Abstract
Background
Germline heterozygous mutations in human STAT1 can cause loss of function (LOF), as in patients with Mendelian susceptibility to mycobacterial diseases (MSMD), or gain of function (GOF), as in patients with chronic mucocutaneous candidiasis (CMC). LOF and GOF mutations are equally rare and can affect the same domains of STAT1, especially the coiled-coil and DNA-binding domains (CCD/DBD). Moreover, 6% of CMC patients with a GOF STAT1 mutation develop mycobacterial disease, obscuring the functional significance of the identified STAT1 mutations. Current computational approaches, such as combined annotation-dependent depletion, do not distinguish LOF and GOF variants
Objective
Estimate variations in CCD/DBD of STAT1
Method
Mutagenized 342 individual wild-type amino acids in CCD/DBD (45.6% of full-length STAT1) to alanine and tested the mutants for STAT1 transcriptional activity.
Results
Of these 342 mutants, 201 were neutral, 30 LOF, and 111 GOF in a luciferase assay. This assay system correctly estimated all previously reported LOF mutations (100%) and slightly fewer GOF mutations (78.1%) in CCD/DBD of STAT1. We found that GOF alanine mutants occurred at the interface of the antiparallel STAT1 dimer, suggesting that they destabilize this dimer. This assay also precisely predicted the impact of two hypomorphic and dominant-negative mutations, E157K and G250E, in CCD of STAT1 that we found in two unrelated MSMD patients.
Conclusion
Systematic alanine-scanning assay is a useful tool to estimate the GOF or LOF status and impact of heterozygous missense mutations in STAT1 identified in patients with severe infectious diseases, including mycobacterial and fungal diseases.
Keywords: STAT1, alanine-scanning, chronic mucocutaneous candidiasis, mendelian susceptibility to mycobacterial disease, reference database
Capsule summary
Reference database based on the alanine-scanning assay is a useful tool for evaluating unknown variations in STAT1. Gain-of-function alanine mutants occurred at the interface of the antiparallel STAT1 dimer, suggesting that they destabilize this dimer.
Introduction
STAT1 is a latent cytoplasmic transcription factor belonging to the signal transducers and activators of transcription (STAT) family. STAT1 is phosphorylated on tyrosine 701 (Y701) by the Janus kinases (JAKs) when a cytokine or growth factor binds to its receptor, allowing the dimerization of STAT1. STAT1 can form a homodimer, known as gamma-interferon activation factor (GAF), after stimulation by interferon (IFN)-γ, IFN-α/β, or interleukin 27 (IL-27). GAF translocates to the nucleus and binds to specific DNA sequences, known as gamma-activating sequences (GAS), in the promoters of IFN-stimulated genes to induce their transcription.1 STAT1 also forms a heterotrimer after stimulation by IFN-α/β or IFN-λ, consisting of STAT1, STAT2, and IRF9, which is known as interferon-stimulated gene factor 3 (ISGF3). ISGF3 binds the interferon-stimulated response element, initiating gene transcription.1 The deactivation of STAT1 is mediated by a nuclear phosphatase thought to be TC45, which dephosphorylates STAT1, allowing its release into the cytoplasm.2,3 In humans, STAT1 plays a nonredundant role in IFN-α/β, IFN-γ, IL-27, and IFN-λ signaling.4
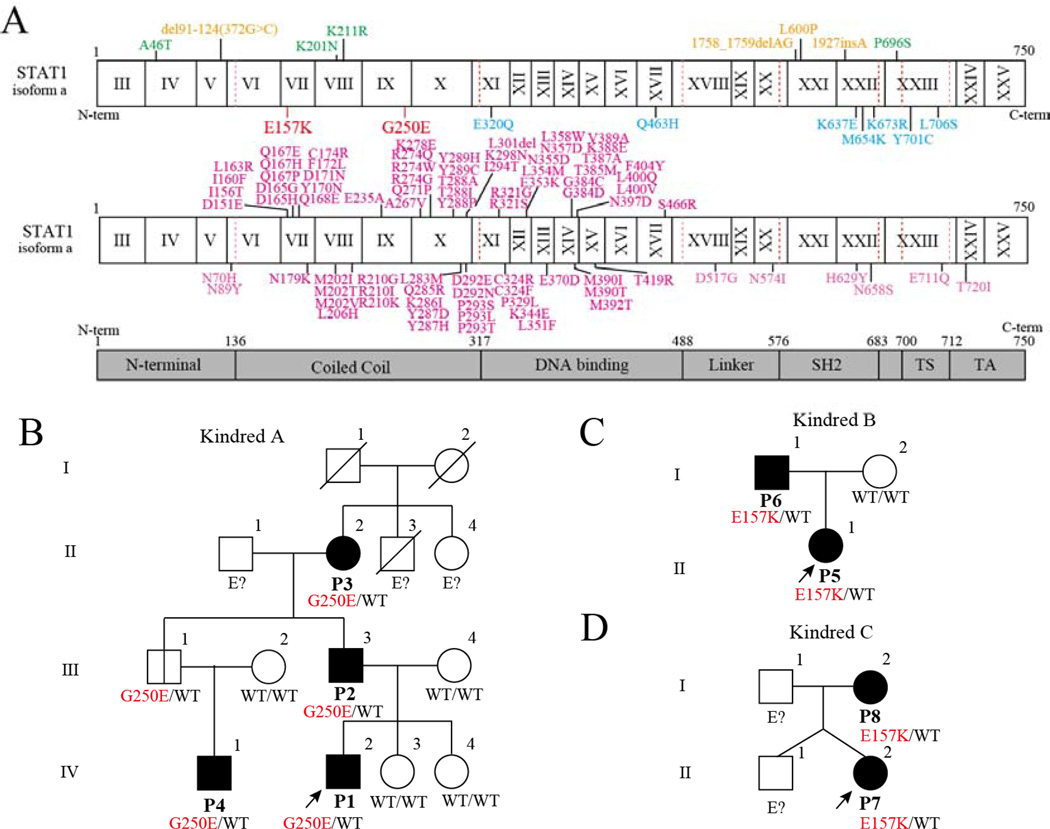
Inborn errors in human STAT1-based immunity cause four types of immune deficiency: i) autosomal recessive (AR) complete STAT1 deficiency; ii) AR partial STAT1 deficiency; iii) autosomal dominant (AD) STAT1 deficiency; and iv) AD STAT1 gain of activity.4 Biallelic loss-of-function (LOF) mutations have been identified in patients with complete and partial AR STAT1 deficiency. These patients suffer from life-threatening viral infections (especially herpes virus infections) because they lack the STAT1-dependent response to IFN-α/β (and perhaps IFN-λ) signaling, and mycobacterial susceptibility because they lack the response to IFN-γ (and perhaps IL-27).5 Partial AR STAT1 deficiency is a milder form, so patients with this disorder suffer from mild viral and mycobacterial diseases and impaired but not abolished responses to IFN-α/β, IFN-γ, IL-27, and IFN-λ signaling.6–8 Heterozygous LOF STAT1 mutations cause Mendelian susceptibility to mycobacterial diseases (MSMD), attributable to the impairment of IFN-γ signaling, and mutations have been identified in the DNA-binding domain (DBD), SH2 domain, and transactivation domain (Figure 1A).9–14 Heterozygous gain-of-function (GOF) mutations underlie chronic mucocutaneous candidiasis (CMC) and have been most commonly identified in the coiled-coil domain (CCD) and DBD (see Figure 1A). 15–47
Figure 1. Family trees and known pathogenic STAT1 mutations.
(A) Human STAT1α isoform with its known pathogenic mutations. Mutations identified in patients with an AR form of complete (orange) or partial (green) STAT1 deficiency are shown in above. Mutations identified in patients with AD MSMD are shown in red (current study) and blue (previously reported). The reported gain-of-function mutations are shown in magenta below the protein. TA, transcriptional activation domain; TS, tail segment. (B, C, D) Family trees.
