Abstract
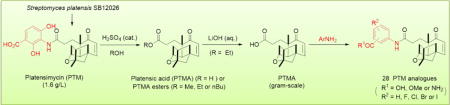
Platensimycin (PTM), produced by several strains of Streptomyces platensis, is a promising drug lead for infectious diseases and diabetes. The recent pilot-scale production of PTM from S. platensis SB12026 has set the stage for the facile semi-synthesis of a focused library of PTM analogues. In this study, gram-quantity of platensic acid (PTMA) was prepared by the sulfuric acid-catalyzed ethanolysis of PTM, followed by a mild hydrolysis in aqueous lithium hydroxide. Three PTMA esters were also obtained in near quantitative yields in a single step, suggesting a facile route to make PTMA aliphatic esters. 1-[Bis(dimethylamino)methylene]-1H-1,2,3-triazolo[4,5-b]pyridinium 3-oxid hexafluorophosphate (HATU)-catalyzed coupling of PTMA and 33 aminobenzoates resulted in the synthesis of 28 substituted aminobenzoate analogues of PTM, among which 26 of them were reported for the first time. Several of the PTM analogues showed weak antibacterial activity against methicillin-resistant Staphylococcus aureus. Our study supported the potential utility to integrate natural product biosynthetic and semi-synthetic approaches for structure diversification.
Keywords: Platensimycin, Platensic acid, Halogen-substituted aminobenzoate, Methicillin-resistant Staphylococcus aureus
1. Introduction
Fatty acids, such as palmitate, are essential building blocks of various lipids, membrane proteins and many intracellular signal molecules for all cells, and play important roles in physiology and diseases.1,2 Fatty acids in bacteria and plants are biosynthesized by type II fatty acid synthases (FASIIs), while multifunctional type I FASs consisting of individual functional domains exist in fungi and animals.3 Due to their importance in de novo fatty acid synthesis, FASs are viable targets for infective diseases, metabolic disorders and cancer.4–7 Therefore there is great interest to identify novel FASs inhibitors with potentially broad pharmacological application.
Platensimycin (PTM), consisting of a 3-amino-2,4-dihydroxy benzoate (ADHBA) and a novel tetracyclic terpene cage moiety (platensic acid, PTMA, 1d), was isolated from Streptomyces platensis by Singh and his co-workers, using an antisense differential screening strategy against an essential enzyme β-Ketoacyl-Acyl Carrier Protein (ACP) Synthase II (FabF) in FASII (Fig. 1).8 It was firstly shown to possess potent antibacterial activity against Gram-positive bacteria including the methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococci, in comparison to clinically used antibiotic linezolid. Later it showed promising antidiabetic activity in mouse models, suggesting its application in metabolic diseases, such as obesity and diabetes.9 Platencin (PTN), a PTM structure analogue with a different tricyclic terpene moiety, was similarly discovered.10 Note that PTN is a dual inhibitor for FabF and β-Ketoacyl-ACP Synthase I (FabH) against many Gram-positive pathogens. The recent discovery of dedicated terpene synthases for PTM and PTN biosynthesis showcased how those elegant molecules were naturally prodcued.11,12
Figure 1.
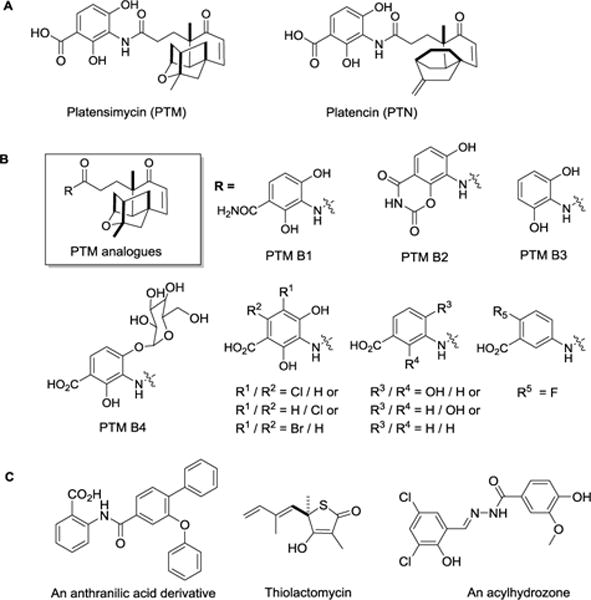
Structures of selected FAS inhibitors and their analogues. (A) Structures of PTM and PTN; (B) PTM aminobenzoate analogues from fermentation broth or semi-synthesis; (C) Structures of an anthranilic acid derivative, thiolactomycin and an acylhydrozone.
