Abstract
Burkholderia pseudomallei produces an extracellular polysaccharide capsule -3)-2-O-acetyl-6-deoxy-β-d-manno-heptopyranose-(1- which has been shown to be an essential virulence determinant. The addition of purified capsule was shown to increase the virulence of a capsule mutant strain in the Syrian hamster model of acute melioidosis. An increase in the number of wild-type B. pseudomallei cells in the blood was seen by 48 h, while the number of capsule mutant cells in the blood declined by 48 h. Capsule expression was shown to be induced in the presence of serum using a lux reporter fusion to the capsule gene wcbB. The addition of purified B. pseudomallei capsule to serum bactericidal assays increased the survival of B. pseudomallei SLR5, a serum-sensitive strain, by 1,000-fold in normal human serum. Capsule production by B. pseudomallei contributed to reduced activation of the complement cascade by reducing the levels of complement factor C3b deposition. An increase in phagocytosis of the capsule mutant compared to the wild type was observed in the presence of normal human serum. These results suggest that the production of this capsule contributes to resistance to phagocytosis by reducing C3b deposition on the surface of the bacterium, thereby contributing to the persistence of bacteria in the blood of the infected host. Continued studies to characterize this capsule are essential for understanding the pathogenesis of B. pseudomallei infections and the development of preventive strategies for treatment of this disease.
Burkholderia pseudomallei is a gram-negative motile bacillus that is the causative agent of melioidosis. The organism is an environmental saprophyte typically found in soil or stagnant water (20). B. pseudomallei is endemic to Southeast Asia and northern Australia; however, the bacterium has been isolated from a number of other areas, predominantly in those countries that lie within 20o north and 20o south of the equator (20, 9). Infection by B. pseudomallei is due to either direct inoculation into wounds and skin abrasions or inhalation of contaminated material (20). The disease melioidosis may present as a variety of clinical manifestations, ranging from acute pneumonia or acute septicemia to chronic and latent infections (16). The acute septicemic form of melioidosis is rapidly fatal, resulting in death within 24 to 48 h after the onset of infection (7). Even with antibiotic therapy, the mortality rate of this form is still 40% (32). The severity of the disease poses a concern for travelers and for military personnel serving in these areas (8).
A number of pathogenic bacteria possess virulence determinants that allow evasion of host defense mechanisms. One of the principal components of innate immunity of the host is the complement cascade. The complement system is comprised of a number of glycoproteins that interact in a series of reactions to promote opsonization and direct cell damage of invading microorganisms (18). To be successful pathogens, microorganisms must avoid destruction by the opsonic, chemotactic, and lytic functions of the complement cascade. Strategies bacteria employ to resist complement attack include impaired complement activation, ineffective opsonization, resistance to complement-mediated lysis, depletion of complement components, and subversion of host complement proteins and receptors to gain access to the intracellular milieu of phagocytes (14, 19). B. pseudomallei has been shown to be resistant to complement-mediated lysis by virtue of the O-polysaccharide (O-PS) moiety of its lipopolysaccharide (LPS) (12, 17, 33). This polysaccharide was originally characterized as type II O-PS with the struc-ture -3)-β-d-glucopyranose-(1-3)-6-deoxy-α-l-talopyranose-(1- but is now referred to as B. pseudomallei O-PS (23, 25).
Previously, subtractive hybridization was carried out between B. pseudomallei and a related avirulent organism, Burkholderia thailandensis, in order to identify putative virulence determinants in B. pseudomallei. This led to the identification of a capsular polysaccharide found to be required for virulence in the Syrian hamster model of acute melioidosis (25). Insertional inactivation of a glycosyltransferase gene resulted in a mutant strain lacking the capsule that was 10,000-fold more attenuated for virulence than the wild-type strain. Sequencing of the genes involved in the biosynthesis of this polysaccharide revealed open reading frames involved in the synthesis and export of capsular polysaccharides, particularly those involved in the production of group 3 capsular polysaccharides, such as the Escherichia coli K-10 capsule, the Haemophilus influenzae group b capsule, and the capsule produced by Neisseria meningitidis serogroup B. The capsule identified by this method was found to be the previously characterized type I O-PS of B. pseudomallei with the structure -3)-2-O-acetyl-6-deoxy-β-d-manno-heptopyranose-(1- (23). However, due to genetic homology, its importance in virulence, and its high molecular mass, it was concluded that the polysaccharide was a capsule (25).
Capsule production has been correlated with virulence in many bacteria, particularly those causing serious invasive infections of humans (2). Although the importance of this capsule in virulence was established previously, the specific contribution of the capsule to the pathogenesis of B. pseudomallei was not defined. The objective of this study was to investigate the role of the capsule in B. pseudomallei pathogenesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. B. pseudomallei, B. thailandensis, and E. coli were grown at 37°C on Luria-Bertani (LB) broth base (Becton Dickinson) agar plates or in LB broth. For animal studies, B. pseudomallei cultures were grown at 37°C in TSBDC medium (4). When appropriate, antibiotics were added at the following concentrations: 50 μg of tetracycline, 100 μg of streptomycin, 25 μg of gentamicin, 25 μg of kanamycin, and 100 μg of polymyxin B per ml for B. pseudomallei and 100 μg of ampicillin, 25 μg of gentamicin, and 25 and 50 μg of kanamycin per ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 endA1 araΔ139 Δ(ara-leu)7697 galU galK λ− rpsL nupG λ−tonA | Invitrogen |
| HB101 | F− Δ(gpt-proA)62 leu supE44 ara14 galK2 lacY1 Δ(mcrC-mrr) rpsL20 (Smr) xyl-5 mtl-1 recA13 | 3 |
| B. pseudomallei | ||
| 1026b | Clinical isolate; Kmr Gmr Smr Pmr Tcs Tps | 11 |
| DD503 | 1026b derivative; allelic-exchange strain; Δ(amrR-oprA) (Kms Gms Sms) rpsL (Smr) | 21 |
| SR1015 | 1026b(pSR1015); Smr Tcr | 25 |
| SLR5 | SR1001 derivative; wcbB::Tn5-OT182 Tcr | 25 |
| SZ210 | DD503 derivative; ΔwcbB | This study |
| SZ210(pSZ219) | SZ210 derivative; pSZ219; Kmr | This study |
| SZ211 | DD503 derivative; pML102; Gmr | This study |
| SZ213 | DD503 derivative; pSZ213; Gmr | This study |
| B. thailandensis | ||
| E264 | Soil isolate; Kmr Gmr Smr Pmr Tcs Tps | 5 |
| Plasmids | ||
| pGSV3-lux | Mobilizable suicide vector containing lux operon from P. luminescens; OriT; Gmr | 22 |
| pBHR1 | Broad-host-range mobilizable cloning vector; Cmr Kmr | MoBiTec |
| pBluescript SK | General cloning vector; ColEI; Apr | Stratagene |
| pKAS46 | Mobilizable allelio-exchange vector; π-dependant R6K replicon; Apr KmrrpsL+ (Sms) | 28 |
| pCR2.1-TOPO | Topoisomerase-mediated cloning vector; Apr Kmr | Invitrogen |
| pRK2013 | Self-transmissible helper plasmid; Kmr | 15 |
| pSZ110 | 1.2-kb BamHI-KpnI fragment containing 600-bp upstream and downstream fragments of wcbB cloned into pBluescript SK; ΔwcbB; Apr | This study |
| pSZ210 | 1.2-kb XbaI-KpnI fragment containing 600-bp upstrcam and downstream fragments of wcbB cloned into pKAS46; ΔwcbB; Apr Kmr | This study |
| pML102 | 670-bp internal region of wcbB gene cloned as an EcoRI fragment into pGSV3-lux; Gmr | This study |
| pSZ212 | 589-bp internal region of wbiA gene cloned into pCR2.1-TOPO; Apr Kmr | This study |
| pSZ213 | 589-bp internal region of wbiA gene cloned as an EcoRI fragment into pGSV3-lux; Gmr | This study |
| pSZ219 | 1.5-kb wcbB gene and promoter region cloned as an NcoI-PinAI fragment into pBHR1; Kmr | This study |
Construction of a B. pseudomallei capsule deletion mutant.
