Abstract
We report three symptomatic children with profound biotinidase deficiency from Sri Lanka. All three children presented with typical clinical features of the disorder. The first is homozygous for a missense mutation in the BTD gene (c.98_104 del7insTCC; p.Cys33PhefsX36) that is commonly seen in the western countries, the second is homozygous for a novel missense mutation (p.Ala439Asp), and the third is the first reported instance of a contiguous gene deletion causing the enzyme deficiency. In addition, this latter finding exemplifies the importance of considering a deletion within the BTD gene for reconciling enzymatic activity with genotype, which can occur in asymptomatic children who are identified by newborn screening.
Keywords: Biotinidase, Biotinidase deficiency, Contiguous gene deletion, Mutation, Enzyme deficiency, Congenital myasthenic syndrome
1. Introduction
Biotinidase (EC 3.5.1.12) is the enzyme responsible for cleaving and recycling biotin from biocytin and from dietary protein-bound sources [1], [2]. Profound biotinidase deficiency (less than 10% of mean normal serum activity) (OMIM #253260) is an autosomal recessively inherited metabolic disorder [3]. Individuals with profound biotinidase deficiency, if untreated, usually exhibit seizures, hypotonia, skin rash, alopecia, vision problems, hearing loss, and developmental delay with accompanying ketolactic acidosis and organic aciduria [2], [3]. Symptoms of the disorder can be successfully improved or prevented with pharmacological doses of oral biotin. However, if untreated, vision or hearing problems and developmental delay occur, they are usually irreversible [3]. Although all states in the United States and many countries screen their newborns for the disorder, some countries do not. Sri Lanka is one of the countries that do not screen for the disorder.
The gene for biotinidase has been isolated and characterized [4], [5] and over 150 mutations causing biotinidase deficiency have been identified [6]. We now report the mutations responsible for profound biotinidase deficiency in three symptomatic children from Sri Lanka. Two of these children are from consanguineous parents, whereas one child was placed in foster care and consanguinity could not be established. One child is homozygous for one of the more common mutations found in children in the United States, a second child is homozygous for a novel missense mutation, and the third child is homozygous for a contiguous gene deletion that encompassed three genes, including BTD. This latter deletion is instructive in that it exemplifies the importance of reconciling enzymatic activity with the genotype especially for confirming if putative positive asymptomatic children by newborn screening actually have biotinidase deficiency.
2. Materials and methods
2.1. Subjects
Individuals with biotinidase deficiency were identified because they developed symptoms that prompted follow-up by measurement of biotinidase activity in blood spots used for newborn screening in Sri Lanka [7]. Subsequently, profound biotinidase deficiency (less than 10% of mean normal activity) was found in all three children. Family and clinical history were obtained with the consent of the families.
2.2. Mutation analysis
Blood for DNA was obtained from these children and from their parents when possible to confirm allelic assignment. Genomic DNA was isolated from peripheral blood lymphocytes using the Gentra Puregene DNA isolation kit (Research Triangle Park, NC) according to the manufacturer's recommendations. The concentration of DNA in each sample was calculated from the optical density at 260 nm and diluted to a concentration of 0.05 μg/μL. DNA sequencing of the biotinidase (BTD) gene was performed by PCR amplification using primers and conditions described previously [8]. All exonic and intron–exon boundaries of the BTD gene were sequenced.
2.3. Deletion studies
DNA labeling was carried out with the Enzo CGH labeling kit for oligo arrays (Enzo Life Sciences, Plymouth Meeting, PA). Array CGH was performed with 0.5 μg of DNA according to the manufacturer's protocol (Agilent Technologies, Santa Clara, CA) using probes for each exon and flanking introns of the genes on the array as previously described [6].
