Abstract
As a desirable agricultural trait, multilocular trait of rapeseed (Brassica rapa; Brassica napus; Brassica juncea), always represents higher yield per plant compared with bilocular plants. We previously isolated a trilocular gene locus, Bjmc1, and identified a set of molecular markers linked to the trilocular gene. With a map-based cloning, we identified that the BjMc1 was located in B genome of Brassica juncea, and it was a CLAVATA1 (CLV1) gene homologue. The insertion of a copia-LTR retrotransposable element 1 (RTE1) into the coding region of BjMc1 interrupted its transcription in rapeseed, leading to the trilocular phenotype. Phylogenetic analysis showed that Mc1 genes were conserved and widespread in land plants. Two amino acid sites had undergone positive selection in the ancestor of Mc1 genes, and then purifying selection was the dominant force after the divergence of dicots and monocots from their common ancestor in the evolutionary process, indicating that Mc1 genes are conserved in modern land plants. Our results provided new insights in molecular regulatory mechanism of multilocularity in rapeseed, and better understanding of molecular mechanism in crop yield improvement.
Introduction
Upon germination, the shoot apical meristem (SAM) produces primary leaves, and after the vegetative-to-reproductive switch, it is transformed into inflorescence meristem, and then floral meristem forms on inflorescence meristem, which generates floral organs, including sepals, petals, stamens and carpels1. In rapeseed (Brassica rapa; Brassica napus; Brassica juncea), morphological studies have shown that the wild-type (bilocular) silique development starts when two carpels initiate from the floral meristem to form a hollow cylinder of cells which grow apically to generate the gynoecium. Simultaneously, two medial ridges grow through directed cell division towards each other to fuse and create the septum. The septum divides the cylinder into two locules corresponding to the original two carpels. However, the multilocular gynoecium consists of more than two carpels and the multilocular silique usually has more than three locules2, 3.
The silique trait of rapeseed directly affects the seed yield, and yield enhancement has always been one of the major goals of rapeseed production and genetic improvement4–6. Multiloculus, leading to a slightly fewer silique per plant but containing much more seeds per silique than biloculus with similar seed size, has been considered a great potential for developing high-yield rapeseed varieties7. However, most rapeseed varieties planted worldwide are bilocular. Classical genetic analysis demonstrated that two recessive nuclear genes controlled the multilocular trait in B. juncea, and one recessive nuclear gene was responsible for the multilocular trait in B. rapa 8, 9. A series of molecular markers linked to the multilocular gene locus had been identified in B. juncea 10, which could promote the utilization of multilocularity in breeding of high-yield cultivars.
The multilocular trait in B. rapa was found to be controlled by a CLAVATA3 (CLV3) gene homologue, and it suggested that the feedback loop involving CLV3 and WUSCHEL (WUS) played a major role in carpel development11. Multilocular trait was also observed and genetically investigated in Arabidopsis 12–15 and tomato16. In Arabidopsis, CLAVATA (CLV) signaling pathway regulating WUS expression is a key component of the network that controls stem cell renewal and differentiation, and the previously described mutants, such as clv1, clv2, clv3 and crn, showed the phenotype of multilocular siliques due to the disturbance of stem cell growth balance17, 18. The sqn mutant showed multiloculus, and SQN was found to be required for the normal accumulation of various miRNAs, indicating that miRNAs might be involved in the regulation of silique trait19. In addition, a recent study showed that another receptor kinase signaling pathway involving ERECTA (ER) regulated the stem cell growth, and the er mutant exhibited a similar silique trait with clv mutants20. In tomato, the mutation of the homologues of both CLV1 and CLV3 resulted in the increased number of fruit locules. Moreover, the mutation of the homologous genes in CLV signaling pathway in maize21, 22 and rice23, such as THICK TASSEL DWARF1, FASCIATED EAR2 and FLORAL ORGAN NUMBER1, could also the increase the seed number per inflorescence, which was similar to multilocular trait in rapeseed.
As a member of CLV signaling pathway that regulates WUS expression, more than 10 alleles of clv1 with multiloculus have been discovered in Arabidopsis, and the mutants exhibited weak, intermediate and strong multilocular phenotypes12, 24, 25. The CLV1 gene encoded a putative receptor kinase26, and mutation at different sites in the gene sequence could lead to different degrees of multilocular phenotype. Moreover, all the clv1 mutants with intermediate and strong multilocular phenotypes were dominant negative, and the mutant of CLV1 homologous gene in tomato was also expected to be dominant-negative16, 27. Similarly, most of the mutants with multiloculus discovered in Brassicas showed variable valve numbers10, 11. However, it has been unknown whether the molecular mechanism controlling multiloculus is similar between Arabidopsis and Brassicas or the dominant-negative character is associated with the instability of multilocular silique trait.
In previous study, we found that the trilocular silique always had three locules, and the trilocular plants had significantly higher yield per plant than the bilocular plants2. The trilocular trait of B. juncea was controlled by two independent recessive nuclear genes, Bjmc1 and Bjmc2. The BjMc1 and BjMc2 gene were isolated from the same plants and mapped by molecular markers28, 29. In present study, we cloned the bilocular gene BjMc1 and trilocular Bjmc1 respectively. A Copia-LTR retrotransposable element 1 (RTE1) inserted in the coding region of Bjmc1 was identified in trilocular plants, which interrupted the transcription of the target gene. We also found that two amino acid sites had undergone positive selection in the ancestor of Mc1 genes, and purifying selection was the dominant force after divergence of dicots and monocots from their common ancestor in the evolutionary process of Mc1 genes, indicating that they were conserved in modern land plants.
Results
Fine mapping of BjMc1 gene
BjMc1 gene was previously mapped to a genomic region between marker EC14MC14 and SC20, which could delimit an interval of 2.7 cM28. Compared with the bilocular siliques in NILs of BjMc1 gene, the trilocular siliques displayed shorter, wider and flatter (Supplementary Fig. 1a,b). But the inflorescence meristem and floral meristem did not show differences between bilocular and trilocular plants in BC6F1 generation (Supplementary Fig. 2a,b). To identify the BjMc1 gene locus, we further screened a BAC library of purple-leaf mustard with the primer C1-1 (Supplementary Table 1). Two positive clones, 26P20 and 83D02, were identified, and four scaffolds (designated as scaffold 1, 2, 3 and 4) (Supplementary Data 1) were obtained by sequencing 83D02. Two sequence-characterized amplified region markers SC40 and SC151 (Supplementary Table 1) were identified according to scaffold 2, and one SSR marker SR52 was identified based on scaffold 4. Subsequently, polymorphic markers SC40, SC151 and SR52 were used to search for the recombinants identifiable between SC13 and SC20. Among the 242 recombinants discovered from the NILs population which consisted of 9,300 individuals, 0, 4 and 8 recombination events were detected for SC40, SC151 and SR52 respectively. Finally, the candidate region of BjMc1 was found to be restricted between SC151 and EC14MC14 at a 1.14-cM region. Twenty-five open reading frames (ORFs) of scaffold 2 in which both the co-segregative SC40 and nearest marker SC151 located were predicted according to: (http://linux1.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind), and were annotated by Arabidopsis genes, including a CLV1 gene (At1g75820) (Fig. 1). For each of the 25 ORFs, we comparatively sequenced the genomic fragments covering the promoter region and the complete coding region from both the bilocular and trilocular plants in BC5F1 population. The results showed that only the predicted gene which was homologous to CLV1 (At1g75820) showed sequence variation. Moreover, the co-segregation molecular marker SC40 was just located in the ORF homologous to CLV1. Thus, the CLV1 homologous gene was selected as the candidate gene for further investigation.
