Abstract
Over the past decade, RNA interference (RNAi) has been ubiquitously utilized to study biological function in vitro, however limitations were associated with its utility in vivo. More recently, small interfering RNA (siRNA) nanoparticles with improved biocompatibility have gained prevalence as a potential therapeutic option for the treatment of various diseases. The adaptability of siRNA nanoparticles enables the delivery of virtually any siRNA, which is especially advantageous for therapeutic applications in heterogeneous diseases that lack unifying molecular features, such as triple negative breast cancer (TNBC). TNBC is an aggressive subtype of breast cancer that is stratified by the lack of estrogen receptor/progesterone receptor expression and HER2 amplification. There are currently no FDA-approved targeted therapies for the treatment of TNBCs, making cytotoxic chemotherapy the only treatment option available to these patients. In this review, we outline the current status of siRNA nanoparticles in clinical trials for cancer treatment, and discuss promising preclinical approaches that have utilized siRNA nanoparticles for TNBC treatment. Next, we address TNBC subtype-specific therapeutic interventions and highlight where and how siRNA nanoparticles fit into these strategies. Lastly, we point out ongoing challenges in the field of siRNA nanoparticle research that, if addressed, would significantly improve the efficacy of siRNA nanoparticles as a therapeutic option for cancer treatment.
Keywords: siRNA nanoparticles, Triple negative breast cancer, Therapeutic strategy
Introduction
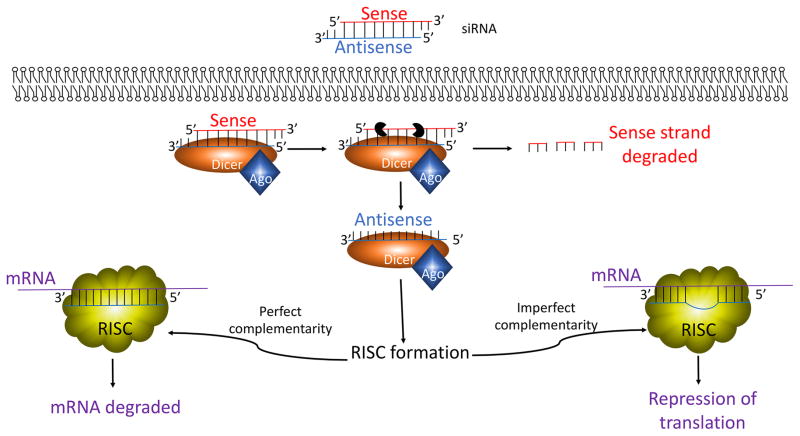
RNA interference (RNAi) is a process where small double-stranded RNAs consisting of approximately 22nt are utilized to repress gene expression. These effects were first observed in nematode worms in 1998, where expression of par-1 mRNA was temporarily depleted following introduction of double-stranded small interfering RNA (siRNA) (Fakhr, et al. 2016; Young, et al. 2016). Further studies identified additional molecular players in the RNAi machinery, including Dicer and the RNA-induced silencing complex (RISC) (Kobayashi and Tomari 2016). Upon uptake of the double-stranded siRNA, Dicer unwinds and cleaves the sense strand (Fig. 1). The antisense strand then acts as a guide for the recognition of complementary mRNAs, and the RISC complex forms (Fig. 1). If the antisense strand is perfectly complement to the mRNA, the mRNA is cleaved by argonaute 2 (Ago2), the catalytic subunit of the RISC complex (Azlan, et al. 2016; Fakhr et al. 2016). Limited complementarity to the target mRNA leads to translational repression, a function that is associated with micro RNAs (miRNAs) (Young et al. 2016). Since its discovery, siRNAs have been widely utilized in vitro to interrogate various cell and molecular biology questions. However, due to limitations associated with its labile nature, proclivity to induce immune responses, and anionic properties, siRNA as a therapeutic strategy requires shielding of the naked siRNA to protect it from adverse serum nucleases and immune cells and to facilitate in vivo uptake.
Figure 1.
The siRNA machinery. Upon entry into the cell, the sense strand of the double-stranded siRNA is cleaved. The antisense siRNA strand then acts as a guide for mRNA complementation. Perfect complementarity between the antisense siRNA strand and the target mRNA leads to mRNA degradation, while imperfect complementarity between the two leads to the mRNA translational repression.
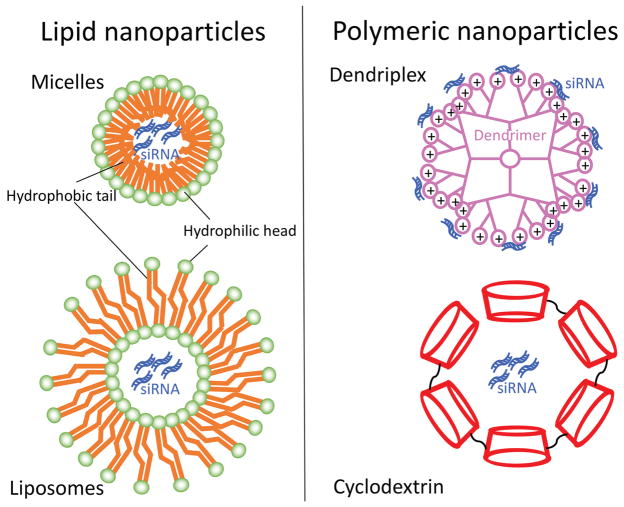
One method of overcoming these limitations is to encapsulate the naked siRNA within nanoparticles. Nanoparticles can be made from various materials (Jeong, et al. 2011; Wang, et al. 2010). Two main categories consist of organic and inorganic nanoparticles (Miele, et al. 2012; Young et al. 2016). Organic nanoparticles are composed of materials that either occur naturally or from a synthesized source (Lopez-Davila, et al. 2012; Young et al. 2016). These include but are not limited to 1) lipid nanoparticles such as micelles and liposomes and 2) polymer-based nanoparticles such as dendriplexes and cyclodextrin nanoparticles (Fig. 2) (Wen and Steinmetz 2016; Young et al. 2016). Inorganic nanoparticles typically consist of non-biodegradable biomaterials including metals, metal oxides, and other carbon materials (Young et al. 2016). Additional complex, hybrid nanoparticles that consist of both organic and inorganic biomaterials also exist. For readers desiring a more in depth understanding of nanoparticle biomaterials as well as advantages and disadvantages associated with each, we recommend several recent comprehensive reviews (Jeong et al. 2011; Miele et al. 2012; Young et al. 2016).
Figure 2.
Select examples of organic nanoparticles. Lipid nanoparticles include micelles and liposomes, which are composed of hydrophobic lipids that contain a hydrophilic head. Polymeric nanoparticles include dendriplexes and cyclodextrin nanoparticles. A dendrimer is a branched amphiphilic polymer and cyclodextrin is an amphiphilic cyclic oligosaccharide.
Nanoparticle size and surface charge significantly affect circulation time, therefore a number of parameters and modifications are adapted to enhance nanoparticle longevity. Three major modes of nanoparticle clearance in vivo include urine excretion through the renal filtration system, biliary excretion through the liver filtration system, and uptake by phagocytic cells of the immune system (Longmire, et al. 2008; Longmire, et al. 2011). Importantly, nanoparticle accumulation and clearance by the kidneys, liver, and blood are significantly dependent on particle size and material (De Jong, et al. 2008; Longmire et al. 2011). First, the glomerulus, or renal filtration functional unit, is capable of excreting nanoparticles with minimal catabolism of the particles. Particles that are excreted by glomerular filtration are typically 6nM or smaller in diameter (Longmire et al. 2008; Longmire et al. 2011; Lorenzer, et al. 2015). In addition to size, particle surface charge can also affect the efficiency of renal excretion. Previous studies demonstrate that amongst particles that are similar in size, positively charged particles are mostly effectively excreted, which is followed by neutral particles and lastly by negatively charged particles (Deen, et al. 2001). These differences in charge selective excretion is partly attributed to the negatively charged glomerular capillary wall that preferentially attracts positively charged particles (Deen et al. 2001). Particles that escape renal excretion may be cleared by the liver. Within the liver, nanoparticles can be internalized by endothelial cells, hepatocytes and Kupffer cells, and different nanomaterials are selectively and differentially internalized by these cells (Bargheer, et al. 2015; Heine, et al. 2014). Hepatocyte and Kupffer cells further mediate enzymatic breakdown of the engulfed particle, which range between 10–20nM in size (Longmire et al. 2008; Longmire et al. 2011). Kupffer cells rely solely on intracellular breakdown of the engulfed material, while hepatocytes-mediated nanoparticle clearance also involve excretion through bile (Longmire et al. 2008; Longmire et al. 2011). Moreover, nanoparticles in circulation are also opsonized, or tagged by serum proteins for phagocytosis by immune cells. Opsonization is affected by various nanoparticle characteristics such as hydrophobicity, size and charge (Alexis, et al. 2008; Lim, et al. 2008). Nanoparticles that are greater than 200nM are readily cleared by phagocytic immune cells (Lorenzer et al. 2015). In addition to size and charge, several additional factors also affect nanoparticle circulation time.
Surface modification with Polyethylene Glycol (PEG), a biocompatible polymer, has also been demonstrated to significantly increase circulation time by 1) reducing charge-based interactions with the glomerular capillaries and 2) shielding nanoparticles from phagocytic cells (Jokerst, et al. 2011; Li and Szoka 2007). The net effect of PEGylation is also associated with decreased cellular uptake of nanoparticles, therefore additional modifications of the nanoparticles are required to improve accumulation within tumor cell. PEGylated nanoparticles are coated with a number of targeting ligands whose receptors are highly expressed on tumor cells to promote receptor-mediated endocytosis. Commonly utilized targeting ligands are summarized in Table 1 (Arosio and Casagrande 2016; Bakrania, et al. 2016; Cao, et al. 2011; Deng, et al. 2013; Feng, et al. 2014; Gu, et al. 2016; Nahta and Esteva 2006; Necela, et al. 2015; Parvani, et al. 2015; Seitz, et al. 2013; Xu, et al. 2016). Importantly, these strategies significantly improve nanoparticle uptake by tumor cells. Various other factors including nanoparticle shape (Geng, et al. 2007; Sadekar, et al. 2011), flexibility (Kobayashi, et al. 2001; Ogawa, et al. 2010), surface-coating (Bargheer et al. 2015; Heine et al. 2014), and cargo (Scholz and Wagner 2012; Zintchenko, et al. 2008) may all play a role in nanoparticle stability and life-span.
Table 1.
