Abstract
IMPORTANCE
Whole-exome sequencing (WES) has the potential to reveal tumor and germline mutations of clinical relevance, but the diagnostic yield for pediatric patients with solid tumors is unknown.
OBJECTIVE
To characterize the diagnostic yield of combined tumor and germline WES for children with solid tumors.
DESIGN
Unselected children with newly diagnosed and previously untreated central nervous system (CNS) and non-CNS solid tumors were prospectively enrolled in the BASIC3 study at a large academic children’s hospital during a 23-month period from August 2012 through June 2014. Blood and tumor samples underwent WES in a certified clinical laboratory with genetic results categorized on the basis of perceived clinical relevance and entered in the electronic health record.
MAIN OUTCOMES AND MEASURES
Clinical categorization of somatic mutations; frequencies of deleterious germline mutations related to patient phenotype and incidental medically-actionable mutations.
RESULTS
Of the first 150 participants (80 boys and 70 girls, mean age, 7.4 years), tumor samples adequate for WES were available from 121 patients (81%). Somatic mutations of established clinical utility (category I) were reported in 4 (3%) of 121 patients, with mutations of potential utility (category II) detected in an additional 29 (24%) of 121 patients. CTNNB1 was the gene most frequently mutated, with recurrent mutations in KIT, TSC2, and MAPK pathway genes (BRAF, KRAS, and NRAS) also identified. Mutations in consensus cancer genes (category III) were found in an additional 24 (20%) of 121 tumors. Fewer than half of somatic mutations identified were in genes known to be recurrently mutated in the tumor type tested. Diagnostic germline findings related to patient phenotype were discovered in 15 (10%) of 150 cases: 13 pathogenic or likely pathogenic dominant mutations in adult and pediatric cancer susceptibility genes (including 2 each in TP53, VHL, and BRCA1), 1 recessive liver disorder with hepatocellular carcinoma (TJP2), and 1 renal diagnosis (CLCN5). Incidental findings were reported in 8 (5%) of 150 patients. Most patients harbored germline uncertain variants in cancer genes (98%), pharmacogenetic variants (89%), and recessive carrier mutations (85%).
CONCLUSIONS AND RELEVANCE
Tumor and germline WES revealed mutations in a broad spectrum of genes previously implicated in both adult and pediatric cancers. Combined reporting of tumor and germline WES identified diagnostic and/or potentially actionable findings in nearly 40% of newly diagnosed pediatric patients with solid tumors.
Genome-scale sequencing methods such as whole-exome sequencing (WES)have provided significant insight into the pathogenesis of cancer as well as a wide spectrum of human diseases.1 Recent studies have begun to establish the yield of WES for the diagnosis of Mendelian disorders and the discovery of incidental (non-phenotype related) medically actionable findings.2–4 However, experience with the use of these tests in the care of patients with cancer remains limited.
Sequencing of tumor and matched normal samples can reveal multiple types of results with implications for clinical practice. The identification of somatic (tumor-specific) mutations has the potential to offer diagnostic and prognostic information and inform selection of therapies, particularly for patients with relapsed or refractory tumors.5 Detection of germline mutations in cancer susceptibility genes may prompt further genetic testing and guide cancer surveillance strategies for both the patient and family members.6 Germline mutations may also explain noncancer phenotypes, predict drug responses, or provide reproductive counseling information for parents (Figure 1).7–11
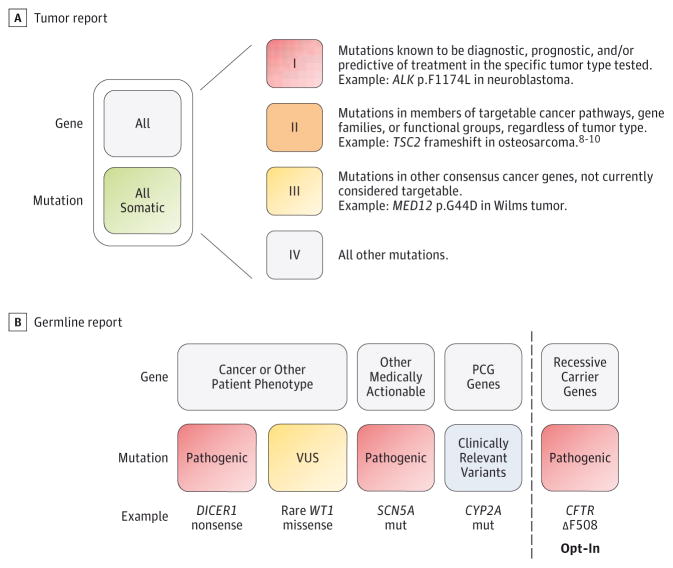
Figure 1. Categories of Mutation Results Included in Clinical Tumor and Germline Whole-Exome Sequencing Reports.
