (variable number of tandem repeats) polymorphisms as predictors of trauma response to mild physical injury. DNA and cell biology. 2010;29(2):71–77. doi: 10.1089/dna.2009.0936. [DOI] [PubMed] [Google Scholar]
- 23.Gressier F, et al. The 5-HTTLPR Polymorphism and Posttraumatic Stress Disorder: A Meta-Analysis. Journal of traumatic stress. 2013;26(6):645–653. doi: 10.1002/jts.21855. [DOI] [PubMed] [Google Scholar]
- 24.Koenen KC, et al. SLC6A4 methylation modifies the effect of the number of traumatic events on risk for posttraumatic stress disorder. Depression and Anxiety. 2011;28(8):639–647. doi: 10.1002/da.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolassa I-T, et al. Association Study of Trauma Load and SLC6A4 Promoter Polymorphism in Posttraumatic Stress Disorder: Evidence From Survivors of the Rwandan Genocide. The Journal of clinical psychiatry. 2010;71(5):543–547. doi: 10.4088/JCP.08m04787blu. [DOI] [PubMed] [Google Scholar]
- 26.Koenen KC, et al. SLC6A4 methylation modifies the effect of the number of traumatic events on risk for posttraumatic stress disorder. Depression and anxiety. 2011;28(8):639–647. doi: 10.1002/da.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.William Tank A, Lee Wong D. Comprehensive Physiology. John Wiley & Sons, Inc; 2011. Peripheral and Central Effects of Circulating Catecholamines. [DOI] [PubMed] [Google Scholar]
- 28.Li L, et al. The Association Between Genetic Variants in the Dopaminergic System and Posttraumatic Stress Disorder: A Meta-Analysis. Medicine. 2016;95(11):e3074. doi: 10.1097/MD.0000000000003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valente NLM, et al. Candidate-gene approach in posttraumatic stress disorder after urban violence: association analysis of the genes encoding serotonin transporter, dopamine transporter, and BDNF. Journal of Molecular Neuroscience. 2011;44(1):59–67. doi: 10.1007/s12031-011-9513-7. [DOI] [PubMed] [Google Scholar]
- 30.Drury SS, Theall KP, Keats BJB, Scheeringa M. The Role of the Dopamine Transporter (DAT) in the Development of PTSD in Preschool Children. Journal of traumatic stress. 2009;22(6):534–539. doi: 10.1002/jts.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang S-C, et al. Molecular Variation at the SLC6A3 Locus Predicts Lifetime Risk of PTSD in the Detroit Neighborhood Health Study. PLoS ONE. 2012;7(6):e39184. doi: 10.1371/journal.pone.0039184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bailey JN, et al. PTSD and dopaminergic genes, DRD2 and DAT, in multigenerational families exposed to the Spitak earthquake. Psychiatry Research. 2010;178(3):507–510. doi: 10.1016/j.psychres.2010.04.043. [DOI] [PubMed] [Google Scholar]
- 33.Voisey J, et al. The DRD2 gene 957C>T polymorphism is associated with Posttraumatic Stress Disorder in war veterans. Depression and Anxiety. 2009;26(1):28–33. doi: 10.1002/da.20517. [DOI] [PubMed] [Google Scholar]
- 34.Young RM, et al. HARMFUL DRINKING IN MILITARY VETERANS WITH POST-TRAUMATIC STRESS DISORDER: ASSOCIATION WITH THE D2 DOPAMINE RECEPTOR A1 ALLELE. Alcohol and Alcoholism. 2002;37(5):451–456. doi: 10.1093/alcalc/37.5.451. [DOI] [PubMed] [Google Scholar]
- 35.Wolf EJ, et al. The Dopamine D(3) Receptor Gene and Posttraumatic Stress Disorder. Journal of traumatic stress. 2014;27(4):379–387. doi: 10.1002/jts.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hemmings SM, et al. BDNF Val66Met and DRD2 Taq1A polymorphisms interact to influence PTSD symptom severity: a preliminary investigation in a South African population. Progress in Neuro-Psychopharmacology and Biological Psychiatry. 2013;40:273–280. doi: 10.1016/j.pnpbp.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Mustapić M, et al. Dopamine beta-hydroxylase (DBH) activity and -1021C/T polymorphism of DBH gene in combat-related post-traumatic stress disorder. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2007;144B(8):1087–1089. doi: 10.1002/ajmg.b.30526. [DOI] [PubMed] [Google Scholar]
- 38.Hamner MB, Gold PB. Plasma dopamine beta-hydroxylase activity in psychotic and non-psychotic post-traumatic stress disorder. Psychiatry Research. 1998;77(3):175–181. doi: 10.1016/s0165-1781(98)00002-x. [DOI] [PubMed] [Google Scholar]
