Abstract
Mutations in B3GALT6, encoding the galactosyltransferase II (GalT-II) involved in the synthesis of the glycosaminoglycan (GAG) linkage region of proteoglycans (PGs), have recently been associated with a spectrum of connective tissue disorders, including spondyloepimetaphyseal dysplasia with joint laxity type 1 (SEMDJL1) and Ehlers–Danlos-like syndrome. Here, we report on two sisters compound heterozygous for two novel B3GALT6 mutations that presented with severe short stature and progressive kyphoscoliosis, joint hypermobility and laxity, hyperextensible skin, platyspondyly, short ilia, and elbow malalignment. Microarray-based transcriptome analysis revealed the differential expression of several genes encoding extracellular matrix (ECM) structural components, including COMP, SPP1, COL5A1, and COL15A1, enzymes involved in GAG synthesis and in ECM remodeling, such as CSGALNACT1, CHPF, LOXL3, and STEAP4, signaling transduction molecules of the TGFβ/BMP pathway, i.e., GDF6, GDF15, and BMPER, and transcription factors of the HOX and LIM families implicated in skeletal and limb development. Immunofluorescence analyses confirmed the down-regulated expression of some of these genes, in particular of the cartilage oligomeric matrix protein and osteopontin, encoded by COMP and SPP1, respectively, and showed the predominant reduction and disassembly of the heparan sulfate specific GAGs, as well as of the PG perlecan and type III and V collagens. The key role of GalT-II in GAG synthesis and the crucial biological functions of PGs are consistent with the perturbation of many physiological functions that are critical for the correct architecture and homeostasis of various connective tissues, including skin, bone, cartilage, tendons, and ligaments, and generates the wide phenotypic spectrum of GalT-II-deficient patients.
Abbreviations: Abs, antibodies; ATCS, adducted-thumb club foot syndrome; BMP, bone morphogenetic proteins; ChPF, chondroitin polymerizing factor; ChSy, chondroitin synthase; COLLs, collagens; COLLI, type I collagen; COLLIII, type III collagen; COLLV, type V collagen; COMP, cartilage oligomeric matrix protein; CS, chondroitin sulfate; CSGALNACT1, chondroitin sulfate N-acetylgalactosaminyltransferase 1; CTDs, connective tissue disorders; C4ST, chondroitin 4-sulfotransferase; C6ST, chondroitin 6-sulfotransferase; DCN, decorin; DEGs, differentially expressed genes; D4ST, dermatan 4 sulfotransferase 1; DS, dermatan sulfate; EDS, Ehlers–Danlos syndrome; ECM, extracellular matrix; FN, fibronectin; GAGs, glycosaminoglycans; Gal, galactose; GalNAc, N-acetylgalactosamine; GalNAcT, β1,4-N-acetylgalactosaminyltransferase; GalNAcT-16, N-acetylgalactosaminyltransferase 16; GalT-I/II, galactosyltransferase I and II; GlcA, glucuronic acid; GlcAT, glucuronosyltransferase; GlcNAc, N-acetylglucosamine; GlcNAcT, α1,4-N-acetylglucosaminyltransferase; GalNAc4S-6ST, GalNAc 4-sulfate 6-O-sulfotransferase; GO, gene ontology; HA, hyaluronic acid; HAS2, hyaluronan synthase 2; Hep, heparin; HOX, homeobox gene family; HPO, human phenotype ontology; HS, heparan sulfate; IdoA, iduronic acid; IF, immunofluorescence microscopy studies; OPN, osteopontin; PGs, proteoglycans; PTC, premature termination codon of translation; qPCR, quantitative polymerase chain reaction; SEMDJL1, spondyloepimetaphyseal dysplasia with joint laxity type 1; TNs, tenascins; Xyl, xylose; XylT, xylosyltransferase
Keywords: B3GALT6, GalT-II deficiency, Spondyloepimetaphyseal dysplasia with joint laxity type 1, Ehlers–Danlos syndrome, Cartilage oligomeric matrix protein, Osteopontin
Highlights
-
•
Clinical features/molecular characterization of two patients with spondyloepimetaphyseal dysplasia with joint laxity type 1
-
•
Identification of two novel B3GALT6 mutations
-
•
First report of transcriptome-wide gene expression profiling on GalT-II-deficient fibroblasts
-
•
Immunofluorescence studies of several ECM structural components in GalT-II-deficient cells
-
•
Enlargement of the knowledge on the GalT-II deficiency’s molecular pathogenesis
1. Introduction
The major connective tissues of the body, such as skin, tendon, ligaments, cartilage and bone, provide the structural and informational framework that is necessary for development. The extracellular matrix (ECM) of connective tissues is a complex interacting network of proteins, glycoproteins and proteoglycans (PGs), which provide the dynamic and essential environment that supports cell maintenance, growth and differentiation. Many mutations have been identified in the genes of either structural ECM components or enzymes involved in their post-translational processing and folding. The molecular basis of how these mutations cause the myriad of connective tissue disorders (CTDs) depends on the function of the gene product, its tissue distribution and the nature of the mutation [1], [2].
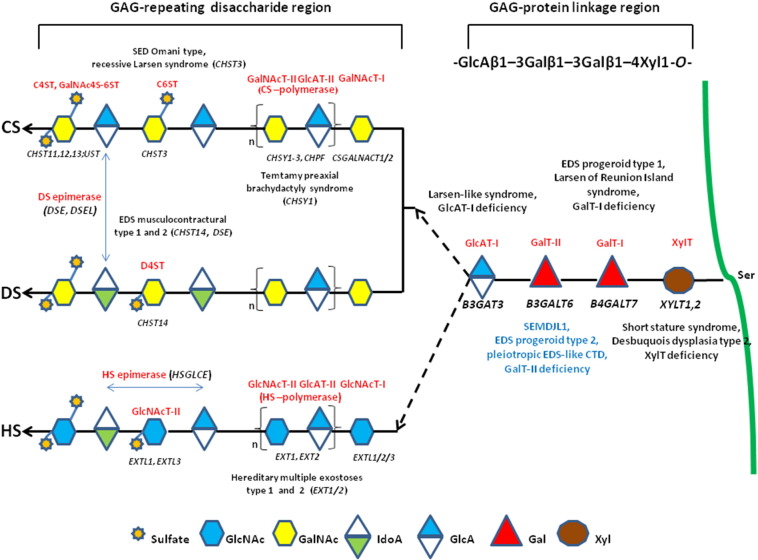
PGs are the major component of the ECM in several tissues, and are composed of core proteins and a variable number of glycosaminoglycan (GAG) side chains that are long, unbranched polysaccharides consisting of repeating disaccharide units of an amino sugar (N-acetylglucosamine [GlcNAc] or N-acetylgalactosamine [GalNAc]) and a uronic acid (glucuronic [GlcA] or iduronic acid [IdoA]). GAGs are distinguished in galactosaminoglycans such as chondroitin sulfate (CS) and dermatan sulfate (DS), and glucosaminoglycans such as hyaluronic acid (HA), keratan sulfate, heparan sulfate (HS), and heparin (Hep) [3]. The PG superfamily is subdivided into two major groups depending on the composition of the disaccharide building block consisting of (GalNAc-GlcA)n or (GlcNAc-GlcA)n to form CS/DS PGs (e.g., versican, decorin) and HS PGs (e.g., perlecan, glypican), respectively [4]. Most GAGs, except for keratan sulfate and HA, are O-glycans that bind to the glycan via an oxygen molecule in the serine of the core protein [5]. GAGs exhibit wide-ranging structural and functional diversity as a result of their complex biosynthetic pathway, as summarized in Fig. 1, which is a tightly regulated process that enables the modified polysaccharide to selectively interact with a large range of ligands [4].
Fig. 1.
Schematic presentation of the synthesis of the GAG backbones of CS/DS or HS chains and related genetic disorders. Each enzyme (glycosyltransferase and/or epimerase, in red) and its coding gene (in black) are described near the sugar symbols. After the synthesis of specific core proteins, the synthesis of the GAG-protein linkage region is initiated by XylT, which transfers [Xyl] to the specific Ser in the endoplasmic reticulum. The synthesis of the linkage region is completed by the consecutive addition of two molecules of [Gal], added by GalT-I/II, followed by the transfer of [GlcA] catalyzed by GlcAT-I in the Golgi. The first [GalNAc] is transferred to the [GlcA] in the linkage region by GalNAcT-I, resulting in the initiation of the synthesis of the repeating disaccharide region of CS/DS chains. Alternatively, the addition of the [GlcNAc] to the linkage region by GlcNAcT-I induces HS biosynthesis. The repeating disaccharide region of the CS/DS chain is elongated by alternate additions of [GlcA] and [GalNAc] catalyzed by CS-GlcAT-II and GalNAcT-II activities of a heterocomplex formed by ChSy and ChPF. The polymerization of the HS chain is catalyzed by HS-GlcAT-II and GlcNAcT-II activities. GAG chains are finally matured by tightly controlled modifications, i.e., epimerization and sulfation. Concerning CS chains, sulfation occurs mainly at positions 4 and 6 of [GalNAc] and position 2 of [GlcA] and is catalyzed by various sulfotransferases. After the formation of the chondroitin backbone, [GlcA] is converted into [IdoA] resulting in the formation of the dermatan backbone, in which position 4 of [GalNAc] is sulfated by D4ST. The unbranched HS chain undergoes final maturation, which consists of a complex series of processing reactions involving [GlcNAc] deacetylation and sulfation, epimerization of [GlcA], and subsequent O-sulfation at different positions (for an extensive review see reference [6]).
The impact of the incorrect synthesis of GAGs is demonstrated by the identification of mutations in several genes encoding key enzymes of this pathway, leading to severe multisystemic disorders [6] (Fig. 1). Diseases caused by defects in the enzymes involved in the synthesis of the so-called GAG linkage region, which is common to the CS/DS and HS/(Hep) chains, are categorized as GAG linkeropathies. The first such disorder to be identified was the progeroid form of Ehlers–Danlos syndrome (EDS), caused by mutations in B4GALT7 (GalT-I deficiency) [7], [8]. The characteristics of this EDS type include an aged appearance, short stature, generalized osteopenia, hypermobile joints, hypotonic muscles, and loose skin [9]. Recently, the recurrent p.Arg270Cys mutation in B4GALT7 was demonstrated to cause “Larsen of Reunion Island syndrome” characterized by dwarfism, hyperlaxity, and multiple dislocations [10]. Mutations in B3GAT3 (GlcAT-I deficiency) were identified in a recessive Larsen-like syndrome characterized by short stature, dysmorphic facies, joint dislocations, and cardiovascular defects [11]. Mutations in XYLT1 were identified in patients with short stature, mild skeletal changes, and moderate intellectual disability [12]. Recently, XYLT1 mutations were demonstrated to cause Desbuquois dysplasia type 2 characterized by dislocations of large joints, severe pre- and post-natal growth retardation, joint laxity, and advanced carpal ossification [13].
Downstream in the biosynthetic pathway, mutations in CHST3 are associated with spondyloepiphyseal dysplasia Omani type, a severe chrondrodysplasia with major involvement of the spine [14]. Furthermore, CHST3 mutations were disclosed in a number of patients who have presented with various diagnoses, i.e., recessive Larsen syndrome, chrondrodysplasia with multiple dislocations, humerospinal dysostosis, and Desbuquois syndrome [15], [16]. Mutations in CHSY1 cause Temtamy pre-axial brachydactyly syndrome [17], and defects in HS synthesis (EXT1 and EXT2) are linked with hereditary multiple exostosis types 1 and 2 [18]. Mutations in CHST14 and DSE were reported in musculocontractural EDS types 1 and 2, respectively [19], [20], and in adducted-thumb club foot syndrome (ATCS) [21]. Musculocontractural EDS and ATCS patients show characteristic craniofacial features, multiple congenital contractures, progressive joint and skin laxity, kyphoscoliosis, and multisystem complications; these features overlap with those of EDS kyphoscoliotic type, which is caused by a deficiency in lysyl hydroxylase encoded by PLOD1[22], [23].
In 2013, two independent research studies identified recessive mutations in B3GALT6 that constitute a novel linkeropathy (GalT-II deficiency) [24], [25]. In particular, Nakajima et al. [24], identified B3GALT6 mutations in 7 Japanese families with spondyloepimetaphyseal dysplasia with joint laxity type 1 (SEMDJL1, or SEMDJL Beighton type). In SEMDJL1, first described in South African patients by Beighton and Kozlowski [26], short stature is associated with progressive spinal malalignment, articular hypermobility especially prominent in the hands, thoracic asymmetry, club feet, dislocation of the radial heads, mild skin extensibility, spatulate terminal phalanges, and lip and palate clefts. Considering the potentially lethal spinal cord compression and pulmonary complications, survival into adulthood is reported to be unusual [26], [27]. In other patients from 3 families with a progeroid form of EDS, without B4GALT7 mutations, Nakajima et al. [24] performed targeted B3GALT6 sequencing and disclosed causal mutations in all of them, defining the progeroid type 2 form of EDS. Furthermore, Malfait et al. [25] identified B3GALT6 mutations in 3 unrelated families with a severe recessive disease, i.e., “pleiotropic EDS-like CTD”, characterized by severe kyphoscoliosis, joint hyperlaxity and contractures, SEMD, skin fragility, intellectual disability, and multiple early-onset fractures. Following the identification of B3GALT6 as the causal gene of SEMDJL1, Vorster et al. [28] identified B3GALT6 mutations in 8 prototype South African families.