LOF and GOF mutations can affect the same domains in STAT1, obscuring the behavior of a particular mutation based solely on its location. Therefore, we systematically investigated the effects of alanine substitutions in CCD and DBD (CCD/DBD) of STAT1, screening 342 alanine mutants (in a total of 750 residues) with a GAS reporter assay after IFN-γ stimulation. All the LOF MSMD-causing mutations and most (78.1%) GOF CMC-related mutations previously identified in CCD/DBD were correctly identified in the alanine mutants, suggesting that this technique can be used to establish a reference library of STAT1 variants. Our results clearly demonstrate that the majority of GOF mutations are located at the interface of the antiparallel STAT1 dimer, probably disrupting the dimerization of antiparallel STAT1 structures. We confirmed our results by identifying two germline heterozygous hypomorphic and dominant-negative mutations in CCD of STAT1 in patients with MSMD.
Materials and Methods
Functional assay based on systematic alanine-scanning mutagenesis
A vector from which to express HaloTag® STAT1 was obtained from the Kazusa cDNA/ORF clone collection (FHC013013). The codons of the vector encoding residues from L136 to F487 of STAT1, except the 10 alanines (A188, A230, A246, A254, A267, A401, A402, A415, A469, and A479), were individually substituted with GCC, the codon most frequently encoding alanine in humans, with site-directed mutagenesis. The activities of the mutants were measured with a luciferase reporter assay with the pGL4.24 vector (Promega Corporation, Madison, WI, USA) driven by five tandem interferon regulatory factor (IRF)-derived GAS elements (TTCCCCGAA) (IRF1 reporter plasmids). Detailed methods of luciferase reporter assay and the other experiments are available in the Supplemental materials and methods.
Case
Detailed case reports of Kindred A, B, and C (see Figure 1) are available in the Supplemental materials and methods. Briefly, 8 patients from 3 unrelated families are included in this study. Six patients received BCG at infancy, and 5 of the 6 developed BCGitis. Multifocal osteomyelitis was observed in 5 patients including 4 patients who developed BCGitis. Two patients who did not receive BCG had a history of tuberculosis. One patient who does not have an obvious history of BCGitis developed intracranial granuloma with M. avium complex detectable by PCR from brain biopsy. The penetrance of STAT1 mutation was not complete. One family member in Kindred A who has no history of BCG vaccination turned out to have STAT1 mutation by familial study.
Results
Functional assay based on systematic alanine-scanning mutagenesis
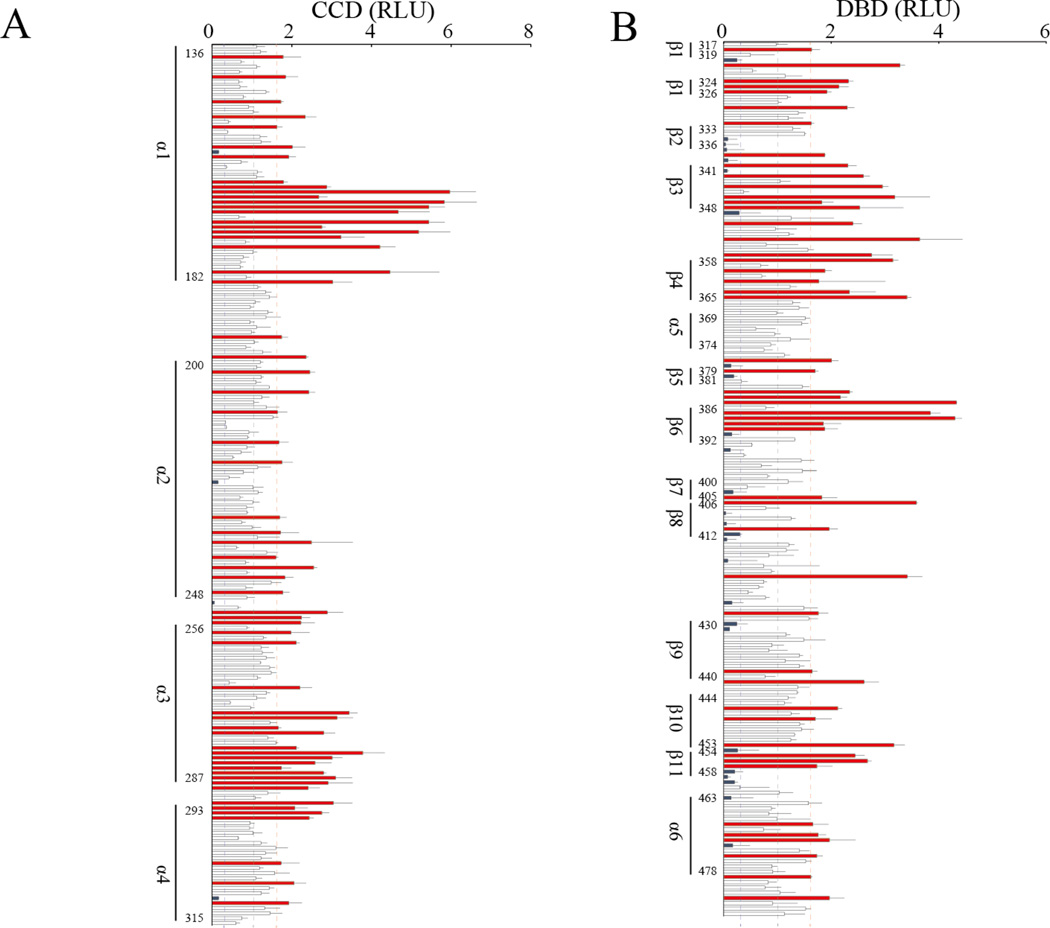
Alanine-scanning mutagenesis is a widely used technique in the determination of the catalytic or functional role of protein residues. We systematically investigated the effects of alanine substitutions in CCD/DBD of STAT1 with a GAS reporter assay after IFN-γ stimulation. We generated 176 alanine mutants in CCD and 166 alanine mutants in DBD. We defined the individual alanine mutants as LOF or GOF if they showed <0.3 times or >1.6 times the GAS transcriptional activity of WT, respectively (see Figure 2A and B). These cutoffs were defined based on the GAS transcriptional activity of known pathogenic mutations. The functional assay based on systematic alanine-scanning mutagenesis (designated the ‘alanine-scanning assay’) showed that 34.7% (61/176) of the alanine mutants in CCD were GOF and 2.2% (4/176) were LOF. In DBD, 30.1% (50/166) of the mutants were GOF and 15.7% (26/166) were LOF. These results are consistent with the clinical observation that most reported STAT1 mutations in CCD/DBD are GOF. Many CMC-related GOF mutations have been identified in CCD/DBD of STAT1 in previous studies. 15–23, 25–44, 48 In contrast, only two MSMD-causing LOF STAT1 mutations (E320Q and Q463H) have been identified in DBD, and no LOF mutation has been reported in CCD.10
Figure 2. GAS transcription assay based on systematic alanine-scanning mutagenesis.
GAS transcriptional activity in response to IFN-γ was measured in 176 STAT1 proteins mutated in CCD (A) and 166 STAT1 proteins mutated in DBD (B). The 34.7% (61/176) and 30.1% (50/166) of alanine mutants were deemed to be GOF, whereas 2.2% (4/176) and 15.7% (26/166) were deemed to be LOF in CCD and DBD, respectively. The experiments were performed in triplicate and the data are expressed in relative luciferase units (RLU).