Common strategies to improve nanoparticle accumulation in tumor cells
| Targeting ligand | Receptor | References |
|---|---|---|
| Vapreotide | Somatostatin receptor | (Feng, et al. 2014) (Seitz, et al. 2013) |
| RGD | MMP2, integrins | (Parvani et al. 2015) (Arosio and Casagrande 2016) |
| Trastuzumab | HER2 | (Nahta and Esteva 2006) (Gu et al. 2016) |
| Folic acid | Folic acid receptor | (Necela, et al. 2015) (Cao, et al. 2011) |
| Hyaluronic acid | HARE, LYVE1, RHAMM, CD44 | (Xu, et al. 2016) (Bakrania, et al. 2016) (Deng et al. 2013) |
There are several advantages to using siRNA nanoparticle platforms as a therapeutic strategy. Recent advances in next generation high throughput sequencing, has revealed extraordinary genetic complexity and heterogeneity in cancer models (Hoelder, et al. 2012). Because siRNAs are readily synthesized and can be optimized to maximize gene silencing (Fakhr et al. 2016), siRNA nanoparticles can be easily adapted to silence virtually any gene in mammalian cells. Various siRNA design software that enable the optimization of siRNA length, specificity, and nucleotide content based on a number of criteria are available online (Fakhr et al. 2016). These tools offer a quick way to focus screening of numerous siRNAs for numerous genes, which contrasts time-consuming traditional ways of searching for small molecule inhibitors against a target (Hoelder et al. 2012). siRNA nanoparticles are also equipped to silence splice variants and transcription factors, which were previously thought of as ‘undruggable’ (Johnston and Carroll 2015). Not surprisingly, several siRNA nanoparticles are currently being tested in clinical trials as cancer therapeutics.
Clinical application of siRNA nanoparticles in cancer treatment
Preclinical studies in various different tumor models have demonstrated siRNA nanoparticles to be effective in inhibiting tumor growth (Parvani et al. 2015; Su, et al. 2015; Zhang, et al. 2016), metastasis (Parvani et al. 2015; Zhao, et al. 2015), angiogenesis (Liu, et al. 2015; Malamas, et al. 2016), and drug resistance (Deng et al. 2013; Zhang et al. 2016). These studies have paved the way for various ongoing clinical trials, which are summarized in Table 2. The efficacies of several different siRNA nanoparticles are currently being evaluated for safety and pharmacokinetics in phase 1 clinical trials, and amongst these, 3 studies have been completed and 1 has been terminated.
Table 2.
siRNA nanoparticles being tested in clinical trials
| Clinical trial phase | Drug name | Nanoparticle material | Targeting strategy | Target | Disease | Company | Stage |
|---|---|---|---|---|---|---|---|
| I | Atu027 | Lipid nanoparticle | None | Protein kinase N3 | Solid tumors | Silence Therapeutics | Completed |
| I | ALN-VSP | Lipid nanoparticle | None | VEGF and KSP | Solid tumors with liver involvement | Alnylam | Completed |
| I | TKM 80301 | Lipid nanoparticle | None | Polo-like kinase 1 | Primary or Secondary liver cancer | NCI | Completed |
| I | CALAA-01 | Polymer (cyclodextrin) nanoparticle | Transferrin | RRM2 | Solid tumors | Calando | Terminated |
| I | siG12D LODER | Polymer (PLGA) nanoparticle | implanted into tumor | KRASG12D | Pancreatic ductal adenocarcinoma | Silenseed Ltd. | Completed |
| I | siRNA-EphA2-DOPC | Lipid nanoparticle | None | EphA2 | Solid tumors | MD Anderson Cancer Ctr | Not yet open |
| II | siG12D LODER | Polymer nanoparticle | implanted into tumor | KRASG12D | Pancreatic ductal adenocarcinoma | Silenseed Ltd. | Not yet open |
| I | TKM 80301 | Lipid nanoparticle | None | Polo-like kinase 1 | Solid tumors | Tekmira | Recruiting |
Atu027 is a lipid nanoparticle with a particle diameter of about 120nm (Santel, et al. 2006), delivering siRNA against protein kinase N3 (PKN3), a downstream effector of the PI3 kinase pathway, for the treatment of various solid tumors (Schultheis, et al. 2014). In preclinical mouse models, silencing PKN3 is associated with decreased tumor growth and metastases (Aleku, et al. 2008). The phase 1 clinical trial evaluating Atu027 consisted of 34 treatment naïve patients harboring a variety of advanced solid tumors (Schultheis et al. 2014). These patients were intravenously infused with 10 escalating doses of Atu027, and results indicated that a) these nanoparticles are well-tolerated in patients up to 0.336mg/kg, and b) 41% of patients exhibited stable disease for at least 8 weeks (Schultheis et al. 2014). These promising results have laid a solid foundation for additional clinical trials.
ALN-VSP is a lipid nanoparticle with a particle diameter of 80–100nm, delivering siRNA against Vascular Endothelial Growth Factor (VEGF) and Kinesin Spindle Protein (KSP) at a 1:1 molar ratio for the treatment of liver cancers (Tabernero, et al. 2013). VEGF is a growth factor that is essential for angiogenesis, endothelial cell permeability, and general preservation of vessel function (Gopal, et al. 2016). KSP is a microtubule interacting protein that is involved in the regulation of mitosis and apoptosis (Naymagon and Abdul-Hay 2016). In a phase 1 clinical trial, patients received intravenous injections of ALN-VSP doses that ranged from 0.4–1.0mg/kg (Tabernero et al. 2013). Tumor biopsies from 12 patients indicated significant accumulation of the siRNA and target mRNA cleavage within the tumor (Tabernero et al. 2013). Overall, ALN-VSP was well-tolerated in patients and exhibited antitumor activity, including the complete regression of liver metastases originating from a primary endometrial tumor. Further development of ALN-VSP is expected.
TKM-080301 is a lipid nanoparticle delivering siRNA against polo-like kinase1 (PLK1) (Liu 2015). PLK1 is a serine-threonine kinase that regulates mitosis, DNA replication, and cellular stress response (Liu 2015). Elevated expression of PLK1 has been observed in various cancers, including but not limited to breast, colorectal, prostate, head and neck cancers (Takai, et al. 2005). Preclinical studies have demonstrated that depletion of PLK1 expression preferentially inhibits the survival of tumor cells in xenograft models (Guan, et al. 2005; Spankuch, et al. 2004); (Liu 2015). A phase 1 trial evaluating the pharmacodynamics of TKM-080301 in patients harboring lymphoma or advanced solid tumors has been completed, however the trial report is currently unavailable.
CALAA-01 is a cyclodextrin-based polymeric nanoparticle with a particle diameter ranging from 60–150nm, delivering siRNA against the ribonucleotide reductase M2 subunit (RRM2) for the treatment of a variety of solid tumors (Zuckerman, et al. 2014). RRM2 is essential for the regulation of nucleotide synthesis, and its expression is frequently upregulated in many cancers (Furuta, et al. 2010). In a phase 1a clinical trial 15 patients were treated with CALAA-01 delivering between 3 to 30mg/m2 of siRNA (Zuckerman et al. 2014). No initial dose-limiting toxicities were observed, however approximately two years after initial treatment, two patients receiving the maximum dose experienced toxicity symptoms. The clinical protocol was subsequently modified to administer patients with a lower initial dose of CALAA-01, with the hypothesis that reduced initial exposure may dampen immunogenicity that was associated with toxicity (Zuckerman et al. 2014). However, the study was terminated when two out of five patients enrolled in the phase 1b clinical trial developed dose-limiting toxicities. The scientific report documenting the termination of the trial has not been published, however preliminary results suggest that the toxicity of CALAA-01 is associated with the delivery vehicle rather than the siRNA (Zuckerman et al. 2014). It is proposed that these toxicities may be neutralized by alternative purification of CALAA-01 after its preparation (Zuckerman et al. 2014).
siG12D LODER is a biodegradable and implantable cylindrical rod (diameter: 0.9mm; length 4mm) that is composed of a copolymer of poly (lactic-co-glyolic) acid (PLGA) (Ramot, et al. 2016). This polymer matrix encapsulates anti-KRAS(12G>D) siRNA (siG12D) to enable sustained local release of siG12D for the treatment of locally advanced pancreatic cancer (Golan, et al. 2015; Ramot et al. 2016; Zorde Khvalevsky, et al. 2013). Greater than 90% of pancreatic ductal adenocarcinomas (PDACs) harbor mutated KRAS, and the majority of these are activating mutations occur within codon 12. The most common codon 12 activating KRAS mutations include 12G>D (61%), 12G>R (18%), and 12G>V (17%) (Rachakonda, et al. 2013). Importantly, previous studies have demonstrated that PDAC cells are addicted to mutant KRAS, such that a reduction in KRAS expression is associated with reduced cell viability (Singh, et al. 2009). These findings have set the foundation to silence mutant KRAS as a therapeutic strategy for PDAC treatment. The phase 1 clinical trial evaluating siG12D LODER consisted of 15 pancreatic cancer divided into 3 dose cohorts. The siG12D LODER implant was inserted into the PDAC lesion via standard biopsy procedures for sustained release of siG12D for 4 months with concomitant weekly intravenous infusion of chemotherapy (Golan et al. 2015). Results indicate that the implant was safe and well-tolerated, with transient adverse effects (Golan et al. 2015). Preliminary CT scans revealed that the majority of patients had stable disease, while 2 patients exhibited partial response (Golan et al. 2015). Moreover, 70% of patients experienced a decrease in the pancreatic tumor marker, CA19-9 (Swords, et al. 2016). A phase 2 clinical trial consisting of a larger cohort of patients to evaluate the response rate of advanced pancreatic cancer patients treated with the chemotherapy and siG12D LODER is underway.
These clinical studies have laid a solid foundation of evaluating siRNA nanoparticles in various solid tumors, and additional trials are currently recruiting patients (Table 2). Lipid nanoparticles appear to be more widely tested than polymer nanoparticles in clinical trials; however, preliminary results from completed trials have demonstrated safety and efficacy using both nanomaterials. Successful evaluation of siRNA nanoparticles could potentially lead to the application of this powerful technology for the future treatment of various cancers, including breast cancer.
Triple Negative Breast Cancer (TNBC) and current standard treatments
Breast cancer is the most common cancer amongst women, with an estimated 246,660 newly diagnosed cases in the United States in 2016 (ACS 2016). Importantly, breast cancer is not a single disease; instead, it is clinically subcategorized into three major subtypes, which include hormone receptor positive, HER2 positive and triple negative breast cancers (TNBCs). This classification system has significant therapeutic and prognostic implications.
As its name suggests, TNBCs are those that lack Estrogen Receptor (ER) expression, Progesterone Receptor (PR) expression, and HER2 amplification as determined by immunohistochemistry (IHC) and fluorescence in-situ hybridization (FISH) analyses. There are currently no biomarkers that positively define TNBCs, therefore TNBCs have been stratified by excluding tumors containing greater than 10% positivity in ER, PR, and HER2. In 2010, the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) updated their guidelines to defining ER and PR negative tumors as those having <1% positivity to better accommodate patients who may benefit from endocrine and HER2-targeted therapies (Hammond, et al. 2010). TNBCs are typically more clinically aggressive, and occur more frequently in younger women and women of African and Hispanic ancestry (Alluri and Newman 2014). Moreover, patients with TNBC are more likely to die from their disease within 5 years of diagnosis in comparison to patients with non-TNBC (Dent, et al. 2007).