Here, we report 2 Moldavian sisters with clinical features reminiscent of the kyphoscoliotic type of EDS, or of the SEMDJL1 skeletal dysplasia. Genetic testing revealed compound heterozygosity for two novel B3GALT6 mutations, leading to the diagnosis of GalT-II deficiency. To identify molecular mechanisms contributing to the etiopathogenesis of GalT-II deficiency, transcriptome-wide expression profiling of skin fibroblasts of the two sisters and immunofluorescence studies were performed.
2. Patients, materials and methods
2.1. Patients
We report on a Moldavian family with two affected sisters, 25 and 21 years old, born from healthy non-consanguineous parents. Both probands were evaluated by clinical geneticists, and dermatologists. Written informed consent was obtained for each proband and parents before sample collection for genetic analyses. Specific consents were obtained for skin biopsy of the patients and for publication of their clinical pictures. This study was approved by the medical ethical committee of the University Hospital Spedali Civili of Brescia and was performed in accordance with the Declaration of Helsinki Principles.
2.2. Cell culture and antibodies
Primary dermal fibroblast cultures from both patients and from 3 sex- and age-matched healthy donors were established from skin biopsies by standard procedures. Fibroblasts were routinely maintained at 37 °C in a 5% CO2 atmosphere in modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg/ml penicillin and streptomycin (Life Technologies, Carlsbad, CA). Fibroblasts were expanded until full confluency was achieved and then harvested by trypsin treatment at the same passage number. A polyclonal rabbit antibody (Ab) against human fibronectin (FN) and monoclonal antibodies (mAbs) against all of the human isoforms of tenascin (TNs) (clone BC-24) and against CS GAGs (clone CS-56) were from Sigma Chemicals (St. Louis, MO). The goat anti-type I collagen (COLLI) Ab and the anti-α5β1 (clone JBS5) and anti-α2β1 (clone BHA.2) integrin mAbs were from Millipore (Billerica, MA). The goat anti-type V collagen (COLLV) and anti-type III collagen (COLLIII) Abs were from LifeSpan BioSciences, Inc. (Seattle, WA). The rabbit anti-versican core protein Ab was from Affinity Bioreagents (Golden, CO), the rabbit anti-perlecan core protein and goat anti-ostepontin (OPN) affinity purified Abs were from Santa Cruz Biotec, Inc. (Heidelberg, Germany), the anti-decorin (DCN) mAb (clone 115402) was from R&D Systems, Inc. (Minneapolis, MN), and the anti-HS GAG mAb was from USBiological Life Sciences (Swampscott, MA). The anti-cartilage oligomeric matrix protein (COMP) RabmAb® EPR6289(2) was from Abcam (Cambridge, UK). Rhodamine-conjugated anti-goat IgG and anti-mouse IgM secondary Abs were from Calbiochem–Novabiochem INTL, and the Alexa Fluor® 488 anti-rabbit and Alexa Fluor® 594 anti-mouse secondary Abs were from Life Technologies.
2.3. Indirect immunofluorescence analysis (IF)
To analyze the FN, COLLI, COLLIII, COLLV, and TNs ECM organization and the α5β1 and α2β1 integrin distribution, fibroblasts were grown, fixed and immunoreacted as previously reported [29]. The HS and CS GAGs, perlecan, versican, and DCN organization were analyzed 48 h from seeding by fixing cells with 3% paraformaldehyde in PBS for 20 min. Subsequently, cells were incubated for 1 h with primary Abs (anti-HS, anti-CS and anti-perlecan at 10 μg/ml; anti-DCN at 25 μg/ml; and anti-versican diluted 1:50 in BSA 1%). The COMP and OPN distribution were analyzed by fixing fibroblasts in cold methanol and reacting cells with 1.5 μg/ml anti-COMP and 4 μg/ml anti-OPN Ab, respectively. After washing in PBS, cells were incubated for 1 h with anti-mouse or anti-rabbit secondary Abs conjugated to Alexa Fluor® 594 and 488, respectively, with the anti-goat IgG or anti-mouse IgM. The IF signals were acquired by a CCD black-and-white TV camera (SensiCam-PCO Computer Optics GmbH, Germany) mounted on a Zeiss fluorescence-Axiovert microscope and digitalized by Image Pro Plus software (Media Cybernetics, Silver Spring, MD). All of the experiments were repeated three times.
2.4. DNA/RNA isolation and mutation analysis
Genomic DNA was purified from blood samples of both patients and their parents using the Wizard Genomic DNA purification Kit (Qiagen, Hilden, Germany) by standard procedures. Total RNA was extracted from dermal fibroblasts of both sisters and 3 unrelated controls using the Qiagen RNeasy kit according to the manufactures' instructions (Qiagen, Hilden, Germany). RNA quality control was assessed on an Agilent 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA, USA).
Primers were designed for all coding exons, including their intron–exon boundaries, using the Primer3 tool. Primer sequences were checked for the absence of known SNPs, based on dbSNP version 139. In particular, all exons and intron flanking regions of the PLOD1 and B3GALT6 genes were amplified by standardized PCR. After enzymatic cleanup of the PCR, all fragments were sequenced in both orientations using the BigDye® Terminator Cycle Sequencing kit protocol followed by capillary electrophoresis on an ABI3130XL Genetic Analyzer (Life Technologies). To detect deletions/duplications, frequently observed in the PLOD1 gene [30], MLPA analysis with the commercially available P359-A1 probemix (MRC-Holland, Amsterdam, The Netherlands) was also performed according to the manufacturer's recommendations. The identified B3GALT6 mutations were not found in 100 control individuals or in the 1000 Genomes Project or NHLBI Exome Variant Server (ESP6500) databases. The mutations were annotated according to HGVS nomenclature. To evaluate causality based on the alteration of the protein structure, the prediction programs Align GVGD, MutationTaster, PolyPhen-2, SIFT and KD4v in the Alamut software version 2.4 were used.
2.5. Microarray hybridization and data analysis
To screen for candidate genes that may contribute to the pathogenesis of GalT-II deficiency, we performed transcriptome-wide expression profiling using the Affymetrix Gene 1.0 ST platform by comparing the gene expression patterns of skin fibroblasts of the two affected sisters with those of three healthy sex and age matched individuals. The microarray analysis was performed starting from 250 ng of total RNA per sample, and labeled targets were prepared using the Ambion® Whole Transcript Expression Kit (Life Technologies) and the GeneChip® WT Terminal Labeling and Controls Kit (Affymetrix UK Ltd, Wycombe La High Wycombe, UK) following the manufacturers' instructions. In brief, total RNA was primed with synthetic primers containing a T7 promoter sequence, reverse transcribed into first-strand cDNA and converted into double-stranded cDNA. Following in vitro transcription, 10 μg of cRNA was reverse transcribed using random primers. Then, 5.5 μg of second-cycle cDNA was fragmented, biotin labeled, and hybridized for 16 h at 45 °C onto the arrays. The chips were then washed in the Affymetrix GeneChip Fluidics station FS 450, scanned using the Affymetrix GeneChip scanner 3000 7G system, and analyzed with the GeneChip® Operating Software. The resulting CEL files were analyzed using Partek® Genomics Suite software version 6.6 (Partek Inc., St. Louis, MO), and gene-level calculation was performed using the Robust Multichip Average algorithm. Gene expression profile analysis was performed using one-way ANOVA. The differentially expressed genes (DEGs) in patients vs controls were selected based on a fold change ± 2.0 and with a P-value < 0.05. Enrichment analysis of Gene Ontology (GO), pathway analysis and functional annotation clustering were performed using the PANTHER (http://www.pantherdb.org/) or DAVID (http://david.abcc.ncifcrf.gov/) databases. To define a genotype–phenotype correlation of the microarray results, we intersected the list of DEGs with the Human Phenotype Ontology (HPO) database using the WebGestalt Toolkit (http://bioinfo.vanderbilt.edu/webgestalt/). All microarray data are MIAME compliant, and the raw data have been deposited in the MIAME compliant GEO database (accession number GSE58312).
2.6. Quantitative real-time PCR (qPCR)
The relative mRNA levels of a series of selected genes identified by array analysis were confirmed by qPCR. Three micrograms of total RNA purified from skin fibroblasts of both patients and 3 unrelated healthy individuals were reverse-transcribed with random primers by a standard procedure. qPCR reactions were performed with SYBR Green qPCR Master Mix using the ABI PRISM 7500 Real-Time PCR System by standard thermal cycling conditions (Life Technologies). The HPRT, ATP5B and CYC1 reference genes were also amplified for normalization of cDNA loading. Relative mRNA expression levels were normalized to the geometric mean of these housekeeping genes and analyzed using the 2−(ΔΔCt) equation. Amplification plots, dissociation curves, and threshold cycle values were generated by ABI Sequence detection system software version 1.3.1. Statistical analyses were performed with the GraphPad Prism software (San Diego, CA, USA). The results were expressed as the mean values of relative quantification ± SEM. Statistical significance between groups was determined using one-way ANOVA. P-values were corrected for multiple testing using Tukey's method (*P < 0.05, **P < 0.01 and ***P < 0.001).
3. Results
3.1. Clinical findings
3.1.1. Patient 1
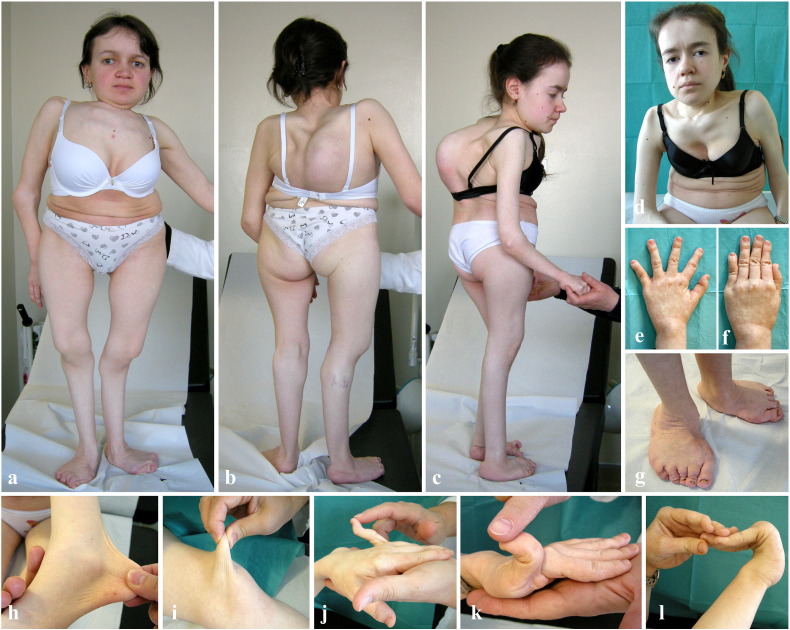
The 25-year-old patient was born following an uneventful pregnancy and delivery. At birth, congenital bilateral hip dislocation and unspecified feet deformities were disclosed. At 4 years of age, the mother reported the first onset of severe and progressive kyphoscoliosis. Other reported clinical data were humeral fracture after minor trauma and episodes of dyspnea at rest. On examination at 25 years of age, we observed the following: flat and oval face, broad nose, long upper lip, light blue sclerae, yellowish teeth and periodontitis, disproportionate short stature (121 cm, lower than the 3rd centile), short trunk (51 cm, upper to lower segment ratio (U/L): 0,36, n.v. 1), thin, doughy and generalized hyperextensible skin without scarring and easy bruising, toenail dystrophy, joint hypermobility according to the Beighton score (6/9) with recurrent dislocations, elbows contracture, severe kyphoscoliosis, genua valga, flat feet, widespread bone deformities and muscular hypotonia (Fig. 2). She required walking aids. Cognitive function and ophthalmologic evaluation were normal.
Fig. 2.
Clinical findings of the two patients. Patient 1 (a, b, g, i) and patient 2 (c–f, h, j–l) show disproportionate short stature, short trunk, severe kyphoscoliosis, generalized bone deformities, genua valga, elbow contracture; dysmorphic signs: frontal bossing, blue sclerae, flat oval face, broad nose (a–d); hand deformities with finger contractures and spatulate terminal phalanges (e, f); foot deformities, i.e., flat foot, hallux valgus, toe contractures (g); hyperextensible skin (h, i); marked hypermobility of small joints (j–l).
3.1.2. Patient 2
In the 21-year-old patient, congenital bilateral hip dislocation, unspecified feet deformities, progressive kyphoscoliosis since 7 years of age, and episodes of dyspnea at rest were reported. On examination at 20 years of age, she presented with a clinical phenotype similar to that of her older sister: flat and oval face, broad nose, long upper lip, light blue sclerae, yellowish teeth, disproportionate short stature (116 cm, below the 3rd centile), short trunk (44 cm, U/L: 0,36, n.v. 1), thin, doughy and generalized hypextensible skin without scarring and easy bruising, toe nail dystrophy, joint hypermobility according to the Beighton score (6/9) with recurrent dislocations, elbow contracture, severe kyphoscoliosis, genua valga, flat feet, widespread bone deformities and muscular hypotonia (Fig. 2). She also required walking aids. Cognitive function and ophthalmologic evaluation were normal.