Joining the growing family of natural FabF or FabH inhibitors, including cerulenin, thiolactomycin and phomallenic acids, PTM and PTN were under intense investigation mainly due to their promising biological activities and unprecedented molecular complexity.13 In order to improve their biological activities or poor pharmacokinetic properties, many PTM and PTN analogues were thus prepared via various approaches. For example, dozens of PTM or PTN congeners have been obtained from its native producers or engineered mutant strains (Fig. 1B).14–19 Many elegant approaches towards the synthesis of their analogues were reported.20–36 These work resulted in a few PTM analogues with improved antibacterial activity, such as 7-phenyl- and 11-methyl 7-phenylplatensimycin,29 as well as PTM D1, a PTM pseudo-dimer.18 Furthermore, Nicolaou and his co-workers prepared a few PTM analogues with various molecular complexities,36 supporting the importance of the ADHBA moiety and the wisdom of making PTM analogues, since there were only a very limited number of natural or synthetic PTM aminobenzoates, especially those halogenated analogues.16,26,27,36 In addition, other molecular scaffolds, such as anthranilic acid derivatives, acylhydrozones or thiolactomycin derivatives, were also potent bacterial FabF or FabH inhibitors by binding to the active sites (Fig. 1C).37–39 Therefore, introduction of diverse functionality via halogen-substituted aminobenzoates to PTM molecular scaffold may improve our understanding of the structure-activity relationship about this important natural product.
2. Results and discussion
(1). Synthesis of PTMA esters (1a – 1c) and PTMA (1d)
With ample PTM in hands from the fermentation of S. platensis SB12026,41 we began our investigation to obtain gram-quantity of PTMA (1d) for semi-synthesis of PTM analogues with varying aminobenzoate moieties (Scheme 1). Singh and his co-workers obtained PTMA via a mild amide hydrolysis through NaNO2/CH3COOH/Ac2O with a moderate yield of about 60%.27 In order to improve the reaction yield, we first discovered that 10 mol% sulfuric acid efficiently converted PTM into corresponding PTMA esters when several alcohols were used as solvents under elevated temperature (Scheme 1A). Methanol (MeOH), ethanol (EtOH) and n-butanol (nBuOH) were all tolerated for this reaction except isopropanol and phenol. The PTMA esters 1a, 1b and 1c were obtained in near quantitative yields. Note that the PTMA methyl ester 1b was further modified to increase the terpene diversity.28 Since many aliphatic alcohols are basic building blocks in organic synthesis, this method may potentially be used to obtain other PTMA esters as well.
Scheme 1.
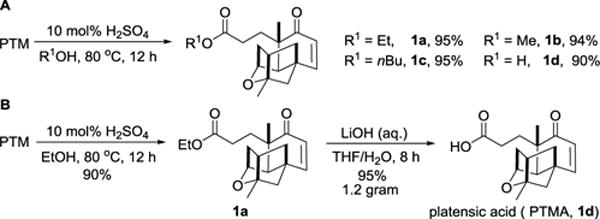
Semi-synthesis of PTMA or PTMA esters from PTM.
The direct hydrolysis was equally effective for this reaction, and PTMA (1d) was obtained in 90% yield under 0.1 mmol scale (Scheme 1A). However, the yield decreased dramatically for a large scale synthesis, which was probably due to the low solubility for PTM under acid conditions.16,41 Therefore, EtOH was used as the solvent to make PTMA ethyl ester 1a, followed by the LiOH-mediated hydrolysis in a large scale synthesis. Thus 1.20 g of PTMA was obtained from 2.11 g of PTM with 85% overall yield in two steps (Scheme 1B). The synthetic PTMA displayed the similar Cotton effect to that of the PTM observed by Circular dichroism (CD), supporting the retention of the PTM-like absolute stereochemistry during the reactions (Supporting Information, Fig. S1). Comparing to the heroic efforts to make PTMA via total synthesis,13 our semi-synthesis approach could take advantage of microbial fermentation and set the stage to rapidly diversify the aminobenzoate moiety of PTM.
