Abstract
Atherosclerotic plaque destabilization is the major determinant of most acute coronary events. Smooth muscle cell (SMC) death contributes to plaque destabilization. Here, we describe a novel antiapoptotic mechanism in vascular SMCs that involves interaction of nuclear glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with apurinic/apyrimidinic endonuclease 1 (Ape1), the major oxidized DNA repair enzyme. GAPDH down-regulation potentiated H2O2-induced DNA damage and SMC apoptosis. Conversely, GAPDH overexpression decreased DNA damage and protected SMCs against apoptosis. Ape1 down-regulation reversed the resistance of GAPDH-overexpressing cells to DNA damage and apoptosis, which indicated that Ape1 is indispensable for GAPDH-dependent protective effects. GAPDH bound Ape1 in the SMC nucleus, and blocking (or oxidation) of GAPDH active site cysteines suppressed GAPDH/Ape1 interaction and potentiated apoptosis. GAPDH up-regulated Ape1 via a transcription factor homeobox protein Hox-A5–dependent mechanism. GAPDH levels were reduced in atherosclerotic plaque SMCs, and this effect correlated with oxidative stress and SMC apoptosis. Thus, we demonstrated that nuclear GAPDH/Ape1 interaction preserved Ape1 activity, reduced DNA damage, and prevented SMC apoptosis. Suppression of SMC apoptosis by maintenance of nuclear GAPDH/Ape1 interactions may be a novel therapy to increase atherosclerotic plaque stability.—Hou, X., Snarski, P., Higashi, Y., Yoshida, T., Jurkevich, A., Delafontaine, P., Sukhanov, S. Nuclear complex of glyceraldehyde-3-phosphate dehydrogenase and DNA repair enzyme apurinic/apyrimidinic endonuclease I protect smooth muscle cells against oxidant-induced cell death.
Keywords: apoptosis, oxidative stress, GAPDH
Atherosclerosis is the principal underlying cause of most cardiovascular disease–related deaths and is a leading cause of death in the Western world (1). Most acute coronary events are related to rupture or erosion of atherosclerotic plaques that are not hemodynamically significant (2). Thus, plaque stability is a critical determinant of clinical events. Apoptosis of vascular smooth muscle cells (SMCs) is the major driving force of atherosclerotic plaque destabilization (3), and preventing plaque SMC apoptosis is a promising antiatherosclerotic strategy (4). DNA-damaging agents are potent inducers of cell death by apoptosis (5), and enhanced DNA damage and/or suppression of DNA repair pathways are now recognized as crucial factors in the initiation and progression of atherosclerosis (6, 7). In the base excision repair pathway, apurinic/apyrimidinic (AP) sites caused by oxidation produce ssDNA breaks and are removed from the genome (8). This process involves AP endonuclease 1/redox factor-1 (Ape1/Ref-1).
Ape1 is a multifunctional protein and plays a crucial role in cell protection from DNA damage. It is also involved in redox regulation of DNA repair (9). Ape1 is the major DNA repair enzyme in human cells (10). The insufficient AP site repair or suppression of Ape1 activity has been shown to promote cell death (9). As reported recently, Ape1-inhibited oxidative stress–induced apoptosis in cardiac progenitor cells (CPCs) and Ape1-overexpressing CPC transplantation in a mouse myocardial infarction model increased CPC survival, restored cardiac function, and reduced cardiac inflammation and fibrosis (11), which suggests an important antiapoptotic function for Ape1. Ape1 is known to bind glyceraldehyde-3-phosphate dehydrogenase (GAPDH; EC 1.2.1.12), and GAPDH reactivates oxidized Ape1 (12).
GAPDH catalyzes the sixth step of glycolysis, and its activity contributes to cell energy production. For a long time, GAPDH was considered to be an important housekeeping molecule, as almost all cells constitutively express GAPDH, and its expression remains relatively constant under stress; however, over the past 2 decades, it has been demonstrated that GAPDH levels change in various models. GAPDH is also implicated in the regulation of multiple non–glycolysis-related processes, including nuclear RNA transport, vesicle transport from ER to Golgi apparatus, DNA replication, and apoptosis (13). We have previously shown that the natural pro-oxidant and key proatherogenic molecule, oxidized LDL (OxLDL), down-regulates GAPDH expression in human vascular SMCs via an H2O2-dependent mechanism (14). OxLDL is associated with apoptotic SMCs in advanced human atherosclerotic plaque (15). It also promotes apoptosis of cultured SMCs (16); therefore, we hypothesized that GAPDH down-regulation mediates oxidant-induced SMC death. Our results show that GAPDH plays a crucial protective role in vascular SMCs, and GAPDH interaction with Ape1 is critical for GAPDH-mediated cell survival. For the first time to our knowledge, we have demonstrated that GAPDH regenerates Ape1 activity by up-regulating Ape1 expression and forming a nuclear GAPDH/Ape1 complex. Our data suggest that both nuclear GAPDH/Ape1 interaction and Ape1 protein up-regulation protect DNA integrity and prevent apoptosis. Our findings provide strong evidence for a predominant role of nuclear GAPDH/Ape1 interaction in the regulation of vascular SMC viability. Targeting of SMC apoptosis via maintenance of nuclear GAPDH/Ape1 interaction could become a novel and potentially beneficial strategy to increase atherosclerotic plaque stability and prevent acute coronary events.
MATERIALS AND METHODS
Materials
Endonuclease IV and DNase I were purchased from Thermo Fisher Scientific (Waltham, MA, USA), human recombinant Ape1 from Sino Biological (Beijing, China), human recombinant GAPDH from Creative Biomart (Shirley, NY, USA), and Alexa Fluor 488 C5 Maleimide from Thermo Fisher Scientific. OxiSelect Oxidative DNA damage ELISA kit was from Cell Biolabs (San Diego, CA, USA) and the NE-PER separation kit was from Pierce Biotechnology (Rockford, IL, USA). RT profiler PCR array was from Qiagen (Valencia, CA, USA). Homeobox protein Hox-A5 (HOXA5) EMSA kit was obtained from Signosis (Santa Clara, CA, USA). 26-mer oligonucleotides (Ape1 substrates) were from Midland Reagent Company (Midland, TX, USA). H2O2 (3% stabilized solution in water) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Koningic (heptelidic) acid (KA) was from BioVision (Milpitas, CA, USA). Mini-ProteanTGX 12% precast protein gels were purchased from Bio-Rad (Hercules, CA, USA), and Sypro Ruby Protein Gel Stain was from Thermo Fisher Scientific.
Cells and animals
Rat aortic SMCs were purchased from Lonza (Allendale, NJ, USA). For experiments, confluent (80–90%) SMCs were grown in SmGM-2 medium (Lonza) that was supplemented with 10% fetal calf serum, antibiotics, 0.5 μg/ml human recombinant epidermal growth factor, 5 μg/ml insulin, and 1 μg/ml human recombinant fibroblast growth factor. To generate SMCs with constitutive overexpression of GAPDH (R3 cells), we used retroviral vector-carrying human GAPDH tagged with V5 (pLZRS-GAPDH; kindly provided by Dr. Douglas Green, St. Jude Children’s Research Hospital, Memphis, TN, USA) (17).
All animal experiments were performed according to protocols approved by the Institutional Committee for Use and Care of Laboratory Animals. ApoE-null mice (age 8 wk; B6.129P2-Apoetm1Unc/J; The Jackson Laboratory, Bar Harbor, ME, USA) were fed a Western-type diet (42% of total calories from fat; 0.15% cholesterol; Harlan-Teklad, Indianapolis, IN, USA) for 12 wk as previously described (18). Under anesthesia, mice were perfused with saline, then 4% buffered paraformaldehyde plus 5% sucrose, and the heart together with aortic arch was dissected, fixed overnight, and embedded in paraffin. Serial sections (6 μm) were taken throughout the entire aortic valve area, per Paigen et al. (19), and used for immunohistochemistry.
Cell death assessment
Cell death was quantified with Cell Death ELISA Plus kit or with In Situ Cell Death Detection TUNEL-TMR kit (both from Roche, Basel, Switzerland) according to manufacturer instructions. Cell apoptosis was defined as TUNEL-positive cell number per 100 DAPI-positive cells.