To construct an in-frame deletion of the wcbB gene, oligodeoxyribonucleotide primers were designed to amplify ∼600 bp of DNA sequence upstream and downstream of the gene. The primers UpwcbBBamHI(+) and WcbBstartHindIII(−) were used to amplify the upstream DNA sequence including the ATG start site (Table 2). The primers WcbBendHindIII (+) and DownwcbBKpnI(−) were used to amplify the region including the stop codon and downstream DNA (Table 2). The primers WcbBstartHindIII(−) and WcbBendHindIII(+) were designed to incorporate a unique HindIII restriction site to facilitate cloning of the upstream and downstream fragments to create a 792-bp in-frame deletion of the wcbB gene. The primers UpwcbBBamHI(+) and DownwcbBKpnI(−) were designed to contain BamHI and KpnI linkers for cloning of the DNA fragments into pBluescript SK. The DNA fragments were amplified from B. pseudomallei 1026b chromosomal DNA via PCR using the QIAGEN HotStarTaq Master Mix kit as previously described (22) and cloned into pBluescript SK to create pSZ110 (Table 1). The assembled in-frame deletion was then cloned as an XbaI/KpnI fragment into pKAS46, an allelic-exchange vector based on rpsL, for counterselection, resulting in the plasmid pSZ210 (Table 1). B. pseudomallei DD503 was the recipient strain used for allelic exchange (Table 1). Allelic exchange was performed as previously described, except that transconjugants were plated on polymyxin B and kanamycin (21, 25). The Pmr Kmr transconjugants were subsequently transferred to plates containing streptomycin to select for the loss of pKAS46. Smr Kms transconjugants were screened by PCR for the in-frame deletion using the primers WcbBstartHindIII(−) and DownwcbBKpnI(−). The mutant strain B. pseudomallei SZ210 contained the expected 792-bp deletion in the wcbB gene relative to wild-type 1026b.
TABLE 2.
Sequence of oligodeoxyribonucleotide primers used in this study
| Primer name | Sequencea |
|---|---|
| UpwcbBBamHI(+) | 5′-GCG GGA TCC GCG CGC CAC TGG CCC CCG AC-3′ |
| WcbBstartHindIII(−) | 5′-GCC AGA AGC TTG TAA TAG CGT GGA AAT TCC ATT CCG CTG TGG CTT G-3′ |
| WcbBendHindIII(+) | 5′-GCC AAG CTT GAC ACG CTG CGC TAA ATG ACC CGA TCC ACC-3′ |
| DownwcbBKpnI(−) | 5′-GCC GGT ACC ACG ATC TCT CGT GCG GGC GAG-3′ |
| WcbB-F2 | 5′-GCC AAT TCC CGA TCG ACG CTG GAA T-3′ |
| WcbB-R2 | 5′-GCA GCG TGT CAA GAA AGG CAT CGA-3′ |
| WbiA-int-F | 5′-GCT TTC TCA TTG CGA AGA GCG G-3′ |
| WbiA-int-R | 5′-GCA AAC GAC GAC AGC AAA CGC T-3′ |
| WcbBfull3NcoI(+) | 5′-GCC CAT GGT GAC ACA CGC GAT CAA AAA CGT-3′ |
| WcbBfull3PinAI(−) | 5′-CGA CCG GTA TGC AGT AAG GTG TTT GAC CA-3′ |
Underlining indicates the restriction enzyme recognition site engineered into the primer.
Complementation of capsule deletion mutant SZ210.
A 1,427-bp DNA fragment containing the full wcbB gene and upstream promoter region was amplified from B. pseudomallei 1026b genomic DNA by PCR using the primers WcbBfullNcoI(+) and WcbBfullPinAI(−) (Table 2). PCR amplification was performed as described above. The PCR product was cloned as an NcoI-PinAI fragment into pBHR1 to create pSZ219 (Table 1). Top10(pSZ219) was conjugated to B. pseudomallei SZ210 using a triparental mating protocol utilizing E. coli HB101(pRK2013) for transfer as previously described (Table 1) (22). Kmr colonies were screened by plasmid DNA isolations and PCR to confirm the presence of pSZ219.
Western blot analysis of capsule production.
Samples for Western blot analysis were prepared as previously described (6). An immunoassay was performed with a 1:500 dilution of the primary antibody, mouse monoclonal antibody 147 raised to B. pseudomallei capsular polysaccharide (CPS). The secondary antibody used was horseradish peroxidase-conjugated goat anti-mouse polyvalent immunoglobulins (immunoglobulin A [IgA], IgM, and IgG).
Animal studies.
The animal model of acute B. pseudomallei infection was previously described (13). To assess the virulence of the capsule mutant strain SZ210 and the complemented strain SZ210(pSZ219) compared to that of wild-type B. pseudomallei 1026b, Syrian hamsters (females; 6 to 8 weeks old) were inoculated intraperitoneally (i.p.) with a number of serial dilutions corresponding to 101 to 105 CFU. At 48 h, the 50% lethal dose (LD50) values were calculated. To determine differences in tissue distribution between the wild type and the capsule mutant, the hamsters were divided into six groups of three animals each and injected i.p. with 102 CFU of either B. pseudomallei 1026b or SR1015. At 12, 24, and 48 h, one group of animals infected with either strain was sacrificed, and the blood, liver, lungs, and spleen were extracted from each animal in the group, homogenized, diluted in sterile phosphate-buffered saline (PBS), and plated on LB agar containing polymyxin B for 1026b or LB plates containing tetracycline for SR1015 for bacterial quantitation. To investigate the effect of purified capsule on virulence in the animal model, hamsters (five per group) were inoculated i.p. with 101 to 104 CFU of bacteria alone or with bacteria and 100 μg of purified B. pseudomallei capsule. After 48 h, the LD50 values were calculated, and the animals were bled. The blood of the infected animals was diluted in sterile PBS and plated on LB plates (for 1026b) or LB plates containing tetracycline (for SR1015) for bacterial quantitation.
Serum bactericidal assays.