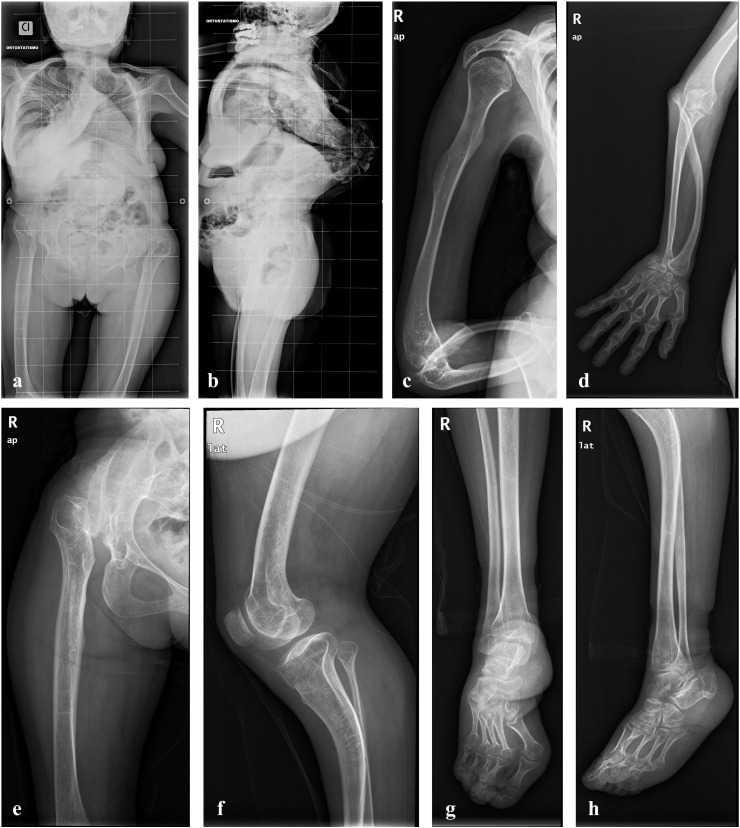
Total skeletal X-ray, heart ultrasound, electrocardiogram and spirometry were performed in both patients after our first evaluation. Skeletal X-ray disclosed in both 4th degree thoraco-lumbar kyphoscoliosis, generalized bone loss and spondyloepimetaphyseal deformities, i.e., platyspondyly, vertebral listhesis, overtubulation of long bones, dislocated and arched radial epiphysis, hypoplastic and dislocated femoral head (Fig. 3). Heart ultrasound revealed mitral valve prolapse in both sisters, and electrocardiography revealed complete right bundle branch block in patient 1. Spirometry disclosed mild restrictive lung disease in both patients.
Fig. 3.
Radiological findings in patient 1 (a, b, e–h) and patient 2 (c, d). a–h: severe bone loss; a, b: thoraco-lumbar kyphoscoliosis (4th degree), platyspondily, and vertebral listhesis; c, d: dislocated and arched radial epiphysis (distal and proximal), arched radial bone, elbow malalignment, humeral overtubulation; e: hypoplastic and dislocated femoral head, femoral overtubulation, and short ileum; f: knee malalignment; g, h: feet deformities with metacarpal shortening (astragalus hypoplasia).
These clinical findings suggested a recessive form of skeletal dysplasia and, in particular, SEMDJL1, for which the genetic basis was not yet known at the time of clinical evaluation, or a recessive form of EDS, i.e., the kyphoscoliotic type.
3.2. Mutation analysis
Initially, mutational screening of PLOD1 was performed with a negative result. Shortly after, B3GALT6 was identified as the causal gene responsible for SEMDJL1 [24], fitting well with the clinical features of our patients. Therefore, sequencing of B3GALT6 was performed in the probands and showed compound heterozygosity for two novel mutations and led to the diagnosis of GalT-II deficiency. In particular, genotyping and segregation analyses revealed the maternal inherited c.227del mutation, leading to a frameshift and the formation of a premature termination codon (PTC) (p.Ile76Thrfs*202), and the paternal c.766C > T transition, resulting in the substitution of a positively charged arginine residue with a larger and neutral tryptophan residue in position 256 (p.Arg256Trp) (Fig. S1). All the in silico prediction algorithms characterized the p.Arg256Trp substitution as disease causing because the mutated and highly conserved residue is located in the glycosyltransferase family 31 domain (IPR002659).
3.3. Transcriptome-wide expression profiling
To investigate the pathomechanisms involved in GalT-II deficiency, we performed a transcriptome-wide expression analysis comparing the gene expression patterns of the two affected patients' skin fibroblasts with those of 3 healthy individuals. A total of 334 DEGs were identified with the Partek Genomic Suite software and by applying a fold change threshold ≥ ± 2 in combination with a P-value ≤ 0.05. In particular, 172 genes were significantly up-regulated, and 162 genes were significantly down-regulated. In Table 1 a selection of up- and down-regulated transcripts, based on fold change, statistical significance, and possible biological relevance related to GalT-II deficiency, is shown (see Table S1 for the full list of DEGs, and Fig. S2 for the scatter plot, hierarchical clustering and GO analysis of the DEGs).
Table 1.
Selection of the most up- and down-regulated DEGs identified in GalT-II-deficient skin fibroblasts.
| Gene description | Gene symbol | Fold-change | P-value |
|---|---|---|---|
| Up-regulated genes | |||
| STEAP family member 4 | STEAP4 | 41.6 | 0.0004 |
| Sema domain, immunoglobulin domain (Ig), short basic domain, secreted 3A | SEMA3A | 15.6 | 0.01 |
| ATP-binding cassette, sub-family A (ABC1), member 8 | ABCA8 | 12.5 | 0.03 |
| Aquaporin 9 | AQP9 | 10.3 | 0.001 |
| Immunoglobulin superfamily, member 10 | IGSF10 | 8.6 | 0.003 |
| Tumor necrosis factor (ligand) superfamily, member 10 | TNFSF10 | 8.6 | 0.007 |
| Elav-like family member 2 | CELF2 | 8.0 | 0.02 |
| Alcohol dehydrogenase 1B (class I), beta polypeptide | ADH1B | 7.6 | 0.01 |
| Insulin-like growth factor binding protein 2 | IGFBP2 | 7.6 | 0.02 |
| Relaxin/insulin-like family peptide receptor 1 | RXFP1 | 6.7 | 0.02 |
| Interleukin 13 receptor, alpha 2 | IL13RA2 | 6.7 | 0.004 |
| Hyaluronan synthase 2 | HAS2 | 6.6 | 0.002 |
| Cytochrome P450, family 19, subfamily A, polypeptide 1 | CYP19A1 | 6.1 | 0.006 |
| Junctional adhesion molecule 2 | JAM2 | 6.0 | 0.007 |
| Collagen and calcium binding EGF domains 1 | CCBE1 | 5.5 | 0.01 |
| Chondroitin sulfate N-acetylgalactosaminyltransferase 1 | CSGALNACT1 | 4.7 | 0.01 |
| Peroxisome proliferator-activated receptor gamma | PPARG | 4.5 | 0.003 |
| Endothelin receptor type B | EDNRB | 4.4 | 0.001 |
| Polypeptide N-acetylgalactosaminyltransferase 16 | GALNTL1 | 3.4 | 0.04 |
| Down-regulated genes | |||
| LIM homeobox 9 | LHX9 | − 14.5 | 0.01 |
| Tumor necrosis factor (ligand) superfamily, member 4 | TNFSF4 | − 11.8 | 0.003 |
| Oxytocin receptor | OXTR | − 10.4 | 0.002 |
| Hairy and enhancer of split 1, (Drosophila) | HES1 | − 9.7 | 0.01 |
| Cartilage oligomeric matrix protein | COMP | − 9.1 | 0.004 |
| Phosphodiesterase 1C, calmodulin-dependent | PDE1C | − 8.9 | 0.004 |
| Nuclear receptor subfamily 4, group A, member 2 | NR4A2 | − 8.0 | 0.01 |
| Potassium voltage-gated channel, KQT-like subfamily, member 5 | KCNQ5 | − 7.9 | 0.02 |
| Mohawk homeobox | MKX | − 7.3 | 0.0007 |
| Cholinergic receptor, muscarinic 2 | CHRM2 | − 6.8 | 0.01 |
| Homeobox D10 | HOXD10 | − 6.8 | 0.02 |
| Cadherin 13, H-cadherin (heart) | CDH13 | − 4.2 | 0.005 |
| Transcription factor AP-2 alpha | TFAP2A | − 4.1 | 0.01 |
| Dickkopf homologue 3 (Xenopus laevis) | DKK3 | − 4.0 | 0.008 |
| Secreted frizzled-related protein 4 | SFRP4 | − 4.0 | 0.03 |
| Lysyl oxidase-like 3 | LOXL3 | − 3.6 | 0.02 |
| Collagen, type XV, alpha 1 | COL15A1 | − 3.6 | 0.005 |
| Matrix metallopeptidase 16 (membrane-inserted) | MMP16 | − 3.5 | 0.02 |
| Short stature homeobox | SHOX | − 3.3 | 0.01 |
| Sparc/osteonectin, cwcv and kazal-like domains proteoglycan | SPOCK1 | − 3.3 | 0.001 |
| Wingless-type MMTV integration site family, member 5A | WNT5A | − 3.3 | 0.03 |
| Homeobox D11 | HOXD11 | − 3.2 | 0.01 |
To identify differentially expressed pathways in GalT-II-deficient skin fibroblasts compared to control cells, a pathway-enrichment analysis was performed using the Pathway-Express algorithm enriched with curated pathways of the KEGG database, with a significance threshold of P-value < 0.05. This approach showed that the “axon guidance,” “cell adhesion molecule,” “cytokine–cytokine receptor interaction,” “calcium signaling,” “ECM–receptor interaction,” “focal adhesion,” and “TGF-beta signaling” pathways were the most affected biological pathways (Table 2).
Table 2.
Top canonical pathways affected in GalT-II-deficient skin fibroblasts.
| Name | P-value | Ratio⁎ | DEGs |
|---|---|---|---|
| Axon guidance | 3.30E-05 | 10/129 | EFNB2, NTN1, UNC5B, EPHA2, SEMA3A, CXCL12, MET, SEMA6A, SEMA5A, PLXNA2 |
| Cell adhesion molecules | 1.72E-04 | 9/34 | NLGN1, NCAM2, NLGN4X, CLDN11, CNTN1, HLA-DPA1, CDH2, JAM2, CD274 |
| Cytokine–cytokine receptor interaction | 8.04E-04 | 12/263 | CCL2, TNFRSF19, PDGFC, TNFSF10, KIT, IL1R1, CXCL6, CXCL1, CXCL12, CCL28, TNFSF4, MET |
| Calcium signaling pathway | 0.002 | 9/182 | GRPR, ADCY4, CHRM2, AVPR1A, AGTR1, ADNRB, PLCD3, OXTR, PDE1C |
| ECM–receptor interaction | 0.009 | 5/84 | CD47, COL5A1, SPP1, ITGA11, COMP |
| Focal adhesion | 0.03 | 7/203 | PDGFD, PDGFC, COL5A1, SPP1, MET, ITGA11, COMP |
| TGF-beta signaling pathway | 0.04 | 5/87 | ID1, GDF6, SMAD7, COMP, BAMBI |
| Neuroactive ligand–receptor interaction | 0.04 | 8/256 | GRPR, CHRM2, GRIK2, AVPR1A, AGTR1, EDNRB, RXFP1, OXTR |
Input genes/genes in pathway, the down-regulated genes are reported in bold.
To identify biological processes that are over- or under-represented in B3GALT6-deficient cells, we classified all up- and down-regulated genes using the Functional Annotation Cluster tool available in the DAVID database, by selecting only those terms with a P-value ≤ 0.05 and a number of genes in each annotation term ≥ 5. This analysis was performed on the 172 up-regulated genes and generated 11 distinct GO clusters. As shown in Table 3 and Table S2, the GO terms were enriched in genes with glycoprotein, leucine-rich repeat, and immunoglobulin-like domains and with functions necessary for chemotaxis, homeostatic processes, ion homeostasis, cell proliferation, migration, and morphogenesis. Interestingly, several up-regulated genes are involved in carbohydrate and GAG binding. Functional analysis of the 162 down-regulated genes resulted in 13 different clusters. From Table 4 and Table S3, it can been observed that the GO terms were particularly enriched in genes with glycoprotein- and DNA-binding domains that play a pivotal role in skeletal system and limb development, embryonic limb and tissue morphogenesis, and cell development. In particular, more than a few genes in the top clusters are essential for cell/tissue homeostasis and encode structural ECM components, such as collagens (COLLs), glycoproteins, and PGs (COL5A1, COL15A1, COL8A1, COMP, and SPP1), enzymes/proteins involved in post-translational processing and folding, and ECM remodeling and/or signaling (LOXL3, MMP16, SULF1, TIMP3, LFNG, WNT5A, SFRP4, GREM2, TNFSF4, PDGFC, CDH13, GDF6, and GDF15). Furthermore, different aspects of gene transcription regulation were affected because genes encoding transcription factors with homeobox protein domains (DLX2, HOXA11, SHOX, MKX, LHX9, HOXD10, and HOXD11) were the most represented.
Table 3.