Confocal and STED microscopy
Cells were plated (1 × 105 cells/ml) onto 35-mm collagen-coated glass-bottom dishes (MatTek Corp., Ashland, MA, USA) and grown for 24 h. Plated cells were then incubated for 6 h with or without 110 µM H2O2 in SmGM-2 growth medium with supplements. After treatment, cells were briefly washed with PBS and fixed in ice-cold methanol for 10 min. Cells were blocked for 1 h at room temperature in PBS that contained 0.1% Triton X-100 and 5% donkey serum, and were incubated with goat polyclonal Ref-1 (Ape1) antibody (Ab) (1:200, sc-9919) and rabbit polyclonal GAPDH Ab (1:200, sc-25778; both from Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h. Ref-1 Ab was detected by biotinylated horse anti-goat Ab (1:200, BA-9500; Vector Laboratories, Burlingame, CA, USA) followed by incubation with streptavidin-Alexa Fluor 594 conjugate (1:500, S32356; Thermo Fisher Scientific). To detect GAPDH Ab, cells were sequentially stained with Atto 647N-labeled goat anti-rabbit Ab (1:400, 40839; Sigma-Aldrich) plus DAPI (Fluoro Pure grade; Thermo Fisher Scientific). To detect oxidized GAPDH, cells were fixed with 4% paraformaldehyde for 5 min, permeabilized with 0.5% Triton X-100, and stained with mouse GAPDH-SO3 mAb (1:100, LF-MA0100, 4A1 clone; Thermo Fisher Scientific). Primary Ab was detected with biotinylated horse anti-mouse Ab (1:200, BA-2000; Vector Laboratories) followed by incubation with streptavidin-Alexa Fluor 594 and costaining with Alexa Fluor 488–conjugated phalloidin (1:100, A12379; Thermo Fisher Scientific) and DAPI. Samples were embedded in ProLong Mountant (P36961; Thermo Fisher Scientific). Confocal and gated stimulated emission depletion (gSTED) super-resolution microscopy was performed with a Leica TCP SP8 STED microscope (Leica Microsystems, Wetzlar, Germany) equipped with a ×100/1.4 NA oil-immersion plan-apochromatic objective lens (HCX PL APO STED; Leica Microsystems), a tunable supercontinuum laser, a 405-nm diode laser, a STED pulsed 775-nm depletion laser, and Leica HyD detectors. Multichannel confocal images were acquired in the sequential scanning mode at a scan speed 600 Hz. Fluorescence of DAPI, Alexa Fluor 488 (phalloidin), Alexa Fluor 594 (Ape1 or GAPDH-SO3), and Atto 647N (GAPDH) were excited at 405, 585, and 650 nm, respectively. Emission band-pass wavelengths were set to 420–480 nm (DAPI), 500–550 nm (Alexa Fluor 488), 592–642 nm (Alexa Fluor 594), and 658–720 nm (Atto 647N). Two-color gSTED imaging of Ape1 and GAPDH in the nuclear compartment was performed in a sequential mode by using a single 755-nm depletion laser. The time-gated detection was limited to a time window from 0.5 to 6.5 ns. Emission band-pass was set to 645–720 nm for Atto 647N and to 551–600 nm for Alexa Fluor 594. Pixel size was 21.6 nm. Confocal and gSTED images were acquired and examined with the LAS X software (Leica Microsystems). STED images were subjected to the colocalization analysis by using the Coloc 2 plugin in Fiji (ImageJ; National Institutes of Health, Bethesda, MD, USA) software. The plugin computed Spearman’s and Pearson’s colocalization indices (20) and generated colocalization pixel maps. We found no detectable Alexa Fluor 594 fluorescence in cells that were stained only with GAPDH Ab, and no Atto 647N fluorescence was detected in Ape1-stained cells showing negligible bleed-through between channels. We used single Ab-stained cells also as a negative control for STED imaging and colocalization analysis. We obtained negligible value of colocalization indices for cells that were stained only with GAPDH Ab (Pearson’s coefficient, 0.005; Spearman’s coefficient, 0.0025) and for cells that were stained only with Ape1 Ab (Pearson’s coefficient, 0.025; Spearman’s coefficient, 0.025).
Surface plasmon resonance
Real-time binding interaction of GAPDH with Ape1 was measured with surface plasmon resonance (SPR) technology by using a BiaCore T100 optical biosensor, NTA sensor chip, and BiaEvaluation software (BiaCore GE Healthcare, Pittsburgh, PA, USA). Recombinant Ape1 (20 µg/ml) was immobilized by amine coupling (BiaCore Amine Coupling Kit) according to manufacturer instructions, and the remaining active carboxylic residues were neutralized with ethanolamine. The sensor chip then was washed and equilibrated in 0.01 M HEPES, pH 7.4, 0.15 M NaCl, 3 mM EDTA, and 0.005% Surfactant P20 (HBS-EP; GE Healthcare) at a flow rate 20 μl/min, and GAPDH was introduced at 8 serial dilutions (1, 5, 10, 25, 100, 250, 500, and 1000 nM) in HBS-EP buffer (72-μl injection volumes at a flow rate of 20 μl/min). Binding was measured at 25°C for 7 min, followed by 10 min dissociation with washing buffer. One minute of sensor chip regeneration was performed with Gentle Ag/Ab elution buffer (Thermo Fisher Scientific). Sensorgram response data were analyzed and kinetic constants were calculated with the BiaEvaluation kinetics package. We ran mouse Ape1 mAb (sc-136057; Santa Cruz Biotechnology) and lactate dehydrogenase as a positive and negative control for SPR, respectively. Ape1 Ab bound Ape1 with high affinity (Kd = 14.4 nM, consistent with the value provided by the manufacturer). No measurable binding with Ape1 was detected for lactate dehydrogenase (data not shown).
Ape1 activity assay
The 5′-endonuclease activity of Ape1 was analyzed by using a quantitative in vitro assay that measured the incision of a 26-mer FAM-labeled oligonucleotide substrate that contained a single synthetic AP site [tetrahydrofuran (F)] (21). To create double-stranded DNA substrates, equal volumes of 200 µM solutions of the respective and complimentary strands [5′-FAM-AATTCACCGGTACG(F)ACTAGAATTCG-3′ and 5′-CGAATTCTAGTCCGTACCGGTGAATT-3, respectively] were mixed in annealing buffer (50 mM Tris, pH 7.5, 25 mM NaCl, 2 mM MgCl2, 1 mM DTT, and 0.01% Tween-20) and incubated at 95°C for 5 min. To assay the endonuclease activity of Ape1, cells were harvested in buffer (50 mM Tris-HCL, pH 7.4, 30 mM KCl, and 10% glycerol) that was supplied with protease inhibitors (Complete Ultra tablets; Roche) and lysed by slow shaking at 4°C for 30 min. Ape1 endonuclease reaction mix contained 300 ng cell lysate and 1 μM FAM-labeled Ape1 substrate in endonuclease IV reaction buffer (Thermo Fisher Scientific) that was supplied with 2 mM MgSO4. Mix was incubated at 37°C for 12 min, and reaction was stopped by adding 2× RNA gel loading buffer (Thermo Fisher Scientific). Samples were heated at 95°C for 5 min to dissociate the labeled oligonucleotide. Reaction products were separated on 15% PAGE/urea gels and imaged on FLA-7000 phosphoimager (Laser 473 nm, filter 520 nm; FujiFilm, Tokyo, Japan) to quantify the FAM signal. Endonuclease activity was calculated as the relative amount of the 13-mer oligo product with the unreacted 26-mer substrate [product/(product + substrate)].
Small interfering RNA
Cells (0.5–1.0 × 106) were harvested and transfected with small interfering RNA (siRNA) at a final concentration of 10 nM with GeneMute siRNA transfection Reagent (SignaGen Laboratories, Rockville, MD, USA) according to manufacturer protocol. siRNA used in this study was Silencer Select positive control GAPDH siRNA and Silencer Select negative control No 1 siRNA (both from Thermo Fisher Scientific), On-Target Plus rat APEX1 siRNA (79116; Dharmacon, Lafayette, CO, USA), nonspecific scrambled siRNA control (On-Target Plus control siRNA – SMARTpool; Dharmacon), and rat HOXA5 siRNA (Thermo Fisher Scientific). Medium was replaced 8 h after transfection and the efficiency of siRNA transfection was confirmed by immunoblotting with Ape1, HOXA5, or GAPDH Ab.
Statistical Analysis
Data are presented as means ± sem. Statistical analysis was performed by using 1-way ANOVA or Student’s t test, with a value of P < 0.05 considered significant.
RESULTS
GAPDH suppressed cell apoptosis and prevented oxidative DNA damage via Ape1 endonuclease
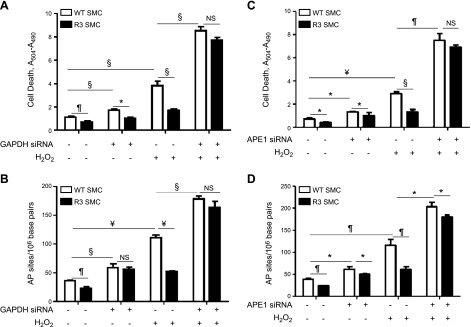
We have previously shown that the proatherogenic molecule, OxLDL, down-regulated GAPDH in SMCs via an H2O2-dependent mechanism (14). Both OxLDL and H2O2 are potent inducers of SMC apoptosis (16, 22). To investigate a potential role for GAPDH in oxidant-induced cell apoptosis, GAPDH expression was targeted by siRNA, or, alternatively, GAPDH was overexpressed in R3 SMCs. We confirmed that GAPDH-targeted siRNA decreased GAPDH expression by >70% in both wild-type (WT) and R3 SMCs, and that R3 cells had a 1.5-fold increase (P < 0.05) in GAPDH protein vs. control cells (Supplemental Fig. 1). H2O2 dose-dependently reduced GAPDH protein levels in WT and R3 cells without changes in the expression level of β-actin (Supplemental Fig. 2). Of importance, R3 cells retained significantly higher GAPDH levels compared with WT SMCs under treatment with H2O2 (110 μM H2O2, R3: 0.71 ± 0.06; WT: 0.34 ± 0.03). H2O2 (110 μM for 16 h) induced significant cell apoptosis in WT cells, and silencing GAPDH potentiated H2O2-induced cell death by ∼2-fold (Fig. 1A); however, H2O2-induced cell apoptosis was significantly blocked in R3 cells, which demonstrated that GAPDH protects SMCs against oxidant-induced cell death.