2.4. Microarray studies
High density SNP microarray studies were performed using Affymetrix Cytoscan HD SNP ArrayTM. Briefly, whole genome PCR amplification is performed on DNA extracted from peripheral blood followed by hybridization, staining, washing and scanning. Data is then analyzed using Affymetrix Chromosome Analysis SuiteTM (ChAS) software (Genome build: hg19). This array contains 2.67 million copy number markers/probes (1.9 million non-polymorphic probes/markers and 750,000 SNP probes/markers) that detect copy number variations (CNVs), loss of heterozygosity (LOH), and segmental or whole chromosome uniparental isodisomy. It has coverage of 12,000 OMIM genes with global resolution of 5–10 kb.
3. Case studies
Patient 1, a male, was the 36 1/2-week product of a normal pregnancy. There were no prenatal complications. The child was given to foster parents at nine days of age. He first developed seizures at seven weeks of age which did not respond to antiepileptic medications. He exhibited screaming episodes, staring gaze, jitteriness and lethargy.
A dried blood spot test revealed elevated 3-hydroxyisovalerylcarnitine and undetectable biotinidase activity (deficient activity is less than 0.029 AU; normal activity is greater than 0.219 AU). Samples were not available from the parents. He improved clinically on biotin therapy and is now asymptomatic at 11 months of age.
Mutation analysis revealed that this boy was homozygous for c.98_104 del7insTCC; p.C33FfsX36 in the BTD gene [6]. This mutation is a relative common mutation in Western European countries and results in a frameshift and the synthesis of a truncated, inactive enzyme protein causing profound biotinidase deficiency.
Patient 2, a male, is the product of first cousin parents. The child was well until two months of age when he exhibited seizures which did not respond to antiepileptic medications. He developed global developmental delay. At two years of age he had increased frequency of the seizures, tachypnea, metabolic acidosis, alopecia and a seborrheic skin rash. He was admitted to the intensive care unit of the hospital. A blood sample for biotinidase activity was sent to the United Kingdom where they found undetectable enzymatic activity. They also found elevations of 3-hydroxyisovalerate, 3-methylcrotonylglycine and methylcitrate in his urine. The child was then treated with biotin and essentially all of his symptoms resolved or markedly improved; however, the child, who is now 11 years old, continues to have global developmental delay.
At the time of his diagnosis, his asymptomatic, seven-month-old sister, was also found to have biotinidase deficiency. Unfortunately, biochemical studies were not performed on her prior to initiating biotin therapy. She was started on biotin and has been continually treated. She is now four years of age and continues to be asymptomatic. She was found to be homozygous for the same mutation as her brother.
The parents are cousins. In addition to a sister, the parents had a prior pregnancy that resulted in a stillborn infant. The cause of the demise is unknown.
Mutation analysis revealed that the proband is homozygous for c.1316T>C; p.A439D and homozygous for the c.1413T>C; p.C471C polymorphism. This missense mutation is a novel alteration and is predicted to be pathogenic by Polyphen 2, SIFT and Mutation Taster analyses. The mother is heterozygous for both the mutation and the polymorphism. The father was deceased.
Patient 3 is a male who exhibited poor head and poor motor coordination and global developmental delay. At 41 days of age he developed uncontrolled, generalized tonic–clonic seizures which initially occurred about once a day, but increased in frequency. Upon presentation to the clinic at eight months of age, he had microcephaly with over-riding sutures, mild hepatomegaly, alopecia, frequent respiratory tract infections and seborrheic dermatitis. His muscle tone and reflexes were normal. He had no ptosis or choking episodes as reported by the mother. He readily improved after biotin therapy was initiated.
A plasma acylcarnitine profile revealed an elevation of 3-hydroxyisovalerylcarnitine. His biotinidase activity in dried blood-soaked filter paper was markedly deficient at 0.017 AU. The child was diagnosed with profound biotinidase deficiency.
Biotin therapy was initiated and his seizures stopped, his skin rash resolved, his motor delays resolved, he has less frequent respiratory tract infections and his growth become normal; however, he still had microcephaly.