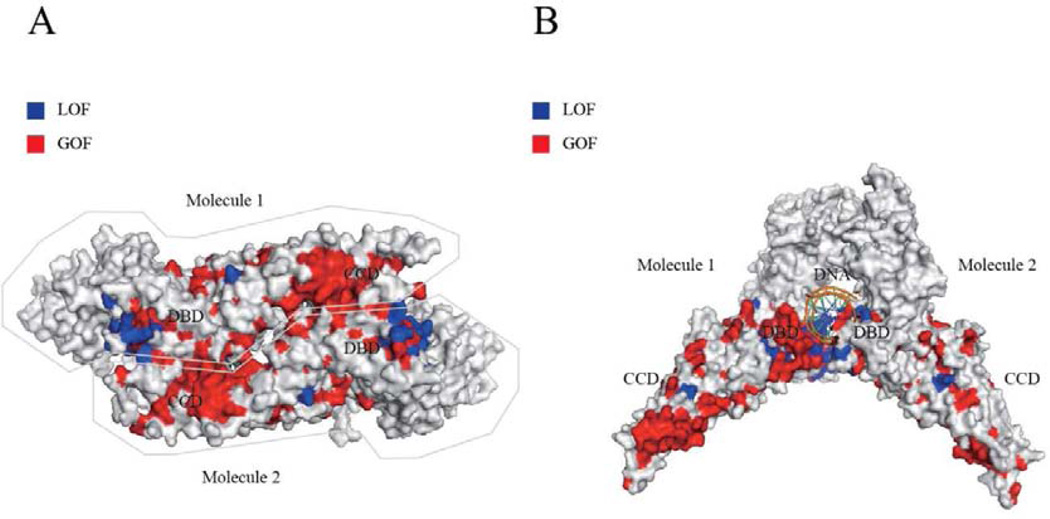
The results of the alanine-scanning assay were compared with previously identified GOF and LOF mutations in STAT1 (see Table SI). The assay allowed us to explain 100% of LOF mutants and 78.1% (86.7% in CCD and 64.3% in DBD) of GOF mutants in CCD/DBD of STAT1. No known LOF mutations were deemed to be GOF. Only one GOF mutation was misidentified as a LOF mutation in the alanine-substituted mutants. Fifteen GOF mutations (20.5%) were falsely identified as neutral. We then evaluated STAT1 variants that are occasionally identified in healthy individuals and are listed in the Single Nucleotide Polymorphism Database (dbSNP), 1000 Genome Projects, and/or the Exome Aggregation Consortium (ExAc) database, with the alanine-scanning assay (see Table SII). This assay estimated 71.1% (27/38), 23.7% (9/38), and 5.4% (2/38) of the variations as neutral, GOF, and LOF, respectively. We next blotted the GOF and LOF alanine mutants in the parallel and antiparallel dimeric STAT1 structures (see Figure 3A and B). Intriguingly, the GOF alanine mutations predominantly localized at the interface of the antiparallel dimer (see Figure 3A). In contrast, several LOF alanine mutations localized close to the DNA-binding site (see Figure 3B). Although the other LOF alanine mutations localized inside the protein, there was no obvious preferential localization.
Figure 3. Distributions of LOF and GOF mutations in antiparallel and parallel STAT1 dimers.
GOF (red) and LOF (blue) alanine substituents were mapped to the antiparallel (A) and parallel dimeric structures (B) of STAT1. (A) GOF alanine mutants predominantly localized at the interface of the antiparallel dimer, whereas LOF alanine mutants localized inside the protein. (B) Many LOF alanine mutants localized close to the DNA-binding site.
Mutation significance cutoff (MSC) impact estimation of STAT1 mutations and variations in CCD/DBD
We evaluated the previously identified mutations with MSC impact estimation based on their combined annotation-dependent depletion (CADD) scores. The CADD scores of all previously identified GOF and LOF mutations, except E235A and R321G, in CCD/DBD of STAT1 were >7.736 (the benign/damaging cutoff specific for STAT1), and were therefore estimated to have a high impact (see Table SI). However, 89.7% (35/39) of natural variations in CCD/DBD of STAT1 listed in the dbSNP, 1000 Genome Project, and ExAc databases were also estimated to have a high impact. We also detected no significant differences in the CADD scores of the LOF and GOF mutations in STAT1.
Identification of STAT1 mutations
The STAT1 mutation in proband (P1) was identified with the candidate gene approach for MSMD. We identified a novel heterozygous mutation, 746G>A (G250E), in STAT1 with Sanger sequencing (see Figure S1). No mutation was identified in IFNGR1, IFNGR2, IKBKG, CYBB, IRF8, or ISG15 in P1. Another novel heterozygous mutation in STAT1, 469G>A (E157K), was identified in unrelated patients (P5 and P7) with WES and confirmed with Sanger sequencing. Neither the E157K nor G250E mutation was found in the NCBI, Ensembl, dbSNPs, or ExAc databases, or in our own in-house database of 3,000 exomes. Nor were they detected in the 1,052 control samples from 52 ethnic groups in the Centre d’Etude du Polymorphisme Humain (CEPH) and Human Genome Diversity (HGD) panels.49, 50 Therefore, they are rare MSMD-causing variants rather than irrelevant polymorphisms. The G250E mutation was identified in P1, P2, P3, and P4, consistent with AD inheritance. The same mutation was also identified in an asymptomatic uncle (A.III.1), who had not been vaccinated with BCG. The E157K mutation was identified in P5 and P6 in kindred B, and in P7 and P8 in kindred C. Thus, we identified two novel heterozygous mutations, E157K and G250E, in CCD of STAT1 in seven patients from three families (Figure 1B–D).
E157K and G250E in STAT1 partially impair its phosphorylation, binding to GAS, and GAS transactivation in response to IFN-γ
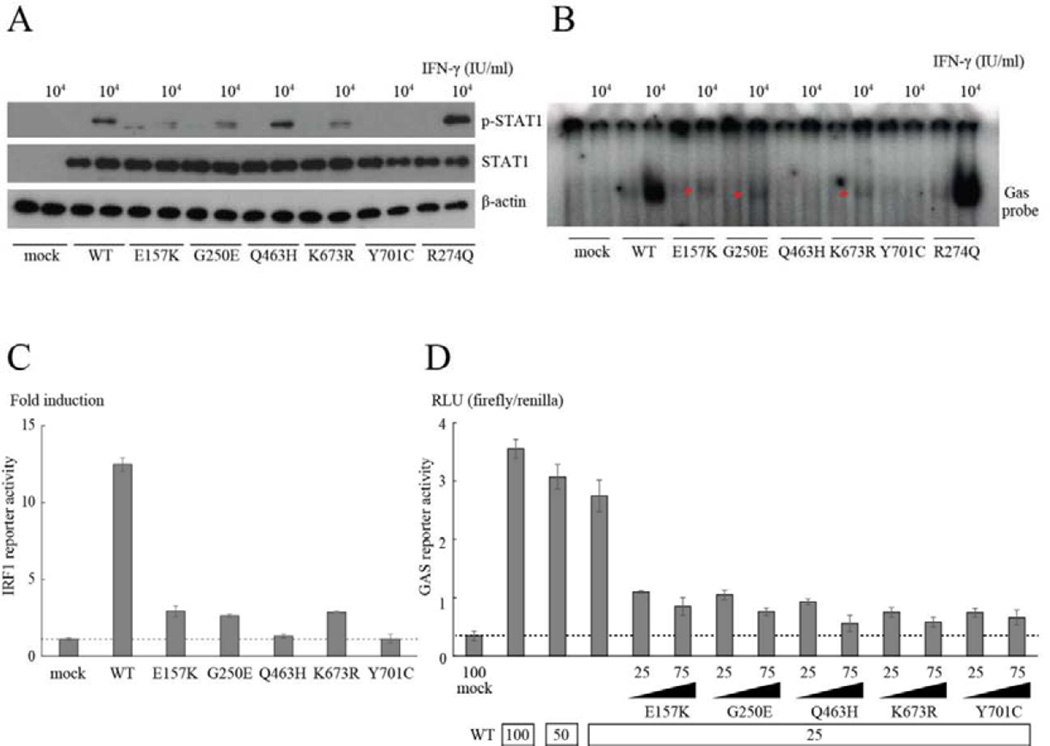
We transiently introduced WT- and/or mutant-STAT1-containing plasmids into U3C cells, a STAT1-null fibrosarcoma cell line, with lipofection, and analyzed the expression and Y701 phosphorylation of STAT1 after IFN-γ stimulation, with immunoblotting (see Figure 4A and S2A). Both E157K and G250E STAT1 and the K673R dominant-negative hypomorphic mutant that causes AD STAT1 deficiency were expressed normally but showed partially impaired phosphorylation after IFN-γ stimulation.12 In contrast, the R274Q mutation, a common GOF mutation in patients with CMC, showed increased phosphorylation in response to IFN-γ.15 Consistent with previous reports, Q463H STAT1 was normally phosphorylated and Y701C abolished STAT1 phosphorylation after IFN-γ stimulation.10, 11 We then analyzed the intrinsic DNA-binding ability of the STAT1 proteins with an electrophoretic mobility shift assay (EMSA). U3C cells expressing WT or mutant STAT1 were stimulated with IFN-γ and subjected to EMSA with a GAS probe. E157K and G250E, as well as K673R STAT1, displayed markedly lower levels of GAF binding to GAS (see Figure 4B and S2B). Y701C STAT1 had no DNA-binding ability, whereas R274Q STAT1 showed increased DNA-binding ability. We next investigated the impact of the E157K and G250E mutations on the transcriptional activity of GAS in assays with GAS reporter plasmids. Like K673R STAT1, both E157K and G250E STAT1 severely impaired STAT1-mediated signaling, but did not abolish it, as Q463H and Y701C STAT1 did, disrupting GAS transcriptional activity after IFN-γ stimulation (see Figure 4C). Both E157K and G250E STAT1, as well as the other previously reported dominant negative STAT1 mutations, had dose-dependent negative effects in cotransfection experiments, suggesting that both mutants exert a dominant-negative effect on IFN-γ-induced WT-STAT1-mediated GAS activation (see Figure 4D and S3).