Figure 1.
GAPDH suppressed cell apoptosis and prevented oxidative DNA damage via Ape1 endonuclease. WT or GAPDH-overexpressing R3 SMCs were transfected with GAPDH-targeted siRNA (A, B), Ape1-targeted siRNA (C, D), or with control (scrambled) siRNA, and cells were exposed to 110 μM H2O2 for 16 h. Cell death was measured with Cell Death ELISA (A, C), and AP site number was quantified by OxiSelect Oxidative DNA damage ELISA (B, D). Data from a representative experiment are shown. Each experiment was repeated at least 3 times. NS, not significant. *P < 0.05, ¶P < 0.01, §P < 0.005, ¥P < 0.001.
Because GAPDH binds and reactivates oxidized Ape1 in human colon carcinoma cells (12), we hypothesized that interaction of GAPDH with Ape1 would mediate the GAPDH-induced antiapoptotic effect. At first, we tested the role of GAPDH in DNA repair by quantifying the number of AP sites (Fig. 1B). GAPDH down-regulation by siRNA significantly increased the basal number of AP sites in WT cells compared with controls, which suggested enhanced DNA damage. GAPDH overexpression in R3 cells reduced the number of AP sites. H2O2 markedly up-regulated AP site formation in WT SMCs; however, this effect was completely blocked in R3 cells (Fig. 1B), which indicated that GAPDH protected DNA against oxidant-induced damage. We next explored a similar approach to test the involvement of Ape1 in the effects of GAPDH on DNA damage and apoptosis. Ape1 siRNA, but not scrambled siRNA (control), significantly decreased Ape1 expression in both WT and R3 cells (Supplemental Fig. 1). Ape1 down-regulation slightly increased basal AP site numbers and induced apoptosis in WT cells (Fig. 1C, D). H2O2 induced a >3-fold increase in AP sites in WT cells and corresponding elevation in apoptosis. Furthermore, cell treatment with Ape1 siRNA potentiated H2O2-induced increase in AP sites and apoptosis. These data show that maintenance of sufficient Ape1 levels is required to prevent oxidative DNA damage and cell death. R3 cells had decreased AP site numbers and reduced cell apoptosis compared with WT control. H2O2 induced approximately 50% fewer AP sites and a corresponding reduction in apoptosis in R3 cells compared with WT cells. These results indicate that GAPDH overexpression in R3 cells suppresses both oxidant-induced DNA damage and apoptosis.
To determine whether Ape1 was responsible for reduced sensitivity of R3 cells to treatment with oxidant, Ape1 was silenced, and cells were exposed to H2O2. Ape1 down-regulation partially blocked the resistance of R3 cells to oxidant-induced DNA damage and completely eliminated R3 cells, resistance to H2O2-elicited apoptosis (Fig. 1C, D). These data demonstrate that GAPDH-induced antiapoptotic effects depend on Ape1 expression.
GAPDH bound Ape1 in vitro, and blocking (or oxidation) of GAPDH active site cysteines reduced GAPDH/Ape1 binding
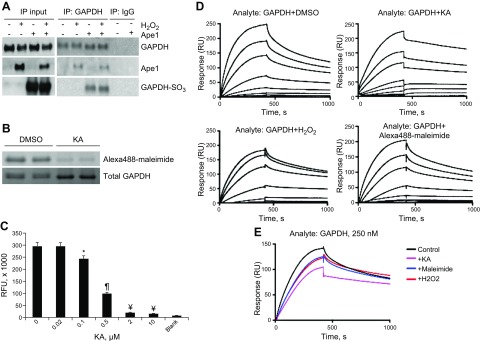
The GAPDH molecule contains 3 cysteines, 2 of which (C149 and C153) are located in the GAPDH active site (13). Oxidative stress induces the transformation of these cysteines from reduced (–SH) to oxidized form (–SOH or –SO3), which results in reduced enzyme activity and function (23–25). Here we determined whether GAPDH binding to Ape1 depends on GAPDH active site cysteines. We incubated 1.35 μM GAPDH with an equimolar amount of Ape1. Protein mixture was exposed to H2O2 (0.5 mM for 15 min) and immunoprecipitated with GAPDH Ab. A strong Ape1 band was detected in the immunoprecipi-tate, showing that GAPDH binds Ape1 (Fig. 2A). To confirm that cell treatment with H2O2 oxidized GAPDH active site cysteines, GAPDH samples were further immunoblotted with Ab generated against oxidized GAPDH active sites (GAPDH-SO3 Ab) (26). H2O2 induced the appearance of a strong GAPDH-SO3 band on immunoblots, which indicated oxidation of GAPDH essenial cysteines. GAPDH/Ape1 binding was also confirmed by standard ELISA method. GAPDH (1–10 μg/ml) strongly bound Ape1 immobilized on the ELISA plate, and vice versa, Ape1 bound to GAPDH immobilized on the plate (Supplemental Fig. 4).
Figure 2.
GAPDH bound Ape1 in vitro, and blocking (or oxidation) of GAPDH active site cysteines reduced GAPDH/Ape1 binding. A) Human recombinant GAPDH was preincubated with an equimolar amount of human recombinant Ape1, protein mixture was exposed to H2O2, and samples were immunoprecipitated (IP) with rabbit GAPDH Ab or with the same amount of rabbit IgG (control). Immunoprecipitate was submitted for immunoblotting with GAPDH, GAPDH-SO3, and Ape1 Ab. Data from a representative immunoprecipitation experiment are shown (n = 3). B) KA bound to GAPDH cysteines. GAPDH was incubated with 2 μg/ml KA or with DMSO (control), and GAPDH cysteines were labeled with 700 μM thiol-reactive probe Alexa Fluor 488-maleimide in PBS for 2 h at room temperature. To visualize Alexa Fluor 488 signal, samples were loaded on 12% mini-protean TGX protein gel and imaged with Gel Doc system. Total GAPDH level was quantified with SYPRO Ruby protein gel stain. Technical replicas are shown. Experiment was repeated 3 times. C) Human recombinant GAPDH (50 μg/ml) was incubated with KA (0–10 ug/ml) in PBS for 2 h, and GAPDH enzyme activity was measured with KD Alert GAPDH assay kit by using the procedure with real-time kinetic measurement. Fluorescence reading was assessed over a 10-min interval with data collected every 1 min. GAPDH activity in each sample was defined as the fluorescence increase over 5 min and was recalculated as activity units using a standard curve obtained with the kit provided with the GAPDH enzyme standard (n = 8 per each KA dose). *P < 0.05, ¶P < 0.01, ¥P < 0.001 vs. no KA control. D) Recombinant Ape1 (20 μg/ml) was immobilized on the BiaCore NTA sensor chip and used as a ligand for SPR experiment. Human recombinant GAPDH was preincubated with H2O2, KA, or DMSO (control) or with Alexa Fluor 488-maleimide, and GAPDH analyte was introduced to sensor chip at 8 serial dilutions (1, 5, 10, 25, 100, 250, 500, and 1000 nM). Sensorgrams for a representative experiment are shown. E) Single sensogram for control and GAPDH-treated samples (250 nM dilution) were combined into 1 graph for comparison. Ape1 immobilization on NTA chip was repeated twice, and all analytes were run over each chip at least 3 times.