DAVID functional annotation clustering of the up-regulated genes.
| Cluster | Enrichment score | Category | Term | P-value |
|---|---|---|---|---|
| 1 | 7.4 | SP_PIR_KEYWORDS | Disulfide bond | 2.99E-09 |
| SP_PIR_KEYWORDS | Signal | 1.68E-08 | ||
| SP_PIR_KEYWORDS | Glycoprotein | 1.81E-06 | ||
| 2 | 5.1 | UP_SEQ_FEATURE | Signal peptide | 2.11E-08 |
| GOTERM_CC_FAT | GO:0005615—extracellular space | 4.80E-05 | ||
| SP_PIR_KEYWORDS | Secreted | 2.88E-04 | ||
| 3 | 3.1 | GOTERM_CC_FAT | GO:0005887—integral to plasma membrane | 5.91E-05 |
| UP_SEQ_FEATURE | Topological domain:extracellular | 1.78E-04 | ||
| GOTERM_CC_FAT | GO:0044459—plasma membrane part | 2.22E-04 | ||
| 4 | 3.0 | INTERPRO | IPR013151:immunoglobulin | 7.32E-05 |
| SP_PIR_KEYWORDS | Immunoglobulin domain | 5.49E-04 | ||
| INTERPRO | IPR013783:immunoglobulin-like fold | 0.001 | ||
| 5 | 2.5 | GOTERM_BP_FAT | GO:0007610—behavior | 4.27E-04 |
| INTERPRO | IPR001811:small chemokine, interleukin-8-like | 4.46E-04 | ||
| GOTERM_BP_FAT | GO:0006935—chemotaxis | 7.39E-04 | ||
| 6 | 2.3 | GOTERM_MF_FAT | GO:0030247—polysaccharide binding | 0.003 |
| GOTERM_MF_FAT | GO:0030246—carbohydrate binding | 0.006 | ||
| GOTERM_MF_FAT | GO:0005539—glycosaminoglycan binding | 0.01 | ||
| 7 | 2.2 | GOTERM_BP_FAT | GO:0048878—chemical homeostasis | 9.66E-04 |
| GOTERM_BP_FAT | GO:0010033—response to organic substance | 0.002 | ||
| GOTERM_BP_FAT | GO:0042592—homeostatic process | 0.01 | ||
| 8 | 2.1 | GOTERM_BP_FAT | GO:0051270—regulation of cell motion | 0.002 |
| GOTERM_BP_FAT | GO:0008283—cell proliferation | 0.007 | ||
| GOTERM_BP_FAT | GO:0030334—regulation of cell migration | 0.004 | ||
| 9 | 1.9 | GOTERM_BP_FAT | GO:0048878—chemical homeostasis | 9.66E-04 |
| GOTERM_BP_FAT | GO:0044057—regulation of system process | 0.002 | ||
| GOTERM_BP_FAT | GO:0050801—ion homeostasis | 0.01 | ||
| 10 | 1.7 | INTERPRO | IPR003591:leucine-rich repeat, typical subtype | 0.005 |
| SMART | SM00013:LRRNT | 0.01 | ||
| 11 | 1.5 | GOTERM_BP_FAT | GO:0000902—cell morphogenesis | 0.006 |
| GOTERM_BP_FAT | GO:0000904—cell morphogenesis involved in differentiation | 0.02 | ||
| GOTERM_BP_FAT | GO:0006928—cell motion | 0.033 |
Table 4.
Selection of DAVID functional annotation clusters obtained with the down-regulated genes.
| Cluster | Enrichment score | Category | Term | P-value |
|---|---|---|---|---|
| 1 | 4.8 | SP_PIR_KEYWORDS | Glycoprotein | 1.35E-10 |
| UP_SEQ_FEATURE | Signal peptide | 3.22E-07 | ||
| GOTERM_CC_FAT | GO:0005576—extracellular region | 2.82E-04 | ||
| 2 | 3.1 | GOTERM_BP_FAT | GO:0001501—skeletal system development | 9.91E-05 |
| GOTERM_BP_FAT | GO:0060173—limb development | 2.66E-04 | ||
| GOTERM_BP_FAT | GO:0048598—embryonic morphogenesis | 0.005 | ||
| 3 | 2.6 | UP_SEQ_FEATURE | Glycosylation site:N-linked (GlcNAc…) | 7.53E-10 |
| GOTERM_CC_FAT | GO:0044459—plasma membrane part | 0.001 | ||
| UP_SEQ_FEATURE | Topological domain:extracellular | 0.005 | ||
| 4 | 2.46 | GOTERM_MF_FAT | GO:0003700—transcription factor activity | 6.84E-04 |
| GOTERM_MF_FAT | GO:0043565—sequence-specific DNA binding | 0.001 | ||
| INTERPRO | IPR001356:homeobox | 0.01 | ||
| 5 | 2.45 | GOTERM_BP_FAT | GO:0060284—regulation of cell development | 7.11E-05 |
| GOTERM_BP_FAT | GO:0045596—negative regulation of cell differentiation | 0.002 | ||
| GOTERM_BP_FAT | GO:0051960—regulation of nervous system development | 0.006 | ||
| 6 | 2.0 | GOTERM_MF_FAT | GO:0030528—transcription regulator activity | 2.91E-04 |
| GOTERM_MF_FAT | GO:0003677—DNA binding | 0.04 | ||
| SP_PIR_KEYWORDS | Transcription regulation | 0.03 | ||
| 7 | 1.98 | GOTERM_BP_FAT | GO:0042325—regulation of phosphorylation | 0.002 |
| GOTERM_BP_FAT | GO:0043549—regulation of kinase activity | 0.03 | ||
| GOTERM_BP_FAT | GO:0031399—regulation of protein modification process | 0.04 | ||
| 8 | 1.96 | GOTERM_BP_FAT | GO:0042981—regulation of apoptosis | 0.009 |
| GOTERM_BP_FAT | GO:0043066—negative regulation of apoptosis | 0.011 | ||
| 9 | 1.87 | GOTERM_BP_FAT | GO:0016481—negative regulation of transcription | 0.006 |
| GOTERM_BP_FAT | GO:0016564—transcription repressor activity | 0.013 | ||
| 10 | 1.54 | GOTERM_BP_FAT | GO:0030182—neuron differentiation | 0.0014 |
| GOTERM_BP_FAT | GO:0048812—neuron projection morphogenesis | 0.037 | ||
| 11 | 1.46 | INTERPRO | IPR000742:EGF-like, type 3 | 0.018 |
| SP_PIR_KEYWORDS | egf-like domain | 0.025 |
To assess whether the differential gene expression identified in GalT-II-deficient skin fibroblasts correlates with the patients' clinical features, we intersected the list of DEGs with the HPO project database, which is integrated in the WebGestalt Toolkit. This analysis indicated that the observed perturbation of gene expression should mainly be related to the phenotypic features that involve the skeletal system organization and the morphology of bone structures, fitting well with the clinical features of SEMDJL1. As shown in Table 5, the HPO terms that were particularly enriched in DEGs comprised of “abnormal morphology of the appendicular skeleton,” “abnormalities of limb bone morphology,” “abnormality of the digits,” “abnormality of the periorbital region,” “abdominal wall defect,” “aplasia/hypoplasia involving hands and bones of the upper limbs,” and “abnormality of spinal cord.” These different classes of phenotypic abnormalities principally involved transcription factors, such as HOXD10, HOXA11, SHOX, TBX3, and TFAP2A and other genes, such as COMP, BMPER, and GDF6, all having a central role either in embryonic or postnatal development. This consistent genotype–phenotype correlation also increases the overall confidence level of our microarray data.
Table 5.
Summary of the HPO analysis.
| HPO terms | DEGs associated with phenotypic abnormalities⁎ |
|---|---|
| Abnormal appendicular skeleton morphology (HP:0011844) and abnormality of limb bone morphology (HP:0002813) | CYP19A1, SHOX, LFNG, ANKH, RIPK4, PTCH1, CBS, HOXA11, CRLF1, DSP, EGR2, PYCR1, WNT5A, CHN1, BMPER, GDF6, CCBE1, TBX3, COL5A1, TFAP2A, EDNRB, RBPJ, SNORD116-1, SEMA3A, KIT, COMP, MASP1, HOXD10, CNTN1 |
| Abnormality of the digits (HP:0011297) | SHOX, LFNG, RIPK4, PTCH1, CBS, HOXA11, CRLF1, DSP, EGR2, WNT5A, CHN1, BMPER, GDF6, CCBE1, TBX3, TFAP2A, EDNRB, RBPJ, SNORD116-1, COMP, MASP1, CNTN1 |
| Abnormality of the periorbital region (HP:0000606) | BDNF, ANKH, LDLR, RIPK4, PTCH1, CDON, DSP, PYCR1, WNT5A, CHN1, BMPER, CCBE1, AFF2, TBX3, COL5A1, TFAP2A, EDNRB, RBPJ, SNORD116-1, KIT, MASP1 |
| Abdominal wall defect (HP:0010866) | BDNF, BMPER, LFNG, CCBE1, TBX3, COL5A1, CBS, ZEB1, PTCH1, MASP1, PYCR1, WNT5A |
| Aplasia/hypoplasia involving hands and bones of the upper limbs (HP:0005927, HP:0006496) | CHN1, SHOX, CCBE1, TBX3, TFAP2A, RIPK4, PTCH1, RBPJ, CRLF1, COMP, MASP1, WNT5A |
| Thoracoabdominal wall defects (HP:0100656) | BDNF, BMPER, LFNG, CCBE1, TBX3, COL5A1, CBS, ZEB1, PTCH1, MASP1, PYCR1, WNT5A |
| Abnormality of spinal cord (HP:0002143) | CHN1, ENPP1, BMPER, GDF6, LFNG, CCBE1, PTCH1, COMP, MASP1 |
| Vertebral segmentation defect (HP: 0003422) | PTCH1, CHN1, BMPER, GDF6, LFNG, WNT5A |
Genes down-regulated in affected sisters are reported in bold.
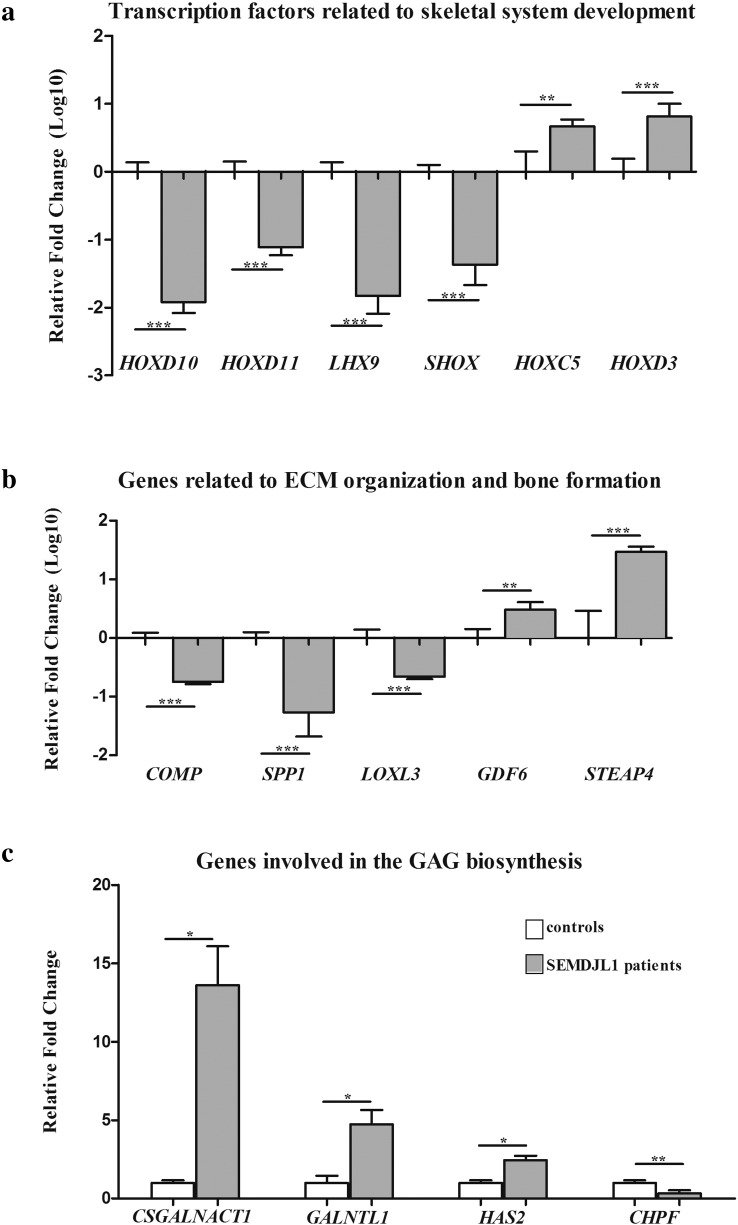
To further confirm the soundness of the gene expression data obtained from microarray analysis, we performed qPCR and verified the differential expression of a subset of DEGs. Genes were prioritized based on fold change, statistical significance, and biological relevance related to GalT-II deficiency. Specifically, we focused on transcription factors associated with skeletal system development, on genes associated with ECM organization and bone morphogenesis, and on genes related to GAG biosynthesis and carbohydrate binding. As shown in Fig. 4A, qPCR confirmed the marked decrease of the transcriptional levels of HOXD10, HOXD11, LHX9 and SHOX as well as the increased expression of HOXC5 and HOXD3. Similarly, the expression levels of COMP and SPP1 were confirmed to be down-regulated. These genes encode the cartilage oligomeric matrix protein (COMP) and osteopontin (OPN) respectively, proteins that are involved in the structural integrity of the ECM and in the bone matrix mineralization. LOXL3, which encodes an extracellular copper-dependent amine oxidase involved in the formation of crosslinks in COLLs and elastin, was also confirmed down-regulated. The highly increased expression of STEAP4, which encodes a member of the six-transmembrane epithelial antigen of prostate protein family with a critical role during osteoclast differentiation, was also validated (Fig. 4B). qPCR also showed the increase of the expression levels of CSGALNACT1, which encodes a subunit of the GalNAcT-1 glycosyltransferase (Fig. 1), of GALNTL1, which encodes a member of the N-acetylgalactosaminyltransferases (GalNAcT-16) involved in the biosynthesis of mucin-type O-glycans, and HAS2, which encodes the hyaluronan synthase 2 involved in the biosynthesis of HA. CHPF, which encodes the chondroitin polymerizing factor (ChPf) that enters the CS polymerase heterocomplex responsible for elongating the repeating disaccharide region of the CS/DS chains (Fig. 1), was confirmed to be down-regulated (Fig. 4C). Overall, qPCR results correlated well with the microarray expression data with the exception of GDF6, which encodes a member of the bone morphogenetic protein (BMP) family. This gene was found to be down-regulated in the microarray analyses (Table S1), whereas qPCR showed an up-regulation.