Figure 4. Functional assays of E157K and G250E STAT1 identified in patients with MSMD.
(A) The levels of pSTAT1 were reduced in the E157K and G250E mutants in response to IFN-γ, to levels similar to that in the K673R mutant. (B, C) E157K and G250E STAT1, as well as the K673R hypomorphic mutant, severely impaired the ability of GAF to bind GAS (B) and induce IRF1 reporter activity after IFN-γ stimulation (C). In contrast, Q463H and Y701C STAT1 completely abolished DNA-binding to GAS (B) and IRF1 activation (C). (D) All the mutants exerted a dose-dependent negative effect on WT-STAT1-mediated GAS activation after IFN-γ stimulation. The amounts of plasmids used are shown below the x-axis.
E157K and G250E in STAT1 partially impair nuclear localization
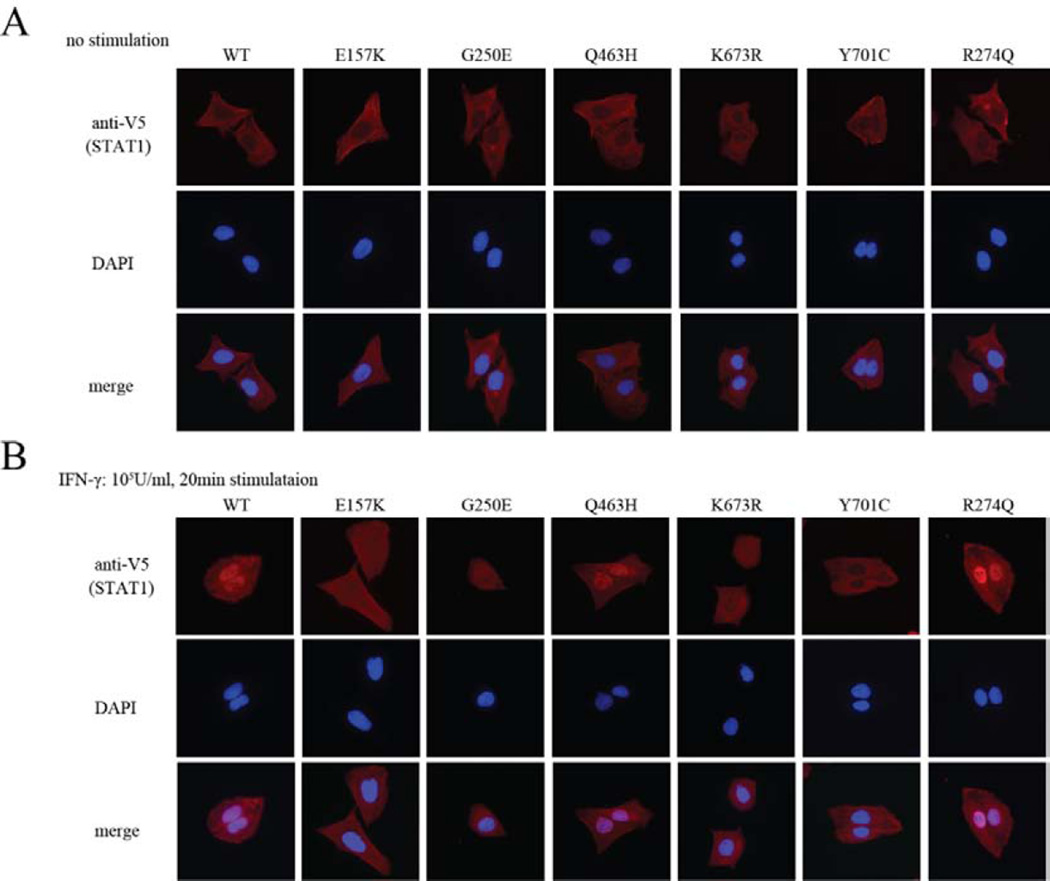
We analyzed the nuclear translocation of STAT1 in U2OS cells stably expressing V5-tagged WT, E157K, G250E, Q463H, K673R, Y701C, or R274Q STAT1 proteins.10–12, 15, 16 STAT1 was mainly observed in the cytoplasm of unstimulated cells (see Figure 5A and S2C). After IFN-γ stimulation, STAT1 was mainly observed in the nuclei of cells expressing WT, Q463H, or R274Q STAT1, whereas Y701C STAT1 remained in the cytoplasm (see Figure 5B and S2D). E157K, G250E, and K673R STAT1 displayed partially impaired nuclear translocation.
Figure 5. Subcellular localization of WT and mutant STAT1.
U2OS cells stably expressed V5-tagged WT or mutant STAT1 was stimulated with IFN-γ for 30 min and subjected to immunostaining. (A) Before IFN-γ stimulation, the WT and all the mutant STAT1 proteins were localized to the cytoplasm. (B) Like the K673R mutant, the nuclear translocation of E157K and G250E STAT1 was partially impaired in cells expressing either protein. In contrast, Y701C completely abolished the nuclear translocation of STAT1 after IFN-γ stimulation.