To further evaluate the importance of GAPDH cysteines in GAPDH/Ape1 interaction, we quantified real-time binding of GAPDH to Ape1 by using SPR technology, with Ape1 immobilized on the SPR sensor chip (Fig. 2D, E). Kd (equilibrium dissociation constant) was calculated to evaluate binding affinity of control (or treated) GAPDH sample with Ape1 (Table 1). Relative increase in Kd indicates reduced binding affinity. GAPDH bound Ape1 with a Kd of 259 ± 20 nM. To determine whether oxidation of GAPDH essential cysteines alters GAPDH/Ape1 binding, GAPDH was incubated with H2O2 at the dose and time shown above to oxidize cysteines, and Kd was measured by SPR. GAPDH with oxidized cysteines had reduced binding affinity to Ape1 (Kd = 349 ± 13 nM) compared with control (Fig. 2E), which indicated that GAPDH/Ape1 binding depends on C149 and C153. To further confirm the role of GAPDH active site cysteines in the formation of the GAPDH/Ape1 complex, GAPDH was preincubated with the thiol-reactive probe Alexa Fluor 488-maleimide or with KA. KA is a specific GAPDH inhibitor that has been reported to bind to GAPDH C149 and C153 (27, 28), and KA is shown to induce DNA fragmentation and apoptosis in neuronal cells (29). To prove that KA directly binds to GAPDH cysteines, recombinant GAPDH was preincubated with 2 μg/ml KA and protein cysteines were labeled with Alexa Fluor 488-maleimide. GAPDH cysteine labeling was decreased by treatment with the inhibitor, which indicated that KA binds GAPDH cysteines (Fig. 2B). We also confirmed that KA dose-dependently suppressed GAPDH activity in SMCs. Cell treatment with 2 μg/ml KA significantly inhibited GAPDH glycolytic activity by 93% (Fig. 2C). GAPDH samples that were preincubated with Alexa488-maleimide or with KA were used for SPR to assess GAPDH-Ape1 binding. Blocking of GAPDH cysteines either by Alexa Fluor 488-maleimide or KA markedly increased Kd, which suggested reduced GAPDH/Ape1 binding affinity. Table 1 summarizes Kd values obtained for Ape1 binding with control GAPDH or GAPDH with blocked cysteines. It is important to note that either GAPDH oxidation or blocking of cysteines decreased the SPR sensogram Rmax, reduced protein association rate, and increased dissociation rate compared with control GAPDH, which indicated decreased surface binding capacity of GAPDH.
TABLE 1.
The equilibrium dissociation constant (Kd) between the control (or treated) GAPDH and Ape1
P < 0.05 vs. control.
GAPDH bound Ape1 in the SMC nucleus, and treatment with H2O2 disrupted the nuclear GAPDH/Ape1 complex
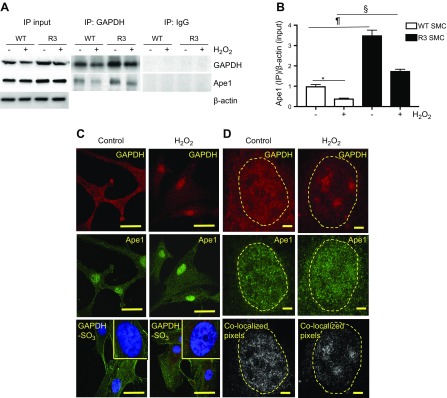
GAPDH is primarily a cytosolic protein; however, it is also found in the nucleus in untreated cells and GAPDH translocated into the nucleus after cell stress (30). In contrast, Ape1 localizes mainly in the nucleus, and some amount is also found in association with mitochondrial DNA (31). Ape1 was coimmunoprecipitated with GAPDH in whole-cell lysates that were obtained from WT and R3 SMCs, which showed that GAPDH binds Ape1 in vivo (Fig. 3A). As expected, GAPDH was down-regulated in H2O2-treated cells. H2O2 also decreased the amount of Ape1 coimmunoprecipitated with GAPDH. Of importance, H2O2-treated R3 SMCs retained a significantly higher Ape1 level in GAPDH immunoprecipitate compared with control cells (Fig. 3A, B).
Figure 3.
GAPDH bound Ape1 in the SMC nucleus, and treatment with H2O2 disrupted nuclear GAPDH/Ape1 complex. A, B) WT and R3 SMCs were grown to confluence and treated with H2O2. A) For immunoprecipitation (IP), total cell lysate was incubated with rabbit GAPDH Ab or with rabbit IgG (control), and immunoprecipitate was immunoblotted with GAPDH and Ape1 Ab. B) Quantitative densitometry data show the Ape1 amount in GAPDH immunoprecipitate (n = 3). *P < 0.05, ¶P < 0.01, §P < 0.005. C, D) Conventional confocal (C) and gSTED super-resolution (D) microscopy. SMCs were incubated with or without 110 μM H2O2 for 6 h, fixed in methanol, permeabilized with 0.5% Triton X-100, and incubated with GAPDH (red) and Ape1 (green) Ab plus DAPI. D) gSTED was performed with a Leica STED microscope equipped with a ×100/1.4 NA oil-immersion plan-apochromatic objective lens. STED images of the nuclear compartment of individual cell without (n = 9) and with H2O2 treatment (n = 13) were used for generation of colocalization pixel map (colocalized pixels shown in white) and for quantification of colocalization coefficients (Pearson’s and Spearman’s) with Fiji software. Dashed line outlines cell nucleus shape. Scale bars: 30 μm (C), 3 μm (D). *P < 0.05 vs. control.
GAPDH and Ape1 localization in the cytoplasmic and nuclear compartments of SMCs after a short-term exposure to H2O2 (110 µM for 6 h) was visualized by conventional confocal microscopy (Fig. 3C) and also by gSTED super-resolution microscopy (Fig. 3D). gSTED microscopy provides more precise detection of colocalized molecules than conventional confocal microscopy (32). In untreated SMCs, immunoreactive GAPDH was detected in the cytosol and nucleus. As expected, H2O2 induced marked GAPDH translocation to the nucleus, where it formed clusters. In control and H2O2-treated cells, immunoreactive Ape1 was localized in the nucleus. Six hours after treatment with H2O2, essential cysteines of GAPDH were strongly oxidized, as was evident by the appearance of multiple GAPDH-SO3 spots in the nucleus and in cytosol (Fig. 3C). GAPDH and Ape1 were colocalized exclusively in the nuclei of untreated and H2O2-treated cells (Fig. 3D). Pearson’s and Spearman’s correlation coefficients (20) were computed to quantify nuclear colocalization of GAPDH and Ape1 (Table 2). Treatment with H2O2 decreased both correlation coefficients by approximately 50%, which suggested a reduction in nuclear GAPDH/Ape1 binding. Overall, microscopy data, along with in vitro results, suggest that GAPDH binds Ape1 in the nucleus and that GAPDH essential cysteines mediate GAPDH/Ape1 binding.
TABLE 2.
Pearson’s and Spearman’s coefficients quantifying nuclear colocalization of GAPDH and Ape1
| Treatment | Pearson’s | Spearman’s |
|---|---|---|
| Control | 0.372 ± 0.024 | 0.363 ± 0.026 |
| H2O2 | 0.247 ± 0.021* | 0.267 ± 0.024* |
P < 0.05 vs. control.
Blocking GAPDH cysteines potentiated cell death
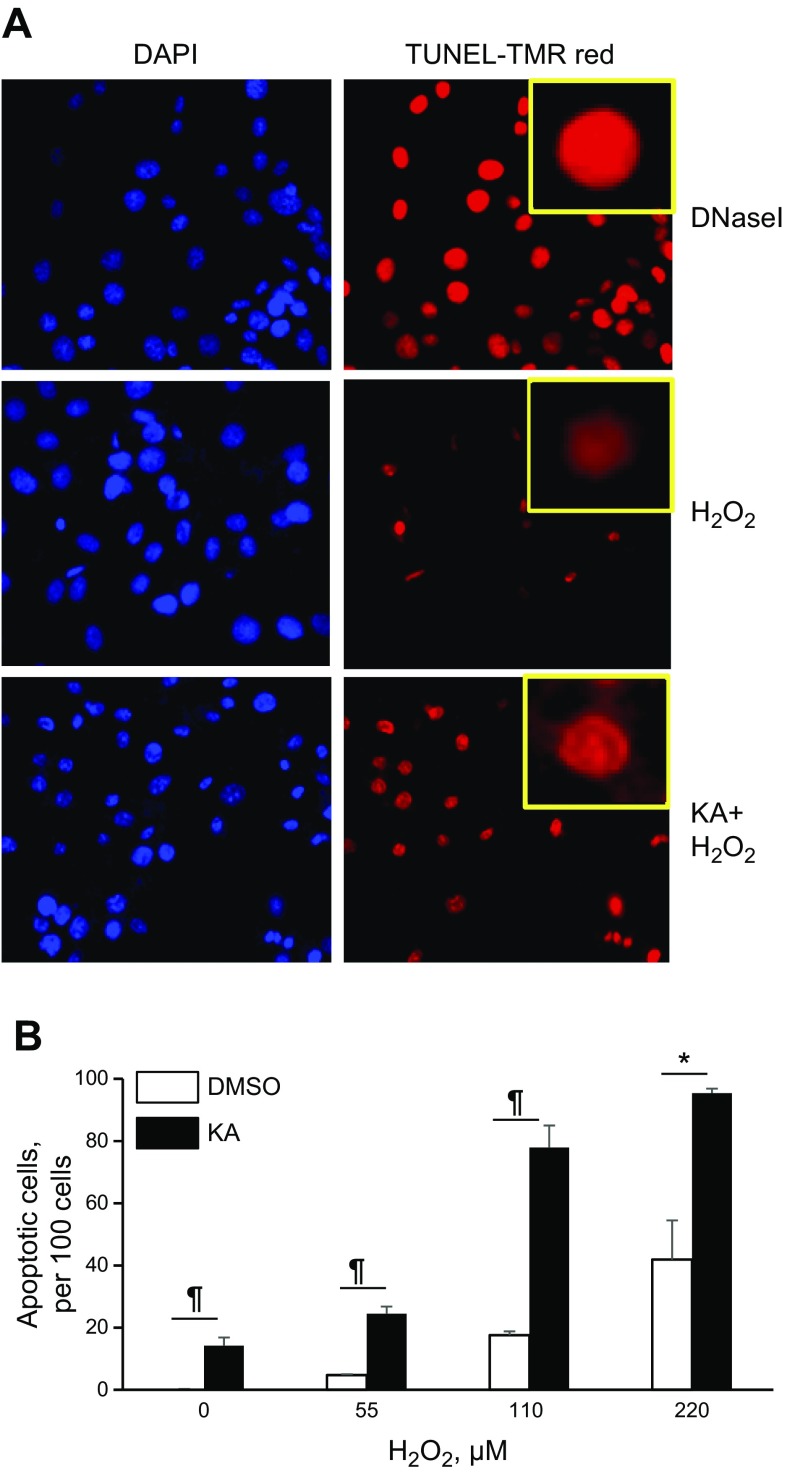
We showed that GAPDH essential cysteines mediate binding with Ape1 and that treatment with KA suppressed their interaction. To determine whether blocking GAPDH cysteines potentiated apoptosis, SMCs were preincubated with KA, and H2O2-induced apoptosis was quantified with TUNEL assay. H2O2 dose-dependently (55–220 μM) increased the number of TUNEL-positive cells, and pretreatment with KA markedly potentiated cell death, which demonstrated the crucial role that GAPDH cysteines play in the prevention of apoptosis (Fig. 4). We noticed that KA-treated SMCs have approximately 5× stronger TUNEL signal per individual cell compared with control cells [KA, TUNEL signal/cell, 4.44 ± 0.10 RU; control (DMSO), 0.86 ± 0.03 RU; P < 0.001]. We observed a similar increase in TUNEL signal/cell after SMC treatment with increasing doses of DNase I (Fig. 4A), an endonuclease that is known to produce single breaks (nicks) in DNA (33). Considering that the TUNEL assay is based on measuring the number of apoptosis-induced nicks in DNA (34), these results suggest that blocking GAPDH cysteines potentiated oxidative DNA damage.