(2). Synthesis of PTM 3-aminobenzoate analogues
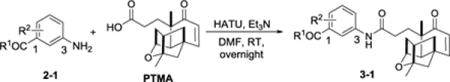
Organofluorine was known to affect the binding affinity and selectivity of small molecules to their targets, as well as to change their physical and pharmacokinetic properties.42 Therefore, several fluorine-containing methyl esters of PTM analogues were first synthesized by coupling of PTMA (1d) with substituted 3-aminobenzoic acid methyl esters (2b-1, 2d-1, 2e-1), with the ratio of methyl esters : PTMA : HATU : Et3N=1.5 : 1.0 :1.2 : 1.5 (entry 2, 4, 5). PTM analogues 3b-1, 3d-1, 3e-1 with different F substitutions on the aromatic rings were obtained with various yields ranging from 34% to 43%. The 4-Cl substituted compound 3c-1 was obtained with a comparable yield of 34% (entry 3). However, these yields were much lower than those for coupling of PTMA with protected ADHBA during the PTM synthesis, typically 50% – 70% under similar coupling conditions.13,36 The possible reason is that the inductive effects of fluoride and chloride reduce the basicity of the 3-amino group, since the coupling of 3-aminobenzoic acid methyl ester to PTMA resulted in a decent yield of 87% (entry 1).
PTM and its analogues were often prepared by the hydrolysis of their methyl esters in the last step of synthesis.13 Due to the low yield for preparing the halogen-substituted PTM aminobenzoic methyl esters (3b-1 – 3e-1), we next tested if unprotected aminobenzoic acids were able to couple to PTMA directly. PTM analogues 3f-1, 3g-1, 3i-1, 3j-1 were obtained with around 80% yield when two equivalents of the aminobenzoates were used, except 3h-1 with a chloride substitution with 46% yield, potentially because of some steric hindrance of chloride (entry 6–10). Very interestingly, the coupling yield between the unsubstituted 3-aminobenzoic acid and the F-substituted acids seemed to be comparable, which was quite different from those of 3-aminobenzoic acid methyl esters. This represented the first directly-coupling of aminobenzoic acids to the PTMA and was a simple method with good yield. It was similar to the synthesis of racemic PTN by the coupling of unprotected aminobenzoate to the platencinic acid.43
(3). Synthesis of the PTM 2-aminobenzoate analogues
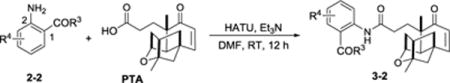
We proposed a convergent route for PTM biosynthesis, by coupling the ADHBA moiety with the putative platensicyl-CoA using amide synthase PtmC.11 It was supported by the successful incorporation of 18 arylamines to the PTM terpene ketolide in a PTM high yield producing mutant strain with one ADHBA biosynthetic gene ptmB1 inactivated.19 We attempted to obtain PTM anthranilic acid analogues through mutasynthesis, while none of the desired products were produced despite five different anthranilic acids were tested. It suggested that PtmC may have very stringent substrate requirement towards 3-aminobenzoates and promoted us to test if 2-aminobenzoates could be employed for the semi-synthesis of PTM analogues (Table 2).
Table 2.
Synthesis of PTM analogues 3-2 from 2-aminobenzoates 2-2 and PTMAa
| |||
|---|---|---|---|
|
| |||
| Entry | R3/R4/2-2 | 3-2 | Yield (%) |
| 1 | OMe/H/2a-2 | 3a-2 | 63 |
| 2 | OMe/5-F/2b-2 | 3b-2 | 52 |
| 3 | OMe/5-Cl/2c-2 | 3c-2 | 27 |
| 4 | OMe/5-Br/2d-2 | 3d-2 | 23 (11) b |
| 5 | OMe/3-F/2e-2 | ---- | trace |
| 6 | OMe/4-F/2f-2 | ---- | trace |
| 7 | OMe/6-F/2g-2 | ---- | trace |
| 8 c | OH/H/2h-2 | 3h-2 | 51 |
| 9 c | OH/5-F/2i-2 | 3i-2 | 61 |
| 10 c | OH/5-Cl/2j-2 | 3j-2 | 45 |
| 11 c | OH/5-Br/2k-2 | 3k-2 | 40 |
| 12 c | OH/5-I/2l-2 | 3l-2 | 38 (38) b |
| 13 c | OH/4-F/2m-2 | 3m-2 | 28 |
| 14 c | OH/3-F/2n-2 | ---- | decomposed |
| 15 c | OH/6-F/2o-2 | 3o-2 | 43 |
| 16 c | OH/4-Cl/2p-2 | 3p-2 | 51 |
| 17 | NH2/H/2q-2 | 3q-2 | 68 |
| 18 | NH2/5-F/2r-2 | 3r-2 | 52 |
| 19 | NH2/5-Cl/2s-2 | 3s-2 | 46 |
| 20 | NH2/5-Br/2t-2 | 3t-2 | 18 |
| 21 | NH2/4-F/2u-2 | 3u-2 | 16 |
| 22 | NH2/3-F/2v-2 | ---- | decomposed |
| 23 | NH2/6-F/2w-2 | 3w-2 | 42 |
On a 0.1 mmol scale, 2-aminobenzoates 2-2 : PTMA : HATU : Et3N = 1.5 : 1.0 :1.2 : 1.5.