E157K STAT1 forms a hydrogen bond with E394 in its antiparallel partner
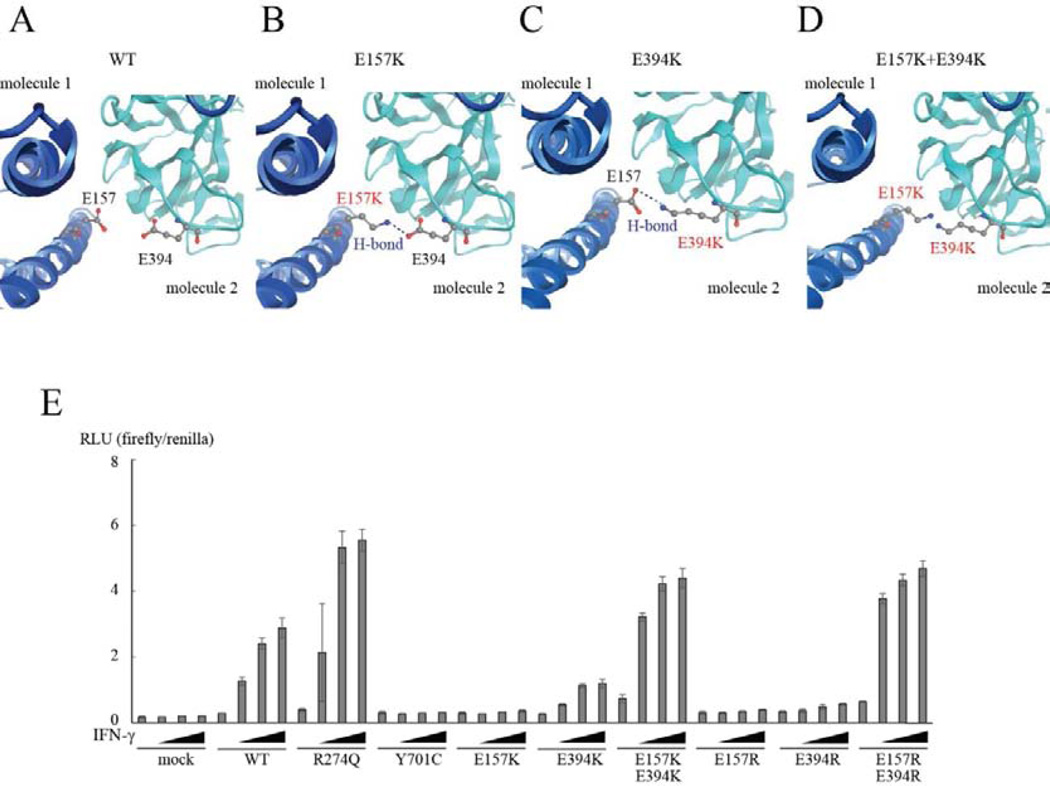
STAT1 forms both parallel and antiparallel dimers.51, 52 Phosphorylated STAT1 forms parallel dimers (active form), whereas unphosphorylated STAT1 preferentially forms antiparallel dimers (inactive form). When STAT1 forms antiparallel dimers, CCD and DBD associate reciprocally.2 We mapped the positions of E157K and G250E, together with six other well-characterized mutations (GOF: D165G and R274Q in CCD, N355D and K388E in DBD; LOF: E320Q and Q463H in DBD) in both the parallel (see Figure S4A) and antiparallel STAT1 dimers (see Figure S4B). In the parallel structure, G250E was located close to the linker domain, which plays an important role in DNA binding.53 This mutation also localized close to E320Q, which impairs STAT1 phosphorylation and the binding of GAF to GAS.10 The Q463H mutation, which specifically impairs DNA binding, was located very close to the DNA-binding site. In contrast, the E157K mutation localized far from the DNA-binding site, and close to the GOF mutations D165G and R274Q in CCD. When we examined these mutations in the antiparallel structure, all four GOF mutations were located at the dimerization interface. The G250E, E320Q, and Q463H mutations were located inside the protein, relatively far from the dimerization interface, in the antiparallel dimer. However, the E157K LOF mutation and other GOF mutations were located at the dimeric interface in the antiparallel structure. We next mapped known pathogenic mutations (35 GOF and four LOF mutations) in CCD/DBD in the parallel (see Figure S5A) and antiparallel dimers (see Figure S5B). As in the alanine-scanning assay, the GOF mutations were preferentially observed at the molecular interface in the antiparallel dimer. These observations suggest that some specific mechanism underlies the LOF of E157K STAT1. Therefore, we analyzed E157K STAT1 in silico using the protein modeling software MOE (Figures 6A and B and S6). The mutant K157 residue was predicted to form a hydrogen bond with E394 of its antiparallel partner, stabilizing the antiparallel STAT1 dimer.
Figure 6. Interaction between E157 and E394 residues affects GAS transcriptional activity.
(A, B) E157K STAT1 was analyzed in silico with MOE. Unlike WT STAT1 (A), the E157K (B) and E394K (C) mutants were predicted to form hydrogen bonds with E394 and E157 in their dimeric partners, respectively. The E157K+E394K double mutant (D) was predicted to allow the access of their side chains, leading to electrostatic repulsion between the two. (E) Both E157K and E394K were LOF mutations, whereas the E157K+E394K double mutant was predicted to be a GOF mutant by reporter assay. Similar results were observed with arginine substitutions.
Strength of interaction between residues E157 and E394 affects GAS transcriptional activity
We then investigated the functional significance of mutation E157K by focusing on the interaction between residues E157 and E394. A MOE analysis predicted that the mutant K394 residue forms a hydrogen bond with E157 (see Figure S6). Therefore, we generated two STAT1 mutants, E394K and E157K+E394K (double mutant), and investigated their functions (see Figure 6C and D). As expected, E394K STAT1 showed LOF in a GAS reporter assay, like the E157K mutant (see Figure 6E). However, surprisingly, the E157K+E394K double mutant showed GOF, with markedly increased GAS transcriptional activity (Figure 6E). Glutamic acid is a negatively charged amino acid, whereas lysine is positively charged. Therefore, we inferred that the electrostatic repulsion between residues K157 and K394 underlies the GOF of the E157K+E394K double mutant (see Figure 6D). To confirm this inference, we substituted residues E157 and E394 with arginine, another positively charged amino acid (see Figure S7A–D), and tested the functional significance of these changes in a GAS reporter assay (see Figure 6E). Like the lysine-substituted mutants, both E157R and E394R STAT1 displayed LOF, whereas the E157R+E394R double mutant displayed GOF.
The alanine-scanning assay showed that E157A STAT1 is a LOF mutant. Therefore, we investigated the effect of A157 with MOE. Although LOF E157A STAT1 formed no hydrogen bond with E394, a conformational change was predicted that allowed amino acid H158 to form a hydrogen bond with E394 (see Figure S8A and B).
Discussion
We systematically evaluated the impact of amino acid substitutions (342 alanine mutants) in CCD/DBD of STAT1 using a GAS reporter assay. The GAS reporter assay is an accurate and practical method with which to assess LOF and GOF STAT1 mutations.11, 12, 15, 19–21 This assay allowed us to explain 100% of known LOF mutations and 78.1% (86.7% in CCD and 64.3% in DBD) of known GOF mutations in CCD/DBD of STAT1 using alanine substituents of the same residues. It showed that 34.7% (61/176) and 30.1% (50/166) of alanine mutants were GOF, whereas 2.2% (4/176) and 15.7% (26/166) were LOF in CCD and DBD, respectively. This is consistent with previous clinical observations that most reported disease-causing mutations in CCD/DBD are GOF and are associated with CMC.4, 15, 16, 22, 36 In contrast, only two AD MSMD-related LOF mutations, E320Q and Q463H, both in DBD, have yet been reported in those domains.10 The alanine-scanning assay revealed that four of 176 alanine substituents (E157A, T224A, G250A, and Q311A) in CCD were LOF. This result was confirmed by the identification of three familial cases of AD MSMD carrying LOF mutations in CCD. The functional significance of the E157K and G250E mutations, newly identified in those patients, were correctly predicted with the alanine-scanning assay, confirming the efficiency and reliability of this assay system.
The alanine-scanning assay also showed that the GOF alanine mutants predominantly localized to the molecular interface of the antiparallel dimer. This assay has contributed to the evaluation of protein–protein interfaces by disturbing the roles of the side-chain functional groups at specific positions and the energetic contributions of individual side-chains to protein binding.54 Therefore, the distribution of the GOF alanine mutants suggests that the disruption of the antiparallel dimer is a key molecular mechanism underlying the GOF STAT1 mutations. In vitro and in silico analyses of two STAT1 residues, E157 and E394, strongly supported this hypothesis. Both E157K and E394K STAT1 were evaluated as LOF with the GAS reporter assay and were predicted to stabilize the antiparallel dimerization of STAT1 by forming an additional hydrogen bond with MOE. In contrast, the E157K+E394K double mutant was identified as a GOF mutant with the GAS reporter assay. These findings were confirmed with arginine substitutions at residues E157 and E394. Because both lysine and arginine are positively charged, the disruption of the antiparallel dimer by the electrostatic repulsion between residues 157 and 394 of the double mutants was suspected to be the molecular basis of GOF. These observations are nicely explained by previous studies in which the antiparallel form of STAT1 was shown to facilitate the access of a phosphatase by presenting pY701 at both ends of the antiparallel dimer for ready dephosphorylation.2, 52 Our results, together with those of previous studies, clearly support the hypothesis that the disturbance of the antiparallel STAT1 dimer affects its activity by altering its phosphorylation at Y701. The mutations that disrupt the antiparallel STAT1 dimer are GOF mutations, whereas the mutations that stabilize the antiparallel dimer are LOF mutations.
