Abstract
The recent introduction of metabolic autopsy in the field of forensic science has made it possible to detect hidden inherited metabolic diseases. Since the next generation sequencing (NGS) has recently become available for use in postmortem examinations, we used NGS to perform metabolic autopsy in 15 sudden unexpected death in infancy cases. Diagnostic results revealed a case of carnitine palmitoyltransferase II deficiency and some cases of fatty acid oxidation-related gene variants. Metabolic autopsy performed with NGS is a useful method, especially when postmortem biochemical testing is not available.
Keywords: Sudden unexpected death in infancy, Metabolic autopsy, Next generation sequencing, Fatty acid oxidation disorder, Carnitine palmitoyltransferase II deficiency
Highlights
-
•
This is the first metabolic autopsy performed with next generation sequencing (NGS).
-
•
We detected one case of CPT II deficiency and three cases of FAOD-related rare variants.
-
•
Some of them had no specific abnormality except for genetic variants.
-
•
These cases would be undetected without NGS.
-
•
We advocate metabolic autopsy performed with NGS.
1. Introduction
Sudden unexpected death in infancy (SUDI) is defined as a term that has been variably used to refer to all cases of sudden and unexpected deaths in infancy, and not just to those where the death has been attributed to sudden infant death syndrome [1]. The common causes of SUDI are accidental death, infection, cardiovascular anomaly, child abuse and metabolic diseases.
Fatty acid oxidation disorder (FAOD) is one type of the inherited metabolic diseases that can lead to patient death during situations such as long fasting or infection. Before the introduction of expanded newborn screening by tandem mass spectrometry, FAODs account for approximately 5% of sudden infant deaths [2], [3], [4], [5], [6], [7] (Table 1). Some patients can now be screened before the symptom is apparent, others still may remain undetected because the screening has just started nationwide in Japan and some disorders including carnitine palmitoyltransferase (CPT) II deficiency are not included in the first target disease [8]. Since FAODs are functional diseases, they are not easy to diagnose, especially during postmortem examinations. Therefore, some cases may remain undiagnosed or may be incorrectly classified as sudden infant death syndrome.
Table 1.
Proportion of inherited metabolic disorders among sudden unexpected death in infancy.
The recent introduction of metabolic autopsy in the field of forensic science has made it possible to detect hidden inherited metabolic diseases [7], [9]. Metabolic autopsy is the autopsy which focuses on the inherited metabolic diseases [10]. The term metabolic autopsy includes microscopic examination of the liver, postmortem blood acylcarnitine analysis and genetic analysis [7]. Since liver steatosis is often seen as a nonspecific change in SUDI and the results of postmortem blood acylcarnitine analysis is often modified by postmortem changes, it is not easy to make a biochemical diagnosis [11]. Although successful genetic analysis has been reported in many cases, there are about 20 FAOD-related genes and thus, surveying all of the genes using Sanger's traditional method of “one gene, one exon at a time” is not an effective way for analyzing these types of cases.
The next generation sequencing (NGS) has recently become available for use in postmortem examinations [12], [13]. This technique makes it possible to examine a much larger number of genes and exons at a lower cost in addition to requiring less time than that for the conventional Sanger's method. However, most of the studies using these new techniques have focused on cardiac diseases and thus, there has yet to be metabolic autopsy performed with NGS [13].
Therefore, the aim of our current study is to detect FAOD-related gene variants among sudden death cases. As far as we know, this is the first metabolic autopsy performed with NGS.
2. Materials and methods
2.1. Subjects and target sequence
A total of 15 SUDI cases, all of which did not have any characteristic appearance and remained undiagnosed after macroscopic examination, were analyzed at the Department of Forensic Pathology at Nagasaki University (Table 2; 11 males, 4 females, age range; 0 days to 11 months). All cases were born before 2014 and none of them were screened by tandem mass spectrometry.
Table 2.
Case summary.
| Case | Age | Sex | Diagnosis | Postmortem acylcarnitine analysis | Fat staining |
|---|---|---|---|---|---|
| 1 | 0 d | F | Unknown | None specific change | Negative |
| 2 | 0 d | M | Unknown | None specific change | Negative |
| 3 | 2 d | M | Pneumonia | Not analyzed | Negative |
| 4 | 22 d | M | Unknown | None specific change | Negative |
| 5 | 2 m | M | Acute respiratory infection | Not analyzed | Negative |
| 6 | 3 m | M | SIDS | None specific change | Negative |
| 7 | 4 m | M | SIDS | None specific change | Moderate |
| 8 | 5 m | M | Pneumonia | Not analyzed | Negative |
| 9 | 5 m | M | Unknown | Not analyzed | Negative |
| 10 | 7 m | M | SIDS | Not analyzed | Negative |
| 11 | 8 m | M | Reye's-like syndrome | Not analyzed | Moderate |
| 12 | 8 m | M | SIDS | Not analyzed | Negative |
| 13 | 10 m | F | Unknown | None specific change | Moderate |
| 14 | 11 m | F | Acute encephalopathy | Not analyzed | Negative |
| 15 | 11 m | F | Reye's-like syndrome | Long-chain fatty acid defect | Distinctive |
SIDS; sudden infant death syndrome, d; day, m; month, M; male, F; female.