Yields are given in parentheses for pyridine instead of Et3N as base.
2.0 eq of aminobenzoate was used.
Firstly, seven methyl esters of 2-aminobenzoic acid with different halogen substitutions were tested. Three PTM 2-aminobenzoic methyl esters with 5-F (3b-2), 5-Cl (3c-2) or 5-Br (3d-2) substitution, along with the unsubstituted ester 3a-2 (entries 1–4) were successfully obtained through HATU-catalyzed coupling in moderate to low yields. Our inability to obtain enough amide products of 3-F (2e-2), 4-F (2f-2), and 6-F (2g-2) (entries 5–7) suggested that the combination effects of the inductive effects from F and the steric hindrance from the nearby methyl ester were detrimental to the amide coupling reaction.
Nine halogen substituted 2-aminobenzoic acids (2h-2 to 2p-2) were directly coupled to PTMA when two equivalents of the aminobenzoic acids were used (entries 8–16). Eight PTM analogues were successfully prepared with moderate yields, while the product of 3-fuloro-2-amino benzoic acid (2n-2) decomposed soon (entry 14). In addition, several halogen-substituted PTM benzoic amides were also synthesized, however the product of 3-fuloro-2-amino benzoic amide (2v-2) also decomposed soon (entries 17–23).
Since the yields for the 2-substituted PTM analogues were generally lower to those of 3-substituted analogues, pyridine was tested as the alternative base instead of triethylamine in the coupling reaction of 2d-2 or 2l-2 to PTMA, according to the published procedure.28 However, no yield improvement was observed (entries 4 and 12). In summary, 15 different 2-aminobenzoate analogues of PTM with diverse halogen substitution were successfully obtained, which greatly complemented our mutasynthesis approach to diversify PTM privileged structure.
(4). The antibacterial activity of semi-synthesized PTM analogues against S. aureus
All of the 32 synthesized compounds, including three PTMA esters and PTMA, ten 3-aminobenzoate PTM analogues and eighteen 3-aminobenzoate PTM analogues, were tested against S. aureus ATCC 29213 and five clinical MRSA strains from local hospitals in central China. The semi-synthesized compounds did not show antibacterial activity when tested in low concentration (10 μg/disk) in the agar diffusion assay, using PTM (5 μg/disk) as a positive control and DMSO as a negative control (data not shown).
Therefore, 100 μg/disk of each compound, along with PTM (5 μg/disk) was next tested. Among the semi-synthesized compounds, only 3f-1, 3j-2, 3k-2 and 3l-2 showed small, but clear zone of inhibition against the tested strains (Supporting Information, Fig. S2). 3j-2 (5-F), 3k-2 (5-Cl) and 3l-2 (5-Br) were the PTM 2-aminobenzoic acid derivatives, while their corresponding methyl esters or amides did not show any activity against S. aureus, which suggested the important role of the carboxylic acid functionality. Interestingly, the position for the halogen substitution in the benzoic acid was also very critical, because none of 3m-2 (4-F), 3o-2 (6-F), 3p-2 (4-Cl) of PTM 2-aminobenzoic acid analogues showed any antibacterial activity. Finally the minimum inhibitory concentrations (MICs) for 3f-1, 3j-2, 3k-2 and 3l-2 were determined to be at least 64 μg/mL against the tested S. aureus strains using agar dilution method, consistent with the reported MIC value (> 82 μg/mL) for 3f-1, which was the unsubstituted PTM 3-aminobenzoic acid.36, 44
Although the new PTM aminobenzoate analogues only showed weak antibacterial activities, it confirmed the importance of the ADHBA moiety for the potent antibacterial activity of PTM.36 In addition, the generation of a focused library of these compounds would be a starting point to discover other interesting application around PTM scaffold, such as anticancer or anti-diabetic agents.