Reported categories of somatic mutations (A) and germline mutations (B) are shown. Germline testing for autosomal recessive carrier mutations was performed if requested by the parents at the time of study enrollment. PCG indicates pharmacogenetic; VUS, variant of uncertain significance. The image in panel B was adapted from the study by Scollon et al,7 an open access article. For more information on the categorization of TSC2 frameshift as a category II mutation, see Perry et al,8 Krueger et al,9 and Iyer et al.10
The BASIC3 (Baylor College of Medicine Advancing Sequencing in Childhood Cancer Care) study is a Clinical Sequencing Exploratory Research (CSER) consortium project funded by the National Genome Human Research Institute (NHGRI) and National Cancer Institute (NCI) focusing on prospective implementation of clinical WES in the pediatric oncology clinic.12 To characterize the diagnostic yield of combined tumor and germline WES for children with newly diagnosed solid tumors, we report here the results for 150 consecutive BASIC3 study patients sequenced in a laboratory certified by the College of American Pathologists (CAP) and Clinical Laboratory Improvement Act (CLIA).
Methods
Study Subjects and Data Collection
The BASIC3 study was approved by the institutional review board of Baylor College of Medicine. Study enrollment was offered to all patients having diagnostic tumor surgery or biopsy at Texas Children’s Hospital for newly diagnosed and previously untreated central nervous system (CNS) and non-CNS solid tumors who had at least 1 English- or Spanish-speaking parent and planned ongoing oncology care at Texas Children’s Cancer Center.7 Written informed consent was obtained for all study participants. Patients with benign non-CNS tumors not requiring ongoing oncology care were excluded.
At study entry, the study medical geneticist or genetic counselor reviewed the age, cancer diagnosis, medical record (including family history), and parental surveys and determined whether clinical genetic testing would be considered as part of routine care, and if so, which specific test(s) would be used. The Texas Children’s Hospital electronic health record was reviewed for all patients as of April 1, 2015, for any germline genetic or molecular tumor testing ordered.
Key Points.
Question
What is the feasibility and diagnostic yield of clinical tumor and germline whole-exome sequencing (WES) in an unselected cohort of children with newly diagnosed solid tumors of both the central nervous system and elsewhere?
Findings
Somatic mutations of established or potential clinical utility were identified in 27% of patients, and pathogenic or likely pathogenic germline mutations in adult and pediatric cancer susceptibility genes were identified in more than 8%of patients. Combined reporting of tumor and germline WES identified diagnostic and/or potentially actionable findings in nearly 40% of newly diagnosed pediatric patients with solid tumors.
Meaning
This study highlights the diversity of tumor and germline results detectable by clinical WES, including many genes not previously associated with the specific pediatric tumor diagnosed in the patient.
Study Samples
Peripheral blood samples were obtained from all study patients and requested from parents. Fresh tumor from pretreatment diagnostic specimens, if available, was snap frozen for the study by the consultant pathologist. A confirmatory pathology review was conducted for each tumor sample. Study genetic counselors completed a clinical WES requisition form for each patient including tumor diagnosis, other medical diagnoses, and family history.
Whole-Exome Sequencing
Whole-exome sequencing was performed in the CAP- and CLIA-certified Whole Genome Laboratory at Baylor College of Medicine following a previously described protocol,2,3 including library construction, exome capture by VCRome, version 2.113 (Roche NimbleGen), and paired-end sequencing on an Illumina HiSeq instrument (Illumina Inc). Tumor and germline library pairs were sequenced on a single lane of a HiSeq 2000/2500, with a mean coverage of 272× and a target base coverage of 20× at 97.3%. Germline-only samples were run 3 per lane.
Variant Calling and Annotation
Data analysis for germline samples was performed as previously described.2,3 Tumor exomes were similarly analyzed using the Mercury v.3 pipeline,14 including variant calling and annotation (eFigure 1 in Supplement 1). A somatic variant list was generated for each patient by subtracting the germline .vcf from the tumor .vcf and applying somatic filters (eTable 1 in Supplement 2) to the merged file with a tumor variant ratio cut-off of 0.05 or greater. Somatic mutations were annotated with information from the Catalog of Somatic Mutations in Cancer (COSMIC) database.15,16 A FamCan (Familial Cancer) variant list was also populated with germline variants in a list of cancer predisposition syndrome genes (eTable 2 in Supplement 2) to allow detection of biallelic mutations (second hits) in the tumor or loss of heterozygosity (LOH) in autosomal dominant cancer genes. An increase in variant allele fraction of 0.20 in the tumor was considered evidence of LOH.17
Data Interpretation, Variant Confirmation, and Clinical Reporting
Germline variants remaining after filtering were classified and interpreted in the context of patient phenotype(s) to generate a germline WES report, as previously described.2,3 For the present study, these reports included variants confirmed by Sanger sequencing in 4 categories (Figure 1):pathogenic or likely pathogenic mutations in disease genes related to cancer susceptibility or other patient phenotype; variants of uncertain significance (VUS) in phenotype-associated disease genes; incidental medically actionable mutations (including the mitochondrial genome); and a limited panel of pharmacogenetic variant alleles.3 Parents were given the option to have their child’s report include carrier status results for any recessive disorder for which genetic testing is currently available. Parental blood samples, if available, underwent Sanger sequencing for specific germline mutations identified in the patient sample. Germline WES reports did not include disease-associated mutations that were not considered actionable in the absence of a phenotype.