Unlike patients harboring luminal and HER2 breast cancers, who benefit from estrogen receptor and HER2 antagonists, there are no FDA-approved targeted therapies for the treatment of TNBC. Current standard local therapies include surgery and radiotherapy, while systemic therapy includes the use of cytotoxic chemotherapy (Yagata, et al. 2011). TNBC lesions are typically unifocal, making breast conserving therapy a plausible option (Yagata et al. 2011). Regional and locoregional recurrences are typically higher in TNBC than in non-TNBC, however radiation therapy after surgery has been demonstrated to decrease recurrence (Kyndi, et al. 2008; Voduc, et al. 2010). Systemic therapy include the use of taxane-anthraycline regimens that most TNBCs patients initially respond to (Chacon and Costanzo 2010; Stover and Winer 2015); however, chemosensitivity is often short-lived and the majority of patients relapse and succumb to metastatic disease (O’Reilly, et al. 2015). Additionally, in comparison to patients harboring non-TNBC, those harboring TNBC also typically experience shorter average time to local recurrence (2.8 years vs. 4.2 years), shorter mean time to distant recurrence (2.6 years vs. 5 years), and increased rate of distant recurrence (33.9% vs. 20.4%) (Dent et al. 2007). Because of these ongoing challenges associated with effective treatments for TNBCs, various novel targeted treatment approaches are currently being explored, including siRNA nanoparticles.
Preclinical studies of siRNA nanoparticles in the treatment of TNBCs
The clinical successes of siRNA nanoparticles for the treatment of various solid cancers have paved the way for its application in TNBC. Several recent preclinical studies have explored different siRNA delivery vehicles to silence an assortment of target genes that are associated with poor prognosis in TNBC. These studies have largely focused on siRNA nanoparticle application in 3 major areas for the treatment of TNBCs: inhibiting components of the cell cycle, inhibiting epithelial-mesenchymal transition (EMT), and improving chemotherapy efficacy.
siRNA silencing of cell cycle regulators
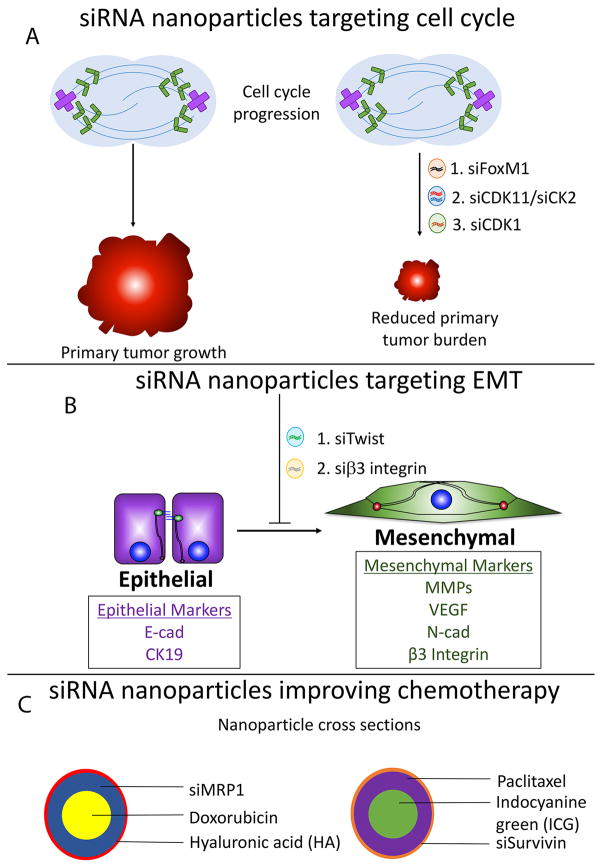
FoxM1 is a transcription factor that functions as a master regulator of the cell cycle by inducing G1→S and G2→M transitions (Saba, et al. 2016). Notably, FoxM1 is overexpressed in many cancers, which has been correlated to metastasis and disease progression (Saba et al. 2016). In a recent study, Hamurcu et al demonstrated that FoxM1 is overexpressed in numerous TNBC cell lines and that depletion of FoxM1 expression inhibits cyclin D1 expression, and Src (Y416) and Erk activation (Hamurcu, et al. 2016). Functionally, these changes coalesced to reduce colony formation, proliferation, invasion, and migration (Hamurcu et al. 2016). Traditionally, transcription factors have commonly been thought of as ‘undruggable’ due to their lack of enzymatic activities (Johnston and Carroll 2015; Yan and Higgins 2013), however these limitations are lifted by siRNA nanoparticles. Two weeks after nude mice were engrafted with MDA-MB-231 tumors, they were administered with control and FoxM1 targeting siRNA encapsulated within liposomal lipid nanoparticles at 0.3mg/kg weekly (Hamurcu et al. 2016). Results demonstrated significant reduction of FoxM1 expression in the primary tumor, which was associated with decreased primary tumor burden (Fig. 3A) (Hamurcu et al. 2016). Future studies should evaluate tumor recurrence following withdrawal of FoxM1 siRNA nanoparticles.
Figure 3.
Current approaches using therapeutic siRNA nanoparticles in the treatment of TNBCs. A) siRNA nanoparticles delivering siRNA against FoxM1, CDK11/CK2, and CDK1 are demonstrated to significantly reduce primary tumor burden. B) siRNA nanoparticles silencing Twist and β3 integrin inhibits EMT. Nanoparticles delivering β3 integrin siRNA also reduce primary tumor burden, primary tumor recurrence, and metastasis in MDA-MB-231 mouse xenograft models. C) Complex nanoparticles utilize siRNAs to improve chemotherapeutic efficacy.
In another study, Cyclin-dependent kinase 11 (CDK11) and casein kinase II (CK2) were evaluated as potential therapeutic targets for the treatment of TNBCs (Kren, et al. 2015). CDK11 and CK2 are two well-established molecular players that mediate cancer cell growth and survival. CDK11 is an atypical CDK that functions in mitosis, transcription, and RNA splicing, and its elevated expression is observed in TNBCs (Zhou, et al. 2016). CK2 is a Ser/Thr protein kinase that phosphorylates numerous substrates that function in cell biology processes such as transcription, cell cycle regulation, apoptosis, and others (Meggio and Pinna 2003). Elevated expression of CK2 in human breast cancers correlates with metastasis (Giusiano, et al. 2011). Kren et al encapsulated CDK11 and CK2 siRNAs into polyamine-based micelles that are coated with tenfibgen (TBG) protein. TBG is specifically recognized by tenascin C, a receptor that is highly expressed in breast cancer stroma (Guttery, et al. 2010). Control TBG-coated nanoparticles or those encapsulating siRNAs against CDK11 were administered via intravenous injection at a dose of 0.01mg/kg, every 3–4 days for 10 days. Mice receiving nanoparticles that delivered CDK11 siRNAs harbored significantly smaller primary tumors (Fig. 3A) (Kren et al. 2015). Future studies should evaluate changes in metastatic potential of the xenografted tumors in response to nanoparticle administration.
In a third study, Liu et al utilized siRNA nanoparticles to target CDK1, a cyclin-dependent kinase that has recently been demonstrated to be synthetic lethal in TNBC models harboring elevated Myc expression (Horiuchi, et al. 2012). Myc is an oncoprotein that is frequently overexpressed in TNBCs. Directly targeting Myc as a therapeutic strategy has not come to fruition because 1) designing a small molecule inhibitor to disrupt protein-protein or protein-DNA interactions necessary for Myc function has proven to be challenging (Horiuchi, et al. 2014) and 2) Myc expression is essential for regeneration and maintenance of stem cells in the bone marrow, skin, and gastrointestinal tract (Soucek, et al. 2008). Using a cationic lipid based PEG-PLA nanoparticle system, Liu et al delivered CDK1 specific siRNA to TNBC cells harboring elevated Myc expression, and observed decreased Myc expression that is accompanied by 1) decreased cell viability, 2) decreased colony formation, and 3) increased apoptosis in vitro (Liu, et al. 2014). In vivo, administering siCDK1 nanoparticles culminated in significant reduction of primary tumor burden (Fig. 3A). Importantly no significant changes in body weight and inflammatory cytokines were noted, indicating no major toxicities (Liu et al. 2014). As with previous studies, changes in metastatic potential were not evaluated.
In summary, various groups have demonstrated the effectiveness of siRNA nanoparticles at reducing primary tumor burden by targeting key cell cycle regulators. Insignificant changes in body weight and immune cytokine production were observed, which suggest that these delivery platforms are safe and well-tolerated. However, since most breast cancer patients undergo surgery to remove the primary tumor, and metastasis is ultimately the factor associated with patient mortality, future studies need to evaluate how tumor recurrence and metastatic potential can be targeted by these siRNA nanoparticles.
siRNA silencing inducers of Epithelial-Mesenchymal Transition
Epithelial-Mesenchymal Transition (EMT) is a normal physiological program that occurs during development, fibrosis, and wound healing (Taylor, et al. 2010). Aberrant activation of EMT engenders its pathophysiological characteristics associated with cancer (Taylor et al. 2010). Over the past decade, the effects of EMT have been expanded to include alterations in cell motility and invasion, acquisition of stem-like characteristics, resistance to chemotherapy, and remodeling of the tumor microenvironment (Morrison, et al. 2013). Given its salient role in malignancy, attention has turned to targeting EMT for the treatment of TNBCs.
Various transcription factors, such as members of the TWIST, Snail, and Zeb families, have previously been characterized as master regulators of EMT (Taylor et al. 2010). Recently, Finlay et al utilized an amphiphilic dendrimer to silence TWIST1 in TNBC cells (Finlay, et al. 2015). The authors verified target cell uptake of the dendriplexes that mediated significant reduction of TWIST expression in SUM1315 TNBC cells (Finlay et al. 2015). In vitro, the functional consequences of silencing TWIST1 coalesced to reduce 1) expression of the EMT markers, N-cadherin and vimentin, and 2) cancer cell migration and invasion (Fig. 3B) (Finlay et al. 2015). Furthermore, the authors confirmed that intratumoral injection of the dendriplexes delivering a fluorescent siRNA effectively induced siRNA uptake by tumor cells, and that the fluorescent siRNA was only minimally internalized by cells in other organs. Future studies will need to characterize the in vivo effects of dendriplexes delivering TWIST1 siRNA to TNBC xenograft models.