Figure 4.
Blocking GAPDH cysteines with KA potentiated cell death. A) SMCs were grown in 8-well chamber slide, preincubated with 2 μg/ml KA, and exposed to H2O2 (0–220 μM, 16 h). For quantification of cell death, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100, and incubated with TUNEL-TMR mix for 45 min. In parallel, fixed and permeabilized SMCs were incubated with DNase I (0.2 U/μl for 20 min at 37°C) and apoptotic cells were identified by incubation with TUNEL-TMR. KA strongly increased nuclear TMR signal per cell similar to the effect of DNase I (insert). TUNEL-positive cell number was measured in 4 viewing fields per well and normalized per 100 DAPI-positive cells. B) Quantitative data are shown for a representative experiment (n = 4). *P < 0.05, ¶P < 0.01.
GAPDH activated Ape1
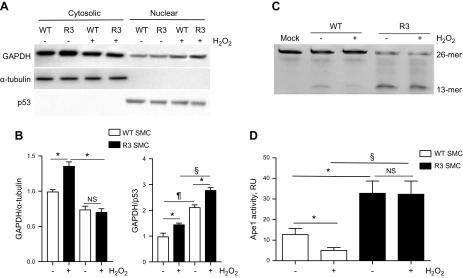
We have shown that H2O2-treated R3 cells had an elevated amount of Ape1 in the GAPDH/Ape1 complex (Fig. 3A, B). To confirm that these cells also had increased nuclear GAPDH levels and up-regulated Ape1 activity, we separated WT and R3 cells, cytosolic and nuclear fractions (Fig. 5). R3 cells had a basal level of nuclear GAPDH similar to WT cells; however, H2O2-treated R3 SMCs had a significantly increased level of nuclear GAPDH compared with H2O2-treated WT SMCs (Fig. 5A, B). It has been shown that GAPDH binding to Ape1 allows GAPDH to convert the oxidized species of Ape1 to the reduced form, thereby reactivating its endonuclease activity to cleave AP sites (12). To directly assess 5′-endonuclease activity of Ape1, we used a quantitative in vitro assay that measures the incision of a 26-mer FAM-labeled oligonucleotide substrate that contains a single synthetic AP site (21). Ape1 activity measurement was performed within linear range of reaction (Supplemental Fig. 3A). Ape1 activity was negligible in cell cytosolic fraction (data not shown). We found that R3 cells had basal 2.6-fold increase in nuclear Ape1 activity (P < 0.05) compared with WT cells (Fig. 5C, D). H2O2 suppressed Ape1 activity by a significant 60% in WT cells; however, H2O2 did not inhibit Ape1 activity in R3 cells. Of importance, Ape1 activity was significantly higher in the nucleus of H2O2-treated GAPDH-overexpressing cells vs. H2O2-treated WT cells, which indicated that GAPDH activated Ape1. Essentially, similar data was obtained when Ape1 activity was assessed in total cellular extracts (Supplemental Fig. 3B). We confirmed that H2O2 (110 μM for 16 h) induced no change in Ape1 expression in both WT and R3 cells (see Fig. 3A, immunoprecipitation input).
Figure 5.
GAPDH activated Ape1. WT and R3 SMCs were grown until confluence and treated with H2O2 (110 μM, 16 h). Cytosolic and nuclear fractions of WT and R3 cells were separated with NE-PER nuclear and cytoplasmic separation kit. A) Samples were immunoblotted with Ab for GAPDH, α-tubulin (cytosolic marker), and p53 (nuclear marker). B) Quantitative data show relative GAPDH amount in cytosolic and nuclear fraction (n = 3). *P < 0.05, ¶P < 0.01, §P < 0.005. C, D) Ape1 activity in nuclear fraction of WT and R3 cells. C) The 5′-endonuclease activity of Ape1 was quantified by measuring the incision of a 26-mer FAM-labeled oligonucleotide substrate that contained a single synthetic AP site. Ape1 endonuclease reaction mix contained 300 ng cell lysate and 1 μM FAM-labeled Ape1 substrate. Products were separated on 15% PAGE/Urea gels. D) Endonuclease activity was calculated as the relative amount of the 13-mer oligo product with the unreacted 26-mer substrate [product/(product + substrate)]. Data from a representative experiment are shown (n = 5). NS, not significant. *P < 0.05, §P < 0.005.
GAPDH up regulated Ape1 expression via a transcription factor HOXA5-dependent mechanism
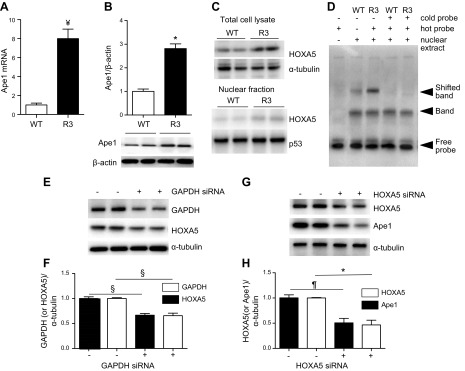
Nuclear GAPDH plays a role in transcriptional regulation of cell-cycle proteins in vivo and in vitro (35). We assessed a potential GAPDH effect on Ape1 protein and mRNA expression in R3 cells. R3 cells had a significant 2.7-fold increase in Ape1 protein levels accompanied by a 7-fold increase in mRNA expression (Fig. 6A, B). GAPDH-targeted siRNA reduced Ape1 protein expression in WT cells, which confirmed Ape1 regulation by GAPDH (data not shown). To identify transcription factors that mediate GAPDH-induced Ape1 up-regulation, we measured relative mRNA levels of 84 transcription factors in R3 vs. WT SMCs. mRNA levels of several transcription factors were significantly altered in GAPDH-overexpressing cells, including Egr1, Hand1, Max, Nfatc2, Smad1, and Tp53 (Supplemental Table 1). We found a dramatic ∼500-fold up-regulation of HOXA5 in R3 cells. R3 cells had increased HOXA5 protein levels in both cytosolic (32% increase vs. control; P < 0.05) and nuclear fractions (63% increase vs. control; P < 0.05), and these cells had a ∼3-fold increase in HOXA5 DNA-binding activity (Fig. 6C, D). To test whether a specific reduction in GAPDH would down-regulate HOXA5 and to investigate whether a decrease in HOXA5 would change Ape1 expression, SMCs were transfected with GAPDH-targeted siRNA or with HOXA5-targeted siRNA, respectively. GAPDH-targeted siRNA significantly decreased HOXA5 levels and HOXA5-targeted siRNA down-regulated Ape1 (Fig. 6E–H), which showed that GAPDH is upstream of HOXA5 and that Ape1 is downstream of HOXA5. Thus, these data suggest that GAPDH up-regulated Ape1 via HOXA5-dependent signaling.
Figure 6.