- 39.Rothbaum BO, et al. Early Intervention Following Trauma May Mitigate Genetic Risk for PTSD in Civilians: A Pilot Prospective Emergency Department Study. The Journal of clinical psychiatry. 2014;75(12):1380–1387. doi: 10.4088/JCP.13m08715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark R, et al. Predicting post-traumatic stress disorder in veterans: Interaction of traumatic load with COMT gene variation. J Psychiatr Res. 2013;47(12):1849–1856. doi: 10.1016/j.jpsychires.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 41.Valente NLM, et al. Catechol-O-methyltransferase (COMT) val158met Polymorphism as a Risk Factor for PTSD After Urban Violence. Journal of Molecular Neuroscience. 2011;43(3):516–523. doi: 10.1007/s12031-010-9474-2. [DOI] [PubMed] [Google Scholar]
- 42.Kolassa I-T, Kolassa S, Ertl V, Papassotiropoulos A, De Quervain DJF. The Risk of Posttraumatic Stress Disorder After Trauma Depends on Traumatic Load and the Catechol-O-Methyltransferase Val158Met Polymorphism. Biological Psychiatry. 2010;67(4):304–308. doi: 10.1016/j.biopsych.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 43.Yehuda R, et al. Enhanced suppression of cortisol following dexamethasone administration in posttraumatic stress disorder. American Journal of Psychiatry. 1993;150:83–83. doi: 10.1176/ajp.150.1.83. [DOI] [PubMed] [Google Scholar]
- 44.Yehuda R, Halligan SL, Golier JA, Grossman R, Bierer LM. Effects of trauma exposure on the cortisol response to dexamethasone administration in PTSD and major depressive disorder. Psychoneuroendocrinology. 2004;29(3):389–404. doi: 10.1016/s0306-4530(03)00052-0. [DOI] [PubMed] [Google Scholar]
- 45.Yehuda R. Status of Glucocorticoid Alterations in Post-traumatic Stress Disorder. Annals of the New York Academy of Sciences. 2009;1179(1):56–69. doi: 10.1111/j.1749-6632.2009.04979.x. [DOI] [PubMed] [Google Scholar]
- 46.Myers B, McKlveen JM, Herman JP. Glucocorticoid actions on synapses, circuits, and behavior: Implications for the energetics of stress. Frontiers in Neuroendocrinology. 2014;35(2):180–196. doi: 10.1016/j.yfrne.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castro-Vale I, van Rossum EFC, Machado JC, Mota-Cardoso R, Carvalho D. Genetics of glucocorticoid regulation and posttraumatic stress disorder — What do we know? Neuroscience & Biobehavioral Reviews. 2016;63:143–157. doi: 10.1016/j.neubiorev.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 48.Yehuda R. Is the glucocorticoid receptor a therapeutic target for the treatment of PTSD. Psychoneuroendocrinology. 2015;61:2. [Google Scholar]
- 49.Hauer D, et al. Relationship of a common polymorphism of the glucocorticoid receptor gene to traumatic memories and posttraumatic stress disorder in patients after intensive care therapy. Critical care medicine. 2011;39(4):643–650. doi: 10.1097/CCM.0b013e318206bae6. [DOI] [PubMed] [Google Scholar]
- 50.Palma-Gudiel H, Córdova-Palomera A, Leza JC, Fañanás L. Glucocorticoid receptor gene (NR3C1) methylation processes as mediators of early adversity in stress-related disorders causality: A critical review. Neuroscience & Biobehavioral Reviews. 2015;55:520–535. doi: 10.1016/j.neubiorev.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 51.Bachmann AW, et al. Glucocorticoid receptor polymorphisms and post-traumatic stress disorder. Psychoneuroendocrinology. 2005;30(3):297–306. doi: 10.1016/j.psyneuen.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 52.Fujii T, et al. Effect of the common functional FKBP5 variant (rs1360780) on the hypothalamic-pituitary-adrenal axis and peripheral blood gene expression. Psychoneuroendocrinology. 2014;42:89–97. doi: 10.1016/j.psyneuen.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 53.van Zuiden M, Kavelaars A, Geuze E, Olff M, Heijnen CJ. Predicting PTSD: Pre-existing vulnerabilities in glucocorticoid-signaling and implications for preventive interventions. Brain, Behavior, and Immunity. 2013;30:12–21. doi: 10.1016/j.bbi.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 54.Xie P, et al. Interaction of FKBP5 with Childhood Adversity on Risk for Post-Traumatic Stress Disorder. Neuropsychopharmacology. 2010;35(8):1684–1692. doi: 10.1038/npp.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Binder EB, et al. Association of FKBP5 Polymorphisms and Childhood Abuse With Risk of Posttraumatic Stress Disorder Symptoms in Adults. JAMA: the journal of the American Medical Association. 