Serum bactericidal assays were performed as previously described (12). In order to test the effect of purified CPS and O-PS on the survival of serum-sensitive strains, purified B. pseudomallei CPS or O-PS was added to the samples to a final concentration of 10, 50, and 100 μg/ml, along with the bacteria and 30% normal human serum (NHS), and the mixture was incubated for 2 h at 37°C. Control samples contained either bacteria and sterile PBS alone or bacteria, 30% NHS, and PBS without the addition of purified CPS or O-PS. In order to assess whether the capsule was affecting the complement cascade, control samples containing NHS heat inactivated at 56°C for 30 min (HI-NHS) were included. In addition, 100 μg of purified CPS or O-PS was preincubated with 30% NHS for 30 min at 37°C (PI-CPS or PI-O-PS) before the addition of bacteria in order to determine the effects of the purified polysaccharides on complement.
Purification of the B. pseudomallei capsule and O-polysaccharide.
The capsular polysaccharide and O-antigenic moiety of the LPS of B. pseudomallei were purified as previously described (23). Protein, nucleic acid, and endotoxin levels were assessed using the BCA protein assay kit (Pierce Chemical Company, Rockford, Ill.) or the LAL assay kit for endotoxin (BioWhittaker, Walkersville, Md.), by photospectroscopy measurements of the optical density at 260 nm (OD260) and OD280 for nucleic acid content, and by nuclear magnetic resonance spectroscopy. Protein, nucleic acid, and endotoxin levels were determined to be negligible by these methods. Purified capsule and O antigen were resuspended overnight in sterile PBS at a concentration of 10 mg/ml for use in serum bactericidal assays and animal experiments.
Detection of complement factor C3b deposition.
Detection of C3b deposition on the bacterial surface was carried out according to a previously described protocol with some modifications (31). Bacteria were grown overnight with the appropriate selection in 2 ml of LB broth at 37°C. The next day, 100 μl of an overnight culture was added to 3 ml of LB broth, and the cultures were grown to late logarithmic phase (∼4 to 5 h). The cultures were diluted in sterile PBS, and 107 organisms were added to pooled NHS at a concentration of either 10 or 30% in a final volume of 1 ml. Control samples included human serum that was heat inactivated at 56°C for 30 min or PBS. Samples were incubated at 37°C, and the reaction of bacteria with complement factors was stopped by the addition of EGTA to a final concentration of 10 mM and incubation in an ice water bath for 1 min. The bacteria were pelleted by centrifugation at 13,000 × g for 5 min at 4°C, and the pellet was washed three times with 500 μl of PBS. For Western blot analysis, the bacterial pellets were resuspended in 40 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample solution (0.3 M Tris HCl [pH 6.8], 2% SDS, 12.5% glycerol, 5% β-mercaptoethanol) and boiled for 5 min. The samples were then separated by SDS-PAGE (10% polyacrylamide). For control of the migration of C3b in the gels, 1 μl of NHS diluted 1:10 in PBS was included. Following electrophoresis, the proteins were transferred onto nitrocellulose filters (Bio-Rad, Mississauga, Canada). An immunoassay was performed with a 1:8 dilution of the primary antibody, AB189, a mouse monoclonal antibody to human complement factor C3b α fragment (Autogen Bioclear). This antibody recognizes a determinant present on native (circulating) C3 and on the fragments C3b and C3c. The antibody is directed to a determinant present on the α chain of the C3 molecule. The secondary antibody used was horseradish peroxidase-conjugated goat anti-mouse IgG (Sigma).
Optical densitometry.
Western blots were scanned using a Hewlett-Packard ScanJet 6100 scanner, and the scanned image was rendered into a TIFF format. Optical densitometry measurements for the area under the curve were made from the TIFF-formatted image using Bio-Rad Quantity One software. Measurements were analyzed using Instat software (GraphPad Software, San Diego, Calif.).
Immunofluorescence microscopy studies of C3b deposition.
To analyze the deposition of C3b on the bacterial surface by immunofluorescence microscopy, the procedure for the analysis of C3b deposition was followed except that 108 bacteria were used. Following the addition of EGTA and washing of the pellets in PBS, the bacterial pellets were resuspended in 1 ml of a 2.5% paraformaldehyde solution and incubated at 37°C for 10 min with gentle agitation. The pellets were centrifuged at 13,000 × g for 5 min, and excess paraformaldehyde was removed by washing the pellets in PBS three times with gentle agitation for 5 min between centrifugations. The pellets were then resuspended in 200 μl of PBS-Tween (PBS-0.05% Tween 20) and incubated for 1 h at 37°C to block nonspecific binding sites. After being blocked, 20 μl of the sample was centrifuged at 13,000 × g for 5 min (107 bacterial cells); the pellet was resuspended in 20 μl of PBS-Tween containing the primary antibody, AB189, at a dilution of 1:20; and the sample was incubated at 37°C for 30 min. The sample was centrifuged, and 5-min washes were carried out in PBS-Tween as described above. Following the washes, the bacteria were centrifuged, and the pellet was resuspended in PBS-Tween containing the secondary antibody, rabbit anti-mouse IgG conjugated to Cy3 (Jackson Laboratories), at a dilution of 1:400 and incubated at 37°C for 30 min. The sample was then centrifuged and washed three times in PBS-Tween as described above. The pellet was finally washed once in sterile distilled H2O for 5 min, and the bacteria were stained for 2 min with DAPI (4′,6′-diamidino-2-phenylindole; 1:2,000 dilution in H2O of a 20 mg/ml solution; Sigma) for visualization of bacterial cells. After centrifugation of the sample, the resulting pellet was resuspended in 20 μl of H2O and centrifuged onto prepared poly-l-lysine-coated coverslips (Sigma) set in a 24-well tissue culture plate at 2,000 rpm for 10 min in a Sorvall T6000B centrifuge (DuPont). Once the samples were allowed to dry, 1 ml of distilled H2O was added to the wells, and the coverslips were gently removed, dried, and mounted on a glass slide. Samples were visualized and photographed with a Leica DMIREB2 inverted microscope and analyzed using compatible imaging software (Openlab; Improvision). Digital images were prepared using Photoshop version 5.0 (Adobe Systems Inc., San Jose, Calif.).
Quantitative radiolabeled phagocytosis assay.
Phagocytosis of B. pseudomallei strains by polymorphonuclear leukocytes (PMNL) was determined by a radiolabeled phagocytosis assay essentially as previously described (14). Briefly, ∼5 × 106 [14C]lysine-labeled bacteria were added to 106 PMNL isolated from a healthy donor in the presence of either 10 or 30% NHS and incubated in a final volume of 500 μl for 30 min at 37°C in a shaking incubator. Control samples contained bacterial cells, PMNL, and PBS alone. For each variable, identical reactions were set up in two polypropylene scintillation vials (Biovials; Beckman, Chicago, Ill.). Following phagocytosis, 3 ml of ACS 11 scintillant (Amersham, Arlington Heights, Ill.) was added to one of the two tubes to represent both cell- and non-cell-associated bacteria. To separate the cell-associated from the non-cell-associated bacteria, 1 ml of ice-cold PBS was added to the other tube, and three differential centrifugation steps (160 × g for 7 min at 4°C) were performed before the final addition of 3 ml of ACS 11 scintillant. All tubes were counted in a Beckman LS 6500 scintillation counter. The percentage of the bacterial population phagocytosed was calculated by the following equation: percent uptake = [counts per minute following differential centrifugation (cell associated)/counts per minute in noncentrifuged tubes (cell and non-cell associated)] × 100. Experiments were performed in triplicate and repeated more than three times to ensure consistency. The purity and viability of the isolated PMNL was assessed by trypan blue exclusion. Statistical significance was determined using GraphPad Instat software.