Fig. 4.
qPCR validation of the microarray expression data. The relative mRNA expression levels of selected genes related to skeletal system development (a), ECM organization and bone formation (b), and GAG biosynthesis (c), were determined with the 2−(ΔΔCt) method normalized with the geometric mean of the HPRT, ATP5B and CYC1 reference genes. Bars represent the mean ratio of target gene expression in patients' fibroblasts compared to three unrelated healthy individuals. qPCR was performed in triplicate, and the results are expressed as means ± SEM. In A and B, the relative mRNA levels of target genes in patients versus controls are expressed as Log10 transformed values. In C, the non-transformed relative fold change is shown. Statistical significance (*P < 0.05, **P < 0.01 and ***P < 0.001) was calculated with one-way ANOVA and the Tukey post hoc test.
Altogether, the results obtained from transcriptome-wide expression profiling indicate that mutations in B3GALT6 lead to the perturbation of many physiological functions, from early morphogenetic processes to postnatal development, allowing for the formation and function of various connective tissues.
3.4. Immunofluorescence microscopy studies
To investigate the pathophysiological consequences of GalT-II deficiency on ECM architecture, the expression of the CS and HS GAGs, the distribution of the PGs versican, perlecan, and DCN, and the organization of COLLI, COLLIII, COLLV, FN, TNs and the α2β1 and α5β1 COLLs and FN integrin receptors were investigated by IF. Furthermore, because the gene expression analysis showed a strong decrease of the expression levels of COMP and SPP1, the organization of the two corresponding proteins, i.e., COMP, and OPN, was also investigated.
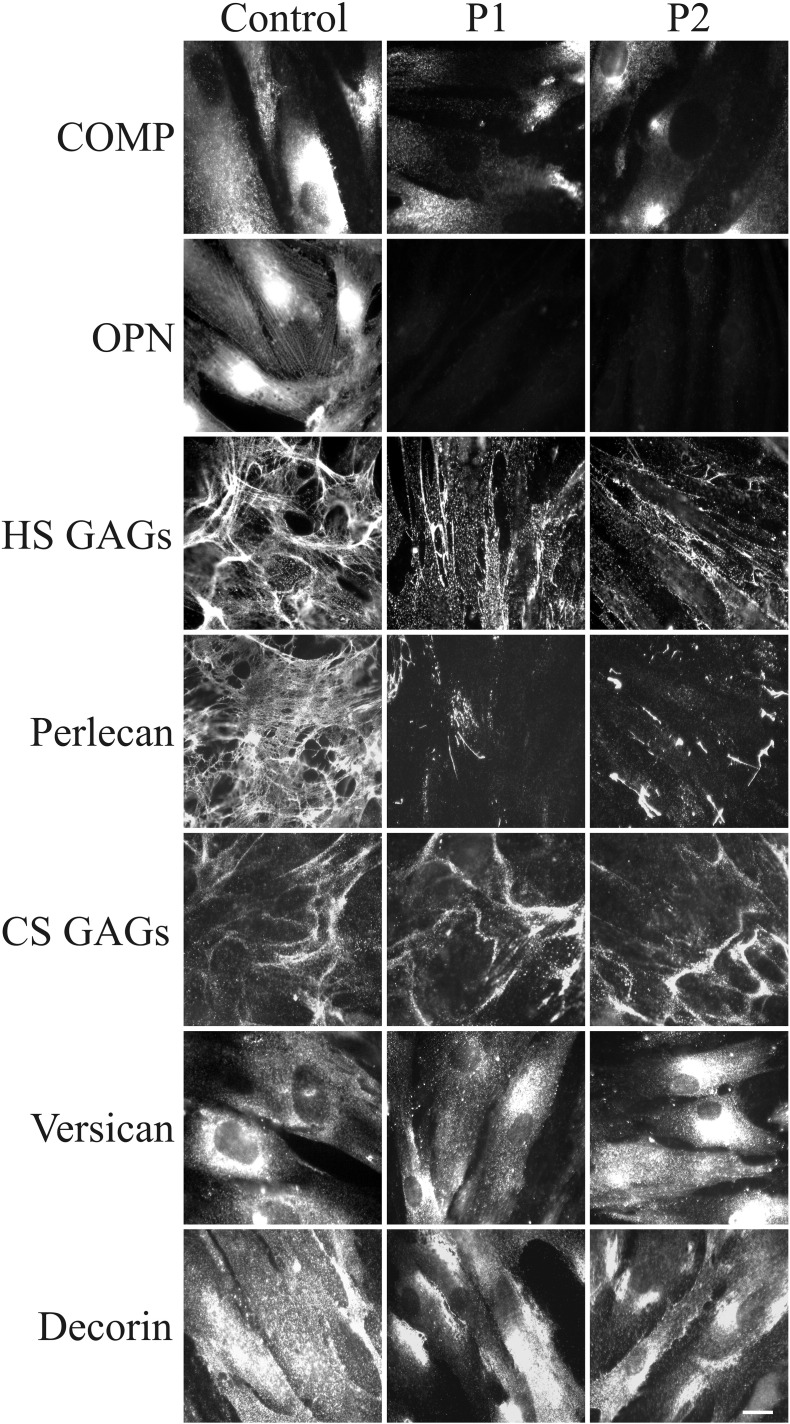
As shown in Fig. 5, either COMP or OPN were expressed by control fibroblasts. In particular, COMP was distributed in the plasma membrane, and large amounts of protein were stored inside the cell; OPN was highly expressed with a perinuclear and cytoskeleton-associated signal. In the patients' fibroblasts, COMP was markedly reduced, and OPN was quite undetectable. These protein data correlate well with the gene expression results.
Fig. 5.
Organization of COMP, OPN, GAGs, and PGs in SEMDJL1 patients' fibroblasts. Control and GalT-II-deficient (patient 1: P1 and patient 2: P2) skin fibroblasts were analyzed with specific Abs directed against COMP, OPN, HS, and CS GAGs chains, and the core proteins of the PGs perlecan, versican and DCN. Scale bar: 10 μm. Experiments were repeated three times. Representative images are shown.
The HS GAGs were organized in a rich fibrillar network covering the control fibroblasts, whereas they decreased in both patients' cells with only a few fibrils localized in the extracellular spaces and in spots associated with the cell surface. The almost HS-specific PG perlecan, organized by control fibroblasts in a fibrillar ECM meshwork and resembling the HS GAGs distribution, was highly reduced in the patients' fibroblasts. On the other hand, the CS GAGs, investigated using a specific Ab that does not recognize the DS GAGs, were organized with a comparable pattern in the ECM of both patients and control cells. The CS/DS-specific PG versican was expressed by both control and patients' cells, with a comparable cytoplasmic distribution. Similarly, the core protein of the hybrid CS/DS PG DCN was detected at comparable levels in the cytoplasm and in the plasma membrane of control and patients' cells.
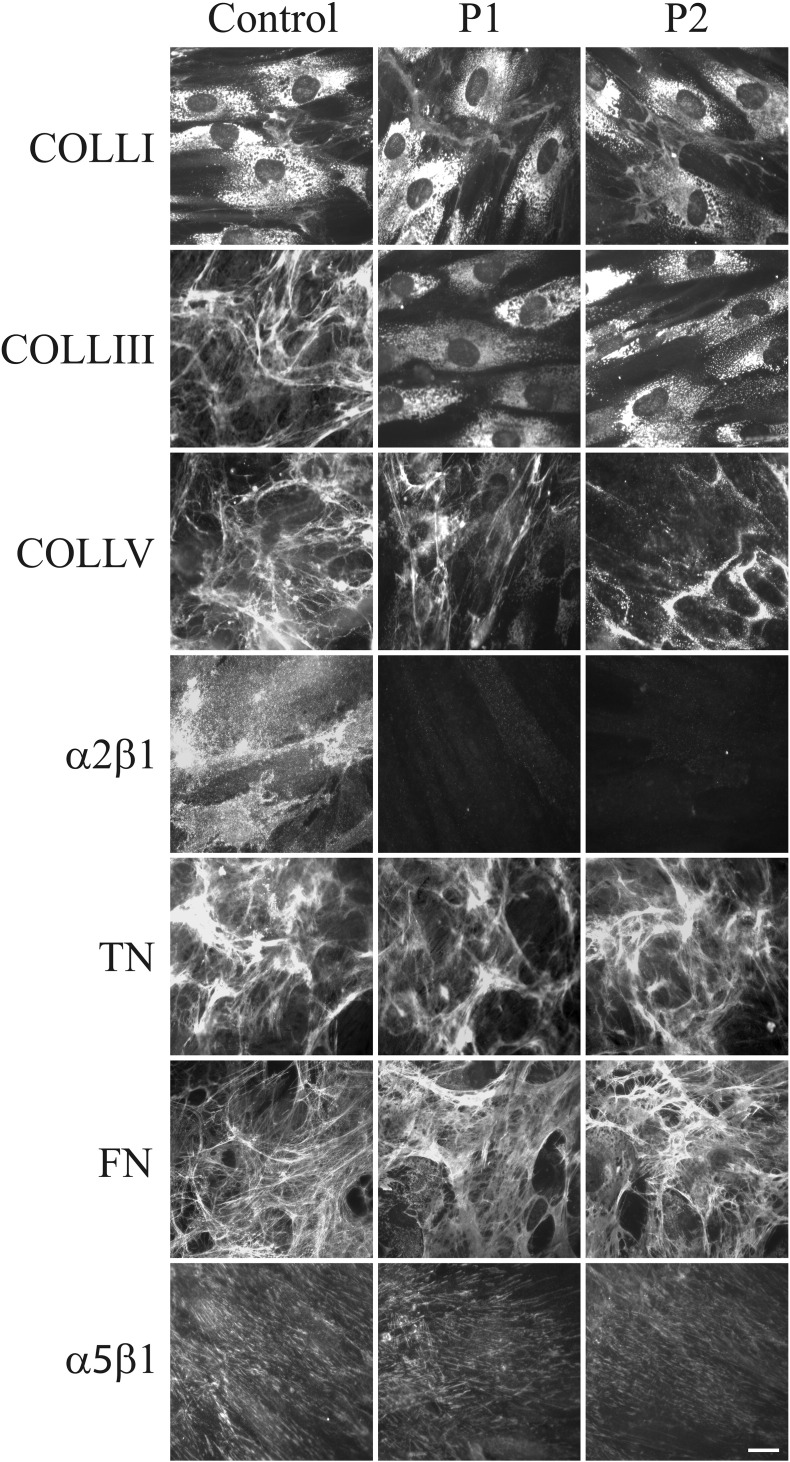
Fig. 6 shows the organization in the ECM of COLLs, glycoproteins and their specific integrin receptors in control and GalT-II-deficient cells. COLLI was mainly stored in cytoplasm and similarly organized in thin ECM fibrils in both control and patients' fibroblasts. COLLIII was assembled in the ECM by control cells, but not by GalT-II-deficient cells, in which the protein was only detected in the cytoplasm. COLLV ECM was organized by control and patient cells; however, in GalT-II-deficient cells, the network was reduced and disorganized. The fibroblast-specific COLL receptor, α2β1 integrin, was organized in thin dots on the control cell surfaces and was not recruited onto the patients' cell membranes. FN and TNs were assembled in fibrillar ECMs in both control and patient fibroblasts in a comparable manner. Similarly, the main receptor for FN, α5β1 integrin, was distributed on both the control and patient cell plasma membranes.
Fig. 6.
Organization of COLLs-, TNs- and FN-ECM, and α2β1 and α5β1 integrin receptors. Control and GalT-II-deficient (P1 and P2) skin fibroblasts were analyzed with specific Abs directed against COLLI, COLLIII, and COLLV. TNs and FN were investigated by labeling the cells with Abs recognizing all TN and FN isoforms. The COLL- and FN-specific receptors, α2β1 and α5β1 integrins, respectively, were analyzed with two specific mAbs recognizing their ligand-binding sites. Scale bar: 10 μm. Experiments were repeated three times. Representative images are shown.