Somatic and FamCan variants were imported into an in house-developed variant review program and subjected to additional filters (eFigure 1 in Supplement 1 and eTable 1 in Supplement 2) followed by inspection of read alignments on the Integrative Genomics Viewer.18 An algorithm for variant ranking based on perceived clinical utility was used to assign somatic mutations to 1 of 4 categories (Figure 1 and eTables 3 to 5 in Supplement 2): established clinical utility (I); potential clinical utility (II); mutations in consensus cancer genes (III); and all other mutations (IV). For a more detailed description of the algorithm see eMethods in Supplement 1. Category I to III somatic mutations were confirmed by Sanger sequencing and reported with accompanying interpretative text.
Statistical Analysis
The Fisher exact test was used to compare the frequencies of category I and II somatic mutations in CNS vs non-CNS tumors and to compare the frequencies of cancer susceptibility variants between study participants and a cohort of 2000 consecutive germline exomes reported from the Baylor College of Medicine Whole Genome Laboratory.3
Results
Patient Characteristics and Samples
Study enrollment began in August 2012. Two hundred eligible patients were identified and offered study enrollment following a previously reported informed consent process,7 of whom 150 (75%) agreed to participate in the study (eFigure 2 in Supplement 1). Patient clinical characteristics and demographics are summarized in eTable 6 in Supplement 2. Enrolled patients were diagnosed with a diverse representation of pediatric CNS (n = 56, 37%) and non-CNS (n = 94, 63%) solid tumors (Table 1). Blood samples were collected from all participants, and adequate tumor samples for WES were obtained from 121 (81%) of the 150 participants, including 40 (71%) of 56 with CNS tumors and 81 (86%) of 94 with non-CNS tumors (eFigure 2 in Supplement 1). The last WES results described here were reported in September 2014 (n = 150 germline reports and n = 121 tumor reports).
Table 1.
Tumor Diagnoses of Study Participants
| Tumor Type | Participants, No. |
|---|---|
| CNS tumors | 56 |
| Medulloblastoma | 11 |
| Low-grade glioma | 11 |
| Ependymoma | 9 |
| Glioneuronal tumor | 5 |
| High-grade glioma | 5 |
| Choroid plexus tumor | 4 |
| Atypical teratoid rhabdoid tumor | 2 |
| Germ cell tumor | 2 |
| Meningioma | 2 |
| Pineoblastoma | 2 |
| Other tumor types | 3 |
| Non-CNS tumors | 94 |
| Neuroblastoma | 19 |
| Wilms tumor | 15 |
| Germ cell tumor | 13 |
| Rhabdomyosarcoma | 9 |
| Soft tissue sarcoma | 7 |
| Ewing sarcoma | 6 |
| Osteosarcoma | 4 |
| Hepatoblastoma | 3 |
| Mucoepidermoid carcinoma | 3 |
| Adrenocortical carcinoma | 2 |
| Hepatocellular carcinoma | 2 |
| Pheochromocytoma | 2 |
| Other tumor types | 9 |
Abbreviation: CNS, central nervous system.
Tumor WES
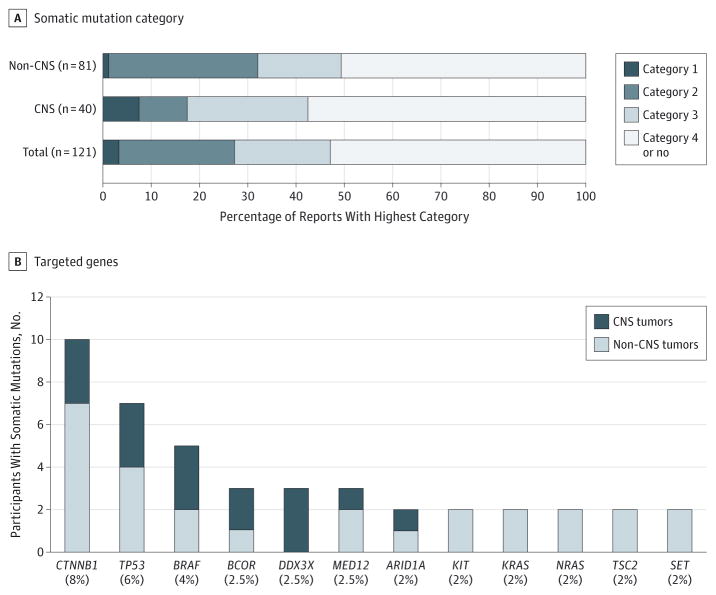
Aggregating the results from 121 tumor WES reports revealed validated somatic mutations that were designated as having established clinical utility (category I) in 4 patients (3%) and potential utility (category II) in an additional 29 patients (24%) (Figure 2A and eTables 7 and 8 in Supplement 2). The fraction of patients with CNS tumors who had category I and II mutations (7 of 40, 17%)was numerically but not statistically lower than that for non-CNS tumors (26 of 81, 32%) (Figure 2A, P = .13). The 37 recurrent category I and II mutations detected included CTNNB1 (in 10 of 121 participants, 8.3%), BRAF (5 of 121, 4.1%), KIT, KRAS, NRAS, and TSC2 (2 of 121 each, 1.7%) (Figure 2B and eTable 7 in Supplement 2). Only 4 (11%) of these 37 mutations were previously detected by routine clinical molecular testing: an ALK hotspot mutation in a neuroblastoma and BRAF V600E mutations in 3 gliomas. An equal number were identified in genes that are now becoming routinely tested in the patient’s tumor type (CTNNB1 in 3 medulloblastomas and H3F3A in a glioblastoma) although not at the time of diagnosis.19 A total of 25 of 37 category I or II mutations (68%) were either oncogene alterations previously reported in the COSMIC database or inactivating mutations in tumor suppressor genes (eTable 7 in Supplement 2).15
Figure 2. Somatic Mutations Detected Among Study Participants.