Although the alanine-scanning assay can be considered a good system for estimating the functional significance of STAT1 mutations, it has some limitations. First, this system cannot evaluate STAT1 mutations involving WT alanine residues, such as A267V, a known GOF mutation. Seconds, the alanine-scanning assay does not always estimate the impact of STAT1 mutations appropriately. Indeed, this assay system misidentified 21.9% of GOF mutations and 28.9% of natural variants. This may have occurred because the alanine mutants cannot mimic the size effects or the hydrophilic or hydrophobic effects exerted by other substituted amino acids. Consequently, the establishment of a reference database of STAT1 variants, that measures the activity of all possible STAT1 substitutions (totaling 14,231 STAT1 mutants), might be a future goal to achieve an accurate evaluation of the functional significance of all STAT1 variants. Next, in order to evaluate the applicability of the alanine-scanning assay, we compared the results of this assay system with the computational impact estimates of The Mutation Significance Cutoffs (MSC) method, in which a CADD benign/damaging cutoff specific for STAT1 was used.55–58 MSC showed a high true-positive estimation rate, with 97.3% (71/73) of known pathogenic GOF and LOF mutations in CCD/DBD identified as deleterious. However, MSC misestimated 89.7% of the natural variants as damaging. The high rate of false-positive estimates, which was recently demonstrated empirically59, is a common problem of current computational impact estimates. Although MSC permits a low false-positive rate56, our results show that its false-positive rate was still 89.7% in evaluating the STAT1 variants. The allele frequencies of the STAT1 variants analyzed in the present study are quite low in the general population (see Table SII), which could explain the high false-positive rate because the allele frequency correlates strongly with the impact estimate.56, 58, 60 Our results stress the difficulty in the computational estimation of the mutations and variations in STAT1 and support the utility of the alanine-scanning assay in their estimation.
The alanine-scanning assay, together with the identification of three familial cases of AD MSMD, have shown that LOF and GOF mutations co-exist in both CCD and DBD of STAT1. AD partial STAT1 deficiency and AD STAT1 gain of activity are distinct primary immune deficiencies based on different molecular mechanisms. However, there is considerable overlap in their clinical manifestations. Although CMC is a prominent clinical manifestation in patients with GOF mutations, nearly 6% of these patients are also susceptible to mycobacterial infection.45 Additionally, many patients with GOF mutations present with broad manifestations associated with bacterial and viral infections.45 Furthermore, some patients with a GOF mutation have been reported to develop combined immunodeficiency disease and immune dysregulation, polyendocrinopathy, enteropathy, or X-linked (IPEX)-like syndrome, with severe and/or atypical clinical manifestations.29, 32 Considering the clinical overlap and phenotypic diversity of their symptoms, we must carefully evaluate any unknown variations in CCD/DBD of STAT1. The current advances in deep sequencing and genotyping methods have furthered the identification of the genetic etiology of many diseases. Simultaneously, the evaluation of unknown variants has become increasingly important in clarifying disease-causing mutations. The current study is the first to establish a reference database for estimating the pathogenesis of naturally-occurring genetic variations in a central disease-causing gene, based on a systematic alanine-scanning assay. This study also demonstrated that the establishment of a reference database based on the alanine-scanning assay would be a useful tool for evaluating unknown STAT1 variations found in genetic study of patients with primary immune deficiency.
Supplementary Material
Key message.
Systematic alanine-scanning assay correctly estimated known LOF mutations (100%) and GOF mutations (78.1%) in CCD/DBD of STAT1
Systematic alanine-scanning assay correctly estimated 71.1% of nonpathogenic natural variants
Alanine scanning assay provided better estimation of STAT1 variants compared with computational estimation.
Acknowledgments
We would like to thank to M. Ciancanelli and T. Le Voyer for helpful discussions. We also thank Y. Nemirovskaya, L. Amar, E. Anderson, M. Courat and T. Nivare for administrative support. The sequence analysis was supported by the Analysis Center of Life Science, Natural Science Center for Basic Research and Development, Hiroshima University.
A declaration of all sources of funding for the research:
This study was supported in part by Grants in Aid for Scientific Research from the Japan Society for the Promotion of Science (22591161 to M.K.; 25713039 to S.O.; 16H05355 to S.O.; and 16K15528 to S.O.) and was supported in part by the Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and development, AMED. This study was also supported in part by Research on Measures for Intractable Diseases funding from the Japanese Ministry of Health, Labour and Welfare (H22-Nanchi-ippan-078 to M.K.), GSK Japan Research Grant 2014, and the Kurozumi Medical Foundation. The Laboratory of Human Genetics of Infectious Diseases is supported by institutional grants from INSERM, University Paris Descartes, the Rockefeller University, the St. Giles Foundation, the US National Institute of Allergy and Infectious Diseases (grant n° R37AI095983 and U01AI109697), and grants from the French National Research Agency (ANR) under the “Investments for the future” program (grant n°ANR-10-IAHU-01).
Abbreviations used
- AD
autosomal dominant
- AR
autosomal recessive
- CADD
combined annotation-dependent depletion
- CEPH
Centre d’Etude du Polymorphisme Humain
- CMC
chronic mucocutaneous candidiasis
- DBD
DNA-binding domain
- dbSNP
Single Nucleotide Polymorphism Database
- EMSA
electrophoretic mobility shift assay
- ExAc
Exome Aggregation Consortium
- GAF
gamma-interferon activation factor
- GOF
gain of function
- HGD
Human Genome Diversity
- IFN
interferon
- IL
interleukin
- IRF
interferon regulatory factor
- ISGF3
interferon-stimulated gene factor 3
- JAKs
Janus kinases
- LOF
loss of function
- MSC
Mutation Significance Cutoffs
- MSMD
Mendelian susceptibility to mycobacterial diseases
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Authorship: Contribution: Reiko Kagawa performed the immunoblotting and immunostaining analyses, and prepared the first draft; Ryoji Fujiki performed the alanine-scanning assay; Miyuki Tsumura., Xiao-Fei Kong., Osamu Hirata., Sonoko Sakata, Shiho Nishimura., and Satoshi Saito generated the mutant STAT1 plasmids, and performed the EMSA and luciferase reporter assay; Zenichiro Kato and Hidenori Ohnishi performed the in silico assay; Maiko Ikeda, Jamila El Baghdadi, Aziz Bousfiha, Kaori Fujiwara, Matias Oleastro, Judith Yancoski, Laura Perez, Silvia Danielian, and Fatima Ailal examined the patients and wrote the clinical reports; Hidetoshi Takada, Toshiro Hara, Stéphanie Boisson-Dupuis, and Jacinta Bustamante analyzed the data; Yuval Itan estimated the functional significance of the STAT1 mutations and variants computationally with the MSC method, based on the CADD method.; Anne Puel, Jean-Laurent Casanova, Osamu Ohara, and Masao Kobayashi supervised the study and revised the manuscript; Satoshi Okada designed and supervised the study, and performed the EMSA and reporter assay.
Disclosure of potential conflict of interest:
The authors have no conflict of interest to declare.