Construction of lux reporter strains SZ211 and SZ213.
A 670-bp internal region of the wcbB gene was amplified by PCR using the primers WcbB-F2 and WcbB-R2 (Table 2). A 589-bp internal fragment of the LPS gene wbiA was amplified by PCR using the primers WbiA-int-F and WbiA-int-R (Table 2). PCR amplification was performed as described above. Both PCR products were cloned into pCR2.1-TOPO (Invitrogen Life Technologies, Burlington, Ontario, Canada) according to the manufacturer's instructions. The 670-bp internal region from wcbB was cloned as an EcoRI fragment into pGSV3-lux, a suicide vector containing a promoterless lux operon as a reporter, to create pML102 (Table 1). The 589-bp internal region from wbiA was cloned as an EcoRI fragment into pGSV3-lux, resulting in the plasmid pSZ213 (Table 1). DH10B(pML102) and DH10B(pSZ213) were conjugated to B. pseudomallei DD503 in a triparental mating protocol as described above (Table 1) (22).
DNA manipulation.
Restriction enzymes and T4 DNA ligase were purchased from Invitrogen Life Technologies and New England Biolabs (Mississauga, Ontario, Canada) and used according to the manufacturers' instructions. Chromosomal DNA was isolated using a Wizard Genomic DNA Purification kit (Promega Corp., Madison, Wis.). Plasmid DNA was isolated using a QIAprep Spin Miniprep kit (QIAGEN). DNA fragments used in cloning procedures were excised from agarose gels and purified using a QIAGEN gel extraction kit. PCR products were cloned into pCR2.1-TOPO using the TOPO TA Cloning kit and chemically competent E. coli Top10 (Invitrogen Life Technologies) according to the manufacturer's instructions.
Bioluminescence assay to assess capsule and LPS gene induction in normal human serum.
The lux reporter fusion strains B. pseudomallei SZ211 and SZ213 were grown in M9 plus 1% glucose, M9 plus 1% glucose plus 30% NHS, or M9 plus 1% glucose plus 30% HI-NHS. Ninety-six-well plates containing 150 μl of medium were inoculated with 5 μl of an overnight culture of either SZ211 or SZ213 (five replicates per condition) and incubated at 37°C with shaking for 18 h. Absorbance readings (OD540) and luminescence (in relative light units) measurements were taken every 2 h. The absorbances of the bacterial cultures were measured using a Bio-Rad Microplate Reader and Microplate Manager version 5.1 software (Bio-Rad). Luminescence was measured with the Reporter Microplate Luminometer (Turner Biosystems, Sunnyvale, Calif.) and Reporter software.
RESULTS
The glycosyltransferase gene wcbB is required for the biosynthesis of B. pseudomallei capsule.
An in-frame deletion was constructed in wcbB, a gene which encodes a glycosyltransferase, resulting in the capsule-minus strain SZ210. To confirm the role of wcbB in the biosynthesis of capsule, SZ210 was complemented by the introduction of a wild-type copy of the wcbB gene cloned into the mobilizable broad-host-range plasmid pBHR1 (Table 1). Western blot analysis of proteinase K-digested whole cells was performed using mouse monoclonal antibody directed to B. pseudomallei capsule to assess capsule production by these strains (Fig. 1). Similar to the capsule-minus strain SR1015 and B. thailandensis E264, which is known to lack this capsule, SZ210 was found to be negative for capsule production, as indicated by the absence of a 200-kDa band that is present for wild-type 1026b. Complementation of SZ210 by providing the wild-type wcbB gene in trans restored capsule production. Whole-cell extracts from the complemented strain SZ210(pSZ219) reacted to the capsule antibody producing the 200-kDa band corresponding to the B. pseudomallei capsule.
FIG. 1.
Western blot analysis of capsule production by capsule-complemented strain B. pseudomallei SZ210(pSZ219). Bacteria were reacted with proteinase K, subjected to SDS-PAGE, electroblotted, and reacted with monoclonal antiserum raised to B. pseudomallei CPS. Lane M, benchmark prestained protein molecular mass standards (Invitrogen Life Technologies); lane 1, B. pseudomallei 1026b; lane 2, B. thailandensis E264; lane 3, B. pseudomallei SR1015; lane 4, B. pseudomallei SZ210; lane 5, blank; lane 6, B. pseudomallei SZ210(pSZ219). The apparent molecular masses of the prestained proteins are indicated.
The B. pseudomallei capsule contributes to virulence in the Syrian hamster model of acute melioidosis.
Syrian golden hamsters were inoculated intraperitoneally with 101 to 105 cells of either wild-type B. pseudomallei 1026b, capsule mutants SR1015 and SZ210, or the complemented strain SZ210(pSZ219). One group of animals inoculated with SR1015 also received 100 μg of purified B. pseudomallei capsule. After 48 h, the LD50 values were calculated, and the blood of the infected animals was diluted and plated for bacterial quantitation. As shown in Table 3 the addition of purified capsule significantly increased the virulence of the capsule mutant strain SR1015. The LD50 value was calculated to be 34 CFU, similar to the LD50 value of wild-type B. pseudomallei 1026b (<10 CFU). In contrast, the LD50 value for SR1015 without the addition of purified capsule was calculated to be 3.5 × 105 CFU, 10,000-fold higher than when capsule was added to the inoculum. In addition, purified capsule enhanced the survival of SR1015 in the blood. Bacteria could not be detected in the blood of hamsters inoculated with SR1015 alone. However, the number of SR1015 CFU recovered from the blood of infected animals was 9.0 × 102 CFU/ml when capsule was added to the inoculum, an almost-1,000-fold increase. This number was comparable to the number of wild-type B. pseudomallei 1026b bacteria recovered from the blood. The addition of capsule was not toxic to the hamsters, as hamsters inoculated with 100 μg of purified capsule alone survived for the duration of the experiment without any ill effects (data not shown).
TABLE 3.
Effects of purified capsule on the virulence of B. pseudomallei SR1015 in the Syrian hamster model of acute melioidosis
| Strain | CPS added | LD50 (CFU) | Blood bacterial count (CFU/ml) |
|---|---|---|---|
| 1026b | No | <10 | 4.1 × 105 |
| SR1015 | No | 3.5 × 105 | 3 |
| SR1015 | Yes | 34 | 9.0 × 102 |
| SZ210 | No | 9.6 × 104 | 10 |
| SZ210(pSZ219) | No | 12 | 4.9 × 105 |
The LD50 value for the capsule mutant strain SZ210 containing an in-frame deletion of the wcbB gene was calculated to be 9.6 × 104 CFU, and the number of bacteria in the blood was determined to be 10 CFU/ml. Complementation of this strain restored virulence in the animal model, resulting in an LD50 value of 12 CFU, comparable to that of wild-type B. pseudomallei 1026b. Furthermore, the number of bacteria in the blood of animals infected with the complemented strain, SZ210(pSZ219), was determined to be 4.9 × 105 CFU/ml, similar to the number of bacteria recovered from animals infected with 1026b.