The parents are first cousins. There are four older siblings. A sister, who exhibited seizures at two months of age that were uncontrollable with anti-epileptic medications, died at six months. Another sister developed seizures on the 45th day of life. She was also unresponsive to medication and she died at two months of age. Post-mortem examinations were not performed. Two other sisters, five and 13 years old, are both healthy.
Mutation analysis was performed on DNA from the proband and his parents. PCR products were obtained and sequenced for exons, 2, 3 and 4 of BTD for the proband. Despite multiple attempts using the routine and alternate primers, PCR amplification of exon 1 was unsuccessful. However, both parents and a control individual produced amplicons of the expected size for all combinations of exon 1 primers used. Sequence analysis was normal for exons 2, 3 and 4 of the proband and parents. In addition, the proband was homozygous for a polymorphism in exon 4 (c.1413T>C; p.C471C), detected in heterozygous form in each of the parents.
At this point we considered the possibility that the child may have a deletion of the BTD gene that involved a region encompassing exon 1 and the PCR primer sites.
Deletion testing was performed by exon-level array comparative genomic hybridization (array CGH) that indicated that the child was homozygous for a deletion involving the entire exon 1 of the BTD gene [9]. The parents were each found to be heterozygous for this deletion. We also determined that the next downstream gene, HACL1, was deleted, but due to available probe placement on the array, the extent of the deletion 5′ to the HACL1 gene could not be determined.
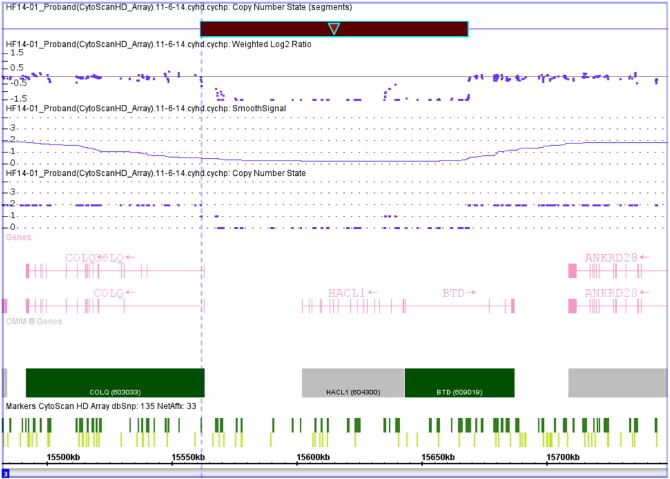
We, therefore, performed further array CGH analysis using a second probe set and confirmed that the deletion encompassed exon 1 of the BTD gene. This analysis also found that the entire HACL1 gene and exon 1 of the adjacent COLQ gene (Fig. 1). The end of the deletion occurs within the intron between exons 1 and 2 of the COLQ gene. The contiguous gene deletion is denoted (arr[hg19] 3p25.1(15,561,697–15,668,594)x0 and is approximately 107 kb. Microarray analysis revealed that both parents are heterozygous for the same deletion.
Fig. 1.
The 107 kb contiguous gene deletion (black bar) in Patient 3. The microarray data indicates that Patient 3 is homozygous for the deletion. The deletion includes exon 1 of the BTD gene, the entire HACL1 gene and a small portion of the 5′-end of the COLQ gene encompassing exon 1. Each of the parents is heterozygous for the identical deletion (not shown).
Since the COLQ gene is associated with congenital myasthenic syndrome, we obtained further medical history and additional physical examination of the child looking for any of the known clinical features of congenital myasthenic syndrome. The proband did not exhibit any of the following: facial, neck, proximal, distal or axial muscle weakness, ptosis, abnormal tendon reflexes, scoliosis, ophthalmoparesis, respiratory crises, fatigability, hyperventilation, waddling gait/ataxia, dysphagia or chewing difficulties, history of poor cry or sucking, scapular winging or slow pupillary light response. He did have delayed milestones, but they improved on the biotin therapy. No electromyelographic, repetitive nerve stimulation, compound muscle action potential or response to acetylcholinesterase inhibitor studies have been performed to date due to availability and costs.