3. Conclusion
We disclosed a facile approach to prepare 28 PTM aminobenzoate analogues bearing various halogen substitutions on the benzene ring. We first developed an efficient procedure to obtain 1.2 g of PTMA, the important biosynthetic intermediate of PTM, through the sulfuric acid-catalyzed ethanolysis of PTM and a mild hydrolysis by aqueous lithium hydroxide. The semi-synthesized PTMA was then used for the synthesis of various PTM aminobenzoate analogues. Finally all the semi-synthesized compounds were tested against S. aureus and five clinical MRSA isolates, a few of which showed weak antibacterial activities. In summary, our approach was rapid and convenient to obtain PTM aminobenzoate analogues, and these synthesized compounds could be further explored for potential pharmacological applications.
4. Experimental section
All commercially available reagents were directly used as received from vendors, unless otherwise stated. N, N-dimethylformamide (DMF) was freshly distilled over calcium hydride prior to use. 1H and 13C NMR spectra were recorded on a Brucker 400 MHz or 500 MHz spectrometer. Chemical shifts were reported in ppm relative to the internal standard tetramethylsilane (δ = 0 ppm) for 1H NMR and CDCl3 (δ = 77.00 ppm) for 13C NMR spectroscopy. The following abbreviations were used to designate chemical shift multiplicities: s = singlet, d =doublet, t = triplet, q = quartet, h = heptet, m = multiplet, br = broad. HRMS spectra were recorded on a LTQ-ORBITRAP-ETD instrument. CD spectra were recorded on J-815 from JASCO.
General Procedures for Compounds (1a-1d)
To a stirred solution of PTM (44.0 mg, 0.10 mmol) in alcohol or H2O (5.0 mL) was added 98% H2SO4 (0.5 μL, 0.01 mmol). The mixture was stirred under reflux conditions for 12 hrs. Thin-layer chromatography (TLC) indicated the reaction was complete. After the reaction mixture was cooled to room temperature, 5.0 mL brine was added. The resulting mixture was extracted with ethylacetate (EtOAc) (3×5.0 mL), and the combined organic portions were dried over anhydrous sodium sulfate. Concentration followed by flash column chromatography (eluent: EtOAc/light petroleum ether = 1:30 ~ 1:5) afforded (1a–1d).
Preparation of PTMA from PTM in Gram-Scale
To a stirred solution of PTM (2.11 g, 4.78 mmol) in EtOH (100 mL) was added 98% H2SO4 (24.5 μL, 0.47 mmol). The mixture was stirred under reflux conditions for 12 hrs. TLC indicated the reaction was complete. After the reaction mixture was cooled to room temperature, 100 mL brine was added. The resulting mixture was extracted with EtOAc (3×100 mL), and the combined organic portions were dried over anhydrous sodium sulfate. Concentration followed by flash column chromatography (eluent: EtOAc/light petroleum ether = 1:30 ~ 1:5) afforded 1a 1.37g, 4.31 mmol in 90% yield.
To a solution of ethyl ester 1a (1.37 g, 4.31 mmol) in THF (50 mL), was added LiOH-H2O (8.6 mL, 2 M). The mixture was stirred overnight at RT. The reaction was quenched with 3N HCl solution and then 10 mL brine was added. The resulting mixture was extracted with CHCl3 (3×50 mL), and the CHCl3 fraction was dried over anhydrous sodium sulfate, and concentrated in vacuo. The crude product was purified by flash column chromatography (eluent: EtOAc/light petroleum ether = 1:5 ~ 1:3) to afford PTMA (1d) (1.20 g, 4.09 mmol) in 95% yield.
General Procedures for Compounds 3
To a solution of PTMA (1d) (29.0 mg, 0.10 mmol) and aminobenzoates 2 (1.5 mmol or 0.20 mmol) in DMF (1.0 mL) at room temperature were added Et3N or pyridine (0.42 mmol) and HATU (76.1 mg, 0.20 mmol). The mixture was stirred at room temperature overnight and monitored by TLC. Then brine (10 mL) was added. The resulting mixture was extracted with EtOAc (3×5.0 mL), and the combined organic portions were dried over anhydrous sodium sulfate and concentrated in vacuo. compounds 3 were finally prepared through flash column chromatography (eluent: EtOAc/light petroleum ether = 1:20 ~ 5:1).