EMT can also be induced in response to microenvironmental regulators. β3 integrin is a transmembrane protein that functions as the vitronectin and fibronectin receptor when it is homodimerized to αv integrin (Missirlis, et al. 2016; Parvani, et al. 2013). Previous studies have demonstrated that β3 integrin plays essential roles in Transforming Growth Factor-beta (TGF-β) mediated EMT (Galliher and Schiemann 2006; Parvani et al. 2013). Using a lipid-based siRNA carrier called ECO, Parvani et al demonstrated that a single 4-hour dose of ECO/siβ3 lipid nanoparticles was sufficient to sustain prolonged silencing of β3 integrin in both mouse and human breast cancer cell lines for up to 7 days (Parvani et al. 2015). Furthermore, silencing β3 integrin in vitro inhibits EMT in TNBC cell lines, in part by the upregulation of epithelial markers, cytokeratin (CK) 19 and E-cadherin and downregulation of mesenchymal markers, N-cadherin and PAI-1 (Fig. 3B) (Parvani et al. 2015). β3 integrin depletion also reduces proliferation and invasion in 2D cultures and inhibits 3D organotypic outgrowth. To improve circulation time and tumor cell uptake, the ECO/siβ3 lipid nanoparticles were further modified with PEG, which is conjugated to an RGD peptide that is recognized by β3 integrin (Iyer, et al. 2013). When administered to nude mice engrafted with MDA-MB-231 tumors, RGD-ECO/siβ3 lipid nanoparticles effectively decreased primary tumor burden, primary tumor recurrence, and metastatic tumor burden (Parvani et al. 2015). The authors verified decreased β3 integrin expression in primary tumors that were surgically removed, and H&E staining of control and RGD-ECO/siβ3 treated tumors indicate reduced tumor associated vasculature in the experimental group (Parvani et al. 2015).
In summary, these studies demonstrated that targeting EMT-inducing factors is an effective way to reduce tumorigenicity of TNBC. Because of the salient role EMT plays in mediating stem-like characteristics and drug resistance, future studies should explore how silencing EMT-inducing factors would affect these phenotypes. Future studies also need to determine which combination of EMT transcription factor, if silenced by siRNA nanoparticles, would be most efficacious at inhibiting TNBC disease progression.
Improving chemotherapeutic efficacy
In 1995, the FDA approved its first nano-drug, Doxil, a PEGylated liposomal formulation of doxorubicin (Fajardo-Ortiz, et al. 2014; Marchal, et al. 2015). Liposomal encapsulation of doxorubicin prevents breakdown of the drug before reaching tumor cells. The effects of this are twofold: 1) increased cytotoxic effect of the drug is achieved since a greater amount of the drug is reaching tumor cells, and 2) decreased side-effects of the drug is also achieved since normal cells are exposed to the active drug less frequently (Lao, et al. 2013). PEGylation of these liposomal formulations further extends circulation time that increases the propensity of the drug to reach tumor cells (Khan, et al. 2015). These improvements have translated to clinical success and Doxil is currently utilized to treat recurrent breast cancer. Despite these successes, limitations associated with Doxil remain. First, although the PEGylated formulation is associated with increased circulation time, the presence of the PEG moiety also creates steric hindrance that limits tumor cell uptake (Khan et al. 2015). Furthermore, doxorubicin inherently exhibits high affinity for extracellular matrix proteins, which further limits tumor cell uptake of the drug (Khan et al. 2015). These limitations are indicative that further modifications of Doxil are required for its optimal use. Deng et al address these deficiencies by constructing a dual-siRNA-chemotherapy co-delivery system, where an siRNA film against multidrug resistant protein 1 (MRP1) was built around a chemotherapy-loaded nanoparticle core (Fig. 3C). Furthermore, these nanoparticles were coated with hyaluronic acid (HA), which has previously been demonstrated to 1) enhance in vivo stability and 2) mediate active uptake by CD44, a receptor that is highly expressed on some TNBC cells (Deng et al. 2013). Nude mice engrafted with MDA-MB-468 tumors were treated with this elegant system every 5 days for 15 days, with each dose of the experimental group consisting of 1mg/kg doxorubicin and 1mg/kg MRP1 siRNA. The control group delivering MRP1 siRNA only did not exhibit tumor inhibitory effects. The experimental group delivering both the siRNA and doxorubicin reduced tumor volume a) by 8-fold in comparison to vehicle-treated group and b) by 4-fold in comparison to the control group delivering doxorubicin only (Deng et al. 2013).
Another group combined photothermal therapy (PTT) with gene therapy and chemotherapy (Su et al. 2015). This “Triple Punch” nanoparticle delivers paclitaxel, siRNA against survivin, a gene associated with poor prognosis in TNBCs, and Indocyanine green (ICG), a PTT agent that has been approved by the FDA as a clinical imaging agent (Fig. 3C) (Su et al. 2015). In response to near infrared (NIR) laser irradiation, hyperthermia produced by ICG mediates deformation of the nanoparticle core structure, which induces the release of paclitaxel and survivin siRNA. MDA-MB-231 tumors were engrafted onto nude mice and treated with 2 doses of triple therapy nanoparticles (NP-IPS) that contain 0.32umol/kg of ICG, 0.54umol/kg of paclitaxel, and 1.5mg/kg of survivin siRNA within 30 days (Su et al. 2015). In comparison to control groups receiving mono or dual therapies, mice receiving NP-IPS had complete growth inhibition of MDA-MB-231 xenograft tumors, without any indication of tumor recurrence. Evaluation of post-mortem histopathology of various organ systems indicates no injury or damage in mice receiving nanoparticles.
TNBC subtype-specific therapeutic strategies: what are the limitations and how do siRNA nanoparticles fit in?
Previous studies have further subcategorized TNBCs into six distinct transcriptional subtypes (Lehmann, et al. 2011). These include two basal-like (BL1 and BL2) subtypes, a mesenchymal (M) subtype, a luminal androgen receptor (LAR) subtype, a mesenchymal stem-like (MSL) subtype, and an immunomodulatory (IM) subtype (Lehmann et al. 2011). Each of these subtypes preferentially respond to different therapeutic agents. Although current standard clinical practice for TNBC treatment do not take these 6 subtypes into account, efforts have been made in ongoing clinical trials to treat patients based on specific molecular subsets (Lehmann, et al. 2015). In this section, we explore how siRNA nanoparticles can be utilized to enhance subtype-specific TNBC treatments.
Basal-like TNBCs and cisplatin
The BL1 and BL2 subtypes are enriched for cell cycle and DNA damage response genes and are preferentially sensitive to cisplatin (Lehmann et al. 2011). Cisplatin is a platinum-containing chemotherapy that exerts its therapeutic effects by binding to DNA, causing DNA cross-linking, which leads to apoptosis (Apps, et al. 2015). Interestingly, BL breast cancers have striking similarities to BRCA1 mutated breast cancers, which exhibit defective DNA homologous recombination. Both BL breast cancers and BRCA1 mutated breast cancers are 1) diagnosed in younger women, have poor prognosis, and high mitotic index 2) characterized by genomic instability and 3) preferentially respond to DNA damaging agents as demonstrated in xenograft mouse models and in clinical trials (Isakoff, et al. 2015; Lehmann et al. 2011; Toft and Cryns 2011). Gene expression analysis indicates that there is a subset of BL breast cancers that harbor BRCA1 mutations, while other BL breast cancers retain wild type (WT) BRCA1 (Prat, et al. 2014). Those that retain WT BRCA1 typically contain mutations or alterations in other molecular players of the homologous recombination pathway, which enhances chemosensitivity to platinum agents (Baker, et al. 2016; Prat et al. 2014; Toft and Cryns 2011). Although BL TNBCs are more responsive to cisplatin, resistance mechanisms have been documented. Mechanisms of cisplatin resistance include secondary reversion mutations in BRCA1/2 that circumvent the therapeutic effects of cisplatin (Dhillon, et al. 2011). Utilizing siRNA nanoparticles to eliminate BRCA1/2 expression in combination with cisplatin may eliminate these resistant mechanisms (Table 3). Should a reversion mutation in BRCA1/2 arise in a region that overlaps with the chosen BRCA1/2 siRNA, alternative siRNA sequences against BRCA1/2 may be utilized. Because functional BRCA1 is essential for DNA damage repair in normal cells, it is imperative that the siRNA delivery platform utilized to deliver BRCA1/2 siRNA exhibit tumor cell specific uptake to reduce complications from off target effects in other organ systems.
Table 3.
siRNA nanoparticle strategies to augment proposed TNBC subtype-specific therapies
| TNBC subtype | Current proposed therapy | Proposed siRNA nanoparticle strategy |
|---|---|---|
| Basal-like | Cisplatin | siBRCA + cisplatin |
| Mesenchymal | PI3K/mTOR inhibitor | sip110β + MEK inhibitor siPIK3CA mutant + chemo siGF combination siEMT combination |
| Luminal androgen receptor | Bicalutamide | siAR |
GF – growth factor; EMT – Epithelial-mesenchymal transition; AR – Androgen receptor
Mesenchymal TNBCs and PI3K/mTOR inhibitors
The mesenchymal subtype of TNBCs harbor elevated growth factor signaling and proteins involved in EMT, and representative cell lines preferentially respond to PI3K/mTOR inhibitors (Lehmann et al. 2011). Clinical trials testing the combination of mTOR inhibitors with a variety of chemotherapies and EGFR-family targeted agents are under way (Massihnia, et al. 2016; Paplomata and O’Regan 2014). Similarly, a phase 2 clinical trial evaluating the efficacy of BKM120, a pan class 1 PI3K inhibitor, is ongoing (Mohamed, et al. 2013; Paplomata and O’Regan 2014). Within the past 2 decades, various modes of PI3K/mTOR and Ras/Erk signaling crosstalk have been elucidated (Mendoza, et al. 2011), and efforts to utilize small molecule inhibitors against both pathways in combination are also being tested in phase 1 clinical trials (Jokinen and Koivunen 2015). Preliminary studies have been disappointing, with a combined overall response rate of 4.7%, suggesting that improved strategies to inhibiting these pathways are necessary (Jokinen and Koivunen 2015).
Class 1 PI3K, consists of p110α, p110β, and p110γ catalytic kinases that exist as heterodimers. Previous studies utilizing several cancer cell lines have demonstrated that small molecule inhibitors against p110β were ineffective, while an siRNA approach that silences p110β effectively inhibits proliferation (Mendoza et al. 2011; Weiss, et al. 2007). Mechanistically, small molecule inhibition of p110β inhibits its kinase activity, which does not alter expression levels of the PI3K regulatory subunit, p85. However, silencing p110β with siRNAs disables its scaffolding functions leading to decreased p85 expression. Alteration of the p85 to p110β ratio induces negative feedback loops that coalesce to inhibit proliferation (Fan, et al. 2006). These results suggest that siRNA nanoparticle silencing of catalytic PI3K kinases in combination with MEK inhibitors may be more effective than the utilization of small molecule inhibitors to both pathways (Table 3) (Jokinen and Koivunen 2015). In addition to the advantages of silencing key players of the PI3K pathway, previous studies have also demonstrated that siRNAs can be adapted to specifically silence point mutations (Fleming, et al. 2005). In fact, silencing mutant KRAS as a therapeutic strategy is currently being explored in clinical trials for the treatment of pancreatic cancers (Table 2). Since PIK3CA mutations are prevalent in TNBCs (Shah, et al. 2012), preferential siRNA depletion of mutant gene expression is desirable, because this strategy is equipped to eliminate dose-limiting toxicities associated with PI3K inhibitors (Table 3). Collectively, these ideas suggest that an siRNA approach to silencing the PI3K pathway may be an effective therapeutic strategy in combination with chemotherapy, because of its versatile effects in downregulating PI3K 1) kinase activity 2) scaffolding activity and 3) mutations.