4. Discussion
GalT-II deficiency is a recently discovered autosomal recessive condition responsible for a spectrum of connective tissue disorders. In particular, mutations in B3GALT6 cause SEMDJL1, a severe skeletal dysplasia first documented by Beighton and Kozlowski in 1980 when the clinical features of 7 affected children in an Afrikaans-speaking community of South Africans were described [26]. In subsequent years, additional patients were identified, and thus far, more than 30 affected cases have been reported in the Afrikaner population [28]. Nakajima et al. [24] described 8 patients with SEMDJL1, all of whom had the typical skeletal abnormalities of the disorder. Four other patients of 3 families also had a range of extra-skeletal and connective tissue abnormalities that overlapped with those observed in the progeroid form of EDS. Furthermore, Malfait et al. [25] described 5 patients with a similar phenotype but with some substantial differences from the SEMDJL1 prototype because these patients also had soft tissue fragility with delayed wound healing, multiple early-onset fractures and intellectual disability. In our patients, the presence of defective growth, joint hypermobility and modeling of the spine and long bones, the lack of tissue fragility and intellectual disability perfectly matched with SEMDJL1. The initial clinical suspicion of the EDS kyphoscoliotic type and the consequent PLOD1 genetic testing were justified not only by the lack of knowledge, at time of clinical evaluation, of the SEMDJL1 causative gene but also by the presence of three major criteria of the EDS kyphoscoliotic type, i.e., marked skin hyperextensibility, progressive kyphoscoliosis and generalized joint laxity.
Altogether, in SEMDJL1, progeroid type 2 of EDS, and “pleiotropic EDS-like CTD”, a total of 17 recessive B3GALT6 mutations have been identified: in particular, 11 (2 recurrent) mutations are missense substitutions of highly conserved amino acids within the catalytic galactosyltransferase domain of GalT-II with a predicted deleterious effect; 5 mutations are small deletions leading to frameshifts and PTC formation, and they are expected to generate truncated proteins because in single-exon genes, such as B3GALT6, nonsense-mediated RNA decay should not be activated, and 1 mutation is a recurrent substitution in the translation initiation codon [[24], [25], [28], this study] (Table S4). Immunocytochemical studies of several mutant Galt-II proteins showed subcellular mislocalization, and activities of all mutant proteins were significantly decreased compared to wild-type proteins, supporting loss-of-function [24], [25]. The concept of different mutations in the same gene causing a spectrum of variable phenotypes is well established in skeletal dysplasias and other disorders. An analogous situation seems to be valid for the B3GALT6 mutations, which are causative in the spectrum of the SEMDJL1 and EDS-like GAG-mediated CTDs.
Concerning the pathological mechanism of GalT-II deficiency some functional studies were performed. In particular, Malfait et al. [25] showed that B3GALT6 mutations resulted in the production of immature DCN, lacking its CS/DS side chain, as well as reduced-to-absent HS chains in dermal patient fibroblasts. The authors also showed, by electron microscopy on a patient's skin biopsy, an abnormal collagen fibril architecture, suggesting that COLL fibrillogenesis is perturbed in GalT-II deficient patients. Nakajima et al. [24], using patients' lymphoblastoid cells, demonstrated a decrease of HS chains and a paradoxical increase of CS/DS chains on the cell surface. The IF analysis performed on our two patients' skin fibroblasts showed a marked reduction of the HS GAG chains ECM organization, and a complete disarray of the perlecan-ECM. Interestingly, the CS GAGs assembled into an ECM very similar to that organized by the control cells, and the core proteins of the hybrid CS/DS PGs DCN and versican showed a control-like distribution. Although the organization of the DS GAGs was not investigated, it is likely that these hybrid PGs should not have a normal composition of their CS/DS side chains, as previously shown for DCN [25].
Perlecan, encoded by HSPG2, is predominantly a HS-specific PG and binds to several ECM components, i.e., COLLI, COLLIV, nidogen, laminin, and FN. It has been demonstrated that the CS/HS form of perlecan, with an expression most likely limited to cartilage, is involved in cartilage COLL ECM homeostasis [31], as also shown in perlecan-null mice that fail to form an adequate cartilage template and therefore display severe skeletal defects [32]. Functional null mutations in HSPG2 result in lethal forms of neonatal short-limbed dwarfism, known as dyssegmental dysplasia Silverman-Handmaker type syndrome, characterized by severe cartilage matrix anomalies [33]. Schwartz–Jampel syndrome type 1 patients with hypomorphic HSPG2 mutations, display a less severe short-stature phenotype accompanied by chondrodysplasias [34].
DCN is one of the major PGs in skin and tendon, and it was shown that the DCN core protein is important for COLL fibril formation, whereas its CS/DS GAGs regulate the space between the COLL fibrils. DCN knockout mice have COLL fibrils varying in dimension with thicker diameters and widely irregular shapes [35]. Interestingly, the biochemical phenotype of these mutant mice overlaps with that observed in musculocontractural EDS type/ATCS due to D4ST deficiency (Fig. 1). In particular, Miyake et al. [19] analyzing fibroblasts of patients with CHST14 mutations, showed that although the DCN core protein was normally organized, the disaccharide composition of its GAG chains consisted only of CS, without DS chains. Therefore, a deficiency of D4ST perturbs normal DS/CS balance, finally leading to disorganization of the COLL network, underlining the pivotal role of DCN in COLL fibrillogenesis.
Altogether, the absence of HS GAG synthesis observed in our patients' fibroblasts, in combination with the disorganization of perlecan and, possibly, of other HS PGs, and the most likely abnormal CS/DS side chain composition of hybrid PGs, such as versican, DCN and probably others, were associated with a defective COLL fibril assembly, as shown by the lack of the COLLV and COLLIII ECM and of their receptor, the α2β1 integrin. In our patients' fibroblasts, the organization into the ECM of TNs, which are involved in the regulation of COLL fibrillogenesis, and of FN, the assembly of which is known to be dependent on COLL deposition, are not affected, and the FN fibril deposition is associated with the recruitment of the FN-specific receptor, the α5β1 integrin. The FN ECM and α5β1 integrin are poorly organized in vitro by EDS fibroblasts [29], [36], [37], independent of the EDS type and gene mutation, indicating that different ECM components organized by skin fibroblasts might distinguish the SEMDJL1 cellular phenotype from the EDS phenotype.
To identify novel molecular mechanisms involved in SEMDJL1 pathogenesis, we performed a transcriptome wide expression profiling on patients' skin fibroblasts. Although the number of patients was limited to two sisters and we are aware that the genetic variation between them is considerably lower than between unrelated individuals, we identified several DEGs and related pathways that should contribute to the etiopathology of GalT-II deficiency, since the obtained results correlate well with the pleiotropic phenotype of GalT-II-deficient patients. In particular, we identified the predominant down-regulation of numerous genes with functions related to the maintenance of ECM structural integrity. For instance, COL5A1 was found to be down-regulated at either the transcriptional or translational level. COLLV plays a central role in COLLs fibrillogenesis and co-assembles with COLLI to form heterotypic fibrils. Haploinsufficiency of COLLV due to mutations in COL5A1 causes classical EDS [38], characterized by widened atrophic scars, skin hyperextensibility, and generalized joint hypermobility; the latter two major criteria of classical EDS were also present in our SEMDJL1 patients. Additionally, COL15A1, encoding the α1 chain of COLLXV, was found to be down-regulated in the array. Interestingly, COLLXV is a human osteoblast-secreted matrix protein, expressed during osteogenic differentiation of bone marrow stromal cells in osteoblasts, and in bone tissue biopsies by osteoblasts forming new bone tissue [39]. Consistent with this evidence, we can speculate that the decreased COL15A1 expression observed in our SEMDJL1 patients, might be associated with an impairment of the osteogenic differentiation and bone remodeling, likely contributing to the clinical phenotype. Among the other down-regulated transcripts, we considered COMP and SPP1 as key genes in the context of SEMDJL1 pathogenesis and analyzed their expression at the protein level as well. IF analyses demonstrated a reduced-to-absent organization of COMP and OPN. COMP, a non-collagenous ECM glycoprotein expressed primarily in cartilage, ligament, tendon, skeletal muscle and growth plate, is essential for the normal development of cartilage and for its conversion to bone [40]. COMP binds to COLLI, COLLII, COLLIX, COLLXII, COLLXIV, FN, and aggregan, thereby accelerating fibrillogenesis through the promotion of matrix component assembly and the maintenance of the stability of the COLLs network of adult tissues [41]. Mutations in COMP produce clinical phenotypes of pseudoachondroplasia and multiple epiphyseal dysplasia, characterized by disproportionate short stature, brachydactyly, joint hypermobility, early onset osteoarthritis, and scoliosis [42]. In a recent transcriptome-wide expression study, COMP was found to be down-regulated in patients with adolescent idiopathic scoliosis, and COMP was proposed as an important biomarker in predicting scoliosis development [43].
OPN is one of the major components of mineralized bone matrix that modulates the osteoblast phenotype, endochondral bone formation, the mineralization of the matrix in physiological or pathological conditions, wound healing and calcification [44]. It has been suggested that OPN facilitates the attachment of the osteoclasts to the mineral matrix of bones and, consequently, promotes the osteoclastic bone resorption essential for bone and mineral homeostasis [45].
Our findings, although obtained in vitro, indicate that B3GALT6 mutations, through the perturbation of the synthesis of the GAG linkage region of HS and CS/DS PGs, lead to a general dysregulation of the deposition of structural ECM proteins, such as COLLs, OPN and COMP, with a pivotal role for a correct ECM architecture and homeostasis of several connective tissues.
Moreover, our analysis showed a differential expression of genes involved in ECM-mediated signal transduction. In particular, the TGFβ/BMP signaling pathway included GDF6 and BMPER, which were up-regulated, and GDF15 which was down-regulated; these genes encode BMPs that are known to regulate tissue differentiation and maintenance acting in complex signaling networks during embryonic bone formation, postnatal growth, remodeling, and regeneration of the skeleton [46]. GDF6 is considered as a potential inhibitor of bone formation because it was shown to inhibit osteogenic differentiation of human bone marrow stromal cells in vitro [47]. BMPER is known to inhibit BMP2- and BMP4-dependent osteoblast differentiation and BMP-dependent differentiation of the chondrogenic cells [48]. GDF15 has a role in bone remodeling and is reported to be involved in human osteoarthritis [49]. Interestingly, a recent genome-wide analysis in a canine model discovered GDF15 and COMP as candidate genes for congenital hip dysplasia, a clinical feature present in both of our SEMDJL1 patients [50]. Additionally, microarray results revealed STEAP4 as the most up-regulated gene in GalT-II-deficient fibroblasts. STEAP4 is an endosomal ferroreductase indispensable for osteoclast development and function, since knockdown of Steap4 in bone marrow macrophages markedly inhibited osteoclast formation by decreasing cellular iron [51]. Overall, our data suggest that GalT-II deficiency leads to an imbalance in bone remodeling caused by increased osteoclastic bone resorption and decreased osteoblastic bone formation. We can hypothesize that the pathological bone phenotype of SEMDJL1 patients, apart from the aberrant PG synthesis, is also consequent to a reduction of osteogenic markers, such as OPN, COLLs and COMP, in combination with an induction of osteogenic differentiation inhibiting factors, such as GDF6 and BMPER, and an increased osteoclast number and/or enhanced activity likely due to increased STEAP4 activity.
Interestingly, our microarray data also indicated the differential expression of several enzymes involved in the synthesis and post-translational processing of ECM components, including genes specifically implicated in GAG biosynthesis. In particular, we found an increased CSGALNACT1 expression; this gene encodes a subunit of GalNAcT-1, a glycosyltransferase that transfers the first GalNAc residue onto the common GAG linkage region, thus initiating CS-specific biosynthesis (Fig. 1). The molecular mechanisms regulating the specific synthesis of CS/DS or HS GAG chains are not well understood. Some studies have indicated that the amino acids of the core protein near the GAG attachment site and the sulfation of the Gal residues in the linkage region, catalyzed by C6ST (encoded by CHST3), may be involved in this differential assembly and/or unknown additional factors, influencing the activities of the glycosyltransferases that continue the synthesis of CS/DS or HS [6]. The enhanced CSGALNACT1 expression observed in our patients' fibroblasts may represent a compensatory mechanism for the perturbed synthesis of the common GAG linkage region due to GalT-II deficiency and could, to some extent, explain the unexpected ECM organization of the CS GAGs by our patients' fibroblasts, as shown by IF. The authentic implication of GalNAcT-1 and other differentially expressed enzymes, such as ChPf, HAS2, GalNAcT-16, LOXL3, and STEAP4, in the molecular mechanisms involved in the etiopathogenesis of GalT-II deficiency, merits future investigation.
Finally, GalT-II deficiency induced the altered expression of genes encoding transcription factors, such as HOXD10, HOXD11, and SHOX, all found significantly down-regulated in SEMDJL1 skin fibroblasts. The majority of these genes are members of the HOX family, known to be principally implicated in the determination of differential genetic programs along the anterior–posterior axis of vertebrates [52]. HOXD10 is related to skeletal muscle tissue development, and its mutations cause congenital vertical talus [53]. Hoxd11 mutant mice exhibit shortened metacarpals and phalanges of forelimbs and hindlimbs with hypoplasia/aplasia of the tibiale mediale [54]. Of particular interest, the SHOX gene encodes a transcription factor that is important for normal bone development and is expressed in growth plate chondrocytes, where it modulates proliferation, differentiation and cell death [55]. SHOX deficiency represents a common congenital form of growth failure and is involved in the etiology of idiopathic short stature and the growth deficits and skeletal anomalies in Leri–Weill dyschondrosteosis, Langer mesomelic dysplasia, and Turner syndrome [55]. In addition to short stature, patients with SHOX deficiency can present a wide variety of skeletal malformations such as shortening and bowing of the forearms, Madelung deformity, and cubitus valgus [56]. Other down-regulated transcription factors identified were LHX9, CRIP2, CSRP2, and TES, which belong to the LIM homeobox family also implicated in skeletal and limb development [57]. Altogether, these data indicate that the altered gene expression of different transcription factors seems to contribute to the skeletal abnormalities of SEMDJL1 patients.