A, Highest category of somatic mutation per participant identified by tumor whole-exome sequencing (WES). Data for the subsets of children with central nervous system (CNS) tumors (n = 40) and non-CNS tumors (n = 81) are shown in addition to the entire patient cohort with tumor samples available (n = 121). I indicates category I mutation; II, category II mutation; III, category III mutation; IV, category IV or no mutation. B, Genes targeted by recurrent somatic mutations in patients for whom tumor WES was performed.
The tumor reports of an additional 24 of 121 patients (20%) included at least 1 somatic mutation in a consensus cancer gene (category III), most frequently TP53 (7 of 121, 6%), BCOR, DDX3X, and MED12 (3 of 121 each, 2.5%), while 64 (53%) of 121 patients had no category I to III mutations reported (Figure 2; eTables 7 and 8 in Supplement 2). Only 22 (59%) of 37 category I and II mutations and 18 (33%) of 54 (33%) category III mutations were in genes known to be mutated at greater than 2% frequency in the tumor type tested; for example, BRAF, JAK3, KRAS, MET, and TSC2 mutations are all rare events in neuroblastoma.15,20,21 A total of 1111 mutations in category IV genes were also identified by WES, but not further validated, in 112 (93%) of 121 patients.
Germline WES
Pathogenic or likely pathogenic mutations underlying the patient’s phenotype were identified in 15 (10%) of 150 patients (Table 2 and eTable 9 in Supplement 2). For 8 (62%) of 13 patients with dominant cancer susceptibility mutations, the gene in which the mutation was reported was known to be associated with childhood cancer risk (VHL × 2, TP53 × 2, DICER1, MSH2, WT1, and KRAS). However, in 3 of these 8 cases, the clinical team caring for the child had not considered genetic testing because (1) the positive family history suggestive of Lynch syndrome was not elicited by the team caring for the child with glioblastoma multiforme and MSH2 mutation; (2) the clinical features had not previously resulted in a Noonan syndrome diagnosis in the child with KRAS mutation; and (3) 1 child with unilateral Wilms tumor and no congenital anomalies or developmental delay was mosaic for WT1 mutation. In addition, the remaining 5 patients with dominant mutations had tumor diagnoses not previously associated with the mutated gene(BRCA1 × 2, BRCA2, SMARCA4, and CHEK2). There was also a single autosomal recessive diagnosis (TJP2 deficiency) in a patient with severe liver disease and hepatocellular carcinoma (HCC). This case (in addition to another concurrent finding at another institution) was the first report of an association of TJP2 deficiency and HCC.22 Finally, 1 patient with previously unexplained proteinuria was found to carry an X-linked CLCN5 mutation. Parental samples were available for testing in 13 of these 15 diagnostic cases. Sanger validation revealed the diagnostic mutation to be inherited from a parent in 11 (85%); the only exceptions were the mosaic WT1 mutation and de novo KRAS mutation.
Table 2.