The B. pseudomallei capsule is responsible for persistence in the blood.
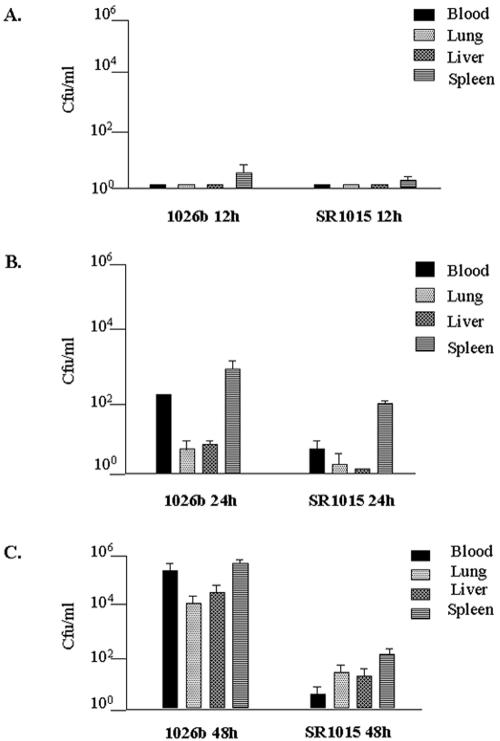
To further demonstrate the role of the capsule in infection by B. pseudomallei, an experiment was designed to investigate differences in tissue distribution between the capsule mutant strain and the wild type in infected hamsters. Animals were inoculated with 102 CFU of either wild-type B. pseudomallei 1026b or the capsule mutant SR1015. At different time points, the animals were sacrificed, and the numbers of bacteria in the blood, liver, lungs, and spleen of each animal were determined. As seen in Fig. 2, the numbers of B. pseudomallei 1026b and SR1015 bacteria were nearly undetectable at 12 h (Fig. 2A). By 24 h, the numbers of 1026b bacteria recovered from the blood, lung, liver, and spleen increased, while SR1015 was detected only in the spleen (Fig. 2B). By 48 h, very high numbers of 1026b bacteria were recovered from all of the organs taken, representing a dramatic increase compared to the inoculum (Fig. 2C). In contrast, all of the organs taken from hamsters infected with SR1015 contained fewer bacteria (Fig. 2C). Of particular interest was the fact that the number of SR1015 bacteria recovered from the blood at 48 h was lower than in the inoculum, suggesting that the capsule mutant was cleared from the blood more effectively than the wild type. The number of SR1015 bacteria recovered from the spleen was higher than the number of SR1015 bacteria in the blood, suggesting that SR1015 was being cleared from the blood and sequestered in the spleen. The difference in virulence between the two strains can be attributed to capsule production, since the capsule mutant strain has a growth rate similar to that of the wild-type strain 1026b (data not shown).
FIG. 2.
Differences in tissue distribution between B. pseudomallei strains 1026b and SR1015 in the Syrian hamster model of acute melioidosis. Female Syrian hamsters (three per group) were inoculated i.p. with 102 CFU of either strain, and at 12, 24, and 48 h, two groups of animals were sacrificed and bacterial quantitation of the tissues was determined. The data represent the average number of bacteria found in each tissue and the standard deviation for a given time point.
Capsule production by B. pseudomallei is responsible for persistence in the blood by inhibiting the effectiveness of the complement cascade.
To define the role of the capsule for persistence in the blood, serum bactericidal assays were performed with the addition of purified capsule to determine if capsule had an effect on the survival of serum-sensitive strains of B. pseudomallei. For these experiments, we utilized a double mutant that we constructed in the laboratory, SLR5, which lacks both capsule and O-polysaccharide, since the capsule mutant SR1015 was previously found to be serum resistant (Table 1) (25). As shown in Fig. 3, the survival of SLR5 was extremely poor when incubated in the presence of 30% NHS. However, the addition of purified capsule increased the survival of SLR5 in NHS. The addition of 50 μg of capsule to the reaction increased the numbers of SLR5 to 5.9 × 101 CFU/ml, and the addition of 100 μg of capsule increased the survival of SLR5 by nearly 1,000-fold to 1.9 × 103 CFU/ml. Furthermore, preincubation of 30% NHS with 100 μg of capsule (PI-CPS) before the addition of bacteria increased the survival of SLR5 100,000-fold to 4.4 × 106 CFU/ml. This was similar to the survival of SLR5 when incubated with serum that was heat inactivated. These effects were found to be specific to capsule, since the addition of 50 or 100 μg of purified B. pseudomallei O-PS or preincubation of the serum with O-PS did not increase the survival of serum-sensitive SLR5 (Fig. 3).
FIG. 3.
Purified B. pseudomallei capsule enhances the survival of B. pseudomallei SLR5 in 30% NHS. Purified B. pseudomallei CPS or O-PS was added at a concentration of either 50 or 100 μg/ml to bacteria and 30% NHS. Purified CPS or O-PS was also incubated with 30% NHS (PI-CPS or PI-O-PS) for 30 min at 37°C before the addition of bacteria. The bars represent the average number of CFU per milliliter and the standard deviation of three separate experiments.
Capsule reduces complement factor C3b deposition in vitro.
Since capsule mutants of B. pseudomallei are serum resistant in that they are not susceptible to lysis by the membrane attack complex (MAC), we postulated that the ability of the capsule to enhance survival in the blood could be due to its ability to inhibit C3b deposition and opsonization. To investigate the effect of capsule on C3b deposition, the amount of C3b deposited on the surfaces of wild-type B. pseudomallei 1026b and the capsule mutant, SR1015, in the presence of serum was determined by Western blot analysis using a mouse monoclonal antibody specific to human complement factor C3b. The deposition of C3b was found to be more pronounced in the capsule mutant SR1015 than in the wild type in both 10 and 30% NHS (Fig. 4A and B). Similar results were observed with the capsule mutant SZ210, a strain containing an in-frame deletion of the wcbB gene. More C3b was detected when SZ210 was incubated in both 10 and 30% NHS than with 1026b (Fig. 4C and D). Optical densitometry measurements were performed in order to quantitate the difference in C3b deposition between the strains. The average amount of C3b deposited on the surfaces of SR1015 and SZ210 bacteria was 3.5-fold higher than for 1026b in 10% NHS and 2.5-fold higher in 30% NHS. In addition, there was a shift in the molecular mass of C3b, which normally runs at 185 kDa, indicating a covalent attachment of the molecule to the bacterial surface (26). The nature of this attachment was not investigated; however, C3b is thought to covalently attach to the bacterial surface through an ester or amide linkage (18).
FIG. 4.