Figure 1.
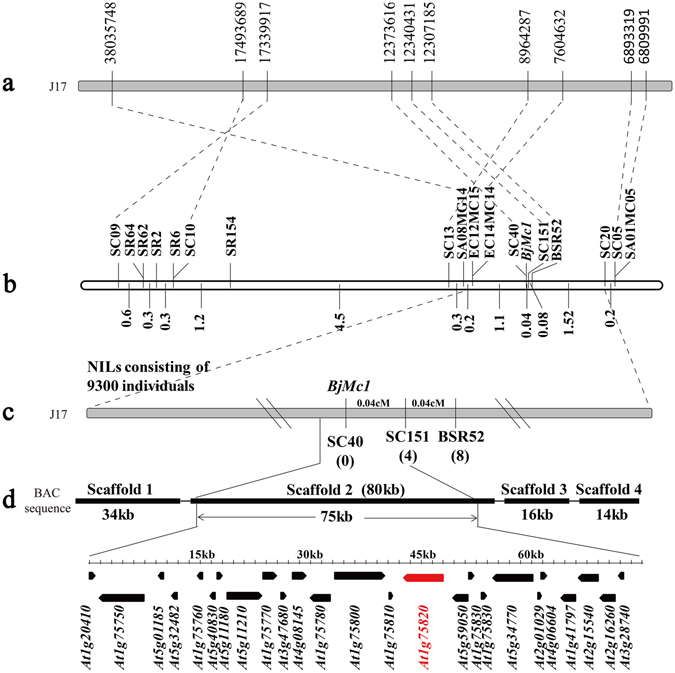
Map-based cloning of BjMc1. The number below the marker indicates the number of recombinants between individual markers and BjMc1 locus. The pentagons represent the predicted genes in the 75-kb target region on chromosome J17 of B. juncea. The candidate gene of BjMc1 is indicated by red color. (a) The physical location of molecular markers of BjMc1 on J17 chromosome. (b) The genetic linkage map of BjMc1. (c) Genetic distance of the three markers identified in this research. (d) the scaffolds of BAC clone 83D02.
Bjmc1 gene transcription of trilocular plants was interrupted by a Copia-LTR RTE1
A pair of primer CV4 (Supplementary Table 1) was designed based on the conserved reference sequences of Bra015812, AT1G75820 and scaffold 2 of 83D02. Two homologous copies, respectively designated as BjCLV1a and BjCLV1b, were detected in bilocular plants of B. juncea, and the sequences were shown in Supplementary Data 2. No sequence variation was detected in BjCLV1a between bilocular and trilocular plants, while an insertion of a 4961 bp Copia-LTR RTE1 was detected in the coding region of BjCLV1b of trilocular plants (Fig. 2a), and the sequence of RTE1 was showed in Supplementary Data 3. The sequence analysis further indicated that the putative Copia-LTR RTE1 had a 3 bp target site duplication (GGC), 364 bp terminal directed repeats and a number of subterminal repeats (ATCAGCGACT) (Fig. 2c). To validate the candidacy of BjCLV1b for BjMc1 locus, two constructs p35S::BjCLV1b and pBjCLV1b:BjCLV1b were transformed into the J163-4 plants. A total of 27 pBjCLV1b:BjCLV1b-transgenetic plants and 20 p35S::BjCLV1b-transgenetic plants were obtained in T0 generation. 18 out of the 27 pBjCLV1b:BjCLV1b-transgenetic plants showed chimeric phenotype, which showed 2–6 bilocular siliques besides the trilocular ones in one plant. By contrast, 16 out of the 20 p35S::BjCLV1b-transgenetic plants showed completely bilocular siliques (Fig. 3a). The phenotypes and genotypes of four pBjCLV1b:BjCLV1b-transgenetic and five p35S::BjCLV1b-transgenetic lines were analyzed in T1 progeny of plants. As shown in Supplementary Table 2, in all five p35S::BjCLV1b-transgenetic lines, the transgenic plants with completely bilocular siliques were observed. By contrast, only one of the four pBjCLV1b:BjCLV1b-transgenetic lines showed a T1 progeny plant with bilocular silique. The expression levels of BjCLV1b in early inflorescence of 4 transgenic lines with pBjCLV1b:BjCLV1b (TGP1-4), 11 transgenic lines with p35S::BjCLV1b (TGP5-15), and the bilocular and trilocular plants of BC5F1 generation were detected by qPCR using the primer DL2. As expected, compared with the bilocular plants of BC5F1 generation, the p35S::BjCLV1b-transgenic plants showed much higher expression level of BjMc1, while the pBjCLV1b:BjCLV1b-transgenic plants exhibited similar or lower expression level (Supplementary Fig. 3a).
Figure 2.
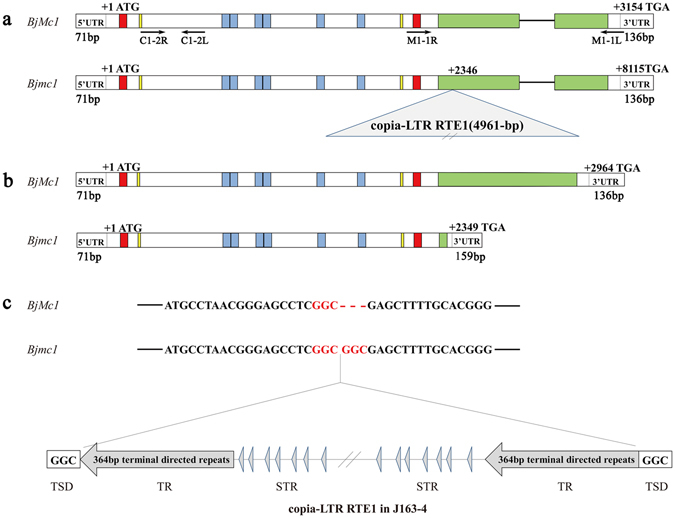
Gene structure of BjMc1 in bilocular and Bjmc1 in trilocular plants. Red box ( ) represents the putative transmembrane domain, yellow box (
) represents the putative transmembrane domain, yellow box ( ) represents the cysteine domain, blue box (
) represents the cysteine domain, blue box ( ) represents the LRR domain, green box (
) represents the LRR domain, green box ( ) represents the serine/threonine protein kinase. (a) Genomic gene structure. The arrows indicate the binding sites of primers (b) cDNA gene structure. (c) Structure of the copia-LTR RTE1 found in the Bjmc1 genomic gene.
) represents the serine/threonine protein kinase. (a) Genomic gene structure. The arrows indicate the binding sites of primers (b) cDNA gene structure. (c) Structure of the copia-LTR RTE1 found in the Bjmc1 genomic gene.
Figure 3.