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Mertens C, Zhong M, Krishnaraj R, Zou W, Chen X, Darnell JE., Jr Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 2006;20:3372–3381. doi: 10.1101/gad.1485406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meyer T, Hendry L, Begitt A, John S, Vinkemeier U. A single residue modulates tyrosine dephosphorylation, oligomerization, and nuclear accumulation of stat transcription factors. J Biol Chem. 2004;279:18998–19007. doi: 10.1074/jbc.M400766200. [DOI] [PubMed] [Google Scholar]
- 4.Boisson-Dupuis S, Kong XF, Okada S, Cypowyj S, Puel A, Abel L, et al. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr Opin Immunol. 2012;24:364–378. doi: 10.1016/j.coi.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 6.Chapgier A, Kong XF, Boisson-Dupuis S, Jouanguy E, Averbuch D, Feinberg J, et al. A partial form of recessive STAT1 deficiency in humans. J Clin Invest. 2009;119:1502–1514. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kong XF, Ciancanelli M, Al-Hajjar S, Alsina L, Zumwalt T, Bustamante J, et al. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood. 2010;116:5895–5906. doi: 10.1182/blood-2010-04-280586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kristensen IA, Veirum JE, Moller BK, Christiansen M. Novel STAT1 alleles in a patient with impaired resistance to mycobacteria. J Clin Immunol. 2011;31:265–271. doi: 10.1007/s10875-010-9480-8. [DOI] [PubMed] [Google Scholar]
- 9.Dupuis S, Dargemont C, Fieschi C, Thomassin N, Rosenzweig S, Harris J, et al. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293:300–303. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 10.Chapgier A, Boisson-Dupuis S, Jouanguy E, Vogt G, Feinberg J, Prochnicka-Chalufour A, et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata O, Okada S, Tsumura M, Kagawa R, Miki M, Kawaguchi H, et al. Heterozygosity for the Y701C STAT1 mutation in a multiplex kindred with multifocal osteomyelitis. Haematologica. 2013;98:1641–1649. doi: 10.3324/haematol.2013.083741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsumura M, Okada S, Sakai H, Yasunaga S, Ohtsubo M, Murata T, et al. Dominant-negative STAT1 SH2 domain mutations in unrelated patients with Mendelian susceptibility to mycobacterial disease. Hum Mutat. 2012;33:1377–1387. doi: 10.1002/humu.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sampaio EP, Bax HI, Hsu AP, Kristosturyan E, Pechacek J, Chandrasekaran P, et al. A novel STAT1 mutation associated with disseminated mycobacterial disease. J Clin Immunol. 2012;32:681–689. doi: 10.1007/s10875-012-9659-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustamante J, Boisson-Dupuis S, Abel L, Casanova JL. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-gamma immunity. Semin Immunol. 2014;26:454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, et al. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, et al. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 17.Toth B, Mehes L, Tasko S, Szalai Z, Tulassay Z, Cypowyj S, et al. Herpes in STAT1 gain-of-function mutation [corrected] Lancet. 2012;379:2500. doi: 10.1016/S0140-6736(12)60365-1. [DOI] [PubMed] [Google Scholar]
- 18.Aldave JC, Cachay E, Nunez L, Chunga A, Murillo S, Cypowyj S, et al. A 1-year-old girl with a gain-of-function STAT1 mutation treated with hematopoietic stem cell transplantation. J Clin Immunol. 2013;33:1273–1275. doi: 10.1007/s10875-013-9947-5. [DOI] [PubMed] [Google Scholar]
- 19.Soltesz B, Toth B, Shabashova N, Bondarenko A, Okada S, Cypowyj S, et al. New and recurrent gain-of-function STAT1 mutations in patients with chronic mucocutaneous candidiasis from Eastern and Central Europe. J Med Genet. 2013;50:567–578. doi: 10.1136/jmedgenet-2013-101570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mekki N, Ben-Mustapha I, Liu L, Boussofara L, Okada S, Cypowyj S, et al. IL-17 T cells’ defective differentiation in vitro despite normal range ex vivo in chronic mucocutaneous candidiasis due to STAT1 mutation. J Invest Dermatol. 2014;134:1155–1157. doi: 10.1038/jid.2013.480. [DOI] [PubMed] [Google Scholar]
- 21.Mizoguchi Y, Tsumura M, Okada S, Hirata O, Minegishi S, Imai K, et al. Simple diagnosis of STAT1 gain-of-function alleles in patients with chronic mucocutaneous candidiasis. J Leukoc Biol. 2014;95:667–676. doi: 10.1189/jlb.0513250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takezaki S, Yamada M, Kato M, Park MJ, Maruyama K, Yamazaki Y, et al. Chronic mucocutaneous candidiasis caused by a gain-of-function mutation in the STAT1 DNA-binding domain. J Immunol. 2012;189:1521–1526. doi: 10.4049/jimmunol.1200926. [DOI] [PubMed] [Google Scholar]
- 23.Yamazaki Y, Yamada M, Kawai T, Morio T, Onodera M, Ueki M, et al. Two novel gain-of-function mutations of STAT1 responsible for chronic mucocutaneous candidiasis disease: impaired production of IL-17A and IL-22, and the presence of anti-IL-17F autoantibody. J Immunol. 2014;193:4880–4887. doi: 10.4049/jimmunol.1401467. [DOI] [PubMed] [Google Scholar]
- 24.Shamriz O, Engelhard D, Rajs AP, Kaidar-Shwartz H, Casanova JL, Averbuch D. Mycobacterium szulgai chronic multifocal osteomyelitis in an adolescent with inherited STAT1 deficiency. Pediatr Infect Dis J. 2013;32:1345–1347. doi: 10.1097/01.inf.0000437067.43859.4c. [DOI] [PubMed] [Google Scholar]
- 25.Frans G, Moens L, Schaballie H, Van Eyck L, Borgers H, Wuyts M, et al. Gain-of-function mutations in signal transducer and activator of transcription 1 (STAT1): chronic mucocutaneous candidiasis accompanied by enamel defects and delayed dental shedding. J Allergy Clin Immunol. 2014;134:1209–1213. e6. doi: 10.1016/j.jaci.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilic SS, Puel A, Casanova JL. Orf Infection in a Patient with Stat1 Gain-of-Function. J Clin Immunol. 2014;35:80–83. doi: 10.1007/s10875-014-0111-7. [DOI] [PubMed] [Google Scholar]
- 27.Kumar N, Hanks ME, Chandrasekaran P, Davis BC, Hsu AP, Van Wagoner NJ, et al. Gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation-related primary immunodeficiency is associated with disseminated mucormycosis. J Allergy Clin Immunol. 2014;134:236–239. doi: 10.1016/j.jaci.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sampaio EP, Hsu AP, Pechacek J, Bax HI, Dias DL, Paulson ML, et al. Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J Allergy Clin Immunol. 2013;131:1624–1634. doi: 10.1016/j.jaci.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharfe N, Nahum A, Newell A, Dadi H, Ngan B, Pereira SL, et al. Fatal combined immunodeficiency associated with heterozygous mutation in STAT1. J Allergy Clin Immunol. 2014;133:807–817. doi: 10.1016/j.jaci.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 30.Higgins E, Al Shehri T, McAleer MA, Conlon N, Feighery C, Lilic D, et al. Use of ruxolitinib to successfully treat chronic mucocutaneous candidiasis caused by gain-of-function signal transducer and activator of transcription 1 (STAT1) mutation. J Allergy Clin Immunol. 2015;135:551–553. doi: 10.1016/j.jaci.2014.12.1867. [DOI] [PubMed] [Google Scholar]