Western blot analysis of decreased complement factor C3b deposition in 10 and 30% normal human serum by B. pseudomallei capsule. B. pseudomallei 1026b, SR1015, and SZ210 were incubated in 10 and 30% NHS, subjected to SDS-polyacrylamide gel electrophoresis, electroblotted, and reacted with a mouse monoclonal antibody to human complement factor C3b (Autogen Bioclear). (A) Lane M, prestained protein molecular mass standards (New England Biolabs); lane 1, B. pseudomallei 1026b incubated in PBS; lane 2, 1026b incubated in 10% NHS for 30 min; lane 3, 1026b incubated in 10% NHS for 60 min; lane 4, B. pseudomallei SR1015 incubated in PBS; lane 5, SR1015 incubated in 10% NHS for 30 min; lane 6, SR1015 incubated in 10% NHS for 60 min; lane 7, C3b positive control. (B) Lane M, prestained protein molecular mass standards; lane 1, B. pseudomallei 1026b incubated in PBS; lane 2, 1026b incubated in 30% NHS for 30 min; lane 3, 1026b incubated in 30% NHS for 60 min; lane 4, B. pseudomallei SR1015 incubated in PBS; lane 5, SR1015 incubated in 30% NHS for 30 min; lane 6, SR1015 incubated in 30% NHS for 60 min; lane 7, C3b positive control. (C) Lane M, prestained protein molecular mass standards; lane 1, B. pseudomallei 1026b incubated in PBS; lane 2, 1026b incubated in 10% NHS for 30 min; lane 3, 1026b incubated in 10% NHS for 60 min; lane 4, B. pseudomallei SZ210 incubated in PBS; lane 5, SZ210 incubated in 10% NHS for 30 min; lane 6, SZ210 incubated in 10% NHS for 60 min; lane 7, C3b positive control. (D) Lane M, prestained protein molecular mass standards; lane 1, B. pseudomallei 1026b incubated in PBS; lane 2, 1026b incubated in 30% NHS for 30 min; lane 3, 1026b incubated in 30% NHS for 60 min; lane 4, B. pseudomallei SZ210 incubated in PBS; lane 5, SZ210 incubated in 30% NHS for 30 min; lane 6, SZ210 incubated in 30% NHS for 60 min; lane 7, C3b positive control. The apparent molecular masses of the prestained proteins are indicated.
Immunofluorescence microscopy analysis was also performed to demonstrate the difference in C3b deposition between the capsule mutant and the wild type. The same experiment described above was performed, and samples were reacted with the mouse monoclonal antibody to human complement factor C3b, except that the samples were reacted with a secondary antibody conjugated to Cy3 and were stained with DAPI for visualization of bacterial cells. As shown in Fig. 5, the B. pseudomallei capsule mutant SR1015 demonstrated more reactivity to the antibody to human C3b in the presence of serum than the wild-type 1026b. This is evident from the red fluorescence that corresponds to the C3b bound to the bacterial surface surrounding the blue DAPI-stained cells seen when the capsule mutant was incubated in the presence of 10% NHS (Fig. 5D to F). In contrast, the amount of red fluorescence surrounding the DAPI-stained wild-type cells was minimal in the presence of 10% NHS (Fig. 5A to C). There was a dramatic difference in the amount of C3b deposited on the surface of the capsule mutant compared to the wild type, which was detectable after only 15 min of incubation of the bacteria with human serum (Fig. 5B and E). By 60 min, there was some C3b deposition on wild-type B. pseudomallei; however, there was still more C3b deposited on the surface of the capsule mutant (Fig. 5C and F). This experiment was not performed with 30% NHS due to excessive clumping of the samples during the fixation process, which resulted in inconsistent and poor staining of the cells.
FIG. 5.
Immunofluorescence microscopy analysis of decreased complement factor C3b deposition in 10% normal human serum by B. pseudomallei capsule. B. pseudomallei 1026b and SR1015 were incubated in 10% normal human serum (NHS), reacted with a mouse monoclonal antibody to human complement factor C3b, reacted with a rabbit anti-mouse IgG conjugated to Cy3 (Jackson Laboratories), and stained with DAPI for visualization of whole bacterial cells (Sigma). (A) B. pseudomallei 1026b incubated in PBS; (B) 1026b incubated in 10% NHS for 15 min; (C) 1026b incubated in 10% NHS for 60 min; (D) B. pseudomallei SR1015 incubated in PBS; (E) SR1015 incubated in 10% NHS for 15 min; (F) SR1015 incubated in 10% NHS for 60 min. The blue fluorescence indicates the DAPI-stained bacteria, and the red fluorescence indicates the binding of complement factor C3b to the bacterial surface.
Western blot analysis was also performed to determine the amount of complement factor C3b deposition on the surface of B. thailandensis E264, a related nonpathogenic organism. The amount of C3b deposition in B. thailandensis E264 was more pronounced than with B. pseudomallei 1026b and was similar to the amount of C3b deposited on the surface of the capsule mutant, B. pseudomallei SR1015, in the presence of human serum (data not shown). The amount of C3b deposition that occurred on the surface of B. thailandensis was expected, since the organism is known to lack this capsule (24, 25).
Capsule production by B. pseudomallei decreases phagocytosis.
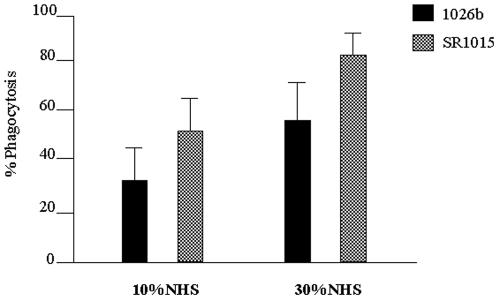
The capsule mutant SR1015 was phagocytosed more significantly by PMNL than the wild-type strain. The proportion of wild-type B. pseudomallei 1026b phagocytosed in the presence of 10% NHS was 35.9%, while the proportion of the capsule mutant SR1015 phagocytosed was 51.7% (P < 0.001) (Fig. 6). When each strain was incubated in the presence of 30% NHS, 59.3% of the wild-type strain 1026b was phagocytosed by the PMNL after 30 min compared to 82.3% for the capsule mutant (P < 0.001) (Fig. 6).
FIG. 6.
Capsule reduces opsonophagocytosis of B. pseudomallei by PMNL. Bacteria labeled with [14C]lysine were incubated with human PMNL in the presence of 10 or 30% NHS and subjected to differential centrifugation. Samples were counted in a Beckman LS 6500 scintillation counter, and the percentage of the bacterial population phagocytosed was calculated by the following equation: percent uptake = [counts per minute following differential centrifugation (cell associated)/counts per minute in noncentrifuged tubes (cell and non-cell associated)] × 100. The data represent the means and the standard deviations from three separate experiments, each performed in triplicate. The difference in opsonophagocytosis between B. pseudomallei 1026b and SR1015 was determined to be significant at both 10% NHS (P < 0.001) and 30% NHS (P < 0.001).