Alternatively, since TNBCs characterized by the M subtype also harbor elevated growth factor signaling, inhibition of growth factors is also a viable therapeutic strategy. In fact, elevated EGFR expression is reported in up to 76% of TNBC patients (Martin, et al. 2012), although inconsistent percentages are reported depending on the method of EGFR detection (Nakai, et al. 2016). Current clinical trials have explored small molecule EGFR inhibitors and anti-EGFR monoclonal antibodies as single agents and in combination with chemotherapy (Nakai et al. 2016). Unfortunately, these trials have been disappointing, owing to activation of alternative signaling pathways that mediate resistance (Nakai et al. 2016). siRNA nanoparticle silencing of a combination of growth factors may present as a feasible approach to circumvent current resistance mechanisms (Table 3), and future studies need to determine what combinations would be most efficacious. Similarly, since mesenchymal subtypes of TNBCs are also characterized with an increase in EMT gene signature, siRNA nanoparticle strategies to silence a combination of EMT transcription factors also presents as a feasible therapeutic strategy (Table 3).
In summary, utilization of siRNA nanoparticles as a therapeutic strategy to inhibit PI3K/mTOR, growth factor, and EMT pathways represents an attractive solution to challenges that are associated with the use of available small molecule PI3K/mTOR inhibitors. Because, functional inhibition of a target using small molecule inhibitors versus using siRNA strategies can have significantly different outcomes (Weiss et al. 2007), future studies need to clearly define what these differences are and which approach exerts the maximal therapeutic efficacy, while limiting toxicities. Future studies should also determine whether delivery of point-mutated siRNA can exert miRNA-like functions on the corresponding WT gene.
Luminal Androgen Receptor expressing TNBCs and bicalutamide
The LAR subtype exhibits elevated androgen receptor (AR) signaling and TNBC cell lines of this subtype preferentially responds to bicalutamide, an AR inhibitor. Approximately 10–43% of TNBCs express AR, however the prognostic value of AR currently remains controversial (Pietri, et al. 2016). Indeed, some studies have demonstrated that AR expression is correlated to increased mortality (Hu, et al. 2011), increased tumor stage and increased lymph node metastasis (McGhan, et al. 2014), while others have shown that AR expressing TNBCs have decreased lymph node metastasis (Rakha, et al. 2007), decreased tumor burden (Luo, et al. 2010; Park, et al. 2011), and increased overall survival (Luo et al. 2010). The reasons behind these contradictory results are currently unclear, although it is possible that differences in scoring systems and utilized reagents could have contributed (McGhan et al. 2014). Recently, a phase 2 clinical trial with 26 patients harboring AR positive and ER/PR negative breast cancers demonstrated that approximately 19% of patients who received bicalutamide exhibited a 6-month clinical benefit (Gucalp, et al. 2013). These results suggest that some patients clearly benefit from AR antagonists, and further testing of next generation AR-targeting agents, including enzalutamide and CYP17 inhibitors are under way (Gucalp et al. 2013). siRNA nanoparticles silencing AR has not been evaluated in the context of TNBCs. Lessons from treating prostate cancer patients with bicalutamide indicates that mutations in the hormone binding pocket of AR can switch anti-androgen antagonists to gaining agonist functions, thereby driving drug resistance (Tian, et al. 2015). It is tempting to speculate that siRNA silencing of AR could circumvent these limitations (Table 3). Future studies need to fully investigate which subsets of AR expressing TNBC patients will benefit from AR-targeted therapies.
TNBCs harboring stromal signatures
Recently, the aforementioned six subtypes of TNBCs have been refined to four distinct transcriptional subtypes with differing clinical characteristics (Lehmann, et al. 2016). These include the BL1, BL2, M, and LAR subtypes, whereas the previously stratified MSL and IM subtypes are now descriptors that indicate the presence of stromal and immune cell within the primary tumor (Lehmann et al. 2016). Indeed, Lehmann et al demonstrated that tumor subtypes changed to MSL when the matched stromal cells were included, suggesting that the MSL gene expression pattern is a characteristic of stromal cells (Lehmann et al. 2016). Prior to the refined 4 transcriptional subtype classification system, MSL TNBC cell lines exhibited in vitro sensitivity to dasatinib, a broad-spectrum tyrosine kinase inhibitor that targets Src family kinases (Lehmann et al. 2011). Not surprisingly, a phase 2 clinical trial evaluating dasatinib for the treatment of TNBC demonstrated only limited activity, and combinations with chemotherapies were subsequently explored (Finn, et al. 2011). Dasatinib is synergistic when treated in combination with cetuximab and cisplatin in vitro (Kim, et al. 2013). However, dasatinib treatment also exerts immunosuppressive effects (Blake, et al. 2008; Fraser, et al. 2009), which may offset the effects of anti-tumor immunity. More recently, a nanoparticle formulation of dasatinib has been utilized in combination with the anti-microtubule chemotherapy, vincristine, and results indicate 1) enhanced tumor cell uptake of dasatinib, 2) increased apoptosis, and 3) decreased vascular mimicry channels (Zeng, et al. 2015). Whether or not nanoparticle delivery of anti-Src siRNA can recapitulate the effects of liposomal dasatinib is unclear. However, the refinement of utilizing MSL as a description to highlight the presence of stromal cells in the microenvironment points at the possibility of utilizing siRNA nanoparticles to target not only tumor cells, but also stromal cells that function in creating a supportive tumor microenvironment (Table 4). Future studies need to determine appropriate therapeutic targets for both the tumor epithelial and stromal components, and localization strategies.
Table 4.
Proposed therapeutic strategies using siRNA nanoparticles to address stromal and immune cells within the primary tumor
| TNBC descriptor | Prosposed siRNA nanoparticle strategy |
|---|---|
| Mesenchymal stem-like | siRNA against stromal cells |
| Immunomodulatory | siPD1/siPD-L1 |
TNBCs harboring immune infiltration
Interestingly, approximately 20% of TNBCs are highly enriched for immune cell infiltration, which is 1) an indicator of good prognosis, 2) a predictor of improved relapse free survival (Lehmann et al. 2016) and pathological complete response (pCR), and 3) suggestive that immunotherapy may be a good therapeutic option (Garcia-Teijido, et al. 2016). Importantly, the anti-PD-1 immune checkpoint inhibiting monoclonal antibody, pembrolizumab, has recently demonstrated clinical activity in a phase 1b clinical trial for the treatment of PD-L1 positive, heavily pretreated, metastatic TNBCs (Nanda, et al. 2016). These preliminary studies demonstrate an overall response rate of 18.5%, and a phase 2 clinical trial is underway. This overall response rate suggests that a subset of patients may be resistant to pembrolizumab and its incorporation for clinical use is currently unclear. Potential mechanisms of pembrolizumab resistance may involve 1) conformational changes or co-receptor expression that shields the therapeutic target from drug binding 2) upregulation of alternative receptors that cooperate to induce hyperactivation of the therapeutic target, 3) activation of downstream effectors that bypass the inhibitory effects of the drug and 4) mutation and constitutive activation of the therapeutic target. Whether or not pembrolizumab resistance actually involves these hypothetical resistance mechanisms is unclear. Interestingly, nanoparticle delivery of anti-HER2 siRNA has been demonstrated to circumvent resistance mechanisms associated with trastuzumab, a humanized monoclonal antibody used for treating HER2 breast cancers (Gu et al. 2016). It is tempting to speculate that silencing PD-1 or PD-L1 expression by siRNA nanoparticles may circumvent pembrolizumab resistance (Table 4), however further testing, including initial studies to determine if siRNA nanoparticles can be adapted to specifically target PD-1 expression on lymphocytes are required.
Ongoing challenges associated with the clinical use of siRNA nanoparticles
Even though preclinical studies and recent clinical trials have established siRNA nanoparticles as a promising therapeutic strategy for cancer treatment, a number of ongoing challenges have limited the technology. First, the percentage of siRNA nanoparticles that are actually taken up by tumor cells is estimated to be only 0.7% of the injected dose (Wilhelm S. 2016). Active targeting strategies such as those summarized in Table 1 improves nanoparticle uptake to ~0.9%, however 0.9% of the injected dose still represents a miniscule amount. While synthesizing sufficient nanoparticles for preclinical mouse models are not limited by these low uptake statistics (the average mouse is 20g), scaling these amounts up for applicability in humans may cause additional complications. Synthesis of nanoparticles in larger amounts may compromise function of the nanoparticles, for example through aggregation (Wilhelm S. 2016). The cost associated with synthesis and quality control could be prohibitively high. Additionally, the large volume of nanoparticles administered into a patient could introduce additional technical challenges and issues with immunity. Lastly, because approximately 99% of the nanoparticles are not taken up by tumor cells, off-target effects may pose serious restrictions. However, the good news is that siRNA silencing of target genes by nanoparticles is a reversible process and withdrawal of the nanoparticle should reverse potential side-effects. Future studies need to clearly dissect the mechanisms of nanoparticle localization and uptake by tumor cells to exploit potential pathways that can improve tumor-specific accumulation.
Conclusion
The success of using siRNA as a tool to dissect molecular pathways has prompted researchers to explore its potential as a therapeutic platform. Indeed, over the past decade various siRNA-nanoparticle strategies have been employed to treat solid tumors in clinical trials. Completed trials have demonstrated both lipid and polymer siRNA nanoparticles to be safe and effective for silencing target gene expression, and preliminary results indicate good therapeutic outcomes in a variety of cancers. These clinical studies have laid a solid foundation for the use of siRNA nanoparticles to address ongoing challenges in cancer treatment, such as the case for TNBC. There is currently a lack of effective FDA-approved targeted therapies for the treatment of TNBCs. Recent studies using high-throughput sequencing has revealed the diverse genetic heterogeneity of TNBC that may underlie the difficulty associated with designing an appropriate targeted therapy for the treatment of this disease. Various ongoing clinical trials are now evaluating TNBC treatment regimens based on distinct molecular characteristics, such as BRCA and LAR. Because TNBC is characterized by a heterogenous genetic landscape, the versatility of selectively targeting different genes by siRNA nanoparticles makes this technology an attractive therapeutic option for TNBC treatment. Various preclinical studies utilizing siRNA nanoparticles to target 1) components of the cell cycle 2) factors that induce EMT, and 3) factors that reduce chemotherapy efficacy have been demonstrated to inhibit TNBC disease progression in mouse models. siRNA nanoparticles may be further adapted to complement proposed therapeutic strategies associated with the recently identified TNBC subtypes. Many of these proposed strategies are currently being tested in clinical trials and the use of siRNA nanoparticles to complement them may further strengthen their therapeutic efficacy. Collectively, siRNA nanotechnology is a promising solution in eliminating the roadblocks to successful development of a targeted therapy for TNBC treatment.