2.2. Extraction of genomic DNA and genetic analysis
Genomic DNA was isolated from blood leukocytes with the QIAamp DNA Blood Mini Kit (Qiagen, Tokyo, Japan) in accordance with the manufacturer's standard methods. A custom-made HaloPlex Target Enrichment System (Agilent Technologies, Santa Clara, CA) was designed to capture the coding exons of the 13 genes targeted to the FAODs (Table 3). The sequencing was performed on an Illumina MiSeq (Illumina, San Diego, CA).
Table 3.
Gene summary.
| Gene | OMIM | Exon | Disease | OMIM |
|---|---|---|---|---|
| SLC22A5 | 603377 | 10 | Primary carnitine deficiency | 212140 |
| CPT1A | 600528 | 19 | CPT I deficiency | 255120 |
| CPT2 | 600650 | 5 | CPT II deficiency | 608836 |
| 600649 | ||||
| 255110 | ||||
| SLC25A20 | 613698 | 9 | CACT deficiency | 212138 |
| ACADVL | 609575 | 20 | VLCAD deficiency | 201475 |
| ACADM | 607008 | 12 | MCAD deficiency | 201450 |
| ACADS | 606885 | 10 | SCAD deficiency | 201470 |
| HADHA | 600890 | 20 | LCHAD, MTP deficiency | 609016 (LCHAD) |
| HADHB | 143450 | 16 | MTP, LCKAT deficiency | 609015 (MTP) |
| HADH | 601609 | 8 | HAD deficiency | 231530 |
| ETFA | 608053 | 12 | MAD deficiency | 231680 |
| ETFB | 130410 | 6 | ||
| ETFDH | 231675 | 13 |
CPT; carnitine palmitoyltransferase, CACT; carnitine-acylcarnitine translocase, VLCAD; very-long-chain acyl-CoA dehydrogenase, MCAD; medium-chain acyl-CoA dehydrogenase, SCAD; short-chain acyl-CoA dehydrogenase, LCHAD; long-chain 3-hydroxyacyl-CoA dehydrogenase, MTP; mitochondrial trifunctional protein, LCKAT; long-chain 3-ketoacyl-CoA thiolase, HAD; 3-hydroxyacyl-CoA dehydrogenase, MAD; multiple acyl-CoA dehydrogenase.
The sequencing reads were mapped on the hg19 human genome sequence using Novoalign version 3.02.12 software (Novocraft, Petaling Jaya, Selangor, Malaysia). Single nucleotide variations and small insertions/deletions were detected using the Genome Analysis Toolkit [14] and annotated using the ANNOVAR software package [15].
Filtering of the variant data was performed as described below. Using the gene information of the GENCODE version 19 [16], single nucleotide variations causing non-synonymous, splice site, or nonsense substitutions and insertions/deletions occurring in the coding regions or the splice sites were retrieved. To identify putatively pathogenic variants, we retained variants with allele frequencies equal to or less than 0.5% in any of the ethnic subgroups found in the various databases. These included variants listed in the Japanese 1200 exomes from the Human Genetic Variation Database (http://www.genome.med.kyoto-u.ac.jp/SnpDB/), the 1000 Genomes Project [17] and the 6500 control exomes from the NHLBI GO Exome Sequencing Project (ESP) (http://evs.gs.washington.edu/EVS/).
The detected variants were confirmed by the traditional Sanger's method. Each of the exons was amplified, and then examined by polymerase chain reaction (PCR). All reactions were performed in a 25-μL volume containing 12.5 μL of PrimeSTAR Max Premix (2 ×) (Takara, Otsu, Japan), 0.4 μM each of the primers and 200 ng of template DNA under the following conditions: 98.0 °C for 1 min, (98.0 °C for 10 s, 54.0 °C for 5 s, 72.0 °C for 30 s) for 30 cycles, and 72.0 °C for 5 min. The products were sequenced using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on Applied Biosystems 3130 DNA Analyzer (Applied Biosystems) in accordance with the manufacturer's instructions. The sequence from both strands was visually inspected in order to confirm the substitution.
2.3. Analysis of the amino acid residues
SIFT (http://sift.jcvi.org/) and PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml/) were used to predict whether each amino acid substitution would affect the function of each protein. These substitutions were also aligned across species (https://genome.ucsc.edu/cgi-bin/hgGateway/).
2.4. Postmortem blood acylcarnitine analysis by tandem mass spectrometry
Whole blood samples were blotted onto one spot on a Guthrie card and then subjected to acylcarnitine analysis by tandem mass spectrometry.