GAPDH up-regulated Ape1 expression via a transcription factor HOXA5-dependent mechanism. A, B) Ape1 mRNA (A) and protein (B) levels in WT and R3 SMCs were quantified by RT-PCR with rat APEX1 gene-specific primers and immunoblotting with Ape1 Ab, respectively. *P < 0.05, ¥P < 0.001 vs. WT cells (n = 3). C) Nuclear fraction of WT and R3 cells was isolated with NE-PER separation kit, and total cell lysate and nuclear fraction were used for immunoblotting with HOXA5 Ab (n = 3). D) Electrophoretic mobility shift assay (EMSA) was performed to determine binding HOXA5 with its corresponding DNA consensus sequences. Nuclear extract of WT and R3 cells was prepared to form transcription factor-DNA complexes. The extent of binding was assayed by gel shift on a 4% nondenaturing PAGE, followed by HOXA5-specific probe labeling on a membrane (n = 3). E–H) SMCs were transfected either with GAPDH-targeted siRNA (E, F) or with HOXA5-targeted siRNA (G, H), and protein level of GAPDH, HOXA5, Ape1, and α-tubulin was measured by immunoblotting (E, G) (n = 3 per each siRNA experiment). Quantitative densitometry data shown (F, H). *P < 0.05, ¶P < 0.01, §P < 0.005.
Reduced GAPDH levels in atherosclerotic plaque SMCs are associated with oxidative stress and apoptosis
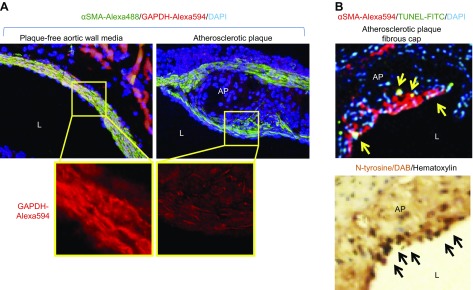
We have shown that oxidant-induced GAPDH down-regulation mediated SMC apoptosis. To test whether these results are relevant to in vivo conditions, such as in atherosclerotic plaque, we obtained aortic valve cross-sections from ApoE-null mice that were fed with high-cholesterol (Western type) diet, a well-established animal model of atherosclerosis (19). We performed immunohistochemistry for GAPDH, N-tyrosine (a marker of oxidative stress), α-smooth muscle actin (a marker of SMC), and visualized apoptotic SMCs by TUNEL assay (Fig. 7). GAPDH immunopositivity was markedly reduced in SMCs in the fibrous plaque cap vs. adjacent SMCs in plaque-free aortic medium. Several SMCs in the plaque cap were apoptotic and had increased levels of N-tyrosine. These data show that GAPDH down-regulation correlated with increased oxidative stress and apoptosis in atherosclerotic plaque SMCs.
Figure 7.
Reduced GAPDH level in atherosclerotic plaque SMCs was associated with oxidative stress and apoptosis. ApoE-null mice (n = 4) were fed a Western-type diet for 12 wk, then humanely killed under anesthesia, and the heart, together with aortic arch, was dissected, fixed, and paraffin embedded. Serial sections (6 μm) were taken throughout the entire aortic valve area and used for immunohistochemistry with GAPDH, α-smooth muscle actin (αSMA; SMC marker), and N-tyrosine (oxidative stress marker) Abs and stained with In Situ Cell Death Detection TUNEL-FITC kit. A) GAPDH immunopositivity (red) was reduced in SMCs from plaque cap (green) vs. adjacent SMCs in plaque-free aortic medium. B) Several SMCs in the atherosclerotic plaque (AP) fibrous cap were apoptotic (yellow arrows) and had increased levels of N-tyrosine (black arrows). L, lumen. Representative data are shown.
DISCUSSION
Here, we described the novel antiapoptotic mechanism that involves the interaction of nuclear GAPDH and Ape1 endonuclease. GAPDH is implicated in numerous cell functions; however, its role in cell death remains controversial and poorly understood (36). Whereas some reports indicate a proapoptotic function for GAPDH (37–39), others demonstrated a protective role (40–43). The current paradigm considers GAPDH as a master switch in the regulation of cell death (44). We have provided the evidence of a novel prosurvival role of GAPDH in vascular SMCs. We have demonstrated that GAPDH protects SMCs against oxidant-induced cell death by preserving genomic DNA integrity. We have identified a mechanism that links GAPDH-dependent DNA protection to an antiapoptotic effect. It involves the direct binding of nuclear GAPDH with Ape1 endonuclease, GAPDH-dependent Ape1 up-regulation, regeneration of Ape1 activity, and stimulation of DNA repair. Our data are in line with reports that suggest an important protective function of nuclear GAPDH. For example, GAPDH has been shown to exert prosurvival effects in HeLa cells by blocking cytochrome C release via a mechanism that involved GAPDH’s nuclear functions (17).
GAPDH is involved in several important nuclear pathways, such as nuclear membrane fusion, maintenance of telomere structure, and transcriptional control of histone expression (13). GAPDH’s nuclear role could be modulated by cellular stress that causes GAPDH translocation (45). In our study, we detected a significant amount of nuclear GAPDH in untreated SMCs by immunoblotting of nuclear fractions and also by super-resolution imaging of the nuclear compartment. We showed that GAPDH binds Ape1 endonuclease in the nuclei of SMCs and that GAPDH enhances 5′-endonuclease activity of Ape1. We found that GAPDH translocates into SMC nucleus upon oxidative stress. As we initially hypothesized, an increase in nuclear GAPDH levels should stimulate its nuclear function and potentially enhance GAPDH/Ape1 binding. To our surprise, nuclear GAPDH/Ape1 colocalization was markedly reduced in H2O2-treated SMCs. To investigate the mechanism, we focused our study on the role of GAPDH active site cysteines in mediating GAPDH/Ape1 interaction and investigated GAPDH effect on Ape1 activity and expression.
Modification of GAPDH active site cysteines directs GAPDH toward the proapoptotic pathway (25, 46). We demonstrated that preserving GAPDH active site cysteines in an unmodified form (i.e., in reduced –SH form) is an important prerequisite for binding nuclear GAPDH with Ape1. Treatment with H2O2 induced GAPDH C149 and C153 oxidation and reduced binding with Ape1, which suggested that essential cysteines are necessary for formation of the GAPDH/Ape1 complex. We noticed the appearance of large GAPDH-SO3 aggregates in H2O2-treated cells, which correlated with decreased nuclear GAPDH/Ape1 colocalization. Nakajima et al. (46) observed similar amyloid-like GAPDH aggregates in human neuroblastoma cells as a result of the formation of S-S bonds between essential GAPDH cysteines. We suggest that either transformation of essential cysteines into –SO3 form or generation of S-S bonds reduce the availability of GAPDH –SH groups to interact with Ape1. We found that KA [a GAPDH blocker that binds to GAPDH active site cysteines (28, 47)] reduced GAPDH binding with Ape1, and that KA also potentiated H2O2-induced SMC apoptosis. Overall, our results reveal a novel and an important role of nuclear GAPDH, namely, enzyme involvement in SMC protection against oxidant-induced cell death via formation of the GAPDH/Ape1 complex.
It has been shown that the preservation of Ape1 activity prevents oxidative DNA damage and suppresses cell apoptosis in vitro and in vivo (48–51). Here, we introduce the innovative hypothesis that the GAPDH/Ape1 complex preserves Ape1 activity, thereby stimulating DNA repair and suppressing cell death. Ape1 down-regulation abolished the resistance of GAPDH-overexpressing R3 cells to DNA damage and apoptosis, which showed that Ape1 is indispensable for GAPDH-dependent protective effects. We described 2 major mechanisms that contribute to GAPDH-dependent Ape1 activation. We now have shown that reduced C149 and C153 mediate GAPDH binding with Ape1 and inhibition of apoptosis. Consistent with our data, Azam et al. (12) have reported that the endonuclease activity of H2O2-inactivated Ape1 was rapidly regenerated by the addition of GAPDH but not by supplementation with antioxidants or sulfhydryl group–recovering agent, such as DTT, which suggested that GAPDH specifically reduces Ape1. We hypothesize that GAPDH essential cysteines directly reduce Ape1 active site residues, which results in its reactivation. We found that GAPDH overexpression increased Ape1 level and that Ape1 up-regulation may also contribute to Ape1 activation. Of interest, H2O2-treated R3 SMCs had a relatively larger amount of Ape1 in the GAPDH/Ape1 complex compared with control; however, it is unknown whether an increase in the nuclear pool of Ape1 would be sufficient to regenerate Ape1 under conditions in which formation of the GAPDH/Ape1 complex is blocked. In fact, we demonstrated that KA blocked formation of the GAPDH/Ape1 complex in vitro and that KA also potentiated cell death, which indicated the predominant role of the GAPDH/Ape1 complex in the prevention of apoptosis.