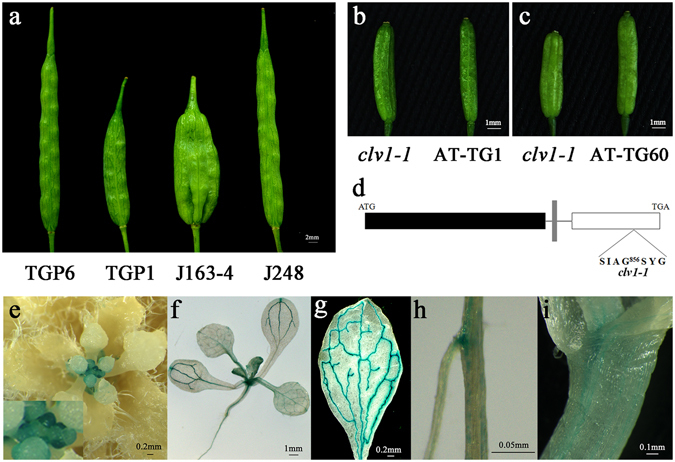
Functional analysis and expression pattern of BjMc1. (a) Silique phenotypes of p35S::BjCLV1b-transgenetic T0 line TGP6, pBjCLV1b:BjCLV1b-transgenetic T0 line TGP1, multilocular parental J163-4 and bilocular parental J248. (b) Silique phenotypes of Arabidopsis mutant clv1-1 and pBjCLV1b:BjCLV1b-transgenetic T0 line AT-TG1. (c) Silique phenotypes of Arabidopsis mutant clv1-1 and 35S::BjCLV1b-transgenetic T0 line AT-TG60. (d) The mutation sites of clv1-1. (e–i) Representative histochemical analysis of GUS expression in tissues under the control of the BjMc1 promoter in the transgenic Arabidopsis T2 plants. (e) Early inflorescence with flower bud meristems. The solid arrow indicates the flower bud meristems on the early inflorescence (f) Seedling 10 d after germination. (g) Rosette leaf. (h) Root. (i) Stem.
In addition, the constructs of p35S::BjCLV1b and pBjCLV1b:BjCLV1b were transformed respectively into Arabidopsis multilocular mutant clv1-1 which showed four valves per silique (Fig. 4c). A total of 56 pBjCLV1b:BjCLV1b-transgenetic plants and 32 p35S::BjCLV1b-transgenetic plants were obtained in T0 generation. 33 out of the 56 pBjCLV1b:BjCLV1b-transgenetic plants showed chimeric phenotype, while 17 out of the 32 p35S::BjCLV1b-transgenetic plants showed completely bilocular phenotype (Fig. 3b,c). Further analysis of phenotypes and genotypes confirmed that the transgenic events were cosegregated with the bilocular trait in T1 progeny.
Figure 4.

Dominant negative phenotype. (a) Silique phenotype of the bilocular plant in BC5F1 generation. (b) Bilocular silique, trilocular silique and trilocular-like silique. (c) Silique phenotypes of Arabidopsis mutant clv1-1 and p35S::bjmc1-transgenetic T0 line AT-TG100.
Bjmc1 encoded a putative truncated protein
Semi-quantitative reverse transcription polymerase chain reaction (RT-PCR) was performed to analyze the transcription levels of BjMc1 and Bjmc1 of early inflorescences of bilocular and trilocular plants in BC5F1 generation. C1-2 and M1-1(Supplementary Table 1) were designed according to different parts of the gene sequence (Fig. 2a). Both C1-2 and M1-1 could identify the transcription of BjMc1 in bilocular plants, while only C1-2 could identify the transcription of Bjmc1 in trilocular plants, indicating that the truncation of transcription has occurred in Bjmc1 gene. Although the transcription of BjMc1 and Bjmc1 could be detected by C1-2, the Bjmc1 expression was down-regulated in trilocular plants (Supplementary Fig. 1i).
To investigate the transcription level of BjMc1 and Bjmc1 in detail, the full-length complementary DNAs (cDNAs) were identified by RACE technology. The cDNA of BjMc1 was 3,155 bp in length, which was composed of two exons and consists of a 71 bp 5′ untranslated region, a 2,964 bp ORF and a 136 bp 3′ untranslated region. The cDNA of Bjmc1, with a full-length of 2,583 bp, comprised a 71 bp 5′ untranslated region which contained the same sequence with the cDNA of BjMc1, a 2,349 bp ORF and a 159 bp 3′ untranslated region (Fig. 2b and Supplementary Data 4). BjMc1 encoded a putative protein of 987 amino acids and contained two putative transmembrane domains, a putative extracellular domain consisting of 6 complete Leu-rich repeats (LRRs) and a putative intracellular domain containing all of the conserved residues found in serine/threonine protein kinase. The LRR region was flanked by a pair of conservatively spaced cysteines (Fig. 2a,b). Sequence analysis indicated that the putative Bjmc1 protein consisting of 782 amino acids contained the extracellular domain of BjMc1, and further analysis showed that Bjmc1 protein was derived from the truncation of BjMc1 gene at Gly782 by Copia-LTR RTE1 (Fig. 2b and Supplementary Fig. 3c), proving that the serine/threonine protein kinase domain of BjMc1 gene was required for the gene to control the development of siliques in B. juncea.
The dominant negative effect of Bjmc1
In back-cross populations, we observed that some bilocular individuals (BjMc1bjmc1bjmc2bjmc2) showed several triloculus-like siliques which showed trilocular shape but with two locules and only a few siliques had three locules (Fig. 4a,b). The silique trait of 219 bilocular plants was investigated in BC8F1 generation, and 92 individuals were found to have triloculus-like siliques.
A previous report has demonstrated that all intermediate and strong clv1 alleles in Aribidopsis are dominant negative27. Similar to the homozygous mutant of CLV1 homologous gene, the heterozygous plants also exhibited weak fasciation in tomato16. Moreover, the Arabidopsis ER, a Leu-rich repeat receptor-like Ser/Thr kinase, regulated organ shape and inflorescence architecture, and a truncated er protein that lacks the cytoplasmic kinase domain confers dominant-negative effects30. In our research, Bjmc1 was shown to be able to encode a truncated protein with a similar structure with er protein, indicating the possibility of dominant negative effect. To verify whether Bjmc1 protein functions in the development process of carpel, the construct p35S::Bjmc1 was transformed into clv1-1 mutant. A total of 62 transgenic plants were obtained and 25 of them showed siliques with five valves (Fig. 4c). The phenotype analysis of T1 progeny showed that the transgenic events could be cosegregated with the phenotype of five valves. The over-expression of Bjmc1 could aggravate the multilocular phenotype of clv1-1. These results further supported the inference that the truncated Bjmc1 protein showed the dominant negative character in controlling carpel development.
Expression pattern and subcellular localization of BjMc1
To investigate the expression pattern of BjMc1, quantitative real-time PCR (qPCR) analysis was performed using the total RNA extracted from various organs of bilocular plants in BC5F1 generation. The results indicated that the highest expression level was detected in early inflorescence, but overall lower and variable expression levels were detected in different developmental periods of siliques. The expression level of BjMc1 was relatively high in SAM, the stem and the root, but it was much lower in the silique peel and the leaf (Supplementary Fig. 3b). To examine BjMc1 activity in greater detail, the expression of GUS in the transgenic plants was detected in the flower bud primordium, the SAM of early inflorescence and mainly in the vascular tissues of other organs, including cotyledons, rosette leaves, stems and roots. But with the development of flower buds, GUS staining faded away in the transgenic plants (Fig. 3e–i). Consistent with qPCR analysis, the flower bud primordium in early inflorescence showed the highest GUS expression. These results proved that the BjMc1 gene was expressed in the flower bud primordium, indicating that the carpel number was determined at the stage of flower bud primordium formation in B. juncea.