Antibacterial activity assay
Bacterial strains used in this study: S. aureus ATCC 29213 was from the American Type Culture Collection. MRSA strains, including MRSA 115 – MRSA 119, were clinical isolates from local hospitals in Hunan Province, China. For agar diffusion assay, the S. aureus 29213 and five MRSA strains were cultivated on Luria-Bertani (LB) agar plates. PTM (5 μg) and various amount of the semi-synthesized PTM analogues (10 μg or 100 μg) were dissolved in DMSO, placed onto 6 mm paper discs and used for the assay. The plates containing the respective bacterial strains were incubated overnight at 37 °C and monitored.
A standard protocol was followed to determine the MIC of tested compounds using agar dilution assay.44 In brief, the S. aureus strains were cultivated on LB broth overnight and diluted. Approximate 104 colony forming units (CFU) of each strain were spotted (2 μL) onto LB agar plates containing different concentration of the tested compounds, ranging from 0.25 μg to 64 μg/mL LB agar. Then the plates were incubated in 37 °C overnight and monitored.
Supplementary Material
Table 1.
Synthesis of PTM analogues 3-1 from 3-aminobenzoates 2-1 and PTMAa
| |||
|---|---|---|---|
|
| |||
| Entry | R1/R2/2-1 | 3-1 | Yield (%) |
| 1 | OMe/H/2a-1 | 3a-1 | 87 |
| 2 | OMe/4-F/2b-1 | 3b-1 | 42 |
| 3 | OMe/4-Cl/2c-1 | 3c-1 | 34 |
| 4 | OMe/6-F/2d-1 | 3d-1 | 43 |
| 5 | OMe/2-F/2e-1 | 3e-1 | 34 |
| 6b | OH/H/2f-1 | 3f-1 | 73 |
| 7b | OH/4-F/2g-1 | 3g-1 | 78 |
| 8b | OH/4-Cl/2h-1 | 3h-1 | 46 |
| 9b | OH/6-F/2i-1 | 3i-1 | 80 |
| 10b | OH/2-F/2j-1 | 3j-1 | 77 |
On a 0.1 mmol scale, 3-aminobenzoate 2-1 : PTMA : HATU : Et3N = 1.5 : 1.0 :1.2 : 1.5.
2.0 eq of aminobenzoate was used.
Acknowledgments
This work was supported in part by National Natural Science Foundation of China 81473124 (to Y.H.), the Chinese Ministry of Education 111 Project B0803420 (to Y.D.), National High Technology Joint Research Program of China grant 2011ZX09401-001 (to Y.D.), National Natural Science Foundation of China 21502238 (to L.Q.), China Postdoctoral Science Foundation 2015M582353 (to L.Q.) and the Postdoctoral Science Foundation of CSU (to L.Q.) and U. S. National Institutes of Health Grants GM114353 (to B.S.). We are grateful to the Center for Advanced Research in CSU, including the High Resolution MS Facility and the NMR facility, for the HRMS and NMR experiments.
Footnotes
Supplementary Data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/.