2008;299(11):1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klengel T, et al. Allele-specific FKBP5 DNA demethylation mediates gene–childhood trauma interactions. Nature neuroscience. 2013;16(1):33–41. doi: 10.1038/nn.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheuer S, et al. FKBP5 polymorphisms moderate the influence of adverse life events on the risk of anxiety and depressive disorders in preschool children. J Psychiatr Res. 2016;72:30–36. doi: 10.1016/j.jpsychires.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 58.Jovanovic T, Ressler KJ. How the Neurocircuitry and Genetics of Fear Inhibition May Inform Our Understanding of PTSD. The American journal of psychiatry. 2010;167(6):648–662. doi: 10.1176/appi.ajp.2009.09071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watkins LE, et al. FKBP5 polymorphisms, childhood abuse, and PTSD symptoms: Results from the National Health and Resilience in Veterans Study. Psychoneuroendocrinology. 2016;69:98–105. doi: 10.1016/j.psyneuen.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Almli LM, et al. ADCYAP1R1 Genotype Associates With Post-Traumatic Stress Symptoms in Highly Traumatized African-American Females. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2013;0(3):262–272. doi: 10.1002/ajmg.b.32145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dias BG, Ressler KJ. PACAP and the PAC1 Receptor in Post-Traumatic Stress Disorder. Neuropsychopharmacology. 2013;38(1):245–246. doi: 10.1038/npp.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ressler KJ, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470(7335):492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Uddin M, et al. ADCYAP1R1 GENOTYPE, POSTTRAUMATIC STRESS DISORDER, AND DEPRESSION AMONG WOMEN EXPOSED TO CHILDHOOD MALTREATMENT. Depression and anxiety. 2013;30(3):251–258. doi: 10.1002/da.22037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang S, et al. No association between ADCYAP1R1 and post-traumatic stress disorder in two independent samples. Molecular psychiatry. 2012;17(3):239. doi: 10.1038/mp.2011.118. [DOI] [PubMed] [Google Scholar]
- 65.Bennett MR, Hatton SN, Lagopoulos J. Stress, trauma and PTSD: translational insights into the core synaptic circuitry and its modulation. Brain Structure and Function. 2016;221(5):2401–2426. doi: 10.1007/s00429-015-1056-1. [DOI] [PubMed] [Google Scholar]
- 66.Laje G, et al. Brain-derived neurotrophic factor Val66Met polymorphism and antidepressant efficacy of ketamine in depressed patients. Biological psychiatry. 2012;72(11):e27. doi: 10.1016/j.biopsych.2012.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang L, et al. PTSD risk is associated with BDNF Val66Met and BDNF overexpression. Molecular psychiatry. 2014;19(1):8. doi: 10.1038/mp.2012.180. [DOI] [PubMed] [Google Scholar]
- 68.Zhang L, Li X-X, Hu X-Z. Post-traumatic stress disorder risk and brain-derived neurotrophic factor Val66Met. World journal of psychiatry. 2016;6(1):1. doi: 10.5498/wjp.v6.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Felmingham KL, Dobson-Stone C, Schofield PR, Quirk GJ, Bryant RA. The Brain-Derived Neurotrophic Factor Val66Met Polymorphism Predicts Response to Exposure Therapy in Posttraumatic Stress Disorder. Biological psychiatry. 2013;73(11):1059–1063. doi: 10.1016/j.biopsych.2012.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maddox SA, Schafe GE, Ressler KJ. Exploring epigenetic regulation of fear memory and biomarkers associated with Post-traumatic stress disorder. Frontiers in Psychiatry. 2013:4. doi: 10.3389/fpsyt.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Frielingsdorf H, et al. Variant brain-derived neurotrophic factor Val66Met endophenotypes: implications for posttraumatic stress disorder. Annals of the New York Academy of Sciences. 2010;1208:150–157. doi: 10.1111/j.1749-6632.2010.05722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bueller JA, et al. BDNF Val66Met allele is associated with reduced hippocampal volume in healthy subjects. Biological psychiatry. 2006;59(9):812–815. doi: 10.1016/j.biopsych.2005.09.022. [DOI] [PubMed] [Google Scholar]