To investigate the subcellular localization of BjMc1, we fused the coding region containing putative transmembrane domains and LRR domains, a without serine/threonine protein kinase of BjMc1 with the coding region of an enhanced GFP driven by a double 35S promoter. This chimeric plasmid was transformed into col-0 Arabidopsis. The result showed that the BjMc1-GFP fusion protein was localized in the plasma membrane (Supplementary Fig. 1c–h), which was consistent with the previous study31 and suggested that BjMc1 could retain some conserved functions of its homologs in other species.
Expression analysis of genes involved in early inflorescence
According to the expression pattern of BjMc1 gene, the homologous genes of CLV singling pathway, including BjCLV3, BjWUS, BjCLV2, BjCRN, BjPOLL, BjPLL1, BjKAPP BjBAM1, BjBAM2 and BjBAM3, and the homologous genes of ABC model of floral organ identity, including BjAP1, BjAP2, BjAP3, BjPI and BjAG, were chosen to perform the qPCR analysis using the total RNA extracted from the early inflorescence of bilocluar and trilocular plants in BC5F1 generation, and primers (Supplementary Table 1) were design according to these homologous gene sequences. The results showed that, in trilocular plants, the A class genes, BjAP1 and BjAP2, were down-regulated significantly, the B class genes, BjAP3 and BjPI were up-regulated significantly, and the C class gene, BjAG, was also down-regulated significantly (Fig. 5). In trilocular plants, the CLV signaling pathway genes, BjCLV2, BjCRN, BjPOLL, BjPLL, BjBAM1, BjBAM2 and BjBAM3, were down-regulated significantly, and BjKAPP was up-regulated. However, as two key components of CLV signaling pathway, BjCLV3 and BjWUS did not show significant variations between bilocular and trilocular plants in early inflorescence (Fig. 5). These results indicated that BjMc1 gene participated in CLV signaling pathway, but it was not the key pathway to control the carpel development in B. juncea. The mutant of Bjmc1 gene in trilocular plants resulted in the significantly expression variation of ABC class genes, indicating that BjMc1 involved in the pathway of flower bud formation to control the carpel development.
Figure 5.
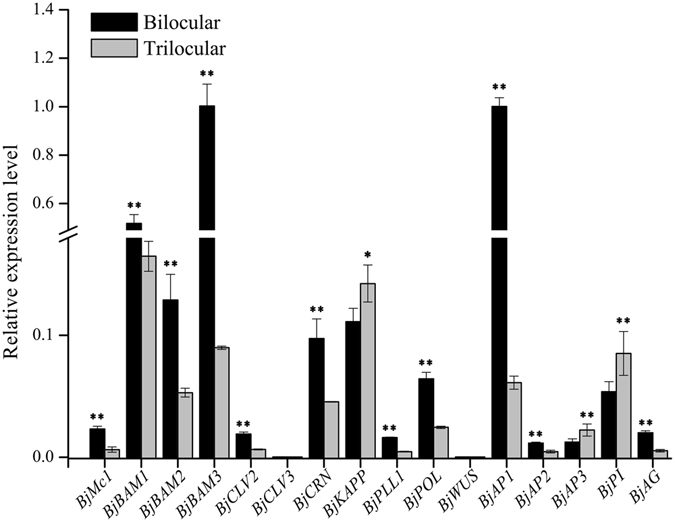
The difference of expression level of related genes between bilocular and trilocular plants during the flower bud differentiation period in BC5F1 generation. *P < 0.05; **P < 0.01.
Mc1 genes being conserved and widespread in land plants
Blast searching against the plant database revealed that Mc1 genes were widespread in land plants and each sequenced land plant genome contained at least one gene encoding BjMc1 homologue. To explore the evolutionary process of BjMc1 in land plants, we characterized CDSs and proteins of Mc1 genes from species representing the main lineages of land plants, including moss Physcomitrella patens, lycophyte Selaginella moellendorffii, Amborellaceae Amborella trichopoda, 9 monocot species and 24 dicot species. The Mc1 genes could be divided into three major branches on the phylogenetic tree (Supplementary Fig. 4), and BjMc1 was in the branch of the Cruciferae species. Structural analysis of Mc1 genes in land plants was performed by comparing the exon-intron organization (http://gsds.cbi.pku.edu.cn/). It was shown that the coding regions of all Mc1 genes of land plants were interrupted by 1–2 introns except for PpaMc1, which contained 8 introns and 9 exons (Supplementary Fig. 5).
Two amino acid sites underwent positive selection in the ancestors of Mc1 genes
In this research, we attempted to further reveal the evolutionary process of BjMc1 homologous genes in land plant species. The LRT of positive selection was applied using codeML method and the codon substitution models32–34, and all Mc1 genes of sampled land plants were tested respectively. First, one-ratio model was used to determine whether there were variations in d N: d S ratio of the codon position for Mc1 genes in land plants. Overall, the maximum likelihood estimates for d N: d S values were close to zero (Supplementary Table 3), suggesting that purifying selection was the predominant force in the evolution of BjMc1 in land plants. Second, the LRTs to compare the data fit to models M1a vs M2a and M7 vs M8 were used to determine whether positive selection promoted the divergence of Mc1 genes in land plants. No amino acid site was influenced by positive selection during the evolution of Mc1 genes in land plants. These results indicated that the primary constraint for Mc1 genes in land plants was purifying selection.
To assess whether the ancestors of BjMc1 gene had undergone a pattern of molecular evolution in the evolutionary process of land plants, branch-model of codeML was performed. Six branches were selected from the phylogenetic tree (branch I, II, III, IV, V and VI) (Supplementary Fig. 4).We found that for branch I and II, the branch-model permitting a class of positively selected codons with d N: d S > 1 had a significantly better fit to the data than the branch-model in which this class of codons were restricted to d N: d S = 1. However, this was not the case for branch III, IV, V and VI (Supplementary Table 4). The results indicated that the evolution process of BjMc1 gene might be influenced by positive selection in its ancestors of branch I and II. Because LRTs suggested that positive selection acted on the ancestral species of BjMc1 gene, the method of Bayes empirical Bayes34, namely branch-site-model of codeML, was used to evaluate the positively selected sites and their posterior probabilities. A total of 32 codons were identified with a >50% posterior probability of d N: d S > 1 along branch I. Of these codons, 2 amino acid sites had a >95% posterior probability of positive selection. Although 2 and 6 codons were identified with a >50% posterior probability of d N: d S > 1 along branch III and branch IV respectively, no amino acid site had a posterior probability ≥95% of positive selection (Supplementary Table 5 and Supplementary Fig. 6). The two positive sites of branch I were located in the first cysteine domain and a key component of serine/threonine protein kinase domain respectively. When dicots and monocots were diverged from Moss, Lycophyte and Amborellaceae, the 139th amino acid tryptophan (W) located in the first cysteine domain was mutated into phenylalanine (F), and the 899th amino acid cysteine (C) located in a component of serine/threonine protein kinase domain was mutated into valine (V) except for in Prunus mume, Eucalyptus grandis and Sesamum indicum (Supplementary Fig. 7). In the long-term evolution process, both the two sites were preserved after mutation, indicating that the two amino acids played vital roles for the gene functions in dicot and monocot plants. The most reasonable explanation for these results was that in the evolutionary process of BjMc1 gene, the ancestor had undergone adaptive evolution during a short period of time before the divergence of dicot and monocot plants from their ancestors on the earth, and then purifying selection was the dominant force for the evolution of these amino acid sites. These results also suggested the adaptability of these conserved Mc1 genes to the land environment today.