2.5. Extraction of genomic DNA and performance of mutational analysis in the other family members
Genomic DNA was purified from buccal cell swabs in accordance with standard methodology. PCR was performed using the previously discussed method. Genomic DNA samples from the parents and the sister were examined for the presence or absence of the CPT2 mutation.
2.6. Ethics
This study was approved by the Ethics Committee of the Nagasaki University Graduate School of Medicine.
3. Case history in Case 15
The patient was a female carried to full-term and delivered via cesarean section. Her Apgar score was 6/9, birth weight was 2870 g, height was 49.0 cm and head circumference was 34.0 cm.
At 11 months of age, she was taken to a doctor's office because of fever and vomiting that had continued for several days. After examination, she was prescribed cephem antibiotics. During the morning of the next day, however, she suddenly lost consciousness and an ambulance was requested. By the time the emergency service personnel arrived, she had already suffered cardiopulmonary arrest. Although cardiopulmonary resuscitation was initiated immediately and continued until she reached the hospital, she was pronounced dead.
The patient had never been ill until the time of her death. Family history was negative for seizures, arrhythmias and sudden death. Her parents were not consanguineous.
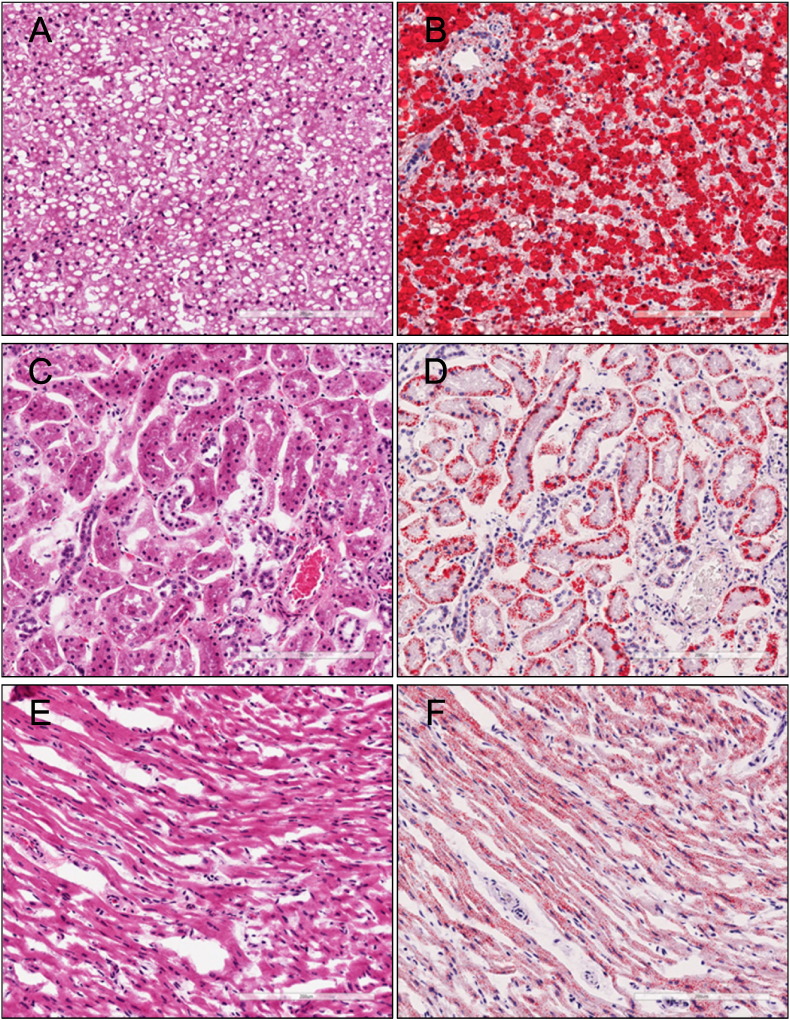
Histological examination of the patient led to the diagnosis of Reye's-like syndrome. The liver showed diffuse and distinctive vacuoles, which Oil-red O staining subsequently confirmed to be the accumulation of fatty acids. Oil-red O-positive vacuoles were also detected in the kidney and heart (Fig. 1).
Fig. 1.
Histological examination of Case 15.
Steatosis was detected in the liver (A; hematoxylin–eosin (H.E.), B; Oil-red-O), kidney (C; H.E., D; Oil-red-O) and heart (E; H.E., F; Oil-red-O).
4. Results
4.1. Target sequence of sudden death cases
Targeted resequencing revealed the mean coverage of the coding sequence in the target genes was 155.4 reads, with an overall average gene level coverage at ≥ 10 reads of 90.9%.
Table 4 shows the detected variants found during the filtering steps. After the filtering, only six variants remained.
Table 4.