- 73.Egan MF, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112(2):257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 74.Soliman F, et al. A genetic variant BDNF polymorphism alters extinction learning in both mouse and human. Science. 2010;327(5967):863–866. doi: 10.1126/science.1181886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olff M, Langeland W, Gersons BP. The psychobiology of PTSD: coping with trauma. Psychoneuroendocrinology. 2005;30(10):974–982. doi: 10.1016/j.psyneuen.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 76.Lee HJ, et al. No association between the brain-derived neurotrophic factor gene Val66Met polymorphism and post-traumatic stress disorder. Stress and Health. 2006;22(2):115–119. [Google Scholar]
- 77.Zhang H, et al. Brain derived neurotrophic factor (BDNF) gene variants and Alzheimer’s disease, affective disorders, posttraumatic stress disorder, schizophrenia, and substance dependence. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2006;141(4):387–393. doi: 10.1002/ajmg.b.30332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nelson EC, et al. Association of childhood trauma exposure and GABRA2 polymorphisms with risk of posttraumatic stress disorder in adults. Molecular Psychiatry. 2009;14(3):234–235. doi: 10.1038/mp.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Amstadter AB, et al. Variant in RGS2 Moderates Posttraumatic Stress Symptoms following Potentially Traumatic Event Exposure. Journal of anxiety disorders. 2009;23(3):369–373. doi: 10.1016/j.janxdis.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dunn EC, et al. Interaction Between Genetic Variants and Exposure to Hurricane Katrina on Post-Traumatic Stress and Post-Traumatic Growth: A Prospective Analysis of Low Income Adults. Journal of affective disorders. 2014;0:10. doi: 10.1016/j.jad.2013.09.018. doi:1016/j.jad.2013.1009.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Badour CL, et al. Exploring the association between a cholecystokinin promoter polymorphism (rs1799923) and posttraumatic stress disorder in combat veterans. Journal of Anxiety Disorders. 2015;36:78–83. doi: 10.1016/j.janxdis.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mota N, et al. The rs1049353 Polymorphism in the CNR1 Gene Interacts With Childhood Abuse to Predict Posttraumatic Threat Symptoms. J Clin Psychiatry. 2015;76(12):E1622–E1623. doi: 10.4088/JCP.15l10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kimbrel NA, et al. Effect of genetic variation in the nicotinic receptor genes on risk for posttraumatic stress disorder. Psychiatry Research. 2015;229(1–2):326–331. doi: 10.1016/j.psychres.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Logue MW, et al. A genome-wide association study of posttraumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Molecular psychiatry. 2013;18(8):937–942. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lowe SR, et al. RORA and posttraumatic stress trajectories: main effects and interactions with childhood physical abuse history. Brain and Behavior. 2015;5(4):e00323. doi: 10.1002/brb3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Amstadter A, et al. Support for association of RORA variant and post traumatic stress symptoms in a population-based study of hurricane exposed adults. Molecular psychiatry. 2013;18(11):1148. doi: 10.1038/mp.2012.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guffanti G, et al. No association between RORA polymorphisms and PTSD in two independent samples. Mol Psychiatry. 2014;19(10):1056–1057. doi: 10.1038/mp.2014.19. [DOI] [PubMed] [Google Scholar]
- 88.Kilaru V, et al. Genome-wide gene-based analysis suggests an association between Neuroligin 1 (NLGN1) and post-traumatic stress disorder. Transl Psychiatry. 2016;6:e820. doi: 10.1038/tp.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lisé M-F, El-Husseini A. The neuroligin and neurexin families: from structure to function at the synapse. Cellular and Molecular Life Sciences CMLS. 2006;63(16):1833–1849. doi: 10.1007/s00018-006-6061-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sindi IA, Dodd PR. New insights into Alzheimer’s disease pathogenesis: the involvement of neuroligins in synaptic malfunction. Neurodegenerative Disease Management. 2015;5(2):137–145. doi: 10.2217/nmt.14.54. [DOI] [PubMed] [Google Scholar]