BjMc1 gene located in B genome of B. juncea originated from Brassica nigro (B. nigro)
To understand the origination and evolution of BjMc1 gene in detail, BLAST analysis was performed in Cruciferae using the protein sequence of BjMc1 as a query. Aside from BjMc1, 13 homologues of BjMc1 were identified from 8 species of Cruciferae, and one homologue was identified and isolated from B. nigro using primer BNI-1 designed based on BjMc1 and CLV1 sequences. Maximum-likelihood analysis using protein sequences classified all BjMc1 homologues into two groups: BjMc1 and its 7 homologues from Brassicas were classified into one group (Group I) and the other 8 homologues of Cruciferae were clustered in the other group (Group II) (Supplementary Fig. 8). Furthermore, BjMc1 was located in the same clade with the homologue BniMc1 of B. nigra, whereas BjCLV1a was located in the same clade with the homologue of B. rapa, indicating that BjMc1 of B. juncea was from B. nigra (BB) and BjCLV1a of B. juncea was from B. rapa (AA). The B. juncea genome sequence information has been released recently35. The 24 AFLP markers obtained in our previous research and the three molecular markers obtained in this research were BLASTed against the B. juncea genome sequence database, and in total 16 molecular markers shared high homology with J17 (Supplementary Table 6). Therefore, it was suggested that the BjMc1 locus derived from B. nigro was located in a genomic region of B genome (J17 chromosome) in B. juncea.
Discussion
In recent years, the multilocular trait, which was thought to possess the potential to increase crop yield, was discovered in rapeseed. In the present study, we cloned the BjMc1 gene controlling the carpel development in B. juncea through fine mapping and transgenic complementation. An insertion of a RTE1 disrupted the transcription of BjMc1 (Fig. 2), leading to the result that the Bjmc1 gene could translate a dominant negative protein and the trilocular silique grows on J163-4 plants. The analysis of the molecular evolutionary process revealed that Mc1 genes had been conserved in modern land plants, and two amino acid sites of this gene located in the first cysteine domain and a key component of serine/threonine protein kinase domain had undergone positive selection during a short period of time before the divergence of dicot and monocot plants from their common ancestors on the earth. Our results also showed that the target gene cloned in this study was from J17 genome of B. juncea.
Although mutilocular phenotype has attracted much research attention in the past few decades, the instability of multilocular trait has always confused researchers. All clv1 mutants in Arabidopsis exhibited weak, intermediate or strong multilocular phenotypes27, and previous studies had shown that most Brassicas plants with multiloculus had variable valve numbers in the siliques of the same plant10, 11. However, in this study, J163-4 planted in both central and northwestern China exhibited four valves stably in trilocular siliques. We transformed the Bjmc1 gene into the clv1-1 allele in Arabidopsis for over-expression, and proved that the truncated Bjmc1 protein had a potential dominant negative effect, which could have played a key role in maintaining the valve number per silique to be four. Moreover, the over-expression of Bjmc1 gene in trilocular plants of NILs is in progress to further prove the dominant negative effect of this mutant gene. The dominant negative model was proposed by previous research: if a protein was monomeric, the activity of a protein was limited by the availability of substrate, then a variant capable of binding substrate without carrying out a subsequent catalytic step could be inhibitory36. In Arabidopsis, clv1-20 showed a weak multilocular phenotype17, while single barely any meristem 1 (bam1) mutant did not display multilocular phenotype37. Double mutants between bam1-3 and clv1-20 exhibited a strongly synergistic defect of extra carpels similar to those of clv1-4 which showed a strong multilocular phenotype and was thought to be a dominant negative mutation, indicating that clv1-4 dominant negative mutation caused inhibitory interaction between clv1 and bam1 protein17. In addition, the BjMc1 was a Leu-rich repeat receptor serine/threonine kinase, which could be bound by the ligand to the extracellular domain. In our study, the truncated Bjmc1 protein contained the putative extracellular domain but without the putative intracellular domain (Supplementary Fig. 3c). Hence, it indicated that although the ligand could normally bind to Bjmc1 protein, the truncated protein had lost the function of serine/threonine domain, and acted as a monomeric variant in the molecular mechanism. Although BjBAM1 interacted synergistically with BjMc1, the BjMc1 protein was the dominant receptor kinase to regulate the carpel development, because it was clv1 but not bam1 that showed multilocular phenotype according to researches in Arabidopsis. The dominant negative molecular model of Bjmc1 protein was shown in Fig. 6.
Figure 6.
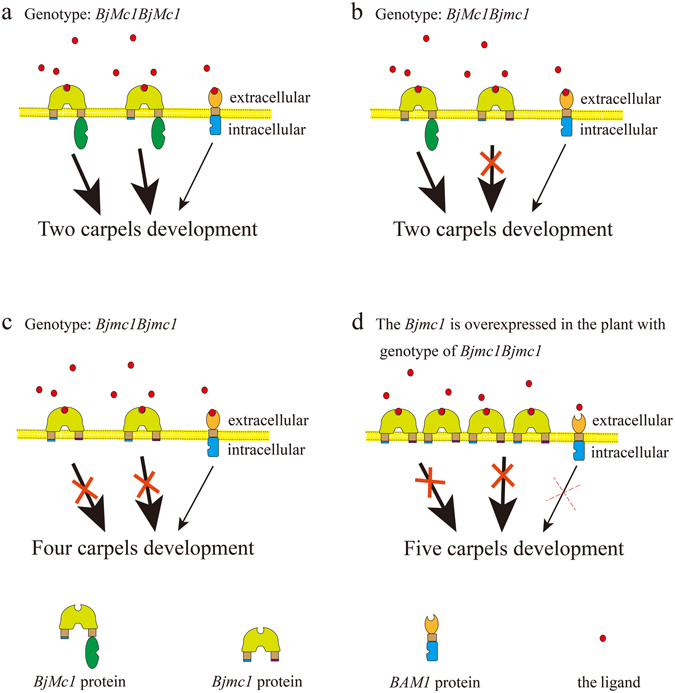
Model of dominant negative receptor action in B. juncea. A putative model for the role of BjMc1 protein, BARELY ANY MERISTEM 1 (BAM1) protein and the ligand protein in regulating the carpel development. Bold arrows indicate that the BjMc1 protein plays a dominant role in regulating the development of carpel. (a) Scenarios for plants with genotype of BjMc1BjMc1. The ligand could bind to both the BjMc1and BjBAM1 protein, but BjMc1protein was the dominant receptor kinase. (b) Scenarios for plants with genotype of BjMc1Bjmc1. The ligand could bind to the BjMc1, Bjmc1and BjBAM1 protein, and the BjMc1protein was the dominant receptor kinase. (c) Scenarios for plants with genotype of Bjmc1Bjmc1. The ligand could bind to the Bjmc1 and BjBAM1 protein, but only the BjBAM1 protein which does not play the dominant role in regulating pathway has the normal function. (d) Scenarios for plants with genotype of Bjmc1Bjmc1 and Bjmc1 overexpression. More ligands are possessed by the Bjmc1, and little ligand could bind to the BjBAM1 protein.