4. Results
Mutations were found in all three individuals with profound biotinidase deficiency. The first child has a relatively common mutation known to cause profound biotinidase deficiency. The second individual was found to be homozygous for a novel mutation that is predicted to be pathogenic and apparently causes profound biotinidase deficiency. In the third child with biotinidase deficiency the disease was caused by a mechanism that has not been reported previously. This individual is homozygous for a contiguous gene deletion that involves three genes. The deletion of exon 1 of the BTD gene results in the failure to synthesize an active enzyme product since exon 1 includes the initiation site and leader signal sequence of the enzyme.
In addition, this deletion includes the HACL1 gene(OMIM #604300). HACL1 encodes for 2-hydroxyphytanoyl-CoA lyase which is involved in alpha-oxidation of phytanic acid and other fatty acids, but no mutations or deletions of the gene have been reported to be pathogenic in humans [10].
Finally, the deletion includes exon 1 of the COLQ gene (OMIM #603033). COLQ encodes the collagen-tail subunit of acetylcholinesterase (Acetylcholinesterase collagenic tail peptide also known as AChE Q subunit, acetylcholinesterase-associated collagen) found in the neuromuscular junction [11]. Defects in COLQ are known to cause congenital myasthenia syndrome (OMIM #603034) [11], [12]. We failed to identify any clinical features of congenital myasthenia syndrome in this child. However, although we were unable to find any reports of individuals with congenital myasthenic syndrome who had a mutation in exon 1 or a deletion of exon 1, we would predict that a homozygous deletion of this exon would have deleterious effects on the gene product and result in the disease state.
The child's symptoms at this time appear to be solely attributable to biotinidase deficiency and he has markedly improved with biotin therapy. However, children with congenital myasthenic syndrome may not exhibit symptoms in the first few years of life [11], [12]. Therefore, we informed the parents about the possibility or likelihood that this child will exhibit symptoms of that disorder. Further electrophysiological studies are required to determine if the child has congenital myasthenic syndrome and is truly at risk of developing symptoms. These studies are currently in progress.
The latter child is the first reported case of biotinidase deficiency due to an exon-level deletion. The fact that the child was the product of consanguineous parents facilitated the discovery of the deletion. If instead a heterozygous deletion had been present, along with a second point mutation elsewhere in the BTD gene, only the normal allele would have been amplified, revealing a normal sequence for exon 1. The proband in this family clearly exhibited symptoms of biotinidase deficiency with biotinidase activity indicative of profound biotinidase deficiency. In addition, the child's symptoms improved rapidly and markedly to biotin treatment. Because of the clinical and enzymatic findings, the enzymatic results were not questioned in this case. However, if this scenario occurred in an asymptomatic infant identified by newborn screening, it would be less obvious how to reconcile the enzymatic data with the identification of a single mutation. In fact, the enzymatic data may have been questioned.
Children have been initially diagnosed with profound biotinidase deficiency by enzymatic activity and subsequently were found to have higher enzymatic activities. This has been attributed to adverse storage of samples [3]. This is precisely why we have recommended that the clinician obtain samples from the proband, parents and an unrelated control for confirmation enzymatic testing [3], [13].
We have three children with profound biotinidase deficiency from Sri Lanka, who are homozygous for distinctly different mutations; two are novel mutations with one caused by a mechanism that has been suspected as a potential cause of the disorder, but has never been reported previously.
Most importantly, the child with the contiguous gene deletion has made it clear that correctly performed enzymatic testing should not be ignored if only a single mutation is identified in an individual suspected of having profound biotinidase deficiency, particularly in cases identified by newborn screening when symptoms may not be clear. The possibility of a deletion encompassing part or all of the BTD gene must be considered in those children having enzymatic deficiency that is not adequately reconciled with the results of mutation analysis.