Detected variants.
| Case | Gene | AA | Substitution | SIFT | PolyPhen-2 | HGVB | EXAC | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 5 | ACADS | P55L | 164C > T | Heterozygote | Damaging | 0.0 | Probably D | 0.993 | 8/8654 | 3/1346 |
| 6 | CPT2 | F383Y | 1148T > A | Heterozygote | Tolerated | 0.61 | Possibly D | 0.932 | 3/8644 | 1/600 |
| 7 | SLC22A5 | D487N | 1459G > A | Heterozygote | Tolerated | 1.0 | Benign | 0.001 | 0/121,410 | 1/600 |
| 14 | ACADS | V84M | 250G > A | Heterozygote | Damaging | 0.02 | Probably D | 0.997 | 0/6496 | 0/600 |
| 15 | CPT2 | F323fs | 968_969 del TC | Heterozygote | ||||||
| 15 | CPT2 | V605L | 1813G > C | Heterozygote | Damaging | 0.004 | Possibly D | 0.885 | 3/8654 | 1/600 |
W; wild type, M; mutation type, AA; amino acid change, probably D; probably damaging, possibly D; possibly damaging, HGVB; Human Genetic Variation Browser, EXAC; The Exome Aggregation Consortium.
4.2. Genetic analysis in Case 15
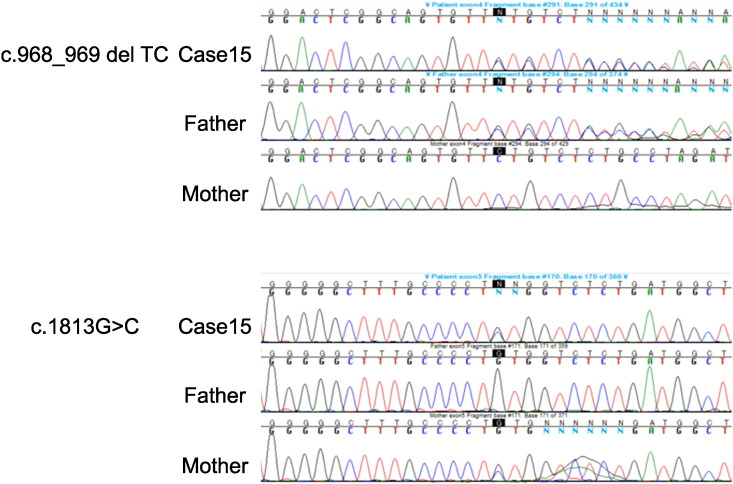
Among the detected variants, only Case 15 was found to have two pairs of heterozygous deletion (c.968_969 del TC, p.F323fs) and substitution (c.1813G > C, p.V605L) in the CPT2 gene, all of which were confirmed by Sanger's sequencing (Fig. 2). Pedigree analysis confirmed that the deletion was transmitted from her father while the substitution was from her mother (Fig. 2). The c.1813G > C (p.V605L) substitution has been previously reported in a Japanese CPT II deficiency patient [21]. This was detected in 3 alleles among the 8654 East Asian control alleles (minor allele frequency: 0.0004, The Exome Aggregation Consortium) and in 1 allele out of the 600 Japanese control alleles (minor allele frequency: 0.002, Human Genetic Variation Browser). Results also predicted there would be an effect on the function of the protein, with a SIFT of 0.004, which indicated damaging and a PolyPhen-2 of 0.885, which indicated possibly damaging. An examination of species ranging from zebrafish to human showed the substitution was conserved (Fig. 3).
Fig. 2.
Sequencing analysis in Case 15.
Case 15 had two pairs of heterozygous deletions (c.968_969 del TC) and substitutions (c.1813G > C) in the CPT2 gene. The father had a heterozygous deletion (c.968_969 del TC) while the mother had a heterozygous substitution (c.1813G > C).
Fig. 3.
Sequence alignment between species.
Except for SLC22A5-D487N, the substitutions were highly conserved between the species.
Analysis also showed there were two homozygous substitutions (c.1055T > G, p.F352C and c.1102G > A, p.V368I) in the CPT2 gene. Since these two substitutions are common genetic polymorphisms [18], [19], [20], [21], [22], they were excluded from the targeted NGS analysis.
4.3. Other genetic substitutions
There were four other substitutions found in four different cases. Case 5 had the substitution ACADS-P55L (c.164C > T), Case 6 had CPT2-F383Y (c.1148T > A), Case 14 had ACADS-V84M (c,250G > A), and Case 7 had SLC22A5-D487N (c.1459G > A) (Table 4). The former three were heterozygous substitutions that according to SIFT or PolyPhen-2 were predicted to affect the function of the protein. An examination of species ranging from zebrafish to human showed the substitution was conserved (Fig. 3).
4.4. Acylcarnitine analysis in Case 15
We performed acylcarnitine analysis of the postmortem whole blood samples using tandem mass spectrometry. Free carnitine was 181.24 μM, which was increased as compared to the normal range (< 60 μM), but which was within the postmortem reference value (< 422 μM). C5-acylcarnitine was increased to 1.51 μM (normal range: < 1.0 μM, postmortem reference value: < 1.5 μM). There was also an increase in the C16, C18, C18:1 and C18:2-acylcarnitine (Table 5). The ratio of [C16 + C18:1] to C2 was as high as 1.83, which suggests that Case 15 had either a CPT II or carnitine–acylcarnitine translocase deficiency.