In this research, although the pBjCLV1b:BjCLV1b-transgenic lines did not display a complete bilocular phenotype, the exogenous gene introduced into J163-4 had worked. Because the J163-4 plants always exhibited trilocular silique but the transgenetic plants showed chimeric phenotype and the exogenous BjMc1 gene expressed (Fig. 3b). There were several explanations for this phenotype. First, the potential dominant negative effect of Bjmc1 gene might make the exogenous BjMc1 gene not working completely. Unless the expression level of the exogenous gene was much higher, the similar or lower expression level to that of endogenous BjMc1 gene could not restore the triloculus; Second, the transgenic receptor material was the trilocular parental line J163-4 rather than the trilocular plants in NILs. Although the genotypes of the trilocular parental line and trilocular plants in NILs were the same, their genetic backgrounds were quite different. In our present research, we are trying to introduce the pBjCLV1b:BjCLV1b construct into the trilocular plants in NILs. Third, BjMc1 gene took part in the development of SAM. In Soybean, Dt2 regulated stem growth, which produced semideterminate plants terminal racemes. When 35S:CDS-Dt2 construct (Dt2 with 35S promoter) and Pro-Dt2:CDS- Dt2 construct (Dt2 with putative endogenous promoter) were introduced into Thorne which displayed the indeterminate stem growth respectively, only 35S:CDS-Dt2-transgenic lines displayed semideterminate stem growth38, and this situation was similar to our research. So, we speculated that the genes involving in the SAM development were controlled by some regulatory mechanism, which could make similar phenotypes in this kind of genes. In our study, although pBjCLV1b:BjCLV1b-transgenic lines did not show a complete bilocular phenotype, 35S::BjCLV1b-transgenic lines showed a complete bilocular phenotype in B. juncea. Similarly, the transgenic lines of Arabidopsis, clv1-1, also showed similar phenotype with J163-4, and Mc1 genes in different species were conservative. Overall, BjCLV1b was the target gene of BjMc1, and the investigation will be carried out to explain the chimeric phenotype of transgenic lines in our following research.
Blast searches revealed that Mc1 genes were widespread in land plants, and analysis of gene structure revealed that Mc1 genes in land plants were conserved, especially at serine/threonine domain. There were two Mc1 copies, BjCLV1a and BjCLV1b, in B. juncea. The evolution analysis revealed that BjCLV1a located in A chromosome was derived from B. rapa while BjCLV1b located in B chromosome was derived from B. nigro, indicating that the evolution of BjMc1 and its homologues in polyploidization followed a regular U-triangle39 evolutionary pattern. In the analysis of evolution process, we found that the dominant driving force for Mc1 genes in land plants was purifying selection, which contributed to functional stabilization. The mutation of BjMc1 homologous genes led to a larger fruit size in tomato, increased kernel row number in maize21, 22 and seed number per silique in rapeseed. The mutation of Mc1 genes always could increase the yields of crops or vegetables. These findings indicated that modulation of the pathways that control fundamental stem cell proliferation had the potential to enhance crop yield. Although Mc1 genes were important to plant growth, their mutations did not exhibit negative impacts on plants, suggesting that they could be an excellent gene resource for breeding of high-yield crops. Therefore, homologous genes of BjMc1 could be edited by genome-editing technology or silenced by RNA interference technology in crops. Moreover, because Bjmc1 was dominant negative and recessive, it could be overexpressed in other crops whose BjMc1 homologous genes had been silenced to obtain the cultivars with consistent high yields. In future work, we will continue to focus on the application of the dominant negative effect to crop breeding for higher yields.
Methods
Plant materials
Homozygous near-isogenic lines (NILs, BC4F1) were constructed using a trilocular line J163-4 and a bilocular line J24828. Based on the NILs, a BC5F1 population (3,700 individuals) and a BC6F1 population (5,600 individuals) were further developed in 2012 to 2014 to fine map Bjmc1 gene. All of the identified individuals with recombination events at Bjmc1 locus were further determined by progeny testing in 2013 to 2015.
Arabidopsis thaliana (Col-0 ecotype) was used to reveal the expression pattern of BjMc1 gene by b-glucuronidase (GUS) staining and subcellular localization. Arabidopsis clv1-1 was purchased from ABRC (http://abrc.osu.edu/), and its mutation site was shown in Fig. 3d.
Availability of data and material
All data generated or analyzed during this study are included in this published article and its additional files.
BAC screening and sequencing
Positive clones from the purple-leaf mustard (Brassica juncea) bacterial artificial chromosome (BAC) library were identified through a two-stage polymerase chain reaction (PCR) screening method with C1-1 primers designed from the sequence of Bra015812 which was located near the homologous sequence of molecular marker SC13 in A07 chromosome of B. rapa. The phenotypes were similar between J163-4 and clv1 mutant in Arabidopsis (Crooijmans et al., 2000). BAC DNA was sequenced as previously described by Yi et al.40.
Genetic mapping and positional cloning
Bulked segregant analysis41 was conducted to screen the molecular markers linked to BjMc1 locus. Simple sequence repeats (SSR) was designed according to the reference sequence retrieved from the positive BAC clone. The parents and bulks were subjected to SSR marker analysis. PCR was performed according to Xu et al.28. PCR products were then separated on 1% polyacrylamide denaturing sequencing gel and shown by silver nitrate staining. Linkage analysis was performed using Joinmap442. All genetic distances were expressed in centiMorgan (cM) using the Kosambi function43.
cDNA preparation and 5′- and 3′- rapid-amplification of cDNA ends (RACE)
Total RNA was extracted from various plant tissues using an RNA extraction kit (RNeasy Plant Mini Kit; QIAGEN). The first-strand cDNA was synthesized using 2 mg of RNA and 200 units of M-MLV reverse transcriptase (Promega Kit) in a volume of 25 ml. The 5′- and 3′- RACE reactions were performed using the SMARTer RACE Amplification Kit (Clontech) according to manufacturer’s instructions.
Constructs and transformation
The genomic fragments of the candidate gene were amplified from the biolocular plants from the NILs using high-fidelity PCR. A 6,613 bp genomic DNA fragment spanning from 2,692 bp upstream the translation start codon to 767 bp downstream the termination codon of BjMc1 using primer E48-2 (Supplementary Table 1) was amplified. The correct fragment confirmed by sequencing was then cloned into Pst I - kpn I site of pCAMBIA2300 vector to construct plasmid pBjMc1:BjMc1. To prepare p35S::BjMc1 construction, we cloned a 3455 bp genomic DNA fragment spanning from start codon to 301 bp downstream the termination codon of BjMc1 using primer E46-1 (Supplementary Table 1) into Pst I - kpn I site of pCAMBIA2300, and double 35S promoter was cloned into Hind III - Pst I site of pCAMBIA2300. To prepare the ProBjMc1-GUS construct, the BjMc1 promoter region (from 1 to 2499 bp) using primer E58-1 (Supplementary Table 1) was amplified. A cassette containing GUS coding region followed by nopaline synthase polyadenylation signal from pBI101 vector was subcloned into the binary vector pCAMBIA 2300 with restriction enzymes Hind III and EcoR I to construct promoter-GUS fusions. The amplified fragments were subcloned into the modified binary vector pCAMBIA 2300 to yield the 2499 bp BjMc1 promoter-GUS construct. To prepare p35S::Bjmc1 construct, a 3036 bp genomic DNA fragment spanning from the start codon to a 687 bp downstream DNA fragment using primer HY-4 (Supplementary Table 1) was amplified. The correct fragment confirmed by sequencing was cloned into Xba I - Sac I site of pMDC83 vector to construct plasmid p35S::Bjmc1. These constructs were introduced into the host cells of Agrobacterium tumefaciens GV3101. The pBjMc1:BjMc1 and p35S::BjMc1 were transformed into the trilocular line J163-4, whereas ProBjMc1-GUS construct was introduced into wild-type Arabidopsis (Col-0 ecotype) and p35S::Bjmc1 was transformed into clv1-1 by floral dipping44. The T2 transgenic plants of ProBjMc1-GUS were grown for GUS staining.