In conclusion, our study revealed important gene expression changes in B3GALT6 mutant fibroblasts, thus providing further insights into potential genes and molecular pathways that should contribute to the etiopathogenesis of GalT-II deficiency and related multisystemic disorders. The key role of GalT-II in GAG synthesis and the crucial biological functions of PGs are consistent with the impairment of many physiological processes, including embryonic and postnatal development and the maintenance of the ECM architecture of various connective tissues, reflecting the pleiotropic phenotype of GalT-II deficient patients. Further studies are needed to better delineate the involvement of some DEGs identified in this study in the pathogenesis of GalT-II deficiency and to search for therapeutic options for this severe CTD.
The following are the supplementary data related to this article.
Mutation screening. In the probands, sequence analysis of B3GALT6 showed compound heterozygosity for two novel mutations: the c.227del one-basepair deletion (p.Ile76Thrfs*202) was inherited from the mother (left panel), and the c.766C > T transition (p.Arg256Trp) was inherited from the father (right panel). For cDNA numbering, + 1 corresponds to the A of the ATG translation initiation codon in the reference sequence (NM_080605.3). For protein numbering, + 1 corresponds to the first translated amino acid (reference sequence NP_542172.2).
Scatter plot, hierarchical clustering, and GO analysis of the DEGs.
A: The volcano plot depicts all statistically significant DEGs identified in GalT-II-deficient cells. The fold-change of DEGs on the x-axis versus the statistical significance (P-value < 0.05) on the y-axis is shown; the up-regulated genes are reported in red, and the down-regulated genes are in blue. B: Hierarchical clustering of the 334 DEGs identified in patients' skin fibroblasts. Although fibroblasts from only 3 control subjects and 2 sisters were analyzed, this analysis showed the presence of two distinct clusters of transcripts that clearly distinguish the patients from the controls, indicating that a remarkable low variability within each group is present, allowing for statistical significance in our findings. The red color represents high gene expression, and blue represents low gene expression. P: patient; C: control.
C: To functionally characterize genes transcriptionally related to GalT-II deficiency, we grouped the DEGs according to the available GO annotations. Analysis on GO Biological Process terms (left panel) showed that cellular process (19.1%, 158/826 involved genes/total process hits), metabolic process (18.5%, 153/826), and developmental process (12.3%, 102/826) were the most enriched terms. Within the cellular process, “cell communication” was the most represented category (65.8%). In the metabolic process category, the majority of DEGs were associated with “primary metabolic process” (88.9%), which includes “nucleobase-containing compound metabolic process” (41.1%), and “protein metabolic process” (36.8%). Concerning the GO developmental process terms, “system development” (68.6%), “ectoderm development” (42.2%) and “mesoderm development” (42.1) were the most enriched, followed by “anatomical structure morphogenesis” (17.6%) and “pattern specification process” (15.7%). Enrichment analysis of GO Molecular Function terms (middle panel) identified binding activity (30.8%), catalytic activity (25.8%), and receptor activity (15.4%) as the top three categories among the affected functions, followed by nucleic acid binding transcription factor activity (10.7%). In the GO Cellular Component terms (right panel), the cell part category was the most enriched (27%), followed by extracellular region (21%), extracellular matrix (17%), macromolecular complex (14%), organelle (13%) and membrane (8%) categories.
DEGs identified in GalT-II-deficient fibroblasts.
DAVID clusters obtained with the 172 up-regulated genes.
DAVID clusters obtained with the 162 down-regulated genes.
Overview of currently known B3GALT6 mutations.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
The authors thank the patients and their family for their kind availability for this study.
References
- 1.Bateman J.F., Boot-Handford R.P., Lamandé S.R. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 2.Karsdal M.A., Nielsen M.J., Sand J.M., Henriksen K., Genovese F., Bay-Jensen A.C., Smith V., Adamkewicz J.I., Christiansen C., Leeming D.J. Extracellular matrix remodeling: the common denominator in connective tissue diseases. Possibilities for evaluation and current understanding of the matrix as more than a passive architecture, but a key player in tissue failure. Assay Drug Dev. Technol. 2013;11:70–92. doi: 10.1089/adt.2012.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulow H.E., Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu. Rev. Cell Dev. Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 4.Esko J.D., Kimata K., Lindahl U. Proteoglycans and sulfated glycosaminoglycans. In: Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etzler M.E., editors. Essentials of Glycobiology. 2nd edn. Cold Spring Harbor Laboratory Press; New York: 2009. (Chapter 16) [Google Scholar]
- 5.Sugahara K., Kitagawa H. Recent advances in the study of the biosynthesis and functions of sulfated glycosaminoglycans. Curr. Opin. Struct. Biol. 2000;10:518–527. doi: 10.1016/s0959-440x(00)00125-1. [DOI] [PubMed] [Google Scholar]
- 6.Mizumoto S., Yamada S., Sugahara K. Human genetic disorders and knockout mice deficient in glycosaminoglycan. Biomed. Res. Int. 2014 doi: 10.1155/2014/495764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Okajima T., Fukumoto S., Furukawa K., Urano T. Molecular basis for the progeroid variant of Ehlers–Danlos syndrome. Identification and characterization of two mutations in galactosyltransferase I gene. J. Biol. Chem. 1999;274:28841–28844. doi: 10.1074/jbc.274.41.28841. [DOI] [PubMed] [Google Scholar]
- 8.Almeida R., Levery S.B., Mandel U., Kresse H., Schwientek T., Bennett E.P., Clausen H. Cloning and expression of a proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I. A seventh member of the human beta4-galactosyltransferase gene family. J. Biol. Chem. 1999;274:26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 9.Bui C., Talhaoui I., Chabel M., Mulliert G., Coughtrie M.W., Ouzzine M., Fournel-Gigleux S. Molecular characterization of b1,4-galactosyltransferase 7 genetic mutations linked to the progeroid form of Ehlers–Danlos syndrome (EDS) FEBS Lett. 2010;584:3962–3968. doi: 10.1016/j.febslet.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 10.Cartault F., Munier P., Jacquemont M.L., Vellayoudom J., Doray B., Payet C., Randrianaivo H., Laville J.M., Arnold Munnich A., Cormier-Daire V. Expanding the clinical spectrum of B4GALT7 deficiency: homozygous p.R270C mutation with founder effect causes Larsen of Reunion Island syndrome. Eur. J. Hum. Genet. 2014 doi: 10.1038/ejhg.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baasanjav S., Al-Gazali L., Hashiguchi T., Mizumoto S., Fischer B., Horn D., Seelow D., Ali B.R., Aziz S.A., Langer R., Saleh A.A.H., Becker C., Nürnberg G., Cantagrel V., Gleeson J.G., Gomez D., Michel J.B., Stricker S., Lindner T.H., Nürnberg P., Sugahara K., Mundlos S., Hoffmann K. Faulty initiation of proteoglycan synthesis causes cardiac and joint defects. Am. J. Hum. Genet. 2011;89:15–27. doi: 10.1016/j.ajhg.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreml J., Durmaz B., Cogulu O., Keupp K., Beleggia F., Pohl E., Milz E., Coker M., Ucar S.K., Nürnberg G., Nürnberg P., Kuhn J., Ozkinay F. The missing “link”: an autosomal recessive short stature syndrome caused by a hypofunctional XYLT1 mutation. Hum. Genet. 2014 doi: 10.1007/s00439-013-1351-y. [DOI] [PubMed] [Google Scholar]
- 13.Bui C., Huber C., Tuysuz B., Alanay Y., Bole-Feysot C., Leroy J.G., Mortier G., Nitschke P., Munnich A., Cormier-Daire V. XYLT1 mutations in desbuquois dysplasia type 2. Am. J. Hum. Genet. 2014;94(3):405–414. doi: 10.1016/j.ajhg.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thiele H., Sakano M., Kitagawa H., Sugahara K., Rajab A., Höhne W., Ritter H., Leschik G., Nȕrnberg P., Mundlos S. Loss of chondroitin 6-O-sulfotransferase-1 function results in severe human chondrodysplasia with progressive spinal involvement. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10155–10160. doi: 10.1073/pnas.0400334101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermanns P., Unger S., Rossi A., Perez-Aytes A., Cortina H., Bonafè L., Boccone L., Setzu V., Dutoit M., Sangiorgi L., Pecora F., Reicherter K. Congenital joint dislocations caused by carbohydrate sulfotransferase 3 deficiency in recessive Larsen syndrome and humero-spinal dysostosis. Am. J. Hum. Genet. 2008;82:1368–1374. doi: 10.1016/j.ajhg.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Unger S., Lausch E., Rossi A., Mègarbanè A., Sillence D., Alcausin M., Aytes A., Mendoza-Londono R., Nampoothiri S., Afroze B., Hall B., Lo I.F., Lam S.T. Phenotypic features of carbohydrate sulfotransferase 3 (CHST3) deficiency in 24 patients: congenital dislocations and vertebral changes as principal diagnostic features. Am. J. Med. Genet. A. 2010;152A:2543–2549. doi: 10.1002/ajmg.a.33641. [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Laue K., Temtamy S., Aglan M., Kotan L.D., Yigit G., Canan H., Pawlik B., Nȕrnberg G., Wakeling E.L., Quarrell O.W., Baessmann L. Temtamy preaxial brachydactyly syndrome is caused by loss-of-function mutations in chondroitin synthase 1, a potential target of BMP signaling. Am. J. Hum. Genet. 2010;87:757–767. doi: 10.1016/j.ajhg.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wuyts W., Van Hul W. Molecular basis of multiple exostoses: mutations in the EXT1 and EXT2 genes. Hum. Mutat. 2000;15:220–227. doi: 10.1002/(SICI)1098-1004(200003)15:3<220::AID-HUMU2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 19.Miyake N., Kosho T., Mizumoto S., Furuichi T., Hatamochi A., Nagashima Y., Arai E., Takahashi K. Loss-of-function mutations of CHST14 in a new type of Ehlers–Danlos syndrome. Hum. Mutat. 2010;31:966–974. doi: 10.1002/humu.21300. [DOI] [PubMed] [Google Scholar]
- 20.Müller T., Mizumoto S., Suresh I., Komatsu Y., Vodopiutz J., Dundar M., Straub V., Lingenhel A., Melmer A., Lechner S., Zschocke J., Sugahara K., Janecke A.R. Loss of dermatan sulfate epimerase (DSE) function results in musculocontractural Ehlers–Danlos syndrome. Hum. Mol. Genet. 2013;22:3761–3772. doi: 10.1093/hmg/ddt227. [DOI] [PubMed] [Google Scholar]
- 21.Dündar M., Muller T., Zhang Q., Pan J., Steinmann B., Vodopiutz J., Gruber R., Sonoda T., Krabichler B., Utermann G., Baenziger J.U., Zhang L., Janecke A.R. Loss of dermatan-4-sulfotransferase 1 function results in adducted thumb-clubfoot syndrome. Am. J. Hum. Genet. 2009;85:873–882. doi: 10.1016/j.ajhg.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinmann B., Royce P.M., Superti-Furga A. The Ehlers–Danlos syndrome. In: Royce P.M., Steinmann B., editors. Connective Tissue and Its Heritable Disorders: Molecular, Genetic, and Medical Aspects. Wiley-Liss; New York: 2002. pp. 351–407. [Google Scholar]
- 23.Malfait F., Syx D., Vlummens P., Symoens S., Nampoothiri S., Hermanns-Lè T., Van Laer L., De Paepe A. Musculocontractural Ehlers–Danlos syndrome (former EDS type VIB) and adducted thumb clubfoot syndrome (ATCS) represent a single clinical entity caused by mutations in the dermatan-4-sulfotransferase 1 encoding CHST14 gene. Hum. Mutat. 2010;31:1233–1239. doi: 10.1002/humu.21355. [DOI] [PubMed] [Google Scholar]
- 24.Nakajima M., Mizumoto S., Miyake N., Kogawa R., Iida A., Ito H., Kitoh H., Hirayama A., Mitsubuchi H., Miyazaki O., Kosaki R., Horikawa R., Lai A. Mutations in B3GALT6, which encodes a glycosaminoglycan linker region enzyme, cause a spectrum of skeletal and connective tissue disorders. Am. J. Hum. Genet. 2013;92:927–934. doi: 10.1016/j.ajhg.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Malfait F., Kariminejad A., Van Damme T., Gauche C., Syx D., Merhi-Soussi F., Gulberti S., Symoens S. Defective initiation of glycosaminoglycan synthesis due to B3GALT6 mutations causes a pleiotropic Ehlers–Danlos-syndrome-like connective tissue disorder. Am. J. Hum. Genet. 2013;92:935–945. doi: 10.1016/j.ajhg.2013.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beighton P., Kozlowski K. Spondylo-epi-metaphyseal dysplasia with joint laxity and severe, progressive kyphoscoliosis. Skeletal Radiol. 1980;5:205–212. doi: 10.1007/BF00580591. [DOI] [PubMed] [Google Scholar]