Acknowledgment
This work was supported by the Safra Research Fund, Henry Ford Hospital.
References
- 1.Pispa J. Animal biotinidase. Ann. Med. Exp. Biol. Fenn. 1965;43(Suppl. 5):1–39. [PubMed] [Google Scholar]
- 2.Wolf B. Disorders of biotin metabolism. In: Scriver C.R., Beaudet A.L., Sly W.S., Valle D., editors. The Metabolic and Molecular Bases of Inherited Disease. McGraw-Hill; New York: 2001. pp. 3935–3962. [Google Scholar]
- 3.Wolf B. Biotinidase deficiency: if you have to have an inherited metabolic disease, this is the one to have. Genet. Med. 2012;14:565–575. doi: 10.1038/gim.2011.6. [DOI] [PubMed] [Google Scholar]
- 4.Cole H., Reynolds T.R., Buck G.B., Lockyer J.M., Denson T., Spence J.E., Hymes J., Wolf B. Human serum biotinidase: cDNA cloning, sequence and characterization. J. Biol. Chem. 1994;269:6566–6570. [PubMed] [Google Scholar]
- 5.Knight H.C., Reynolds T.R., Meyers G.A., Pomponio R.J., Buck G.A., Wolf B. Structure of the human biotinidase gene. Mamm. Genome. 1998;9:327–330. doi: 10.1007/s003359900760. [DOI] [PubMed] [Google Scholar]
- 6.Procter M., Wolf B., Crockett D.K., Mao R. The biotinidase variants registry: a paradigm public database. Genes Genomics Genet. 2015 doi: 10.1534/g3.113.005835. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heard G.S., Secor McVoy J.R., Wolf B. A screening method for biotinidase deficiency in newborns. Clin. Chem. 1984;30:125–127. [PubMed] [Google Scholar]
- 8.Pomponio R.J., Hymes J., Reynolds T.R., Meyers G.A., Fleischhauer K., Buck G.A., Wolf B. Mutations in the human biotinidase gene that cause profound biotinidase deficiency in symptomatic children: molecular, biochemical and clinical analysis. Pediatr. Res. 1997;42:840–848. doi: 10.1203/00006450-199712000-00020. [DOI] [PubMed] [Google Scholar]
- 9.Aradhya S., Lewis R., Bonaga T., Nwokekeh N., Stafford A., Boggs B., Hrusha K., Smaoul N., Compton J.G., Richard G., Suchy S. Exon-level array CGH in a large clinical cohort demonstrates increased sensitivity of diagnostic testing for Mendelian disorders. Genet. Med. 2012;14:594–603. doi: 10.1038/gim.2011.65. [DOI] [PubMed] [Google Scholar]
- 10.HACL1 . 2015. 2-hydroxyacyl-CoA lyase 1.http://www.ncbi.nlm.nih.gov/gene/26061 [Google Scholar]
- 11.Abicht A., Muller J., Lochmuller H. Congenital myasthenic syndromes. Gene Rev. 2014 [Google Scholar]
- 12.Mihaylova V., Muller J.S., Vilchez J.J., Salih M.A., Kabiraj M.M., D'Amico A., Bertini E., Wolfle J., Scheiner F., Kurlemann G., Rasic V.M., Siskova D., Colomer J., Herczegfalvi A., Fabricova K., Weschke B., Scola R., Hoellen F., Schara U., Abicht A., Lochmuller H. Clinical and molecular genetic findings in COLQ-mutant congenital myasthenic syndromes. Brain. 2008;131:747–759. doi: 10.1093/brain/awm325. [DOI] [PubMed] [Google Scholar]
- 13.Wolf B. Clinical issues and frequent questions about biotinidase deficiency. Mol. Genet. Metab. 2013;100:6–13. doi: 10.1016/j.ymgme.2010.01.003. [DOI] [PubMed] [Google Scholar]