Acknowledgments
Funding
Research support was provided in part by the Department of Defense to JGP (BC133808), and by the National Institutes of Health to MWJ (R01 CA138421).
Members of the Jackson laboratory are thanked for providing helpful comments and suggestions.
Footnotes
Declaration of interest
There is no conflict of interest that could be perceived as prejudicing the impartiality of the perspectives reported.
References
- ACS. 2016 Cancer Facts and Figures 2016. American Cancer Society; [Google Scholar]
- Aleku M, Schulz P, Keil O, Santel A, Schaeper U, Dieckhoff B, Janke O, Endruschat J, Durieux B, Roder N, et al. Atu027, a liposomal small interfering RNA formulation targeting protein kinase N3, inhibits cancer progression. Cancer Res. 2008;68:9788–9798. doi: 10.1158/0008-5472.CAN-08-2428. [DOI] [PubMed] [Google Scholar]
- Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alluri P, Newman LA. Basal-like and triple-negative breast cancers: searching for positives among many negatives. Surg Oncol Clin N Am. 2014;23:567–577. doi: 10.1016/j.soc.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apps MG, Choi EH, Wheate NJ. The state-of-play and future of platinum drugs. Endocr Relat Cancer. 2015;22:R219–233. doi: 10.1530/ERC-15-0237. [DOI] [PubMed] [Google Scholar]
- Arosio D, Casagrande C. Advancement in integrin facilitated drug delivery. Adv Drug Deliv Rev. 2016;97:111–143. doi: 10.1016/j.addr.2015.12.001. [DOI] [PubMed] [Google Scholar]
- Azlan A, Dzaki N, Azzam G. Argonaute: The executor of small RNA function. J Genet Genomics. 2016;43:481–494. doi: 10.1016/j.jgg.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Baker LA, Holliday H, Swarbrick A. ID4 controls luminal lineage commitment in normal mammary epithelium and inhibits BRCA1 function in basal-like breast cancer. Endocr Relat Cancer. 2016;23:R381–392. doi: 10.1530/ERC-16-0196. [DOI] [PubMed] [Google Scholar]
- Bakrania AK, Variya BC, Patel SS. Novel targets for paclitaxel nano formulations: Hopes and hypes in triple negative breast cancer. Pharmacol Res. 2016;111:577–591. doi: 10.1016/j.phrs.2016.07.023. [DOI] [PubMed] [Google Scholar]
- Bargheer D, Giemsa A, Freund B, Heine M, Waurisch C, Stachowski GM, Hickey SG, Eychmuller A, Heeren J, Nielsen P. The distribution and degradation of radiolabeled superparamagnetic iron oxide nanoparticles and quantum dots in mice. Beilstein J Nanotechnol. 2015;6:111–123. doi: 10.3762/bjnano.6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake SJ, Bruce Lyons A, Fraser CK, Hayball JD, Hughes TP. Dasatinib suppresses in vitro natural killer cell cytotoxicity. Blood. 2008;111:4415–4416. doi: 10.1182/blood-2008-02-138701. [DOI] [PubMed] [Google Scholar]
- Cao N, Cheng D, Zou S, Ai H, Gao J, Shuai X. The synergistic effect of hierarchical assemblies of siRNA and chemotherapeutic drugs co-delivered into hepatic cancer cells. Biomaterials. 2011;32:2222–2232. doi: 10.1016/j.biomaterials.2010.11.061. [DOI] [PubMed] [Google Scholar]
- Chacon RD, Costanzo MV. Triple-negative breast cancer. Breast Cancer Res. 2010;12(Suppl 2):S3. doi: 10.1186/bcr2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong WH, Hagens WI, Krystek P, Burger MC, Sips AJ, Geertsma RE. Particle size-dependent organ distribution of gold nanoparticles after intravenous administration. Biomaterials. 2008;29:1912–1919. doi: 10.1016/j.biomaterials.2007.12.037. [DOI] [PubMed] [Google Scholar]
- Deen WM, Lazzara MJ, Myers BD. Structural determinants of glomerular permeability. Am J Physiol Renal Physiol. 2001;281:F579–596. doi: 10.1152/ajprenal.2001.281.4.F579. [DOI] [PubMed] [Google Scholar]
- Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT. Layer-by-layer nanoparticles for systemic codelivery of an anticancer drug and siRNA for potential triple-negative breast cancer treatment. ACS Nano. 2013;7:9571–9584. doi: 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429–4434. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- Dhillon KK, Swisher EM, Taniguchi T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011;102:663–669. doi: 10.1111/j.1349-7006.2010.01840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo-Ortiz D, Duran L, Moreno L, Ochoa H, Castano VM. Mapping knowledge translation and innovation processes in Cancer Drug Development: the case of liposomal doxorubicin. J Transl Med. 2014;12:227. doi: 10.1186/s12967-014-0227-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhr E, Zare F, Teimoori-Toolabi L. Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther. 2016;23:73–82. doi: 10.1038/cgt.2016.4. [DOI] [PubMed] [Google Scholar]
- Fan QW, Knight ZA, Goldenberg DD, Yu W, Mostov KE, Stokoe D, Shokat KM, Weiss WA. A dual PI3 kinase/mTOR inhibitor reveals emergent efficacy in glioma. Cancer Cell. 2006;9:341–349. doi: 10.1016/j.ccr.2006.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Yu MZ, Wang JC, Hou WJ, Gao LY, Ma XF, Pei XW, Niu YJ, Liu XY, Qiu C, et al. Synergistic inhibition of breast cancer by co-delivery of VEGF siRNA and paclitaxel via vapreotide-modified core-shell nanoparticles. Biomaterials. 2014;35:5028–5038. doi: 10.1016/j.biomaterials.2014.03.012. [DOI] [PubMed] [Google Scholar]
- Finlay J, Roberts CM, Lowe G, Loeza J, Rossi JJ, Glackin CA. RNA-based TWIST1 inhibition via dendrimer complex to reduce breast cancer cell metastasis. Biomed Res Int. 2015;2015:382745. doi: 10.1155/2015/382745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RS, Bengala C, Ibrahim N, Roche H, Sparano J, Strauss LC, Fairchild J, Sy O, Goldstein LJ. Dasatinib as a single agent in triple-negative breast cancer: results of an open-label phase 2 study. Clin Cancer Res. 2011;17:6905–6913. doi: 10.1158/1078-0432.CCR-11-0288. [DOI] [PubMed] [Google Scholar]
- Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3:413–423. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- Fraser CK, Blake SJ, Diener KR, Lyons AB, Brown MP, Hughes TP, Hayball JD. Dasatinib inhibits recombinant viral antigen-specific murine CD4+ and CD8+ T-cell responses and NK-cell cytolytic activity in vitro and in vivo. Exp Hematol. 2009;37:256–265. doi: 10.1016/j.exphem.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Furuta E, Okuda H, Kobayashi A, Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galliher AJ, Schiemann WP. Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res. 2006;8:R42. doi: 10.1186/bcr1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Teijido P, Cabal ML, Fernandez IP, Perez YF. Tumor-Infiltrating Lymphocytes in Triple Negative Breast Cancer: The Future of Immune Targeting. Clin Med Insights Oncol. 2016;10:31–39. doi: 10.4137/CMO.S34540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusiano S, Cochet C, Filhol O, Duchemin-Pelletier E, Secq V, Bonnier P, Carcopino X, Boubli L, Birnbaum D, Garcia S, et al. Protein kinase CK2alpha subunit over-expression correlates with metastatic risk in breast carcinomas: quantitative immunohistochemistry in tissue microarrays. Eur J Cancer. 2011;47:792–801. doi: 10.1016/j.ejca.2010.11.028. [DOI] [PubMed] [Google Scholar]
- Golan T, Khvalevsky EZ, Hubert A, Gabai RM, Hen N, Segal A, Domb A, Harari G, David EB, Raskin S, et al. RNAi therapy targeting KRAS in combination with chemotherapy for locally advanced pancreatic cancer patients. Oncotarget. 2015;6:24560–24570. doi: 10.18632/oncotarget.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal S, Miller KB, Jaffe IZ. Molecular mechanisms for vascular complications of targeted cancer therapies. Clin Sci (Lond) 2016;130:1763–1779. doi: 10.1042/CS20160246. [DOI] [PubMed] [Google Scholar]
- Gu S, Hu Z, Ngamcherdtrakul W, Castro DJ, Morry J, Reda MM, Gray JW, Yantasee W. Therapeutic siRNA for drug-resistant HER2-positive breast cancer. Oncotarget. 2016;7:14727–14741. doi: 10.18632/oncotarget.7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R, Tapang P, Leverson JD, Albert D, Giranda VL, Luo Y. Small interfering RNA-mediated Polo-like kinase 1 depletion preferentially reduces the survival of p53-defective, oncogenic transformed cells and inhibits tumor growth in animals. Cancer Res. 2005;65:2698–2704. doi: 10.1158/0008-5472.CAN-04-2131. [DOI] [PubMed] [Google Scholar]
- Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA, Blackwell K, Rugo H, Nabell L, Forero A, et al. Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res. 2013;19:5505–5512. doi: 10.1158/1078-0432.CCR-12-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttery DS, Shaw JA, Lloyd K, Pringle JH, Walker RA. Expression of tenascin-C and its isoforms in the breast. Cancer Metastasis Rev. 2010;29:595–606. doi: 10.1007/s10555-010-9249-9. [DOI] [PubMed] [Google Scholar]
- Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, Fitzgibbons PL, Francis G, Goldstein NS, Hayes M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28:2784–2795. doi: 10.1200/JCO.2009.25.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamurcu Z, Ashour A, Kahraman N, Ozpolat B. FOXM1 regulates expression of eukaryotic elongation factor 2 kinase and promotes proliferation, invasion and tumorgenesis of human triple negative breast cancer cells. Oncotarget. 2016;7:16619–16635. doi: 10.18632/oncotarget.7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine M, Bartelt A, Bruns OT, Bargheer D, Giemsa A, Freund B, Scheja L, Waurisch C, Eychmuller A, Reimer R, et al. The cell-type specific uptake of polymer-coated or micelle-embedded QDs and SPIOs does not provoke an acute pro-inflammatory response in the liver. Beilstein J Nanotechnol. 2014;5:1432–1440. doi: 10.3762/bjnano.5.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoelder S, Clarke PA, Workman P. Discovery of small molecule cancer drugs: successes, challenges and opportunities. Mol Oncol. 2012;6:155–176. doi: 10.1016/j.molonc.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi D, Anderton B, Goga A. Taking on challenging targets: making MYC druggable. Am Soc Clin Oncol Educ Book. 2014:e497–502. doi: 10.14694/EdBook_AM.2014.34.e497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi D, Kusdra L, Huskey NE, Chandriani S, Lenburg ME, Gonzalez-Angulo AM, Creasman KJ, Bazarov AV, Smyth JW, Davis SE, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med. 2012;209:679–696. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R, Dawood S, Holmes MD, Collins LC, Schnitt SJ, Cole K, Marotti JD, Hankinson SE, Colditz GA, Tamimi RM. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res. 2011;17:1867–1874. doi: 10.1158/1078-0432.CCR-10-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakoff SJ, Mayer EL, He L, Traina TA, Carey LA, Krag KJ, Rugo HS, Liu MC, Stearns V, Come SE, et al. TBCRC009: A Multicenter Phase II Clinical Trial of Platinum Monotherapy With Biomarker Assessment in Metastatic Triple-Negative Breast Cancer. J Clin Oncol. 2015;33:1902–1909. doi: 10.1200/JCO.2014.57.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer AK, Singh A, Ganta S, Amiji MM. Role of integrated cancer nanomedicine in overcoming drug resistance. Adv Drug Deliv Rev. 2013;65:1784–1802. doi: 10.1016/j.addr.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Jeong JH, Park TG, Kim SH. Self-assembled and nanostructured siRNA delivery systems. Pharm Res. 2011;28:2072–2085. doi: 10.1007/s11095-011-0412-y. [DOI] [PubMed] [Google Scholar]