Diagnostic Germline Findings Related to Patient Phenotype
| Gene Mutation | Tumor Diagnosis and Relevant Medical History | Family History at Study Entry | Inherited From Parent | Tumor LOH |
|---|---|---|---|---|
| Heterozygous Mutation in Autosomal Dominant Disorder Associated With the Specific Childhood Cancer (n = 8) | ||||
|
WT1 (mosaic) c.865_867delinsAA, p.Y289fs |
Wilms tumor | None | No | Yes |
|
DICER1 c.2062C>T, p.R688X |
Pulmonary pleuroblastoma | Multinodular thyroid disease | Yes | ND |
|
VHL c.499C>T, p.R167W |
Pheochromocytoma | None | Yes | Yes |
|
VHL c.499C>T, p.R167W |
Pheochromocytoma | Von-Hippel-Lindau | Yes | No |
|
MSH2 c.1697delA, p.K566fs |
Glioblastoma | Lynch syndrome tumors | Yes | ND |
|
TP53 c.743G>A, p.R248Q |
Neuroblastoma | Li-Fraumeni syndrome | Yes | Yes |
|
TP53 (likely pathogenic) c.470T>C, p.V157A |
Adrenocortical carcinoma | Adult cancers | ND | Yes |
|
KRAS (likely pathogenic) c.194G>T, p.S65I |
Plexiform neurofibroma; multiple anomalies | None | No | No |
| Heterozygous Mutation in Autosomal Dominant Disorder Not Previously Associated With the Specific Childhood Cancer (n = 5) | ||||
|
SMARCA4 c.1156_1157del, p.E386fs |
Neuroblastoma | None | ND | Yes |
|
BRCA1 c.68_69delAG, p.E23fs |
Neuroblastoma | Breast cancer | Yes | No |
|
BRCA1 c.697_698del, p.V233fs |
Anaplastic medulloblastoma | Breast cancer | Yes | No |
|
BRCA2 c.1278delA, p.D427fs |
Ewing sarcoma; short stature, anemia | Breast and ovarian cancer | Yes | No |
|
CHEK2 c.1100delC, p.T367fs |
Wilms tumor | None | Yes | No |
| Autosomal Recessive Diagnosis Not Previously Associated With the Specific Childhood Cancer (n = 1) | ||||
|
TJP2 (homozygous) c.817delG, p.A273fsa |
Hepatocellular cancer, severe liver disease | None | Yes | No |
| Mutation in X-Linked Recessive Disorder Associated With Nononcologic Disorder (n = 1) | ||||
|
CLCN5 c.1466G>A, p.W489X |
Ependymoma; proteinuria | Renal disease, hypertension | Yes | ND |
Abbreviations: HCC, hepatocellular carcinoma; LOH, loss of heterozygosity; ND, not determined.
Additional clinical description of this patient with liver disease and HCC is reported by Zhou et Al.22
An additional 10 participants (6%) were found to have a single pathogenic mutation in a gene associated with an autosomal recessive cancer syndrome(eTable 10 in Supplement 2), including several genes in the Fanconi anemia pathway, with no further evidence of the disorder noted by the oncologist or found in the medical record. Of this group, only 1 participant, with Wilms tumor and a single DIS3L2 mutation, had the tumor type most commonly associated with the recessive condition, Perlman syndrome.23
We further explored the possibility of second events, either somatic mutation or LOH in the tumor WES data of patients with germline findings (Table 2; eTables 9 and 10 in Supplement 2). Nosomatic mutations in the same gene were detected. However, tumor LOH was documented in 6 patients. Of note, there was no evidence of LOH in the 3 tumors with germline BRCA1 or BRCA2 mutations or any of the single recessive cancer mutations other than LOH of DIS3L2 in the Wilms tumor sample.
Finally, we performed comparisons of some of the recurring mutations found in the BASIC3 cohort with 2000 clinical exomes reported3 by our same testing laboratory in a primarily pediatric noncancer clinical population. There was only a modest increased rate of BRCA1/2 mutations in the BASIC3 cohort (3 of 150, 2% vs 14 of 2000, 0.7%; P = .10). Summing across all the 14 Fanconi genes, we found no evidence that putative loss-of-function mutations were enriched in the BASIC3 cohort (P > .99). However, 1 recurrent FANCL frameshift mutation was found in 0.5% of the 2000 exome data set vs 3 of 150 among the BASIC3 participants (P = .19); thus, evaluation of the significance of this finding requires a larger data set.
Twelve patients had clinical germline gene testing outside of WES (the majority being TP53 comprehensive analysis) with no additional sequence-based mutations reported. The WES report does not include deletions, and a large deletion of the SMARCB1 gene was reported in a single patient with an atypical teratoid/rhabdoid tumor of the CNS. In addition, WES revealed the germline nature of a TP53 mutation previously reported from a tumor mutation panel.
Medically actionable incidental findings reported included those genes recommended by the American College of Medical Genetics and Genomics (ACMG)24 and additional findings (including mitochondrial DNA), as recently described.2,3 Eight (5%) of 150 patients had incidental findings, including 5 in genes on the ACMG list (eTable 11 in Supplement 2). Two findings were specifically reported as being actionable for an oncology patient: an immunodeficiency allele and a mitochondrial DNA allele associated with aminoglycoside toxic effects. The remaining patient was diagnosed incidentally with the MELAS mitochondrial disorder (mitochondrial encephalomyopathy, lactic acidosis, and strokelike episodes).
The germline exome report includes 3 additional categories. The laboratory reports VUS in genes related to the patient’s phenotype, which included the cancer susceptibility genes listed in eTable 2 in Supplement 2. One hundred forty-four (96%) of 150 patients had at least 1 (median of 3) VUS on their report, which is consistent with previous reports of 2VUS per study participants using a much smaller gene panel.25 In addition, the WES reports include the common variants recommended by the US Food and Drug Administration for dosing in VKORC1/CYP2C9 (altered warfarin metabolism) and CYP2C19 (altered Plavix metabolism), with 133 (89%)of 150 patients having at least 1 pharmacogenetic variant reported (median of 1). Finally, parents are provided the option of reporting of carrier status (pathogenic mutations only) for the very large number of autosomal recessive disorders currently available for single gene testing. Of the 137 probands whose parents requested reporting (91%), 115 (84%) had at least 1 recessive carrier finding (median of 2).