References and notes
- 1.Muro E, Atilla-Gokcumen GE, Eggert US. Mol Biol Cell. 2014;25:1819. doi: 10.1091/mbc.E13-09-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niphakis MJ, Lum KM, Cognetta AB, Iii, Correia BE, Ichu T-A, Olucha J, Brown SJ, Kundu S, Piscitelli F, Rosen H, Cravatt BF. Cell. 2015;161:1668. doi: 10.1016/j.cell.2015.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wakil SJ. Biochemistry. 1989;28:4523. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 4.Parsons JB, Rock CO. Curr Opin Microbiol. 2011;14:544. doi: 10.1016/j.mib.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardwicke MA, Rendina AR, Williams SP, Moore ML, Wang L, Krueger JA, Plant RN, Totoritis RD, Zhang G, Briand J, Burkhart WA, Brown KK, Parrish CA. Nat Chem Biol. 2014;10:774. doi: 10.1038/nchembio.1603. [DOI] [PubMed] [Google Scholar]
- 6.Ventura R, Mordec K, Waszczuk J, Wang Z, Lai J, Fridlib M, Buckley D, Kemble G, Heuer TS. EBioMedicine. 2015;2:808. doi: 10.1016/j.ebiom.2015.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alwarawrah Y, Hughes P, Loiselle D, Carlson David A, Darr David B, Jordan Jamie L, Xiong J, Hunter Lucas M, Dubois Laura G, Thompson JW, Kulkarni Manjusha M, Ratcliff Annette N, Kwiek Jesse J, Haystead Timothy AJ. Cell Chem Biol. 2016;23:678. doi: 10.1016/j.chembiol.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang CW, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. Nature. 2006;441:358. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 9.Wu M, Singh SB, Wang J, Chung CC, Salituro G, Karanam BV, Lee SH, Powles M, Ellsworth KP, Lassman ME, Miller C, Myers RW, Tota MR, Zhang BB, Li C. Proc Natl Acad Sci USA. 2011;108:5378. doi: 10.1073/pnas.1002588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Kodali S, Lee SH, Galgoci A, Painter R, Dorso K, Racine F, Motyl M, Hernandez L, Tinney E, Colletti SL, Herath K, Cummings R, Salazar O, Gonzalez I, Basilio A, Vicente F, Genilloud O, Pelaez F, Jayasuriya H, Young K, Cully DF, Singh SB. Proc Natl Acad Sci USA. 2007;104:7612. doi: 10.1073/pnas.0700746104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smanski MJ, Yu Z, Casper J, Lin S, Peterson RM, Chen Y, Wendt-Pienkowski E, Rajski SR, Shen B. Proc Natl Acad Sci USA. 2011;108:13498. doi: 10.1073/pnas.1106919108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudolf JD, Dong L-B, Cao H, Hatzos-Skintges C, Osipiuk J, Endres M, Chang C-Y, Ma M, Babnigg G, Joachimiak A, Phillips GN, Shen B. J Am Chem Soc. 2016;138:10905. doi: 10.1021/jacs.6b04317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicolaou KC, Chen JS, Edmonds DJ, Estrada AA. Angew Chem Int Ed. 2009;48:660. doi: 10.1002/anie.200801695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herath KB, Zhang C, Jayasuriya H, Ondeyka JG, Zink DL, Burgess B, Wang J, Singh SB. Org Lett. 2008;10:1699. doi: 10.1021/ol800251v. [DOI] [PubMed] [Google Scholar]
- 15.Yu Z, Smanski MJ, Peterson RM, Marchillo K, Andes D, Rajski SR, Shen B. Org Lett. 2010;12:1744. doi: 10.1021/ol100342m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Ondeyka J, Herath K, Jayasuriya H, Guan Z, Zink DL, Dietrich L, Burgess B, Ha SN, Wang J, Singh SB. J Nat Prod. 2011;74:329. doi: 10.1021/np100635f. [DOI] [PubMed] [Google Scholar]
- 17.Rudolf JD, Dong L-B, Huang T, Shen B. Mol Biosyst. 2015;11:2717. doi: 10.1039/c5mb00303b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong L-B, Rudolf JD, Shen B. Bioorg Med Chem. 2016;24:6348. doi: 10.1016/j.bmc.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong L-B, Rudolf JD, Shen B. Org Lett. 2016;18:4606. doi: 10.1021/acs.orglett.6b02248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudolf JD, Dong LB, Shen B. Biochem Pharm. 2016 doi: 10.1016/j.bcp.2016.11.013. [DOI] [Google Scholar]