Table 5.
Acylcarnitine profile in Case 15.
| C0 | C2 | C5 | C16 | C18 | C18:1 | C18:2 | |
|---|---|---|---|---|---|---|---|
| Case 15 | 181.24 | 4.65 | 1.51 | 4.13 | 2.3 | 4.39 | 0.97 |
| PRV | 422.59 | 147.13 | 1.5 | 3.495 | 2.495 | 3.095 | 0.925 |
| NR | 60.0 | 45.0 | 1.0 | 7.0 | 2.1 | 3.2 | 0.8 |
PRV; postmortem reference value, NR; normal range.
5. Discussion
After many research studies and case reports examined postmortem samples [2], [3], [4], [23], [24], [25], [26], [27], [28], in 2001, Chace et al. performed a large-scale study of FAODs in which they analyzed postmortem blood samples by tandem mass spectrometry [5]. In our previous study, we also performed postmortem acylcarnitine analyses, which subsequently led to our discovery of other FAODs cases [7]. Due to postmortem changes, the interpretation of the postmortem acylcarnitine analysis is sometimes not easy. Table 6 shows our in-house data for our postmortem acylcarnitine analysis. Based on our analytical findings, it was possible to definitively diagnose a CPT II deficiency case, which we reported in our previously published study [7]. In the two false-positive cases that we observed, we found there were increases in the long-chain acylcarnitine in the absence of any genetic abnormality. The C16, C18, C18:1 and C18:2-acylcarnitine values in the present case were much lower than that of not only our definitively diagnosed case, but also the two false-positive cases.
Table 6.
The comparison with other postmortem samples.
| C0 | C2 | C5 | C16 | C18 | C18:1 | C18:2 | |
|---|---|---|---|---|---|---|---|
| Case 15 | 181.24 | 4.65 | 1.51 | 4.13 | 2.3 | 4.39 | 0.97 |
| DD | 69.46 | 8.5 | 0.13 | 7.41 | 5.29 | 6.79 | 1.64 |
| FP 1 | 147.82 | 96.45 | 4.29 | 13.65 | 6.74 | 8.99 | 1.23 |
| FP 2 | 127.4 | 35.25 | 1.97 | 6.13 | 3.74 | 3.87 | 1.55 |
DD, definitively diagnosed case, FP; false-positive case.
Definite diagnoses of FAODs can be made by either a genetic or an enzyme analysis. The relationship between SUDI and FAODs was first discussed in detail in the 1990's [24], [29], [30]. After Bennett and Powell reported finding SUDI cases with an A985G mutation of the medium-chain acyl-CoA dehydrogenase (MCAD) deficiency, they recommended genetic analysis for SUDI be performed [2]. Unfortunately, this study only targeted one particular substitution and did not cover any of the other candidate genes. However, it should be noted that the number of candidate genes for FAODs is approximately 20 genes and thus, postmortem genetic screening is not really practical.
Recently, NGS has been developed and is more widely used, especially in the field of sudden cardiac death [12], [13]. This technique makes it possible to examine a larger number of genes and exons at a lower cost, in addition to requiring a smaller amount of time compared to the conventional Sanger's method [13]. Unlike that for clinical patients, there is a lack of information and no definitive clinical symptoms for sudden death cases. Thus, there are numerous candidate genes that could be responsible for sudden death cases.
In our current study, we used NGS to perform target exon sequencing, in addition to conducting postmortem comprehensive genetic screening for FAODs. As far as we know, this is the first study to examine metabolic autopsy performed with NGS.
In most of the cases, the sequence covered more than 90% of the targeted exons. False-positives and false-negatives were rare. After excluding the variants that have been reported elsewhere, there were six substitutions and deletions remaining. Because the majority of the FAODs are inherited in an autosomal recessive manner, we chose the one case that had at least two pairs of the heterozygous variant (Case 15). This patient had a compound heterozygous deletion (c.968_969 del TC, p.F323fs) and substitution (c.1813G > C, p.V605L) in the CPT2 gene. Sanger's method was used to confirm these variants. The former variant was inherited from her father and caused a frameshift, thereby resulting in an immature protein. The latter variant was inherited from her mother and has been previously found and reported in a Japanese CPT II deficiency patient [21]. SIFT and PolyPhen-2 classified the function of the protein as damaging. Our analyses additionally showed that the amino acid was conserved between the species examined. Thus, when taken together, these genetic analyses demonstrated that this patient had a CPT II deficiency.