An additional novel finding from our report is that GAPDH increased Ape1 expression via a HOXA5-dependent mechanism. HOXA5 transcription factor has been reported to play important roles in embryogenesis, hematopoiesis, and tumorigenesis (52, 53), and HOXA5 gene is implicated in 5-aza-2'deoxycytidine–induced inhibition of atherosclerosis (54). Recently, it has been shown that HOXA5-specific siRNA induced apoptosis and cell-cycle arrest in Jurkat cells (55); however, the underlying mechanisms were not identified. Our demonstration that HOXA5-specific siRNA suppressed Ape1 expression provides a potential mechanism. We believe that the identification of HOXA5 as a novel molecule that is involved in the prevention of cell death could have further clinical implications.
Elevated DNA damage is involved in the induction of cell apoptosis and progression of atherosclerosis (7). Here, we report that GAPDH reduced oxidative DNA damage and protected cultured SMCs against oxidative stress–induced apoptosis. We also found decreased GAPDH levels in the atherosclerotic plaque fibrous cap and that GAPDH down-regulation correlated with increased plaque SMC apoptosis and oxidative stress. Fibrous cap thinning and loss of viable SMCs are hallmarks of human vulnerable plaques (56), and, therefore, suppression of SMC apoptosis is a potential clinically relevant atheroprotective strategy. Our data suggest that Ape1 activation via preservation of the nuclear GAPDH/Ape1 interaction and/or via a HOXA5-dependent mechanism is a potential approach to improve atherosclerotic plaque stability. Increased levels of oxidative DNA damage have also been implicated in various age-related diseases, such as Parkinson’s disease and Alzheimer’s disease, as well as in the general aging process (57, 58), and GAPDH levels are reported to be decreased by aging (59). Of interest, it has been reported that aging differentially regulates nuclear GAPDH vs. perinuclear/cytosolic GAPDH protein (60). Reduced Ape1 activity and increased DNA damage were associated with increased cell death in neuronal cells in senescence-accelerated mice, a well-established model of age-related neurodegeneration (61). Taken together, these reports suggest that developing a therapy on the basis of preserving the GAPDH/Ape1 interaction may be beneficial to slow aging and to treat aging-associated diseases.
In summary, we have described a novel antiapoptotic mechanism that protects vascular SMCs against oxidative stress–induced DNA damage and cell apoptosis. We found that GAPDH interacts with Ape1 in the SMC nucleus and that GAPDH/Ape1 binding depends on GAPDH active site cysteines. GAPDH up-regulates Ape1 via a HOXA5-dependent mechanism, and Ape1 up-regulation as well as formation of the nuclear GAPDH/Ape1 complex contribute to regeneration of oxidized Ape1. We found reduced GAPDH levels in the atherosclerotic plaque fibrous cap in association with increased oxidative stress and cell apoptosis. We suggest that preserving the nuclear GAPDH/Ape1 complex could be a novel beneficial strategy to suppress DNA damage, reduce SMC apoptosis, and ultimately enhance atherosclerotic plaque stability.
ACKNOWLEDGMENTS
This work was supported by U.S. National Institutes of Health (NIH) National Heart, Lung, and Blood Institute Grants R01-HL070241, R01-HL080682 (to P.D.), and R21-HL113705 (to S.S.), as well as American Heart Association Grant-in-Aid 13GRNT17230069 (to S.S.). The project has received funds from COBRE Cardiovascular Research Program (NIH National Institute of General Medical Sciences P20-RR018766; principal investigator, Dr. Daniel Kapusta, Louisiana Health Sciences Center, New Orleans, LA, USA). All authors had full access to all of the data in the study and had final responsibility for the decision to submit for publication. The authors thank to Dr. Daniel Green (St. Jude Children’s Research Hospital, Memphis, TN, USA) for providing GAPDH retroviral expression vector (pLZRS-GAPDH). The authors also thank to Dr. Karen Kirby (Charles W. Gehrke Proteomics Center, University of Missouri) for assistance with SPR experiments. The authors are grateful for the support provided by the Molecular Cytology Core of University of Missouri.
Glossary
- Ab
antibody
- AP
apurinic/apyrimidinic
- Ape1
apurinic/apyrimidinic endonuclease 1
- CPC
cardiac progenitor cell
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- gSTED
gated stimulated emission depletion
- HOXA5
homeobox protein Hox-A5
- KA
koningic acid
- OxLDL
oxidized LDL
- siRNA
small interfering RNA
- SMC
smooth muscle cell
- SPR
surface plasmon resonance
- WT
wild type
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
AUTHOR CONTRIBUTIONS
X. Hou and S. Sukhanov conducted most of the experiments, analyzed the results, and wrote most of the paper; P. Snarski conducted animal experiments; A. Jurkevich and S. Sukhanov conducted confocal and gSTED microscopy imaging; and Y. Higashi, T. Yoshida, and P. Delafontaine conceived the idea for the project, assisted with critical discussion of results, and contributed to the writing of manuscript.
REFERENCES
- 1.Mozaffarian D., Benjamin E. J., Go A. S., Arnett D. K., Blaha M. J., Cushman M., Das S. R., de Ferranti S., Després J. P., Fullerton H. J., Howard V. J., Huffman M. D., Isasi C. R., Jiménez M. C., Judd S. E., Kissela B. M., Lichtman J. H., Lisabeth L. D., Liu S., Mackey R. H., Magid D. J., McGuire D. K., Mohler E. R. III, Moy C. S., Muntner P., Mussolino M. E., Nasir K., Neumar R. W., Nichol G., Palaniappan L., Pandey D. K., Reeves M. J., Rodriguez C. J., Rosamond W., Sorlie P. D., Stein J., Towfighi A., Turan T. N., Virani S. S., Woo D., Yeh R. W., Turner M. B.; Writing Group Members; American Heart Association Statistics Committee; Stroke Statistics Subcommittee (2016) Heart disease and stroke statistics-2016 update: a report from the American Heart Association. Circulation 133, e38–e360 [DOI] [PubMed] [Google Scholar]
- 2.Ambrose J. A., Tannenbaum M. A., Alexopoulos D., Hjemdahl-Monsen C. E., Leavy J., Weiss M., Borrico S., Gorlin R., Fuster V. (1988) Angiographic progression of coronary artery disease and the development of myocardial infarction. J. Am. Coll. Cardiol. 12, 56–62 [DOI] [PubMed] [Google Scholar]
- 3.Littlewood T. D., Bennett M. R. (2003) Apoptotic cell death in atherosclerosis. Curr. Opin. Lipidol. 14, 469–475 [DOI] [PubMed] [Google Scholar]
- 4.Martinet W., Schrijvers D. M., De Meyer G. R. (2011) Pharmacological modulation of cell death in atherosclerosis: a promising approach towards plaque stabilization? Br. J. Pharmacol. 164, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roos W. P., Kaina B. (2013) DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 332, 237–248 [DOI] [PubMed] [Google Scholar]
- 6.Cervelli T., Borghini A., Galli A., Andreassi M. G. (2012) DNA damage and repair in atherosclerosis: current insights and future perspectives. Int. J. Mol. Sci. 13, 16929–16944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shah N. R., Mahmoudi M. (2015) The role of DNA damage and repair in atherosclerosis: a review. J. Mol. Cell. Cardiol. 86, 147–157 [DOI] [PubMed] [Google Scholar]
- 8.Schraufstätter I., Hyslop P. A., Jackson J. H., Cochrane C. G. (1988) Oxidant-induced DNA damage of target cells. J. Clin. Invest. 82, 1040–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demple B., Sung J. S. (2005) Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst.) 4, 1442–1449 [DOI] [PubMed] [Google Scholar]
- 10.Blackstock C. D., Higashi Y., Sukhanov S., Shai S. Y., Stefanovic B., Tabony A. M., Yoshida T., Delafontaine P. (2014) Insulin-like growth factor-1 increases synthesis of collagen type I via induction of the mRNA-binding protein LARP6 expression and binding to the 5′ stem-loop of COL1a1 and COL1a2 mRNA. J. Biol. Chem. 289, 7264–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aonuma T., Takehara N., Maruyama K., Kabara M., Matsuki M., Yamauchi A., Kawabe J., Hasebe N. (2016) Apoptosis-resistant cardiac progenitor cells modified with apurinic/apyrimidinic endonuclease/redox factor 1 gene overexpression regulate cardiac repair after myocardial infarction. Stem Cells Transl. Med. 5, 1067–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azam S., Jouvet N., Jilani A., Vongsamphanh R., Yang X., Yang S., Ramotar D. (2008) Human glyceraldehyde-3-phosphate dehydrogenase plays a direct role in reactivating oxidized forms of the DNA repair enzyme APE1. J. Biol. Chem. 283, 30632–30641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sirover M. A. (2005) New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell. Biochem. 95, 45–52 [DOI] [PubMed] [Google Scholar]