Clinical Yield of Tumor and Germline Whole Exome Sequencing
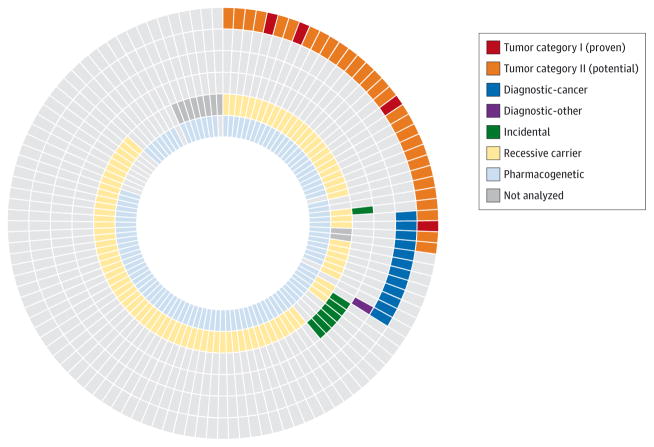
In combination, tumor and germline WES revealed potentially clinically relevant alterations (category I or II somatic mutations, pathogenic germline mutations in genes related to cancer or other patient phenotype [excluding single recessive mutations], or medically actionable incidental germline mutations) in 47 (39%) of 121 patients (Figure 3).
Figure 3. Combined Yield of Tumor and Germline Whole-Exome Sequencing (WES).
Mutations with known or potential clinical implications identified by tumor and germline WES of children with newly diagnosed solid tumors. Each slice represents a different patient undergoing both tumor and germline sequencing (n = 121), and each concentric ring a category of somatic or germline mutation.
Discussion
Here we report that clinical tumor WES in children with newly diagnosed solid tumors revealed somatic mutations of established or potential clinical utility (category I and II) in approximately one-quarter of patients and mutations in cancer genes not classified as targetable (category III) in a similar number (Figure 2A and eTable 8 in Supplement 2). The vast majority of these mutations had not been detected by targeted molecular testing as part of routine clinical care. Consistent with this current practice, tumor WES revealed somatic mutations of established clinical utility (category I) in only a small minority of patients, with far more tumors harboring mutations of potential clinical utility (category II). To our knowledge, this is the first study to investigate the diagnostic yield of WES in the wide spectrum of childhood solid tumors routinely seen in a large children’s hospital. Further studies in larger cohorts of patients will be required to evaluate the yield of such testing for specific tumor types.
Our tumor WES data highlight several points regarding the use of genomic tests for children with solid tumors. First, despite being less frequently mutated than most adult cancers, pediatric solid tumors harbor potentially attractive targets for therapy, prominently including mitogen-activated protein kinase (MAPK) signaling and MTOR/PI3K pathway mutations (eTable 7 in Supplement 2). As expected, given that children with cancer are typically initially treated on clinical trials or following established treatment regimens, the mutations detected did not alter the management of the newly diagnosed cancer in this cohort. Further studies will be required to assess their potential relevance in the context of refractory and relapsed cancers.
Second, although pediatric solid tumors share common genetic alterations with adult epithelial cancers, mutations in developmental pathways such as Wnt/Wingless signaling (eg, CTNNB1, the most frequently mutated gene in this unselected cohort) are particularly prominent in pediatric solid tumors. Accordingly, both pediatric cancer-specific mutation panels and novel therapeutic strategies for targeting these key pediatric cancer pathways are needed.
Third, 56% (51/91) of mutations were found in genes not known to be recurrently mutated in that specific tumor type, demonstrating the potential value of testing strategies and clinical trials based on molecular alterations rather than tumor histology.26
Fourth, to maximize the yield of such strategies, genomic testing to detect alterations outside of known hotspots is necessary, such as the inactivating TSC2 mutations observed in this cohort. The clinical relevance of noncanonical mutations in oncogenes, such as the detected missense kinase domain mutations in JAK2 and FGFR1, is currently unclear and will require further study.
Fifth, many potentially actionable mutations were found in a small fraction of patients (eg, mutation of FGFR1 in <1%), indicating that effective pediatric precision oncology approaches will require access to a large number of targeted agents.
The clinical categorization of mutations in tumor WES reports was based on expert opinion, using knowledge of mutations, genes, and availability of molecularly targeted agents. For example, the reported category III mutations include variants with diagnostic or prognostic utility (eg, DDX3X mutations in Wnt subtype medulloblastoma)27,28 and others that may be predictive of drug response, such as mutations in BRCA1 and BRCA2 and poly-ADP ribose polymerase (PARP) inhibitors.29 The development of standardized definitions for clinical categorization of somatic mutations will be critical to allow comparative analyses between different genomic testing platforms and patient populations.