B. pseudomallei capsule expression is elevated in the presence of 30% normal human serum.
The lux reporter strain B. pseudomallei SZ211 was constructed by cloning an internal fragment of the wcbB gene into pGSV3-lux, a suicide vector containing the lux operon from Photorhabdus luminescens (Table 1). Absorbance (OD540) and luminescence (in relative light units) measurements were taken every 2 h. As shown in Fig. 7A, capsule expression was higher in the presence of M9 plus 1% glucose plus 30% NHS and M9 plus 1% glucose plus 30% HI-NHS than was growth in M9 plus 1% glucose alone. The increase in light production of SZ211 in the presence of serum supports the requirement for capsule for survival in serum.
FIG. 7.
B. pseudomallei capsule and LPS expression is elevated in the presence of 30% normal human serum. B. pseudomallei lux reporter strains SZ211 (A) and SZ213 (B) were grown in M9 plus 1% glucose, in M9 plus 1% glucose plus 30% NHS, or in M9 plus 1% glucose plus 30% HI-NHS. Absorbance (OD540) and luminescence (in relative light units [rlu]) were measured at each time point. Capsule (SZ211) and LPS (SZ213) expression was determined by normalizing light production for growth (rlu/OD540) of each strain. The data represent the average amount of lux reporter activity normalized for growth and standard deviation from three separate experiments.
The strain B. pseudomallei SZ213 was constructed by cloning an internal region of the wbiA gene, which encodes an O-acetyltransferase required for O-acetylation of the O-PS component of B. pseudomallei LPS (12). Light production of this strain was measured under the same conditions to determine whether LPS expression was induced in the presence of serum. Similar to the capsule, LPS expression was elevated in the presence of both 30% NHS and 30% HI-NHS (Fig. 7B). The levels of expression of both capsule and LPS were not significantly different in NHS and HI-NHS, suggesting that the environment of the serum may be required for induction of gene expression rather than complement.
DISCUSSION
We have previously demonstrated that a capsular polysaccharide of B. pseudomallei is essential for the pathogenesis of B. pseudomallei (25). In this study, we have provided evidence that this capsule is important for B. pseudomallei survival in the host, as it limits the effectiveness of opsonization and phagocytosis, leading to decreased clearance of the organism from the blood.
Capsule production by B. pseudomallei contributes to persistence of the organisms in the blood by promoting evasion of the complement cascade. The addition of purified capsule was shown to increase the virulence of a capsule-minus mutant strain in the Syrian hamster model of acute melioidosis. The numbers of wild-type B. pseudomallei bacteria were considerably higher in the blood of infected hamsters than those of capsule-minus mutant strain bacteria by 48 h. Capsule expression was shown to be induced in 30% normal human serum. The addition of purified capsule to serum bactericidal assays showed that the capsule contributes to increased resistance of serum-sensitive strains lacking the O-PS moiety of LPS to the bactericidal effects of normal human serum. The addition of purified capsule increased the survival of a B. pseudomallei serum-sensitive strain by 1,000-fold. Incubation of 30% NHS with 100 μg of capsule for 30 min before the addition of bacteria resulted in a 100,000-fold increase in survival of the serum-sensitive strain. The survival of the serum-sensitive strain under these conditions was similar to its survival in heat-inactivated serum, suggesting that the capsule was affecting the complement cascade. This effect was found to be specific to capsule, since the addition of purified B. pseudomallei O-PS did not increase the survival of the serum-sensitive strain. In addition, serum bactericidal assay results showed that double mutants in capsule and O-PS were more serum sensitive than mutants lacking only O-PS, indicating the importance of capsule in preventing the bactericidal effects of complement (data not shown).
Capsular polysaccharides are often shed during the growth of the organism, as well as during complement attack, which serves to prevent complement attack at the bacterial surface. In addition, surface molecules released into the environment may act to deploy complement activation away from the intact organism (10, 19). Shedding of the capsule may have a role in preventing complement attack against invading B. pseudomallei bacteria. There is evidence to suggest that the 200-kDa capsular polysaccharide of B. pseudomallei is shed into the culture medium during growth (1). Immunofluorescence microscopy studies performed by our laboratory using polyclonal rabbit sera specific to the capsule demonstrated that when purified B. pseudomallei capsule is added exogenously, it does not reassociate with the bacterial cell (data not shown). This could explain the ability of exogenous capsule to inhibit complement attack and enhance the survival of the serum-sensitive strain in 30% normal human serum.
Since capsule mutants of B. pseudomallei were previously found to be serum resistant, we postulated that the capsule enhanced B. pseudomallei survival in the blood by inhibiting C3b deposition and opsonization. Both Western blot analysis and immunofluorescence microscopy experiments using a mouse monoclonal antibody to human C3b demonstrated decreased C3b deposition by the B. pseudomallei capsule. In both experiments, C3b deposition was more pronounced on the surface of the capsule mutant strains than on the wild type. Also evident from Western blot analysis and immunofluorescence microscopy was the fact that some C3b deposition occurred in wild-type B. pseudomallei. These results indicate that the B. pseudomallei capsule does not completely prevent the association of C3b with the bacterial surface but that the presence of the capsule reduces the amount of C3b deposited. This can be expected, since bacterial capsules are known to allow the diffusion of some C3b molecules through to the bacterial surface (27). Furthermore, previous studies have shown that B. pseudomallei is capable of activating the alternative pathway of complement, and this results in the assembly of complement factors C5b to C9, the MAC, on the surface of the invading organism (14, 12). The MAC can still form on a bacterial surface that is covered by a capsule due to the diffusion of some complement proteins. Therefore, capsule production by B. pseudomallei does not completely prevent complement activation, but it reduces the effects of complement through the inhibition of C3b deposition and opsonization of the organisms for effective clearance. This would explain the increased clearance of capsule mutants from the blood of infected animals compared to clearance of the wild type. This conclusion is supported by the fact that B. thailandensis, a related nonpathogenic organism which lacks the capsule, has been shown to be serum resistant but is not capable of establishing bacteremia in the Syrian hamster model of acute melioidosis (4, 12, 25).
Effective opsonization of invading bacteria results in enhanced phagocytosis and clearance of the organisms from the blood of the infected host (18). Quantitative radiolabeled phagocytosis assays were performed to establish a correlation between opsonization of the bacteria and phagocytosis by polymorphonuclear leukocytes. In the presence of serum, the capsule mutant was phagocytosed more effectively than wild-type B. pseudomallei 1026b. From these experiments, we were able to conclude that the B. pseudomallei capsule is antiphagocytic in that it inhibits opsonization for effective phagocytosis of the bacteria. In addition, the capsule may act as a barrier, blocking access of the CR1 receptor on phagocytes to the C3b deposited on the bacterial surface (10). Verbrugh et al. showed that a capsule-producing strain of Staphylococcus aureus incubated with normal human serum had C3 localized on the cell wall beneath the capsular layer (30). These researchers determined that the capsule was antiphagocytic because it interfered with the recognition of cell wall-bound C3b and iC3b molecules by phagocytic cell receptors. Further experiments are necessary to determine whether the B. pseudomallei capsule has a dual function of blocking phagocytosis through the CR1 or CR3 receptors, as well as inhibiting opsonization of the bacteria.