Expression analysis
Total RNA was extracted using Trizol reagent (Invitrogen). The tissues of the early inflorescence of trilocular plants and SAM, early inflorescence, 1–4 mm ovary, 4–7 mm ovary, 7–10 mm ovary, silique peel, leaves, stem and root of bilocular plants in BC5F1 generation were selected, and 3 mg of total RNA from each tissue was treated with RNase-free DNase I to remove contaminated DNA respectively, then reverse transcribed into the first-strand complementary DNA (cDNA) with M-MLV reverse transcriptase (Fermentas, Vilnius, Lithuania) using oligo d(T)25 primer. The reverse-transcribed products from various tissues were used as templates for qPCR assay using the Bj3 primer (Supplementary Table 1) which could particularly amplify BjMc1 copy to examine the expression of BjMc1 gene. The qPCR was conducted according to Li et al.4. The measurements were obtained using the relative quantification method45. ACTIN2 gene was used as the internal control for B. juncea 46. All expression level data obtained by qPCR were based on three biological samples and three replicates for each sample.
The cDNA of BjMc1 and Bjmc1 were detected from the reverse-transcribed products from early inflorescence of trilocular and bilocular plants in BC5F1 generation using the semi-quantitative RT-PCR by C1-2 and M1-1. The ACTIN2 gene was used as the control. Semi-quantitative RT-PCR was performed as following: 94 °C for 3 min; twenty-five cycles of 94 °C for 30 s, 55 °C for 30 s and 72 °C for 45 s; and a final 10-min elongation step.
Histochemical GUS staining
Twelve independent T2 transgenic lines of ProBjMc1-GUS were subjected to histochemical GUS assays. Seedlings of 10 d and various organs of the transgenic plants were incubated at 37 °C overnight in 5-bromo-4-chloro-3-indolyl-b-glucuronic acid solution and then cleaned in 75% (v/v) ethanol. The treated tissues were observed on an Olympus IX-70 Microscope equipped with Nomarski Optics4.
Subcellular localization of BjMc1
The BjMc1 coding sequence without the termination codon (TAA) was amplified from the biolocular plants in BC5F1 generation by PCR using YXB4 primer (Supplementary Table 1). The amplified cDNA fragments were inserted downstream of the double 35S promoter through Xba I - BamH I site in frame with GFP in pMDC83 vector. This plasmid was transformed into Arabidopsis. The roots of transgenic plants in T2 generation were incubated with 10 μM FM4-64 for at least 5 min before observation. The emission light was dispersed and recorded at 500–540 nm for GFP. Confocal images were taken with a Nikon Eclipse80i fluorescence microscope equipped a water-immersed ×40 lens with an excitation wavelength of 488 nm and the following detection wavelengths: 500–540 nm for GFP and at >650 nm for FM4-64 (Nikon, Japan). All fluorescence experiments were independently repeated at least three times.
Identification of Mc1 genes from Cruciferae and other land plant species
To identify Mc1 homologous genes in Cruciferae, BLAST analysis using protein sequence of BjMc1 as a query was performed in Cruciferae (http://brassicadb.org/brad/, http://www.arabidopsis.org/, http://122.205.95.67/blast/blast.php).
To identify Mc1 homologous genes in the land plants, the coding sequence (CDS) of BjMc1 was used as query to search the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/), Ensembl Plants (http://plants.ensembl.org/index.html) and the Arabidopsis Information Resource database (http://www.arabidopsis.org/). The most highly similar sequence was selected from each species. The deduced nucleotide and protein sequences of land plant Mc1 genes identified in this analysis were used for further analysis.
Phylogenetic analyses and detection of positive selection
Using the MEGA547, Cruciferae Mc1 amino acid sequences were aligned by ClastalW and land plant Mc1 CDSs were aligned by ClastalW condons, and finally the FASTA formats were exported. The maximum-likelihood approach was used for the phylogenetic analysis of Cruciferae and all the land plants, respectively.
To test the selective pressure of Mc1 genes during the long period of evolution in land plants, the value of d N: d S ratio (or ω) for Mc1 genes was calculated with the program codeML from Phylogenetic Analysis by Maximum Likelihood (PAML) v4.432. In this study, three likelihood ratio tests (LRTs), M0, M1a vs M2a and M7 vs M8, were used to examine the selective pressure. M0 was used to calculate the average ω value of all codon sites, and the other two LRTs were used to detect the role of positive selection. For one LRT, the differences of log likelihood of the two models were compared using chi-squared (χ2) statistics. In our analyses, the degree of freedom was 1 for M1a/M2a and M7/M8 tests33, 48.
The improved branch model and branch site model49 were also used to detect the role of positive selection on the land plant Mc1 genes. In these two models, six branches were selected from the phylogenetic tree (branch I, II, III, IV, V and VI); and when any one of the branches served as the foreground branch, the remaining branches were background branches (Supplementary Fig. 4). For the analysis of branch site model, we compared the null hypothesis (ω fixed to 1) with the alternative hypothesis (free ω) to test whether positive selection affected the evolution of land plant Mc1 genes. The Bayes empirical Bayes procedure in codeML 34 was used to calculate the posterior probability that each site in the foreground branch was subjected to positive selection.
Electronic supplementary material
Revised Supplementary Figures and Tables
Acknowledgements
The authors are grateful to Zuoxiong Liu from English Department of Huazhong Agricultural University in China and Dr. Cilla Luo in New Zealand for critically reading the manuscript. This work was financially supported by the National Natural Science Foundation of China (NSFC, grant No. 31571698), the National Key Research and Development Program of China (grant No. 2016YFD0101300) and the Program for Modern Agricultural Industrial Technology System of China (grant No. nycytx-00501).