- 27.Beighton P. Spondyloepimetaphyseal dysplasia with joint laxity (SEMDJL) J. Med. Genet. 1994;31:136–140. doi: 10.1136/jmg.31.2.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vorster A.A., Beighton P., Ramesar R.S. Spondyloepimetaphyseal dysplasia with joint laxity (beighton type); mutation analysis in 8 affected South African families. Clin. Genet. 2014 doi: 10.1111/cge.12413. [DOI] [PubMed] [Google Scholar]
- 29.Zoppi N., Gardella R., De Paepe A., Barlati S., Colombi M. Human fibroblasts with mutations in COL5A1 and COL3A1 genes do not organize collagens and fibronectin in the extracellular matrix, down-regulate alpha2beta1 integrin, and recruit alphavbeta3 Instead of alpha5beta1 integrin. J. Biol. Chem. 2004;279:18157–18168. doi: 10.1074/jbc.M312609200. [DOI] [PubMed] [Google Scholar]
- 30.Giunta C., Randolph A., Steinmann B. Mutation analysis of the PLOD1 gene: an efficient multistep approach to the molecular diagnosis of the kyphoscoliotic type of Ehlers–Danlos syndrome (EDS VIA) Mol. Genet. Metab. 2005;86:269–276. doi: 10.1016/j.ymgme.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Kvist A.J., Johnson A.E., Mörgelin M., Gustafsson E., Bengtsson E., Lindblom K., Aszódi A., Fässler R., Sasaki T., Timpl R., Aspberg A. Chondroitin sulfate perlecan enhances collagen fibril formation. Implications for perlecan chondrodysplasias. J. Biol. Chem. 2006;281:33127–33139. doi: 10.1074/jbc.M607892200. [DOI] [PubMed] [Google Scholar]
- 32.Arikawa-Hirasawa E., Watanabe H., Takami H., Hassell J.R., Yamada Y. Perlecan is essential for cartilage and cephalic development. Nat. Genet. 1999;23:354–358. doi: 10.1038/15537. [DOI] [PubMed] [Google Scholar]
- 33.Arikawa-Hirasawa E., Wilcox W.R., Le A.H., Silverman N., Govindraj P., Hassell J.R., Yamada Y. Dyssegmental dysplasia, Silverman-Handmaker type, is caused by functional null mutations of the perlecan gene. Nat. Genet. 2001;27:431–434. doi: 10.1038/86941. [DOI] [PubMed] [Google Scholar]
- 34.Nicole S., Davoine C.S., Topaloglu H., Cattolico L., Barral D., Beighton P., Ben Hamida C., Hammouda H., Cruaud C., White P.S., Samson D., Urtizberea J.A. Perlecan, the major proteoglycan of basement membranes, is altered in patients with Schwartz–Jampel syndrome (chondrodystrophic myotonia) Nat. Genet. 2000;26:480–483. doi: 10.1038/82638. [DOI] [PubMed] [Google Scholar]
- 35.Danielson K.G., Baribault H., Holmes D.F., Graham H., Kadler K.E., Iozzo R.V. Targeted disruption of decorin leads to abnormal collagen fibril morphology and skin fragility. J. Cell Biol. 1997;136:729–743. doi: 10.1083/jcb.136.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumann M., Giunta C., Krabichler B., Ruschendorf F., Zoppi N., Colombi M., Bittner R.E., Quijano-Roy S., Muntoni F. Mutations in FKBP14 cause a variant of Ehlers–Danlos syndrome with progressive kyphoscoliosis, myopathy, and hearing loss. Am. J. Hum. Genet. 2012;90:201–216. doi: 10.1016/j.ajhg.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burkitt Wright E.M., Spencer H.L., Daly S.B., Manson F.D., Zeef L.A., Urquhart J., Zoppi N., Bonshek R. Mutations in PRDM5 in brittle cornea syndrome identify a pathway regulating extracellular matrix development and maintenance. Am. J. Hum. Genet. 2011;88:767–777. doi: 10.1016/j.ajhg.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritelli M., Dordoni C., Venturini M., Chiarelli N., Quinzani S., Traversa M., Zoppi N., Vascellaro A., Wischmeijer A., Manfredini E., Garavelli L., Calzavara-Pinton P., Colombi M. Clinical and molecular characterization of 40 patients with classic Ehlers–Danlos syndrome: identification of 18 COL5A1 and 2 COL5A2 novel mutations. Orphanet J. Rare Dis. 2013;8:58. doi: 10.1186/1750-1172-8-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lisignoli G., Codeluppi K., Todoerti K., Manferdini C., Piacentini A., Zini N., Grassi F., Cattini L., Piva R., Rizzoli V., Facchini A., Giuliani N., Neri A. Gene array profile identifies collagen type XV as a novel human osteoblast-secreted matrix protein. J. Cell. Physiol. 2009;220:401–409. doi: 10.1002/jcp.21779. [DOI] [PubMed] [Google Scholar]
- 40.Koelling S., Clauditz T.S., Kaste M., Miosge N. Cartilage oligomeric matrix protein is involved in human limb development and in the pathogenesis of osteoarthritis. Arthritis Res. Ther. 2006;8:R56. doi: 10.1186/ar1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agarwal P., Zwolanek D., Keene D.R., Schulz J.N., Blumbach K., Heinegård D., Zaucke F., Paulsson M., Krieg T., Koch M., Eckes B. Collagen XII and XIV, new partners of cartilage oligomeric matrix protein in the skin extracellular matrix suprastructure. J. Biol. Chem. 2012;287:22549–22559. doi: 10.1074/jbc.M111.335935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briggs M.D., Hoffman S.M., King L.M., Olsen A.S., Mohrenweiser H., Leroy J.G., Mortier G.R., Rimoin D.L., Lachman R.S., Gaines E.S., Cekleniak J.A., Knowlton R.G., Cohn D.H. Pseudoachondroplasia and multiple epiphyseal dysplasia due to mutations in the cartilage oligomeric matrix protein gene. Nat. Genet. 1995;10:330–336. doi: 10.1038/ng0795-330. [DOI] [PubMed] [Google Scholar]
- 43.Fendri K., Patten S.A., Kaufman G.N., Zaouter C., Parent S., Grimard G., Edery P., Moldovan F. Microarray expression profiling identifies genes with altered expression in adolescent idiopathic scoliosis. Eur. Spine J. 2013;22:1300–1311. doi: 10.1007/s00586-013-2728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alford A.I., Hankenson K.D. Matricellular proteins: extracellular modulators of bone development, remodeling, and regeneration. Bone. 2006;38:749–757. doi: 10.1016/j.bone.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 45.Reinholt F.P., Hultenby K., Oldberg A., Heinegård D. Osteopontin—a possible anchor of osteoclasts to bone. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4473–4475. doi: 10.1073/pnas.87.12.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen D., Zhao M., Mundy G.R. Bone morphogenetic proteins. Growth Factors. 2004;22:233–241. doi: 10.1080/08977190412331279890. [DOI] [PubMed] [Google Scholar]
- 47.Shen B., Bhargav D., Wei A., Williams L.A., Tao H., Ma D.D., Diwan A.D. BMP-13 emerges as a potential inhibitor of bone formation. Int. J. Biol. Sci. 2009;5:192–200. doi: 10.7150/ijbs.5.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binnerts M.E., Wen X., Canté-Barrett K., Bright J., Chen H.T., Asundi V., Sattari P., Tang T., Boyle B., Funk W., Rupp F. Human crossveinless-2 is a novel inhibitor of bone morphogenetic proteins. Biochem. Biophys. Res. Commun. 2004;315:272–280. doi: 10.1016/j.bbrc.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 49.Hinoi E., Ochi H., Takarada T., Nakatani E., Iezaki T., Nakajima H., Fujita H., Takahata Y., Hidano S., Kobayashi T., Takeda S., Yoneda Y. Positive regulation of osteoclastic differentiation by growth differentiation factor 15 up-regulated in osteocytic cells under hypoxia. J. Bone Miner. Res. 2012;27:938–949. doi: 10.1002/jbmr.1538. [DOI] [PubMed] [Google Scholar]
- 50.Lavrijsen I.C., Leegwater P.A., Martin A.J., Harris S.J., Tryfonidou M.A., Heuven H.C., Hazewinkel H.A. Genome wide analysis indicates genes for basement membrane and cartilage matrix proteins as candidates for hip dysplasia in Labrador Retrievers. PLoS One. 2014;9:e87735. doi: 10.1371/journal.pone.0087735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J., Ye S., Fujiwara T., Manolagas S.C., Zhao H. Steap4 plays a critical role in osteoclastogenesis in vitro by regulating cellular iron/reactive oxygen species (ROS) levels and cAMP response element-binding protein (CREB) activation. J. Biol. Chem. 2013;288:30064–30074. doi: 10.1074/jbc.M113.478750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinonez S.C., Innis J.W. Human HOX gene disorders. Mol. Genet. Metab. 2014;111:4–15. doi: 10.1016/j.ymgme.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 53.Shrimpton A.E., Levinsohn E.M., Yozawitz J.M., Packard D.S., Jr., Cady R.B., Middleton F.A., Persico A.M., Hootnick D.R. A HOX gene mutation in a family with isolated congenital vertical talus and Charcot–Marie–Tooth disease. Am. J. Hum. Genet. 2004;75:92–96. doi: 10.1086/422015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Davis A.P., Capecchi M.R. Axial homeosis and appendicular skeleton defects in mice with a targeted disruption of hoxd-11. Development. 1994;120:2187–2198. doi: 10.1242/dev.120.8.2187. [DOI] [PubMed] [Google Scholar]
- 55.Marchini A., Häcker B., Marttila T., Hesse V., Emons J., Weiss B., Karperien M., Rappold G. BNP is a transcriptional target of the short stature homeobox gene SHOX. Hum. Mol. Genet. 2007;16:3081–3087. doi: 10.1093/hmg/ddm266. [DOI] [PubMed] [Google Scholar]
- 56.Ross J.L., Scott C., Jr., Marttila P., Kowal K., Nass A., Papenhausen P., Abboudi J., Osterman L., Kushner H., Carter P., Ezaki M., Elder F., Wei F., Chen H., Zinn A.R. Phenotypes associated with SHOX deficiency. J. Clin. Endocrinol. Metab. 2001;86:5674–5680. doi: 10.1210/jcem.86.12.8125. [DOI] [PubMed] [Google Scholar]
- 57.Tzchori I., Day T.F., Carolan P.J., Zhao Y., Wassif C.A., Li L., Lewandoski M., Gorivodsky M., Love P.E., Porter F.D., Westphal H., Yang Y. LIM homeobox transcription factors integrate signaling events that control three-dimensional limb patterning and growth. Development. 2009;136:1375–1385. doi: 10.1242/dev.026476. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mutation screening. In the probands, sequence analysis of B3GALT6 showed compound heterozygosity for two novel mutations: the c.227del one-basepair deletion (p.Ile76Thrfs*202) was inherited from the mother (left panel), and the c.766C > T transition (p.Arg256Trp) was inherited from the father (right panel). For cDNA numbering, + 1 corresponds to the A of the ATG translation initiation codon in the reference sequence (NM_080605.3). For protein numbering, + 1 corresponds to the first translated amino acid (reference sequence NP_542172.2).
Scatter plot, hierarchical clustering, and GO analysis of the DEGs.
A: The volcano plot depicts all statistically significant DEGs identified in GalT-II-deficient cells. The fold-change of DEGs on the x-axis versus the statistical significance (P-value < 0.05) on the y-axis is shown; the up-regulated genes are reported in red, and the down-regulated genes are in blue. B: Hierarchical clustering of the 334 DEGs identified in patients' skin fibroblasts. Although fibroblasts from only 3 control subjects and 2 sisters were analyzed, this analysis showed the presence of two distinct clusters of transcripts that clearly distinguish the patients from the controls, indicating that a remarkable low variability within each group is present, allowing for statistical significance in our findings. The red color represents high gene expression, and blue represents low gene expression. P: patient; C: control.
C: To functionally characterize genes transcriptionally related to GalT-II deficiency, we grouped the DEGs according to the available GO annotations. Analysis on GO Biological Process terms (left panel) showed that cellular process (19.1%, 158/826 involved genes/total process hits), metabolic process (18.5%, 153/826), and developmental process (12.3%, 102/826) were the most enriched terms. Within the cellular process, “cell communication” was the most represented category (65.8%). In the metabolic process category, the majority of DEGs were associated with “primary metabolic process” (88.9%), which includes “nucleobase-containing compound metabolic process” (41.1%), and “protein metabolic process” (36.8%). Concerning the GO developmental process terms, “system development” (68.6%), “ectoderm development” (42.2%) and “mesoderm development” (42.1) were the most enriched, followed by “anatomical structure morphogenesis” (17.6%) and “pattern specification process” (15.7%). Enrichment analysis of GO Molecular Function terms (middle panel) identified binding activity (30.8%), catalytic activity (25.8%), and receptor activity (15.4%) as the top three categories among the affected functions, followed by nucleic acid binding transcription factor activity (10.7%). In the GO Cellular Component terms (right panel), the cell part category was the most enriched (27%), followed by extracellular region (21%), extracellular matrix (17%), macromolecular complex (14%), organelle (13%) and membrane (8%) categories.
DEGs identified in GalT-II-deficient fibroblasts.
DAVID clusters obtained with the 172 up-regulated genes.
DAVID clusters obtained with the 162 down-regulated genes.
Overview of currently known B3GALT6 mutations.