- 31.Romberg N, Morbach H, Lawrence MG, Kim S, Kang I, Holland SM, et al. Gain-of-function STAT1 mutations are associated with PD-L1 overexpression and a defect in B-cell survival. J Allergy Clin Immunol. 2013;131:1691–1693. doi: 10.1016/j.jaci.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uzel G, Sampaio EP, Lawrence MG, Hsu AP, Hackett M, Dorsey MJ, et al. Dominant gain-of-function STAT1 mutations in FOXP3 wild-type immune dysregulation-polyendocrinopathy-enteropathy-X-linked-like syndrome. J Allergy Clin Immunol. 2013;131:1611–1623. doi: 10.1016/j.jaci.2012.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hori T, Ohnishi H, Teramoto T, Tsubouchi K, Naiki T, Hirose Y, et al. Autosomal-dominant chronic mucocutaneous candidiasis with STAT1-mutation can be complicated with chronic active hepatitis and hypothyroidism. J Clin Immunol. 2012;32:1213–1220. doi: 10.1007/s10875-012-9744-6. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Lin Z, Gao L, Wang A, Wan Z, Chen W, et al. Exome sequencing reveals a signal transducer and activator of transcription 1 (STAT1) mutation in a child with recalcitrant cutaneous fusariosis. J Allergy Clin Immunol. 2013;131:1242–1243. doi: 10.1016/j.jaci.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Lee PP, Mao H, Yang W, Chan KW, Ho MH, Lee TL, et al. Penicillium marneffei infection and impaired IFN-gamma immunity in humans with autosomal-dominant gain-of-phosphorylation STAT1 mutations. J Allergy Clin Immunol. 2014;133:894–896. e5. doi: 10.1016/j.jaci.2013.08.051. [DOI] [PubMed] [Google Scholar]
- 36.Depner M, Fuchs S, Raabe J, Frede N, Glocker C, Doffinger R, et al. The Extended Clinical Phenotype of 26 Patients with Chronic Mucocutaneous Candidiasis due to Gain-of-Function Mutations in STAT1. J Clin Immunol. 2016;36:73–84. doi: 10.1007/s10875-015-0214-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhalla F, Fox H, Davenport EE, Sadler R, Anzilotti C, van Schouwenburg PA, et al. Chronic mucocutaneous candidiasis: characterization of a family with STAT-1 gain-of-function and development of an ex-vivo assay for Th17 deficiency of diagnostic utility. Clin Exp Immunol. 2016;184:216–227. doi: 10.1111/cei.12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giardino G, Somma D, Cirillo E, Ruggiero G, Terrazzano G, Rubino V, et al. Novel STAT1 gain-of-function mutation and suppurative infections. Pediatr Allergy Immunol. 2016;27:220–223. doi: 10.1111/pai.12496. [DOI] [PubMed] [Google Scholar]
- 39.Kataoka S, Muramatsu H, Okuno Y, Hayashi Y, Mizoguchi Y, Tsumura M, et al. Extrapulmonary tuberculosis mimicking Mendelian susceptibility to mycobacterial disease in a patient with signal transducer and activator of transcription 1 (STAT1) gain-of-function mutation. J Allergy Clin Immunol. 2016;137:619–622. e1. doi: 10.1016/j.jaci.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Martinez L, Martinez-Saavedra MT, Fuentes-Prior P, Barnadas M, Rubiales MV, Noda J, et al. A novel gain-of-function STAT1 mutation resulting in basal phosphorylation of STAT1 and increased distal IFN-gamma-mediated responses in chronic mucocutaneous candidiasis. Mol Immunol. 2015;68:597–605. doi: 10.1016/j.molimm.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 41.Nielsen J, Kofod-Olsen E, Spaun E, Larsen CS, Christiansen M, Mogensen TH. A STAT1-gain-of-function mutation causing Th17 deficiency with chronic mucocutaneous candidiasis, psoriasiform hyperkeratosis and dermatophytosis. BMJ Case Rep. 2015;2015 doi: 10.1136/bcr-2015-211372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanimura M, Dohi K, Hirayama M, Sato Y, Sugiura E, Nakajima H, et al. Recurrent inflammatory aortic aneurysms in chronic mucocutaneous candidiasis with a gain-of-function STAT1 mutation. Int J Cardiol. 2015;196:88–90. doi: 10.1016/j.ijcard.2015.05.183. [DOI] [PubMed] [Google Scholar]
- 43.Zheng J, van de Veerdonk FL, Crossland KL, Smeekens SP, Chan CM, Al Shehri T, et al. Gain-of-function STAT1 mutations impair STAT3 activity in patients with chronic mucocutaneous candidiasis (CMC) Eur J Immunol. 2015;45:2834–2846. doi: 10.1002/eji.201445344. [DOI] [PubMed] [Google Scholar]
- 44.Zerbe CS, Marciano BE, Katial RK, Santos CB, Adamo N, Hsu AP, et al. Progressive Multifocal Leukoencephalopathy in Primary Immune Deficiencies: Stat1 Gain of Function and Review of the Literature. Clin Infect Dis. 2016;62:986–994. doi: 10.1093/cid/civ1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Toubiana J, Okada S, Hiller J, Oleastro M, Lagos Gomez M, Aldave Becerra JC, et al. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 2016;127:3154–3164. doi: 10.1182/blood-2015-11-679902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baris S, Alroqi F, Kiykim A, Karakoc-Aydiner E, Ogulur I, Ozen A, et al. Severe Early-Onset Combined Immunodeficiency due to Heterozygous Gain-of-Function Mutations in STAT1. J Clin Immunol. 2016 doi: 10.1007/s10875-016-0312-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dotta L, Scomodon O, Padoan R, Timpano S, Plebani A, Soresina A, et al. Clinical heterogeneity of dominant chronic mucocutaneous candidiasis disease: presenting as treatment-resistant candidiasis and chronic lung disease. Clin Immunol. 2016;164:1–9. doi: 10.1016/j.clim.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 48.Shamriz O, Engelhard D, Rajs AP, Kaidar-Shwartz H, Casanova JL, Averbuch D. Mycobacterium szulgai Chronic Multifocal Osteomyelitis in an Adolescent with Inherited STAT1 Deficiency. Pediatr Infect Dis J. 2013 doi: 10.1097/01.inf.0000437067.43859.4c. [DOI] [PubMed] [Google Scholar]
- 49.Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, Piouffre L, et al. A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 50.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 51.Mao X, Ren Z, Parker GN, Sondermann H, Pastorello MA, Wang W, et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 52.Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, et al. Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci U S A. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 54.Moreira IS, Fernandes PA, Ramos MJ. Hot spots--a review of the protein-protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 55.Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Itan Y, Shang L, Boisson B, Ciancanelli MJ, Markle JG, Martinez-Barricarte R, et al. The mutation significance cutoff: gene-level thresholds for variant predictions. Nat Methods. 2016;13:109–110. doi: 10.1038/nmeth.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Itan Y, Casanova JL. Can the impact of human genetic variations be predicted? Proc Natl Acad Sci U S A. 2015;112:11426–11427. doi: 10.1073/pnas.1515057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Itan Y, Shang L, Boisson B, Patin E, Bolze A, Moncada-Velez M, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc Natl Acad Sci U S A. 2015;112:13615–13620. doi: 10.1073/pnas.1518646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miosge LA, Field MA, Sontani Y, Cho V, Johnson S, Palkova A, et al. Comparison of predicted and actual consequences of missense mutations. Proc Natl Acad Sci U S A. 2015;112:E5189–E5198. doi: 10.1073/pnas.1511585112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gussow AB, Petrovski S, Wang Q, Allen AS, Goldstein DB. The intolerance to functional genetic variation of protein domains predicts the localization of pathogenic mutations within genes. Genome Biol. 2016;17:9. doi: 10.1186/s13059-016-0869-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smeekens SP, Plantinga TS, van de Veerdonk FL, Heinhuis B, Hoischen A, Joosten LA, et al. STAT1 hyperphosphorylation and defective IL12R/IL23R signaling underlie defective immunity in autosomal dominant chronic mucocutaneous candidiasis. PLoS One. 2011;6:e29248. doi: 10.1371/journal.pone.0029248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Al Rushood M, McCusker C, Mazer B, Alizadehfar R, Grimbacher B, Depner M, et al. Autosomal dominant cases of chronic mucocutaneous candidiasis segregates with mutations of signal transducer and activator of transcription 1, but not of Toll-like receptor 3. J Pediatr. 2013;163:277–279. doi: 10.1016/j.jpeds.2013.02.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.