There were four cases with other variants, each of which had a heterozygous substitution. The substitution in Case 5 was ACADS-P55L (c.164C > T), in Case 6 was CPT2-F383Y (c.1148T > A), in Case 14 was ACADS-V84M (c.259G > A) and in Case 7 was SLC22A5-D487N (c.1459G > A). Because the substitution of CPT2-F383Y has also been shown to cause decreases in the CPT II activity and been reported in a CPT II deficiency patient [8], [19], [20], [21], [31], even in the heterozygous F383Y mutation state [21], [32], [33], the heterozygous CPT2-F383Y mutation could be a cause of sudden death. While the postmortem acylcarnitine analysis in our current case did not suggest any CPT II deficiency, it is possible that Case 6 might have been affected by the mutation.
The substitutions of ACADS are controversial. ACADS-P55L was detected in 8 alleles among the 8654 East Asian control alleles (minor allele frequency: 0.0009, The Exome Aggregation Consortium) and in 3 alleles out of the 1346 Japanese control alleles that were examined (minor allele frequency: 0.002, Human Genetic Variation Browser). ACADS-V84M was neither detected in the 6496 East Asian control alleles (The Exome Aggregation Consortium) nor in the 600 Japanese control alleles (Human Genetic Variation Browser). The SIFT scores for these two ACADS substitutions were classified as damaging, with both substitutions conserved between species. It has been reported that ACADS-P55L decreases the enzyme activity of SCAD [34]. Although the final diagnoses in Case 14 and Case 5 were acute encephalopathy and respiratory infection, respectively, it cannot be denied that these substitutions could have potentially affected the enzyme activity to a greater or lesser extent. Thus, the possibility exists that individuals with rare variants are susceptible to environmental stress. However, MCAD can compensate for the SCAD enzyme deficiency because of the overlap in the substrate specificity [35] and the substitution may not have a major effect to the death. Further accumulation of the genetic data will be necessary in order to link these rare variants and SUDI.
Even though SLC22A5-D487N was not detected in the 121,410 control alleles (The Exome Aggregation Consortium), it was detected in 1 allele out of the 600 Japanese control alleles (minor allele frequency: 0.002, Human Genetic Variation Browser). The SIFT score was classified as tolerated while the PolyPhen-2 was classified as benign. Since amino acid position 487 is asparagine in X. tropicalis and zebrafish, this substitution might not affect the activity of this gene.
In the current study, our use of NGS led to finding a case of compound heterozygote CPT II deficiency. Furthermore, we additionally found a case of heterozygote CPT II deficiency-related gene variant and two cases of SCAD deficiency-related variants. It is true that the cost of NGS is still higher than postmortem blood acylcarnitine analysis, but these variants would not have been detected without the use of NGS. Thus, metabolic autopsy performed with NGS is a useful method, especially when postmortem blood acylcarnitine analysis is not available. These findings suggest that metabolic autopsy should be performed in all cases of sudden death.
Acknowledgments
We would like to thank Norimasa Kageyama, Fumi Kuriiwa, Yuko Moriyama and Yutaka Araki for their technical advice. This study was supported in part by a grant from the Ministry of Health, Labour and Welfare of Japan, and grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (25860490).
References
- 1.Blair P.S., Byard R.W., Fleming P.J. Sudden unexpected death in infancy (SUDI): suggested classification and applications to facilitate research activity. Forensic Sci. Med. Pathol. 2012;8:312–315. doi: 10.1007/s12024-011-9294-x. [DOI] [PubMed] [Google Scholar]
- 2.Bennett M.J., Powell S. Metabolic disease and sudden, unexpected death in infancy. Hum. Pathol. 1994;25:742–746. doi: 10.1016/0046-8177(94)90241-0. [DOI] [PubMed] [Google Scholar]
- 3.Lundemose J.B., Kolvraa S., Gregersen N., Christensen E., Gregersen M. Fatty acid oxidation disorders as primary cause of sudden and unexpected death in infants and young children: an investigation performed on cultured fibroblasts from 79 children who died aged between 0–4 years. Mol. Pathol. 1997;50:212–217. doi: 10.1136/mp.50.4.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boles R.G., Buck E.A., Blitzer M.G., Platt M.S., Cowan T.M., Martin S.K., Yoon H., Madsen J.A., Reyes-Mugica M., Rinaldo P. Retrospective biochemical screening of fatty acid oxidation disorders in postmortem livers of 418 cases of sudden death in the first year of life. J. Pediatr. 1998;132:924–933. doi: 10.1016/s0022-3476(98)70385-3. [DOI] [PubMed] [Google Scholar]