- Johnston SJ, Carroll JS. Transcription factors and chromatin proteins as therapeutic targets in cancer. Biochim Biophys Acta. 2015;1855:183–192. doi: 10.1016/j.bbcan.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanoparticle PEGylation for imaging and therapy. Nanomedicine (Lond) 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokinen E, Koivunen JP. MEK and PI3K inhibition in solid tumors: rationale and evidence to date. Ther Adv Med Oncol. 2015;7:170–180. doi: 10.1177/1758834015571111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan DR, Webb MN, Cadotte TH, Gavette MN. Use of Targeted Liposome-based Chemotherapeutics to Treat Breast Cancer. Breast Cancer (Auckl) 2015;9:1–5. doi: 10.4137/BCBCR.S29421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EM, Mueller K, Gartner E, Boerner J. Dasatinib is synergistic with cetuximab and cisplatin in triple-negative breast cancer cells. J Surg Res. 2013;185:231–239. doi: 10.1016/j.jss.2013.06.041. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sato N, Kawamoto S, Saga T, Hiraga A, Haque TL, Ishimori T, Konishi J, Togashi K, Brechbiel MW. Comparison of the macromolecular MR contrast agents with ethylenediamine-core versus ammonia-core generation-6 polyamidoamine dendrimer. Bioconjug Chem. 2001;12:100–107. doi: 10.1021/bc000075s. [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Tomari Y. RISC assembly: Coordination between small RNAs and Argonaute proteins. Biochim Biophys Acta. 2016;1859:71–81. doi: 10.1016/j.bbagrm.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Kren BT, Unger GM, Abedin MJ, Vogel RI, Henzler CM, Ahmed K, Trembley JH. Preclinical evaluation of cyclin dependent kinase 11 and casein kinase 2 survival kinases as RNA interference targets for triple negative breast cancer therapy. Breast Cancer Res. 2015;17:19. doi: 10.1186/s13058-015-0524-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyndi M, Sorensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J Danish Breast Cancer Cooperative G. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol. 2008;26:1419–1426. doi: 10.1200/JCO.2007.14.5565. [DOI] [PubMed] [Google Scholar]
- Lao J, Madani J, Puertolas T, Alvarez M, Hernandez A, Pazo-Cid R, Artal A, Anton Torres A. Liposomal Doxorubicin in the treatment of breast cancer patients: a review. J Drug Deliv. 2013;2013:456409. doi: 10.1155/2013/456409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, Moses HL, Sanders ME, Pietenpol JA. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One. 2016;11:e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, Pietenpol JA, Tan AR. Triple-negative breast cancer: molecular subtypes and new targets for therapy. Am Soc Clin Oncol Educ Book. 2015:e31–39. doi: 10.14694/EdBook_AM.2015.35.e31. [DOI] [PubMed] [Google Scholar]
- Li W, Szoka FC., Jr Lipid-based nanoparticles for nucleic acid delivery. Pharm Res. 2007;24:438–449. doi: 10.1007/s11095-006-9180-5. [DOI] [PubMed] [Google Scholar]
- Lim J, Guo Y, Rostollan CL, Stanfield J, Hsieh JT, Sun X, Simanek EE. The role of the size and number of polyethylene glycol chains in the biodistribution and tumor localization of triazine dendrimers. Mol Pharm. 2008;5:540–547. doi: 10.1021/mp8000292. [DOI] [PubMed] [Google Scholar]
- Liu JY, Chiang T, Liu CH, Chern GG, Lin Ts T, Gao DY, Chen Y. Delivery of siRNA Using CXCR4-targeted Nanoparticles Modulates Tumor Microenvironment and Achieves a Potent Antitumor Response in Liver Cancer. Mol Ther. 2015;23:1772–1782. doi: 10.1038/mt.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Targeting Polo-Like Kinases: A Promising Therapeutic Approach for Cancer Treatment. Transl Oncol. 2015;8:185–195. doi: 10.1016/j.tranon.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhu YH, Mao CQ, Dou S, Shen S, Tan ZB, Wang J. Triple negative breast cancer therapy with CDK1 siRNA delivered by cationic lipid assisted PEG-PLA nanoparticles. J Control Release. 2014;192:114–121. doi: 10.1016/j.jconrel.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Longmire M, Choyke PL, Kobayashi H. Clearance properties of nano-sized particles and molecules as imaging agents: considerations and caveats. Nanomedicine (Lond) 2008;3:703–717. doi: 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire MR, Ogawa M, Choyke PL, Kobayashi H. Biologically optimized nanosized molecules and particles: more than just size. Bioconjug Chem. 2011;22:993–1000. doi: 10.1021/bc200111p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Davila V, Seifalian AM, Loizidou M. Organic nanocarriers for cancer drug delivery. Curr Opin Pharmacol. 2012;12:414–419. doi: 10.1016/j.coph.2012.02.011. [DOI] [PubMed] [Google Scholar]
- Lorenzer C, Dirin M, Winkler AM, Baumann V, Winkler J. Going beyond the liver: progress and challenges of targeted delivery of siRNA therapeutics. J Control Release. 2015;203:1–15. doi: 10.1016/j.jconrel.2015.02.003. [DOI] [PubMed] [Google Scholar]
- Luo X, Shi YX, Li ZM, Jiang WQ. Expression and clinical significance of androgen receptor in triple negative breast cancer. Chin J Cancer. 2010;29:585–590. doi: 10.5732/cjc.009.10673. [DOI] [PubMed] [Google Scholar]
- Malamas AS, Jin E, Gujrati M, Lu ZR. Dynamic Contrast Enhanced MRI Assessing the Antiangiogenic Effect of Silencing HIF-1alpha with Targeted Multifunctional ECO/siRNA Nanoparticles. Mol Pharm. 2016;13:2497–2506. doi: 10.1021/acs.molpharmaceut.6b00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal S, El Hor A, Millard M, Gillon V, Bezdetnaya L. Anticancer Drug Delivery: An Update on Clinically Applied Nanotherapeutics. Drugs. 2015;75:1601–1611. doi: 10.1007/s40265-015-0453-3. [DOI] [PubMed] [Google Scholar]
- Martin V, Botta F, Zanellato E, Molinari F, Crippa S, Mazzucchelli L, Frattini M. Molecular characterization of EGFR and EGFR-downstream pathways in triple negative breast carcinomas with basal like features. Histol Histopathol. 2012;27:785–792. doi: 10.14670/HH-27.785. [DOI] [PubMed] [Google Scholar]
- Massihnia D, Galvano A, Fanale D, Perez A, Castiglia M, Incorvaia L, Listi A, Rizzo S, Cicero G, Bazan V, et al. Triple negative breast cancer: shedding light onto the role of pi3k/akt/mtor pathway. Oncotarget. 2016 doi: 10.18632/oncotarget.10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhan LJ, McCullough AE, Protheroe CA, Dueck AC, Lee JJ, Nunez-Nateras R, Castle EP, Gray RJ, Wasif N, Goetz MP, et al. Androgen receptor-positive triple negative breast cancer: a unique breast cancer subtype. Ann Surg Oncol. 2014;21:361–367. doi: 10.1245/s10434-013-3260-7. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36:320–328. doi: 10.1016/j.tibs.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele E, Spinelli GP, Miele E, Di Fabrizio E, Ferretti E, Tomao S, Gulino A. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. Int J Nanomedicine. 2012;7:3637–3657. doi: 10.2147/IJN.S23696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missirlis D, Haraszti T, Scheele C, Wiegand T, Diaz C, Neubauer S, Rechenmacher F, Kessler H, Spatz JP. Substrate engagement of integrins alpha5beta1 and alphavbeta3 is necessary, but not sufficient, for high directional persistence in migration on fibronectin. Sci Rep. 2016;6:23258. doi: 10.1038/srep23258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed A, Krajewski K, Cakar B, Ma CX. Targeted therapy for breast cancer. Am J Pathol. 2013;183:1096–1112. doi: 10.1016/j.ajpath.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Morrison CD, Parvani JG, Schiemann WP. The relevance of the TGF-beta Paradox to EMT-MET programs. Cancer Lett. 2013;341:30–40. doi: 10.1016/j.canlet.2013.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahta R, Esteva FJ. HER2 therapy: molecular mechanisms of trastuzumab resistance. Breast Cancer Res. 2006;8:215. doi: 10.1186/bcr1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Hung MC, Yamaguchi H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res. 2016;6:1609–1623. [PMC free article] [PubMed] [Google Scholar]
- Nanda R, Chow LQ, Dees EC, Berger R, Gupta S, Geva R, Pusztai L, Pathiraja K, Aktan G, Cheng JD, et al. Pembrolizumab in Patients With Advanced Triple-Negative Breast Cancer: Phase Ib KEYNOTE-012 Study. J Clin Oncol. 2016;34:2460–2467. doi: 10.1200/JCO.2015.64.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naymagon L, Abdul-Hay M. Novel agents in the treatment of multiple myeloma: a review about the future. J Hematol Oncol. 2016;9:52. doi: 10.1186/s13045-016-0282-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necela BM, Crozier JA, Andorfer CA, Lewis-Tuffin L, Kachergus JM, Geiger XJ, Kalari KR, Serie DJ, Sun Z, Moreno-Aspitia A, et al. Folate receptor-alpha (FOLR1) expression and function in triple negative tumors. PLoS One. 2015;10:e0122209. doi: 10.1371/journal.pone.0122209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly EA, Gubbins L, Sharma S, Tully R, Guang MH, Weiner-Gorzel K, McCaffrey J, Harrison M, Furlong F, Kell M, et al. The fate of chemoresistance in triple negative breast cancer (TNBC) BBA Clin. 2015;3:257–275. doi: 10.1016/j.bbacli.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Regino CA, Marcelino B, Williams M, Kosaka N, Bryant LH, Jr, Choyke PL, Kobayashi H. New nanosized biocompatible MR contrast agents based on lysine-dendri-graft macromolecules. Bioconjug Chem. 2010;21:955–960. doi: 10.1021/bc9005442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paplomata E, O’Regan R. The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers. Ther Adv Med Oncol. 2014;6:154–166. doi: 10.1177/1758834014530023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Koo JS, Kim MS, Park HS, Lee JS, Lee JS, Kim SI, Park BW, Lee KS. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Ann Oncol. 2011;22:1755–1762. doi: 10.1093/annonc/mdq678. [DOI] [PubMed] [Google Scholar]