Capsule production by B. pseudomallei contributes to the persistence of the organism in the blood through the reduction of complement factor C3b deposition and phagocytosis. The presence of the capsule enables B. pseudomallei to survive in the blood and spread to other organs, which may result in the overwhelming septicemia that is common in culture-positive melioidosis patients (29). Therefore capsule production is important for the pathogenesis of acute septicemic melioidosis. Further characterization of the B. pseudomallei capsule is essential for understanding the pathogenesis of B. pseudomallei infections and the development of preventive strategies for the treatment of melioidosis.
Acknowledgments
This work was supported by the Canadian Institutes for Health Research MOP 36343 and the Canadian Bacterial Diseases Networks of Centers of Excellence.
We thank Paul Brett for preparing the B. pseudomallei capsular polysaccharide and O-polysaccharide preparations and Marina Tom for excellent technical assistance.
Editor: J. T. Barbieri
REFERENCES
- 1.Anuntagool, N., T. Panichakul, P. Aramsri, and S. Sirisinha. 2000. Shedding of lipopolysaccharide and 200-kDa surface antigen during the in vitro growth of virulent Ara− and avirulent Ara+ Burkholderia pseudomallei. Acta Trop. 74:221-228. [DOI] [PubMed] [Google Scholar]
- 2.Boulnois, G. J., and I. S. Roberts. 1990. Genetics of capsular polysaccharide production in bacteria. Curr. Top. Microbiol. Immunol. 150:1-18. [DOI] [PubMed] [Google Scholar]
- 3.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 4.Brett, P. J., D. DeShazer, and D. E. Woods. 1997. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol. Infect. 118:137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., description of a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, L. E., S. Wong, D. E. Woods, D. A. Dance, and W. Chaowagul. 1994. Passive protection of diabetic rats with antisera specific for the polysaccharide portion of the lipopolysaccharide isolated from Pseudomonas pseudomallei. Can. J. Infect. Dis. 5:170-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaowagul, W., N. J. White, D. A. Dance, Y. Wattanagoon, P. Naigowit, T. M. Davis, S. Looareesuwan, and N. Pitakwatchara. 1989. Melioidosis: a major cause of community-aquired septicemia in northeastern Thailand. J. Infect. Dis. 159:890-899. [DOI] [PubMed] [Google Scholar]
- 8.Dance, D. A. B. 1990. Melioidosis. Rev. Med. Microbiol. 1:143-150. [Google Scholar]
- 9.Dance, D. A. B. 1991. Melioidosis: the tip of the iceberg? Clin. Microbiol. Rev. 4:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Densen, P. 1995. Complement, p. 58-78. In G. L. Mandell, J. E. Bennett, and R. Dolin (ed.), Principles and practice of infectious diseases. Churchill Livingstone, New York, N.Y.
- 11.DeShazer, D., P. J. Brett, R. Carylon, and D. E. Woods. 1997. Mutagenesis of Burkholderia pseudomallei with Tn5-OT182: isolation of motility mutants and molecular characterization of the flagellin structural gene. J. Bacteriol. 179:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeShazer, D., P. J. Brett, and D. E. Woods. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30:1081-1100. [DOI] [PubMed] [Google Scholar]
- 13.DeShazer, D., and D. E. Woods. 1999. Animal models of melioidosis, p. 199-203. In O. Zak and M. Sande (ed.), Handbook of animal models of infection. Academic Press, London, England.
- 14.Egan, A. M., and D. L. Gordon. 1996. Burkholderia pseudomallei activates complement and is ingested but not killed by polymorphonuclear leukocytes. Infect. Immun. 64:4952-4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Figurski, D. H., and D. R. Helsinki. 1979. Replication of an origin-containing derivative of plasmid RK2 dependant on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ip, M., L. G. Osterberg, P. Y. Chau, and T. A. Raffin. 1995. Pulmonary melioidosis. Chest 108:1420-1424. [DOI] [PubMed] [Google Scholar]
- 17.Ismail, G., N. Razak, R. Mohamed, N. Embi, and O. Omar. 1988. Resistance of Pseudomonas pseudomallei to normal human serum bactericidal action. Microbiol. Immunol. 32:645-652. [DOI] [PubMed] [Google Scholar]
- 18.Joiner, K. A., E. J. Brown, and M. M. Frank. 1984. Complement and bacteria: chemistry and biology in host defense. Annu. Rev. Microbiol. 2:461-491. [DOI] [PubMed] [Google Scholar]
- 19.Joiner, K. A. 1988. Complement evasion by bacteria and parasites. Annu. Rev. Microbiol. 42:201-230. [DOI] [PubMed] [Google Scholar]
- 20.Leelarasamee, A., and S. Bovornkitti. 1989. Melioidosis: review and update. Rev. Infect. Dis. 11:413-425. [DOI] [PubMed] [Google Scholar]
- 21.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. The contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perry, M. B., L. L. MacLean, T. Schollaardt, L. E. Bryan, and M. Ho. 1995. Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect. Immun. 63:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perry, M. Personal communication.
- 25.Reckseidler, S. L., D. DeShazer, P. A. Sokol, and D. E. Woods. 2001. Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69:34-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosse, W. F. 1988. The complement system, p. 204-210. In W. K. Jolik, H. P. Willett, D. E. Amos, and C. M. Wilfert (ed.), Zinsser microbiology. Appleton & Lange, Norwalk, Conn.
- 27.Salyers, A. A., and D. D. Whitt. 2002. Bacterial pathogenesis: a molecular approach, 2nd ed., p. 115-130. ASM Press, Washington, D.C.
- 28.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 29.Suputtamongkol, Y., A. Rajchanuwong, W. Chaowagul, D. A. B. Dance, M. D. Smith, V. Wuthiekanun, A. L. Walsh, S. Pukrittayakamee, and N. J. White. 1994. Ceftazadime vs. amoxicillin/clavulanate in the treatment of severe melioidosis. Clin. Infect. Dis. 19:846-853. [DOI] [PubMed] [Google Scholar]
- 30.Verbrugh, H. A., P. K. Peterson, B.-Y. T. Nguyen, S. P. Sisson, and Y. Kim. 1982. Opsonization of encapsulated Staphylococcus aureus: the role of specific antibody and complement. J. Immunol. 129:1681-1687. [PubMed] [Google Scholar]
- 31.Vogel, U., A. Weinberger, R. Frank, A. Müller, J. Köhl, J. P. Atkinson, and M. Frosch. 1997. Complement factor C3 deposition and serum resistance in isogenic capsule and lipooligosaccharide sialic acid mutants of serogroup B Neisseria meningitidis. Infect. Immun. 65:4022-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White, N. J., D. A. B. Dance, W. Chaowagul, Y. Wattanagoon, V. Wuthiekanun, and N. Pitakwatchara. 1989. Halving of mortality of severe melioidosis by ceftazadime. Lancet 2:697-701. [DOI] [PubMed] [Google Scholar]
- 33.Woods, D. E., A. L. Jones, and P. J. Hill. 1993. Interaction of insulin with Pseudomonas pseudomallei. Infect. Immun. 61:4045-4050. [DOI] [PMC free article] [PubMed] [Google Scholar]