- 5.Chace D.H., DiPerna J.C., Mitchell B.L., Sgroi B., Hofman L.F., Naylor E.W. Electrospray tandem mass spectrometry for analysis of acylcarnitines in dried postmortem blood specimens collected at autopsy from infants with unexplained cause of death. Clin. Chem. 2001;47:1166–1182. [PubMed] [Google Scholar]
- 6.Wilcox R.L., Nelson C.C., Stenzel P., Steiner R.D. Postmortem screening for fatty acid oxidation disorders by analysis of Guthrie cards with tandem mass spectrometry in sudden unexpected death in infancy. J. Pediatr. 2002;141:833–836. doi: 10.1067/mpd.2002.130259. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto T., Tanaka H., Kobayashi H., Okamura K., Tanaka T., Emoto Y., Sugimoto K., Nakatome M., Sakai N., Kuroki H., Yamaguchi S., Matoba R. Retrospective review of Japanese sudden unexpected death in infancy: the importance of metabolic autopsy and expanded newborn screening. Mol. Genet. Metab. 2011;102:399–406. doi: 10.1016/j.ymgme.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Kuhara T. Present status of expanded newborn screening project for inborn errors of metabolism by tandem mass spectrometry. Nihon Eiseigaku Zasshi. 2014;69:60–74. doi: 10.1265/jjh.69.60. [in Japanese] [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto T., Emoto Y., Murayama K., Tanaka H., Kuriu Y., Ohtake A., Matoba R. Metabolic autopsy with postmortem cultured fibroblasts in sudden unexpected death in infancy: diagnosis of mitochondrial respiratory chain disorders. Mol. Genet. Metab. 2012;106:474–477. doi: 10.1016/j.ymgme.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Bennett M.J., Rinaldo P. The metabolic autopsy comes of age. Clin. Chem. 2001;47:1145–1146. [PubMed] [Google Scholar]
- 11.Pryce J.W., Weber M.A., Heales S., Malone M., Sebire N.J. Tandem mass spectrometry findings at autopsy for detection of metabolic disease in infant deaths: postmortem changes and confounding factors. J. Clin. Pathol. 2011;64:1005–1009. doi: 10.1136/jclinpath-2011-200218. [DOI] [PubMed] [Google Scholar]
- 12.Bagnall R.D., Das K.J., Duflou J., Semsarian C. Exome analysis-based molecular autopsy in cases of sudden unexplained death in the young. Heart Rhythm. 2014;11:655–662. doi: 10.1016/j.hrthm.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 13.Narula N., Tester D.J., Paulmichl A., Maleszewski J.J., Ackerman M.J. Post-mortem whole exome sequencing with gene-specific analysis for autopsy-negative sudden unexplained death in the young: a case series. Pediatr. Cardiol. 2015;36:768–778. doi: 10.1007/s00246-014-1082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., Mukherjee G., Rajan J., Despacio-Reyes G., Saunders G., Steward C., Harte R., Lin M., Howald C., Tanzer A., Derrien T., Chrast J., Walters N., Balasubramanian S., Pei B., Tress M., Rodriguez J.M., Ezkurdia I., van Baren J., Brent M., Haussler D., Kellis M., Valencia A., Reymond A., Gerstein M., Guigo R., Hubbard T.J. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taroni F., Verderio E., Fiorucci S., Cavadini P., Finocchiaro G., Uziel G., Lamantea E., Gellera C., DiDonato S. Molecular characterization of inherited carnitine palmitoyltransferase II deficiency. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8429–8433. doi: 10.1073/pnas.89.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamamoto S., Abe H., Kohgo T., Ogawa A., Ohtake A., Hayashibe H., Sakuraba H., Suzuki Y., Aramaki S., Takayanagi M., Hasegawa S., Niimi H. Two novel gene mutations (Glu174 → Lys, Phe383 → Tyr) causing the “hepatic” form of carnitine palmitoyltransferase II deficiency. Hum. Genet. 1996;98:116–118. doi: 10.1007/s004390050170. [DOI] [PubMed] [Google Scholar]
- 20.Wataya K., Akanuma J., Cavadini P., Aoki Y., Kure S., Invernizzi F., Yoshida I., Kira J., Taroni F., Matsubara Y., Narisawa K. Two CPT2 mutations in three Japanese patients with carnitine palmitoyltransferase II deficiency: functional analysis and association with polymorphic haplotypes and two clinical phenotypes. Hum. Mutat. 1998;11:377–386. doi: 10.1002/(SICI)1098-1004(1998)11:5<377::AID-HUMU5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 21.Yasuno T., Kaneoka H., Tokuyasu T., Aoki J., Yoshida S., Takayanagi M., Ohtake A., Kanazawa M., Ogawa A., Tojo K., Saito T. Mutations of carnitine palmitoyltransferase II (CPT II) in Japanese patients with CPT II deficiency. Clin. Genet. 2008;73:496–501. doi: 10.1111/j.1399-0004.2008.00986.x. [DOI] [PubMed] [Google Scholar]