- Parvani JG, Galliher-Beckley AJ, Schiemann BJ, Schiemann WP. Targeted inactivation of beta1 integrin induces beta3 integrin switching, which drives breast cancer metastasis by TGF-beta. Mol Biol Cell. 2013;24:3449–3459. doi: 10.1091/mbc.E12-10-0776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvani JG, Gujrati MD, Mack MA, Schiemann WP, Lu ZR. Silencing beta3 Integrin by Targeted ECO/siRNA Nanoparticles Inhibits EMT and Metastasis of Triple-Negative Breast Cancer. Cancer Res. 2015;75:2316–2325. doi: 10.1158/0008-5472.CAN-14-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri E, Conteduca V, Andreis D, Massa I, Melegari E, Sarti S, Cecconetto L, Schirone A, Bravaccini S, Serra P, et al. Androgen receptor signaling pathways as a target for breast cancer treatment. Endocr Relat Cancer. 2016;23:R485–498. doi: 10.1530/ERC-16-0190. [DOI] [PubMed] [Google Scholar]
- Prat A, Cruz C, Hoadley KA, Diez O, Perou CM, Balmana J. Molecular features of the basal-like breast cancer subtype based on BRCA1 mutation status. Breast Cancer Res Treat. 2014;147:185–191. doi: 10.1007/s10549-014-3056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachakonda PS, Bauer AS, Xie H, Campa D, Rizzato C, Canzian F, Beghelli S, Greenhalf W, Costello E, Schanne M, et al. Somatic mutations in exocrine pancreatic tumors: association with patient survival. PLoS One. 2013;8:e60870. doi: 10.1371/journal.pone.0060870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer. 2007;109:25–32. doi: 10.1002/cncr.22381. [DOI] [PubMed] [Google Scholar]
- Ramot Y, Rotkopf S, Gabai RM, Zorde Khvalevsky E, Muravnik S, Marzoli GA, Domb AJ, Shemi A, Nyska A. Preclinical Safety Evaluation in Rats of a Polymeric Matrix Containing an siRNA Drug Used as a Local and Prolonged Delivery System for Pancreatic Cancer Therapy. Toxicol Pathol. 2016;44:856–865. doi: 10.1177/0192623316645860. [DOI] [PubMed] [Google Scholar]
- Saba R, Alsayed A, Zacny JP, Dudek AZ. The Role of Forkhead Box Protein M1 in Breast Cancer Progression and Resistance to Therapy. Int J Breast Cancer. 2016;2016:9768183. doi: 10.1155/2016/9768183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadekar S, Ray A, Janat-Amsbury M, Peterson CM, Ghandehari H. Comparative biodistribution of PAMAM dendrimers and HPMA copolymers in ovarian-tumor-bearing mice. Biomacromolecules. 2011;12:88–96. doi: 10.1021/bm101046d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santel A, Aleku M, Keil O, Endruschat J, Esche V, Fisch G, Dames S, Loffler K, Fechtner M, Arnold W, et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- Scholz C, Wagner E. Therapeutic plasmid DNA versus siRNA delivery: common and different tasks for synthetic carriers. J Control Release. 2012;161:554–565. doi: 10.1016/j.jconrel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Schultheis B, Strumberg D, Santel A, Vank C, Gebhardt F, Keil O, Lange C, Giese K, Kaufmann J, Khan M, et al. First-in-human phase I study of the liposomal RNA interference therapeutic Atu027 in patients with advanced solid tumors. J Clin Oncol. 2014;32:4141–4148. doi: 10.1200/JCO.2013.55.0376. [DOI] [PubMed] [Google Scholar]
- Seitz S, Buchholz S, Schally AV, Jayakumar AR, Weber F, Papadia A, Rick FG, Szalontay L, Treszl A, Koster F, et al. Targeting triple-negative breast cancer through the somatostatin receptor with the new cytotoxic somatostatin analogue AN-162 [AEZS-124] Anticancer Drugs. 2013;24:150–157. doi: 10.1097/CAD.0b013e32835a7e29. [DOI] [PubMed] [Google Scholar]
- Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N, Settleman J. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, Karnezis AN, Swigart LB, Nasi S, Evan GI. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spankuch B, Matthess Y, Knecht R, Zimmer B, Kaufmann M, Strebhardt K. Cancer inhibition in nude mice after systemic application of U6 promoter-driven short hairpin RNAs against PLK1. J Natl Cancer Inst. 2004;96:862–872. doi: 10.1093/jnci/djh146. [DOI] [PubMed] [Google Scholar]
- Stover DG, Winer EP. Tailoring adjuvant chemotherapy regimens for patients with triple negative breast cancer. Breast. 2015;24(Suppl 2):S132–135. doi: 10.1016/j.breast.2015.07.032. [DOI] [PubMed] [Google Scholar]
- Su S, Tian Y, Li Y, Ding Y, Ji T, Wu M, Wu Y, Nie G. “Triple-punch” strategy for triple negative breast cancer therapy with minimized drug dosage and improved antitumor efficacy. ACS Nano. 2015;9:1367–1378. doi: 10.1021/nn505729m. [DOI] [PubMed] [Google Scholar]
- Swords DS, Firpo MA, Scaife CL, Mulvihill SJ. Biomarkers in pancreatic adenocarcinoma: current perspectives. Onco Targets Ther. 2016;9:7459–7467. doi: 10.2147/OTT.S100510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, Paz-Ares L, Cho DC, Infante JR, Alsina M, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene. 2005;24:287–291. doi: 10.1038/sj.onc.1208272. [DOI] [PubMed] [Google Scholar]
- Taylor MA, Parvani JG, Schiemann WP. The pathophysiology of epithelial-mesenchymal transition induced by transforming growth factor-beta in normal and malignant mammary epithelial cells. J Mammary Gland Biol Neoplasia. 2010;15:169–190. doi: 10.1007/s10911-010-9181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, He Y, Zhou J. Progress in antiandrogen design targeting hormone binding pocket to circumvent mutation based resistance. Front Pharmacol. 2015;6:57. doi: 10.3389/fphar.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft DJ, Cryns VL. Minireview: Basal-like breast cancer: from molecular profiles to targeted therapies. Mol Endocrinol. 2011;25:199–211. doi: 10.1210/me.2010-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voduc KD, Cheang MC, Tyldesley S, Gelmon K, Nielsen TO, Kennecke H. Breast cancer subtypes and the risk of local and regional relapse. J Clin Oncol. 2010;28:1684–1691. doi: 10.1200/JCO.2009.24.9284. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, Han Y, Liang LH, Ji A. Nanoparticle-based delivery system for application of siRNA in vivo. Curr Drug Metab. 2010;11:182–196. doi: 10.2174/138920010791110863. [DOI] [PubMed] [Google Scholar]
- Weiss WA, Taylor SS, Shokat KM. Recognizing and exploiting differences between RNAi and small-molecule inhibitors. Nat Chem Biol. 2007;3:739–744. doi: 10.1038/nchembio1207-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen AM, Steinmetz NF. Design of virus-based nanomaterials for medicine, biotechnology, and energy. Chem Soc Rev. 2016;45:4074–4126. doi: 10.1039/c5cs00287g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm STAJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW. Analysis of nanoparticle delivery to tumors. Nature Reviews Materials. 2016:1. [Google Scholar]
- Xu H, Tian Y, Yuan X, Liu Y, Wu H, Liu Q, Wu GS, Wu K. Enrichment of CD44 in basal-type breast cancer correlates with EMT, cancer stem cell gene profile, and prognosis. Onco Targets Ther. 2016;9:431–444. doi: 10.2147/OTT.S97192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagata H, Kajiura Y, Yamauchi H. Current strategy for triple-negative breast cancer: appropriate combination of surgery, radiation, and chemotherapy. Breast Cancer. 2011;18:165–173. doi: 10.1007/s12282-011-0254-9. [DOI] [PubMed] [Google Scholar]
- Yan C, Higgins PJ. Drugging the undruggable: transcription therapy for cancer. Biochim Biophys Acta. 2013;1835:76–85. doi: 10.1016/j.bbcan.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SW, Stenzel M, Yang JL. Nanoparticle-siRNA: A potential cancer therapy? Crit Rev Oncol Hematol. 2016;98:159–169. doi: 10.1016/j.critrevonc.2015.10.015. [DOI] [PubMed] [Google Scholar]
- Zeng F, Ju RJ, Liu L, Xie HJ, Mu LM, Zhao Y, Yan Y, Hu YJ, Wu JS, Lu WL. Application of functional vincristine plus dasatinib liposomes to deletion of vasculogenic mimicry channels in triple-negative breast cancer. Oncotarget. 2015;6:36625–36642. doi: 10.18632/oncotarget.5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Q, Qin L, Fu H, Fang Y, Han B, Duan Y. EGF-modified mPEG-PLGA-PLL nanoparticle for delivering doxorubicin combined with Bcl-2 siRNA as a potential treatment strategy for lung cancer. Drug Deliv. 2016:1–10. doi: 10.3109/10717544.2015.1126769. [DOI] [PubMed] [Google Scholar]
- Zhao X, Li F, Li Y, Wang H, Ren H, Chen J, Nie G, Hao J. Co-delivery of HIF1alpha siRNA and gemcitabine via biocompatible lipid-polymer hybrid nanoparticles for effective treatment of pancreatic cancer. Biomaterials. 2015;46:13–25. doi: 10.1016/j.biomaterials.2014.12.028. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Shen JK, Hornicek FJ, Kan Q, Duan Z. The emerging roles and therapeutic potential of cyclin-dependent kinase 11 (CDK11) in human cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem. 2008;19:1448–1455. doi: 10.1021/bc800065f. [DOI] [PubMed] [Google Scholar]
- Zorde Khvalevsky E, Gabai R, Rachmut IH, Horwitz E, Brunschwig Z, Orbach A, Shemi A, Golan T, Domb AJ, Yavin E, et al. Mutant KRAS is a druggable target for pancreatic cancer. Proc Natl Acad Sci U S A. 2013;110:20723–20728. doi: 10.1073/pnas.1314307110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerman JE, Gritli I, Tolcher A, Heidel JD, Lim D, Morgan R, Chmielowski B, Ribas A, Davis ME, Yen Y. Correlating animal and human phase Ia/Ib clinical data with CALAA-01, a targeted, polymer-based nanoparticle containing siRNA. Proc Natl Acad Sci U S A. 2014;111:11449–11454. doi: 10.1073/pnas.1411393111. [DOI] [PMC free article] [PubMed] [Google Scholar]