We determined the yield of germline WES and whether those findings would have been revealed by a genetic evaluation. The gene most frequently considered for testing by the BASIC3 genetics study team was TP53 (n = 23), although only 2 TP53 germline mutations were identified in this cohort. Substantial heterogeneity was observed in the distribution of cancer susceptibility mutations, with most genes mutated only in a single case. Overall, 5 of the 15 genetic diagnoses in this cohort had specifically been considered for testing by the teams caring for the patient. For many of the remaining diagnoses, the mutations occurred in genes not previously associated with the child’s cancer diagnosis. This may partially relate to recently described disease genes with limited data, eg SMARCA4, which was first described in 2010 in rhabdoid tumors30 and identified in a child with neuroblastoma in this study. Consistent with the somatic mutation analysis, our data suggest that a pediatric hereditary cancer gene panel would also need to be broadly constituted to be an effective test platform. In addition, cancer risk may be influenced by gene-by-gene interactions, as may be the case for the Wilms tumor found in the patient with heterozygous CHEK2, FANCC, and DIS3L2 mutations. In addition, the transmission of cancer susceptibility mutations in these families suggests that the population genetics of childhood cancer susceptibility may be distinct from that of other disease populations; for example, 85% of patients in our study inherited the cancer susceptibility mutation from a parent, with only 1 de novo and 1 mosaic mutation detected in contrast to a 70% de novo rate observed in children with neurodevelopmental disorders.3 This high rate of inheritance has also resulted in rapid targeted testing of at-risk siblings, with cancer screening initiated for those found to carry the identified disease mutation.
A number of children carried pathogenic mutations in adult-onset cancer genes, in particular 3 (2%) of 150 participants with BRCA1 or BRCA2 mutations. Polymorphisms in BARD1, which encodes a BRCA1-interacting protein, have been associated with neuroblastoma risk,31 but 1 study reported no increase in childhood cancer diagnosis in breast cancer kindreds with BRCA1/2 mutations.32 The 3 different tumor types, the absence of LOH, and no statistically significant increase in BRCA1/2 mutations compared with a nonpediatric cancer cohort would suggest that this may be an incidental finding. The discovery here of both germline (n = 3) and somatic (n = 2) BRCA1/2 mutations by WES, as well as the clarification that a TP53 mutation initially identified on a tumor mutation panel was germline, demonstrates the value of sequencing matched normal samples as part of tumor genomic testing.33 The integration of paired tumor and normal sequencing analyses can also be useful for interpretation of germline data, including evaluation of tumor LOH in cases with germline mutations in cancer susceptibility genes, such as our patient with a likely pathogenic TP53 mutation.
Conclusions
Combined reporting of tumor and germline WES results in an unselected cohort of 150 children with newly diagnosed solid tumors revealed potentially actionable findings in nearly 40% of patients across a spectrum of genes previously implicated in both pediatric and adult cancers. The further integration of methods to detect copy number abnormalities and fusion genes will increase the diagnostic yield of these tests, particularly for tumor types that are known to harbor few mutations detectable by WES. Strategies to effectively and responsibly use these diverse results are required to incorporate WES and other genomic tests into childhood cancer care and clinical trials.
Supplementary Material
Acknowledgments
Funding/Support: This study is a Clinical Sequencing Exploratory Research (CSER) program project supported by the National Human Genome Research Institute and the National Cancer Institute grant U01HG006485 (Drs Parsons and Plon).
Footnotes
Open Access: This article is published under the JAMA Oncology open access model and is free to read on the day of publication.
Conflict of Interest Disclosures: As of February 2015 Baylor College of Medicine and Miraca Holdings Inc have formed a joint venture with shared ownership and governance of the Baylor Miraca Genetics Laboratories, which performs exome sequencing. Dr Gibbs is the Chief Scientific Officer; Dr Eng is the Vice President and Executive Laboratory Director; and Dr Plon is a member of the scientific advisory board of Baylor Miraca Genetics Laboratories. No other conflicts are reported.
Role of the Funder/Sponsor: The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: The authors are grateful to the patients and families, pediatric oncologists, and clinical staff at Texas Children’s Cancer Center for their participation in the study. The authors also thank Dr Ramamurthy’s team from the Dan L. Duncan Institute for Clinical and Translational Research, who developed and maintained the BASIC3 study database: Medha Naik, MS, Xingquan Lu, MS, and Vivek Ramanathan, MS. They received no compensation beyond their salaries as full-time Baylor College of Medicine employees.
Author Contributions: Drs Parsons and Plon had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs Parsons and Plon served as co–senior authors, each with equal contribution to this report.
Study concept and design: Parsons, Monzon, Hicks, McGuire, Muzny, Berg, Gibbs, Plon.
Acquisition, analysis, or interpretation of data: All authors.
Drafting of the manuscript: Parsons, Roy, Bergstrom, Kerstein, Petersen, Hicks, Eldin, Ramamurthy, Berg, Plon.
Critical revision of the manuscript for important intellectual content: Parsons, Roy, Yang, Wang, Scollon, Bergstrom, Gutierrez, Bavle, Lin, López-Terrada, Monzon, Hicks, Quintanilla, Adesina, Mohila, Whitehead, Jea, Vasudevan, Nuchtern, McGuire, Hilsenbeck, Reid, Muzny, Wheeler, Berg, Chintagumpala, Eng, Gibbs, Plon.