Author Contributions
J.S., P.X., Z.L., J.W., B.Y., C.M., J.T. and T.F. planned and designed the research; P.X., S.C., X.W., W.H. and G.W. performed experiments, conducted fieldwork; K.H., P.X., X.W. and S.C. analyzed the data; P.X., J.S., X.W. and K.H. wrote the manuscript; All the authors have commented, read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03755-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Barton MK. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010;341:95–113. doi: 10.1016/j.ydbio.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 2.Lv ZW, et al. Primary study on anatomic and genetic analyses of multi-loculus in Brassica juncea. Chin. J. Oil Crop Sci. 2012;34:461–466. [Google Scholar]
- 3.He YT, et al. Anatomic and genetic studies on multicapsular character in Brassica campestris L. Chin. J. Oil Crop Sci. 2003;25:1–4. [Google Scholar]
- 4.Li SP, et al. BnaC9. SMG7b functions as a positive regulator of the number of seeds per silique in Brassica napus by regulating the formation of functional female gametophytes. Plant Physiol. 2015;169:2744–2760. doi: 10.1104/pp.15.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, et al. Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc. Natl. Acad. Sci. USA. 2015;112:5123–5132. doi: 10.1073/pnas.1502160112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang LW, Li SP, Chen L, Yang GS. Identification and mapping of a major dominant quantitative trait locus controlling seeds per silique as a single Mendelian factor in Brassica napus L. Theor. Appl. Genet. 2012;125:695–705. doi: 10.1007/s00122-012-1861-3. [DOI] [PubMed] [Google Scholar]
- 7.Zhao HC, et al. Performances in main characteristic of multilocular B. juncea. Acta. Agric. Boreali-Occident Sinica. 2003;12:62–64. [Google Scholar]
- 8.Choudhary BR, Solanki ZS. Inheritance of silique locule number and seed coat colour in Brassica juncea. Plant Breed. 2007;126:104–106. doi: 10.1111/j.1439-0523.2007.01245.x. [DOI] [Google Scholar]
- 9.Yadava SK, et al. Tetralocular ovary and high silique width in yellow sarson lines of Brassica rapa (subspecies trilocularis) are due to a mutation in Bra034340 gene, a homologue of CLAVATA3 in Arabidopsis. Theor. Appl. Genet. 2014;127:2359–2369. doi: 10.1007/s00122-014-2382-z. [DOI] [PubMed] [Google Scholar]
- 10.Xiao L, et al. Genetic and physical fine mapping of a multilocular gene Bjln1 in Brassica juncea to a 208-kb region. Mol. Breed. 2013;32:373–383. doi: 10.1007/s11032-013-9877-1. [DOI] [Google Scholar]
- 11.Fan CC, et al. A novel single-nucleotide mutation in a CLAVATA3 gene homologue controls a multilocular silique trait in Brassica rapa L. Mol. Plant. 2014;7:1788–1792. doi: 10.1093/mp/ssu090. [DOI] [PubMed] [Google Scholar]
- 12.Clark SE, Running MP, Meyerowitz EM. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- 13.Muller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forstheofel NR, Wu Y, Schulz B, Bennett MJ, Feldmann K. T-DNA insertion mutagenesis in Arabidopsis: prospects and perspectives. Aust. J. Plant Physiol. 1992;10:353–366. doi: 10.1071/PP9920353. [DOI] [Google Scholar]
- 15.Kayes JM, Clark SE. CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development. 1998;125:3843–3851. doi: 10.1242/dev.125.19.3843. [DOI] [PubMed] [Google Scholar]
- 16.Xu C, et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015;47:784–792. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- 17.Durbak AD, Tax FE. CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics. 2011;189:177–194. doi: 10.1534/genetics.111.130930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng ZP, Yang ZN, Zhang S. CLV1 interacts with UFO in modulation of gynoecium development in Arabidopsis thaliana. J. Plant Biol. 2013;56:13–23. doi: 10.1007/s12374-012-0325-2. [DOI] [Google Scholar]
- 19.Prunet N, et al. SQUINT promotes stem cell homeostasis and floral meristem termination in Arabidopsis through APETALA2 and CLAVATA signaling. J. Exp. Bot. 2015;66:6905–6916. doi: 10.1093/jxb/erv394. [DOI] [PubMed] [Google Scholar]
- 20.Mandel T, et al. The ERECTA receptor kinase regulates Arabidopsis shoot apical meristem size, phyllotaxy and floral meristem identity. Development. 2014;141:830–841. doi: 10.1242/dev.104687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bommert P, et al. thick tassel dwarf 1 encodes a putative maize ortholog of the Arabidopsis CLAVATA1 leucine-rich repeat receptor-like kinase. Development. 2004;132:1235–1245. doi: 10.1242/dev.01671. [DOI] [PubMed] [Google Scholar]
- 22.Bommert P, Nagasawa NS, Jackson D. Quantitative variation in maize kernel row number is controlled by the FASCIATED EAR2 locus. Nat. Genet. 2013;45:334–337. doi: 10.1038/ng.2534. [DOI] [PubMed] [Google Scholar]
- 23.Suzaki T, et al. The gene FLORAL ORGAN NUMBER1 regulates floral meristem size in rice and encodes a leucine-rich repeat receptor kinase orthologous to Arabidopsis CLAVATA1. Development. 2004;131:5649–5657. doi: 10.1242/dev.01441. [DOI] [PubMed] [Google Scholar]
- 24.Medford JI, Behringer FJ, Callos JD, Feldmann KA. Normal and abnormal development in the Arabidopsis vegetative shoot apex. Plant Cell. 1992;4:631–643. doi: 10.1105/tpc.4.6.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pogany JA, et al. Identifying novel regulators of shoot meristem development. J. Plant Res. 1998;111:307–313. doi: 10.1007/BF02512189. [DOI] [Google Scholar]
- 26.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 27.Diévart A, et al. CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell. 2003;15:1198–1211. doi: 10.1105/tpc.010504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu P, et al. Identification of molecular markers linked to trilocular gene (mc1) in Brassica juncea L. Mol. Breed. 2014;33:425–434. doi: 10.1007/s11032-013-9960-7. [DOI] [Google Scholar]
- 29.Wang G, et al. Fine mapping of polycyetic gene (Bjmc2) in Brassica juncea L. Acta. Agronomica Sinica. 2016;42:1735–1742. [Google Scholar]
- 30.Shpak ED, Lakeman MB, Torii KU. Dominant-Negative Receptor Uncovers Redundancy in the Arabidopsis ERECTA Leucine-Rich Repeat Receptor–Like Kinase Signaling Pathway That Regulates Organ Shape. Plant Cell. 2003;15:1095–1110. doi: 10.1105/tpc.010413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl Y, et al. Moderation of Arabidopsis root stemness by CLAVATA1 and ARABIDOPSIS CRINKLY4 receptor kinase complexes. Curr. Biol. 2013;23:362–371. doi: 10.1016/j.cub.2013.01.045. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Z, Wong WS, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- 35.Yang JH, et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016;48:1225–1232. doi: 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- 36.Herskowitz I. Functional inactivation of genes by dominant negative mutation. Nature. 1987;329:219–222. doi: 10.1038/329219a0. [DOI] [PubMed] [Google Scholar]
- 37.DeYoung BJ, et al. The CLAVATA1-related BAM1, BAM2, and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 38.Ping JQ, et al. Dt2 is a gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell. 2014;26:2831–2842. doi: 10.1105/tpc.114.126938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagaharu U. Genome analysis in Brassica with special reference to then experimental formation of B. napus and peculiar mode of fertilization. Jpn. J. Bot. 1935;7:389–452. [Google Scholar]
- 40.Yi B, et al. Two duplicate CYP704B1 homologous genes BnMs1 and BnMs2 are required for pollen exine formation and tapetal development in Brassica napus. Plant J. 2010;63:925–938. doi: 10.1111/j.1365-313X.2010.04289.x. [DOI] [PubMed] [Google Scholar]
- 41.Michelmore RW, Paran I, Kesseli R. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA. 1991;88:9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Ooijen, J. Joinmap®: software for the calculation of genetic linkage maps in experimental populations, version 4. Wageningen, the Netherlands: Kyazma BV (2006).
- 43.Kosambi D. The estimation of map distances from recombination values. Ann. Hum. Genet. 1943;12:172–175. [Google Scholar]
- 44.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Ame´lie AK, Eve S, Stephen JP, Smita K, Peter JE. Suppression of the SUGAR-DEPENDENT1 triacylglycerollipase family during seed development enhances oil yield in oilseed rape (Brassica napus L.) Plant Biotechnol. J. 2013;11:355–361. doi: 10.1111/pbi.12021. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong WS, Yang Z, Goldman N, Nielsen R. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics. 2004;168:1041–1051. doi: 10.1534/genetics.104.031153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Revised Supplementary Figures and Tables