- 21.Shang R, Liang J, Yi Y, Liu Y, Wang J. Molecules. 2015;20:16127. doi: 10.3390/molecules200916127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saleem M, Hussain H, Ahmed I, van Ree T, Krohn K. Nat Prod Rep. 2011;28:1534. doi: 10.1039/c1np00010a. [DOI] [PubMed] [Google Scholar]
- 23.Martens E, Demain AL. J Antibiot. 2011;64:705. doi: 10.1038/ja.2011.80. [DOI] [PubMed] [Google Scholar]
- 24.Lu X, You Q. Curr Med Chem. 2010;17:1139. doi: 10.2174/092986710790827852. [DOI] [PubMed] [Google Scholar]
- 25.Palanichamy K, Kaliappan KP. Chem Asia J. 2010;5:668. doi: 10.1002/asia.200900423. [DOI] [PubMed] [Google Scholar]
- 26.Singh SB, Jayasuriya H, Ondeyka JG, Herath KB, Zhang C, Zink DL, Tsou NN, Ball RG, Basilio A, Genilloud O, Diez MT, Vicente F, Pelaez F, Young K, Wang J. J Am Chem Soc. 2006;128:11916. doi: 10.1021/ja062232p. [DOI] [PubMed] [Google Scholar]
- 27.Singh SB, Herath KB, Wang J, Tsou N, Ball RG. Tetrahedron Lett. 2007;48:5429. [Google Scholar]
- 28.Shen HC, Ding F-X, Singh SB, Parthasarathy G, Soisson SM, Ha SN, Chen X, Kodali S, Wang J, Dorso K, Tata JR, Hammond ML, MacCoss M, Colletti SL. Bioorg Med Chem Lett. 2009;19:1623. doi: 10.1016/j.bmcl.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Jang KP, Kim CH, Na SW, Jang DS, Kim H, Kang H, Lee E. Bioorg Med Chem Lett. 2010;20:215. doi: 10.1016/j.bmcl.2010.02.037. [DOI] [PubMed] [Google Scholar]
- 30.Jang KP, Kim CH, Na SW, Kim H, Kang H, Lee E. Bioorg Med Chem Lett. 2009;19:4601. doi: 10.1016/j.bmcl.2009.06.092. [DOI] [PubMed] [Google Scholar]
- 31.Fisher M, Basak R, Kalverda AP, Fishwick CWG, Turnbull WB, Nelson A. Org Biomol Chem. 2014;12:486. doi: 10.1039/c3ob41975d. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Sintim HO. Chem Eur J. 2011;17:3352. doi: 10.1002/chem.201002410. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Lee V, Sintim HO. Chem Eur J. 2009;15:2747. doi: 10.1002/chem.200802568. [DOI] [PubMed] [Google Scholar]
- 34.Waalboer DCJ, Leenders SHAM, Schulin-Casonato T, van Delft FL, Rutjes FPJT. Chem Eur J. 2010;16:11233. doi: 10.1002/chem.201001744. [DOI] [PubMed] [Google Scholar]
- 35.Yeung Y-Y, Corey EJ. Org Lett. 2008;10:3877. doi: 10.1021/ol801400a. [DOI] [PubMed] [Google Scholar]
- 36.Nicolaou KC, Stepan AF, Lister T, Li A, Montero A, Tria GS, Turner CI, Tang Y, Wang J, Denton RM, Edmonds DJ. J Am Chem Soc. 2008;130:13110. doi: 10.1021/ja8044376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie Z, Perretta C, Lu J, Su Y, Margosiak S, Gajiwala KS, Cortez J, Nikulin V, Yager KM, Appelt K, Chu S. J Med Chem. 2005;48:1596. doi: 10.1021/jm049141s. [DOI] [PubMed] [Google Scholar]
- 38.Wang X-L, Zhang Y-B, Tang J-F, Yang Y-S, Chen R-Q, Zhang F, Zhu H-L. Eur J Med Chem. 2012;57:373. doi: 10.1016/j.ejmech.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Bommineni GR, Kapilashrami K, Cummings JE, Lu Y, Knudson SE, Gu C, Walker SG, Slayden RA, Tonge PJ. J Med Chem. 2016;59:5377. doi: 10.1021/acs.jmedchem.6b00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hindra, Huang T, Yang D, Rudolf JD, Xie P, Xie G, Teng Q, Lohman JR, Zhu X, Huang Y, Zhao L-X, Jiang Y, Duan Y, Shen B. J Nat Prod. 2014;77:2296. doi: 10.1021/np5006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi J, Pan J, Liu L, Yang D, Lu S, Zhu X, Shen B, Duan Y, Huang Y. J Ind Microbiol Biotechnol. 2016;43:1027. doi: 10.1007/s10295-016-1769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Müller K, Faeh C, Diederich F. Science. 2007;317:1881. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 43.Hayashida J, Rawal VH. Angew Chem Int Ed. 2008;47:4373. doi: 10.1002/anie.200800756. [DOI] [PubMed] [Google Scholar]
- 44.Wiegand I, Hilpert K, Hancock REW. Nat Protocols. 2008;3:163. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