- 91.Apte SS, Parks WC. Metalloproteinases: A parade of functions in matrix biology and an outlook for the future. Matrix Biology. 2015;44–46:1–6. doi: 10.1016/j.matbio.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 92.Xie P, et al. Genome-wide Association Study Identifies New Susceptibility Loci for Posttraumatic Stress Disorder. Biological Psychiatry. 2013;74(9):656–663. doi: 10.1016/j.biopsych.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ashley-Koch AE, et al. Genome-wide association study of posttraumatic stress disorder in a cohort of Iraq–Afghanistan era veterans. Journal of Affective Disorders. 2015;184:225–234. doi: 10.1016/j.jad.2015.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Montesinos ML. Roles for DSCAM and DSCAML1 in Central Nervous System Development and Disease. In: Berezin V, Walmod SP, editors. Cell Adhesion Molecules: Implications in Neurological Diseases. Springer; New York, New York, NY: 2014. pp. 249–270. [DOI] [PubMed] [Google Scholar]
- 95.Kaur C, Sivakumar V, Yip GW, Ling EA. Expression of syndecan-2 in the amoeboid microglial cells and its involvement in inflammation in the hypoxic developing brain. Glia. 2009;57(3):336–349. doi: 10.1002/glia.20764. [DOI] [PubMed] [Google Scholar]
- 96.Guardado P, et al. Altered gene expression of the innate immune, neuroendocrine, and nuclear factor-kappa B (NF-κB) systems is associated with posttraumatic stress disorder in military personnel. Journal of Anxiety Disorders. 2016;38:9–20. doi: 10.1016/j.janxdis.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 97.Stein MB, Chen C, Ursano RJ, et al. Genome-wide association studies of posttraumatic stress disorder in 2 cohorts of us army soldiers. JAMA Psychiatry. 2016;73(7):695–704. doi: 10.1001/jamapsychiatry.2016.0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nievergelt CM, et al. Genomic predictors of combat stress vulnerability and resilience in U.S. Marines: A genome-wide association study across multiple ancestries implicates PRTFDC1 as a potential PTSD gene. Psychoneuroendocrinology. 2015;51:459–471. doi: 10.1016/j.psyneuen.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 99.Suzuki E, et al. PRTFDC1, a possible tumor-suppressor gene, is frequently silenced in oral squamous-cell carcinomas by aberrant promoter hypermethylation. Oncogene. 2007;26(57):7921–7932. doi: 10.1038/sj.onc.1210589. [DOI] [PubMed] [Google Scholar]
- 100.Logue MW, et al. The Psychiatric Genomics Consortium Posttraumatic Stress Disorder Workgroup: Posttraumatic Stress Disorder Enters the Age of Large-Scale Genomic Collaboration. Neuropsychopharmacology. 2015;40(10):2287–2297. doi: 10.1038/npp.2015.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lebois LAM, Wolff JD, Ressler KJ. Neuroimaging genetic approaches to Posttraumatic Stress Disorder. Experimental neurology. doi: 10.1016/j.expneurol.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Block SR, Liberzon I. Attentional processes in posttraumatic stress disorder and the associated changes in neural functioning. Experimental neurology. doi: 10.1016/j.expneurol.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 103.Morey RA, et al. Serotonin transporter gene polymorphisms and brain function during emotional distraction from cognitive processing in posttraumatic stress disorder. BMC Psychiatry. 2011;11:76–76. doi: 10.1186/1471-244X-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Andero R, et al. Amygdala-Dependent Fear Is Regulated by Oprl1 in Mice and Humans with PTSD. Science translational medicine. 2013;5(188):188ra173–188ra173. doi: 10.1126/scitranslmed.3005656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Holbrook TL, Galarneau MR, Dye JL, Quinn K, Dougherty AL. Morphine use after combat injury in Iraq and post-traumatic stress disorder. New England Journal of Medicine. 2010;362(2):110–117. doi: 10.1056/NEJMoa0903326. [DOI] [PubMed] [Google Scholar]
- 106.Gee DG, et al. Individual differences in frontolimbic circuitry and anxiety emerge with adolescent changes in endocannabinoid signaling across species. Proceedings of the National Academy of Sciences. 2016;113(16):4500–4505. doi: 10.1073/pnas.1600013113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dincheva I, et al. FAAH genetic variation enhances fronto-amygdala function in mouse and human. Nature communications. 2015:6. doi: 10.1038/ncomms7395. [DOI] [PMC free article] [PubMed] [Google Scholar]