- 22.Yamamoto T., Tanaka H., Emoto Y., Umehara T., Fukahori Y., Kuriu Y., Matoba R., Ikematsu K. Carnitine palmitoyltransferase 2 gene polymorphism is a genetic risk factor for sudden unexpected death in infancy. Brain Dev. 2014;36:479–483. doi: 10.1016/j.braindev.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Bennett M.J., Ragni M.C., Hood I., Hale D.E. Comparison of postmortem urinary and vitreous-humor organic-acids. Ann. Clin. Biochem. 1992;29:541–545. doi: 10.1177/000456329202900509. [DOI] [PubMed] [Google Scholar]
- 24.Boles R.G., Martin S.K., Blitzer M.G., Rinaldo P. Biochemical diagnosis of fatty acid oxidation disorders by metabolite analysis of postmortem liver. Hum. Pathol. 1994;25:735–741. doi: 10.1016/0046-8177(94)90240-2. [DOI] [PubMed] [Google Scholar]
- 25.Rashed M.S., Ozand P.T., Bennett M.J., Barnard J.J., Govindaraju D.R., Rinaldo P. Inborn errors of metabolism diagnosed in sudden death cases by acylcarnitine analysis of postmortem bile. Clin. Chem. 1995;41:1109–1114. [PubMed] [Google Scholar]
- 26.Kemp P.M., Little B.B., Bost R.O., Dawson D.B. Whole blood levels of dodecanoic acid, a routinely detectable forensic marker for a genetic disease often misdiagnosed as sudden infant death syndrome (SIDS): MCAD deficiency. Am. J. Forensic Med. Pathol. 1996;17:79–82. doi: 10.1097/00000433-199603000-00015. [DOI] [PubMed] [Google Scholar]
- 27.Rinaldo P., Stanley C.A., Hsu B.Y., Sanchez L.A., Stern H.J. Sudden neonatal death in carnitine transporter deficiency. J. Pediatr. 1997;131:304–305. doi: 10.1016/s0022-3476(97)70171-9. [DOI] [PubMed] [Google Scholar]
- 28.Rinaldo P., Yoon H.R., Yu C., Raymond K., Tiozzo C., Giordano G. Sudden and unexpected neonatal death: a protocol for the postmortem diagnosis of fatty acid oxidation disorders. Semin. Perinatol. 1999;23:204–210. doi: 10.1016/s0146-0005(99)80052-4. [DOI] [PubMed] [Google Scholar]
- 29.Miller M.E., Brooks J.G., Forbes N., Insel R. Frequency of medium-chain acyl-CoA dehydrogenase deficiency G-985 mutation in sudden infant death syndrome. Pediatr. Res. 1992;31:305–307. doi: 10.1203/00006450-199204000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Arens R., Gozal D., Jain K., Muscati S., Heuser E.T., Williams J.C., Keens T.G., Ward S.L. Prevalence of medium-chain acyl-coenzyme A dehydrogenase deficiency in the sudden infant death syndrome. J. Pediatr. 1993;122:715–718. doi: 10.1016/s0022-3476(06)80010-7. [DOI] [PubMed] [Google Scholar]
- 31.Aoki J., Yasuno T., Sugie H., Kido H., Nishino I., Shigematsu Y., Kanazawa M., Takayanagi M., Kumami M., Endo K., Kaneoka H., Yamaguchi M., Fukuda T., Yamamoto T. A Japanese adult form of CPT II deficiency associated with a homozygous F383Y mutation. Neurology. 2007;69:804–806. doi: 10.1212/01.wnl.0000267665.44477.85. [DOI] [PubMed] [Google Scholar]
- 32.Olpin S.E., Afifi A., Clark S., Manning N.J., Bonham J.R., Dalton A., Leonard J.V., Land J.M., Andresen B.S., Morris A.A., Muntoni F., Turnbull D., Pourfarzam M., Rahman S., Pollitt R.J. Mutation and biochemical analysis in carnitine palmitoyltransferase type II (CPT II) deficiency. J. Inherit. Metab. Dis. 2003;26:543–557. doi: 10.1023/a:1025947930752. [DOI] [PubMed] [Google Scholar]
- 33.Joshi P.R., Deschauer M., Zierz S. Clinically symptomatic heterozygous carnitine palmitoyltransferase II (CPT II) deficiency. Wien. Klin. Wochenschr. 2012;124:851–854. doi: 10.1007/s00508-012-0296-9. [DOI] [PubMed] [Google Scholar]
- 34.Shirao K., Okada S., Tajima G., Tsumura M., Hara K., Yasunaga S., Ohtsubo M., Hata I., Sakura N., Shigematsu Y., Takihara Y., Kobayashi M. Molecular pathogenesis of a novel mutation, G108D, in short-chain acyl-CoA dehydrogenase identified in subjects with short-chain acyl-CoA dehydrogenase deficiency. Hum. Genet. 2010;127:619–628. doi: 10.1007/s00439-010-0822-7. [DOI] [PubMed] [Google Scholar]
- 35.Jethva R., Bennett M.J., Vockley J. Short-chain acyl-coenzyme A dehydrogenase deficiency. Mol. Genet. Metab. 2008;95:195–200. doi: 10.1016/j.ymgme.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]