Statistical analysis: Wang, Petersen, Hilsenbeck.
Obtained funding: Parsons, Berg, Gibbs, Plon.
Administrative, technical, or material support: Parsons, Roy, Yang, Bergstrom, Kerstein, Gutierrez, Lin, López-Terrada, Monzon, Hicks, Adesina, Jea, Nuchtern, Ramamurthy, Reid, Muzny, Chintagumpala, Eng, Gibbs.
Study supervision: Parsons, López-Terrada, Monzon, Hicks, Wheeler, Chintagumpala, Plon.
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369(16):1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Muzny DM, Xia F, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312(18):1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dorschner MO, Amendola LM, Turner EH, et al. National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93(4):631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garraway LA. Genomics-driven oncology: framework for an emerging paradigm. J Clin Oncol. 2013;31(15):1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 6.American Society of Clinical Oncology. American Society of Clinical Oncology policy statement update: genetic testing for cancer susceptibility. J Clin Oncol. 2003;21(12):2397–2406. doi: 10.1200/JCO.2003.03.189. [DOI] [PubMed] [Google Scholar]
- 7.Scollon S, Bergstrom K, Kerstein RA, et al. Obtaining informed consent for clinical tumor and germline exome sequencing of newly diagnosed childhood cancer patients. Genome Med. 2014;6(9):69. doi: 10.1186/s13073-014-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perry JA, Kiezun A, Tonzi P, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111(51):E5564–E5573. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krueger DA, Care MM, Holland K, et al. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med. 2010;363(19):1801–1811. doi: 10.1056/NEJMoa1001671. [DOI] [PubMed] [Google Scholar]
- 10.Iyer G, Hanrahan AJ, Milowsky MI, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338(6104):221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;370(25):2418–2425. doi: 10.1056/NEJMra1312543. [DOI] [PubMed] [Google Scholar]
- 12. [accessed July 1, 2015];Clinical Sequencing Exploratory Research program. HTTPs://cser-consortium.org.
- 13.Bainbridge MN, Wang M, Wu Y, et al. Targeted enrichment beyond the consensus coding DNA sequence exome reveals exons with higher variant densities. Genome Biol. 2011;12(7):R68. doi: 10.1186/gb-2011-12-7-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid JG, Carroll A, Veeraraghavan N, et al. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.COSMIC. [accessed April 1, 2015];catalogue of somatic mutations in cancer. Http://cancer.sanger.ac.uk/cosmic.
- 16.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanchi KL, Johnson KJ, Lu C, et al. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. 2014;5:3156. doi: 10.1038/ncomms4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gajjar A, Bowers DC, Karajannis MA, Leary S, Witt H, Gottardo NG. Pediatric brain tumors: Innovative genomic information is transforming the diagnostic and clinical landscape. J Clin Oncol. 2015;33(27):2986–2998. doi: 10.1200/JCO.2014.59.9217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugh TJ, Morozova O, Attiyeh EF, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45(3):279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sausen M, Leary RJ, Jones S, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2013;45(1):12–17. doi: 10.1038/ng.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou S, Hertel PM, Finegold MJ, et al. Hepatocellular carcinoma associated with tight-junction protein 2 deficiency. Hepatology. 2015;62(6):1914–1916. doi: 10.1002/Hep.27872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Astuti D, Morris MR, Cooper WN, et al. Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet. 2012;44(3):277–284. doi: 10.1038/ng.1071. [DOI] [PubMed] [Google Scholar]
- 24.Green RC, Berg JS, Grody WW, et al. American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15(7):565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32(19):2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redig AJ, Jänne PA. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J Clin Oncol. 2015;33(9):975–977. doi: 10.1200/JCO.2014.59.8433. [DOI] [PubMed] [Google Scholar]
- 27.Pugh TJ, Weeraratne SD, Archer TC, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488(7409):106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson G, Parker M, Kranenburg TA, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488(7409):43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sandhu SK, Schelman WR, Wilding G, et al. The poly(ADP-ribose) polymerase inhibitor niraparib (MK4827) in BRCA mutation carriers and patients with sporadic cancer: a phase 1 dose-escalation trial. Lancet Oncol. 2013;14(9):882–892. doi: 10.1016/S1470-2045(13)70240-7. [DOI] [PubMed] [Google Scholar]
- 30.Schneppenheim R, Frühwald MC, Gesk S, et al. Germline nonsense mutation and somatic inactivation of SMARCA4/BRG1 in a family with rhabdoid tumor predisposition syndrome. Am J Hum Genet. 2010;86(2):279–284. doi: 10.1016/j.ajhg.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41(6):718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brooks GA, Stopfer JE, Erlichman J, Davidson R, Nathanson KL, Domchek SM. Childhood cancer in families with and without BRCA1 or BRCA2 mutations ascertained at a high-risk breast cancer clinic. Cancer Biol Ther. 2006;5(9):1098–1102. doi: 10.4161/cbt.5.9.3167. [DOI] [PubMed] [Google Scholar]
- 33.Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7(283):283Ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.