- 14.Sukhanov S., Higashi Y., Shai S. Y., Itabe H., Ono K., Parthasarathy S., Delafontaine P. (2006) Novel effect of oxidized low-density lipoprotein: cellular ATP depletion via downregulation of glyceraldehyde-3-phosphate dehydrogenase. Circ. Res. 99, 191–200 [DOI] [PubMed] [Google Scholar]
- 15.Okura Y., Brink M., Itabe H., Scheidegger K. J., Kalangos A., Delafontaine P. (2000) Oxidized low-density lipoprotein is associated with apoptosis of vascular smooth muscle cells in human atherosclerotic plaques. Circulation 102, 2680–2686 [DOI] [PubMed] [Google Scholar]
- 16.Li Y., Higashi Y., Itabe H., Song Y. H., Du J., Delafontaine P. (2003) Insulin-like growth factor-1 receptor activation inhibits oxidized LDL-induced cytochrome C release and apoptosis via the phosphatidylinositol 3 kinase/Akt signaling pathway. Arterioscler. Thromb. Vasc. Biol. 23, 2178–2184 [DOI] [PubMed] [Google Scholar]
- 17.Colell A., Ricci J. E., Tait S., Milasta S., Maurer U., Bouchier-Hayes L., Fitzgerald P., Guio-Carrion A., Waterhouse N. J., Li C. W., Mari B., Barbry P., Newmeyer D. D., Beere H. M., Green D. R. (2007) GAPDH and autophagy preserve survival after apoptotic cytochrome c release in the absence of caspase activation. Cell 129, 983–997 [DOI] [PubMed] [Google Scholar]
- 18.Sukhanov S., Higashi Y., Shai S. Y., Blackstock C., Galvez S., Vaughn C., Titterington J., Delafontaine P. (2011) Differential requirement for nitric oxide in IGF-1-induced anti-apoptotic, anti-oxidant and anti-atherosclerotic effects. FEBS Lett. 585, 3065–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paigen B., Morrow A., Holmes P. A., Mitchell D., Williams R. A. (1987) Quantitative assessment of atherosclerotic lesions in mice. Atherosclerosis 68, 231–240 [DOI] [PubMed] [Google Scholar]
- 20.Dunn K. W., Kamocka M. M., McDonald J. H. (2011) A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shatilla A., Ramotar D. (2002) Embryonic extracts derived from the nematode Caenorhabditis elegans remove uracil from DNA by the sequential action of uracil-DNA glycosylase and AP (apurinic/apyrimidinic) endonuclease. Biochem. J. 365, 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park W. H. (2016) Exogenous H2O2 induces growth inhibition and cell death of human pulmonary artery smooth muscle cells via glutathione depletion. Mol. Med. Rep. 14, 936–942 [DOI] [PubMed] [Google Scholar]
- 23.Jarosz A. P., Wei W., Gauld J. W., Auld J., Özcan F., Aslan M., Mutus B. (2015) Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is inactivated by S-sulfuration in vitro. Free Radic. Biol. Med. 89, 512–521 [DOI] [PubMed] [Google Scholar]
- 24.Hwang N. R., Yim S. H., Kim Y. M., Jeong J., Song E. J., Lee Y., Lee J. H., Choi S., Lee K. J. (2009) Oxidative modifications of glyceraldehyde-3-phosphate dehydrogenase play a key role in its multiple cellular functions. Biochem. J. 423, 253–264 [DOI] [PubMed] [Google Scholar]
- 25.Batthyany C., Schopfer F. J., Baker P. R., Durán R., Baker L. M., Huang Y., Cerveñansky C., Branchaud B. P., Freeman B. A. (2006) Reversible post-translational modification of proteins by nitrated fatty acids in vivo. J. Biol. Chem. 281, 20450–20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hwang I. K., Yoo K. Y., Kim D. W., Choi J. H., Lee I. S., Won M. H. (2007) Hyperoxidized peroxiredoxins and glyceraldehyde-3-phosphate dehydrogenase immunoreactivity and protein levels are changed in the gerbil hippocampal CA1 region after transient forebrain ischemia. Neurochem. Res. 32, 1530–1538 [DOI] [PubMed] [Google Scholar]
- 27.Endo A., Hasumi K., Sakai K., Kanbe T. (1985) Specific inhibition of glyceraldehyde-3-phosphate dehydrogenase by koningic acid (heptelidic acid). J. Antibiot. 38, 920–925 [DOI] [PubMed] [Google Scholar]
- 28.Kato M., Sakai K., Endo A. (1992) Koningic acid (heptelidic acid) inhibition of glyceraldehyde-3-phosphate dehydrogenases from various sources. Biochim. Biophys. Acta 1120, 113–116 [DOI] [PubMed] [Google Scholar]
- 29.Nakazawa M., Uehara T., Nomura Y. (1997) Koningic acid (a potent glyceraldehyde-3-phosphate dehydrogenase inhibitor)-induced fragmentation and condensation of DNA in NG108-15 cells. J. Neurochem. 68, 2493–2499 [DOI] [PubMed] [Google Scholar]
- 30.Tristan C., Shahani N., Sedlak T. W., Sawa A. (2011) The diverse functions of GAPDH: views from different subcellular compartments. Cell. Signal. 23, 317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masuda Y., Bennett R. A., Demple B. (1998) Rapid dissociation of human apurinic endonuclease (Ape1) from incised DNA induced by magnesium. J. Biol. Chem. 273, 30360–30365 [DOI] [PubMed] [Google Scholar]
- 32.MacDonald L., Baldini G., Storrie B. (2015) Does super-resolution fluorescence microscopy obsolete previous microscopic approaches to protein co-localization? Methods Mol. Biol. 1270, 255–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samejima K., Earnshaw W. C. (2005) Trashing the genome: the role of nucleases during apoptosis. Nat. Rev. Mol. Cell Biol. 6, 677–688 [DOI] [PubMed] [Google Scholar]
- 34.Gavrieli Y., Sherman Y., Ben-Sasson S. A. (1992) Identification of programmed cell death in situ via specific labeling of nuclear DNA fragmentation. J. Cell Biol. 119, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng L., Roeder R. G., Luo Y. (2003) S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 114, 255–266 [DOI] [PubMed] [Google Scholar]
- 36.Colell A., Green D. R., Ricci J. E. (2009) Novel roles for GAPDH in cell death and carcinogenesis. Cell Death Differ. 16, 1573–1581 [DOI] [PubMed] [Google Scholar]
- 37.Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 38.Suarez S., McCollum G. W., Jayagopal A., Penn J. S. (2015) High glucose-induced retinal pericyte apoptosis depends on association of GAPDH and Siah1. J. Biol. Chem. 290, 28311–28320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.You B., Huang S., Qin Q., Yi B., Yuan Y., Xu Z., Sun J. (2013) Glyceraldehyde-3-phosphate dehydrogenase interacts with proapoptotic kinase mst1 to promote cardiomyocyte apoptosis. PLoS One 8, e58697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai R., Xue W., Liu S., Petersen R. B., Huang K., Zheng L. (2015) Overexpression of glyceraldehyde 3-phosphate dehydrogenase prevents neurovascular degeneration after retinal injury. FASEB J. 29, 2749–2758 [DOI] [PubMed] [Google Scholar]
- 41.Takaoka Y., Goto S., Nakano T., Tseng H. P., Yang S. M., Kawamoto S., Ono K., Chen C. L. (2014) Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) prevents lipopolysaccharide (LPS)-induced, sepsis-related severe acute lung injury in mice. Sci. Rep. 4, 5204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacquin M. A., Chiche J., Zunino B., Bénéteau M., Meynet O., Pradelli L. A., Marchetti S., Cornille A., Carles M., Ricci J. E. (2013) GAPDH binds to active Akt, leading to Bcl-xL increase and escape from caspase-independent cell death. Cell Death Differ. 20, 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu H., McElwee-Witmer S., Perrone M., Clark K. L., Zilberstein A. (2000) Phenylephrine protects neonatal rat cardiomyocytes from hypoxia and serum deprivation-induced apoptosis. Cell Death Differ. 7, 773–784 [DOI] [PubMed] [Google Scholar]
- 44.Zhang J. Y., Zhang F., Hong C. Q., Giuliano A. E., Cui X. J., Zhou G. J., Zhang G. J., Cui Y. K. (2015) Critical protein GAPDH and its regulatory mechanisms in cancer cells. Cancer Biol. Med. 12, 10–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dastoor Z., Dreyer J. L. (2001) Potential role of nuclear translocation of glyceraldehyde-3-phosphate dehydrogenase in apoptosis and oxidative stress. J. Cell Sci. 114, 1643–1653 [DOI] [PubMed] [Google Scholar]
- 46.Nakajima H., Amano W., Fujita A., Fukuhara A., Azuma Y. T., Hata F., Inui T., Takeuchi T. (2007) The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J. Biol. Chem. 282, 26562–26574 [DOI] [PubMed] [Google Scholar]
- 47.Sakai K., Hasumi K., Endo A. (1988) Inactivation of rabbit muscle glyceraldehyde-3-phosphate dehydrogenase by koningic acid. Biochim. Biophys. Acta 952, 297–303 [DOI] [PubMed] [Google Scholar]
- 48.Li M. X., Shan J. L., Wang D., He Y., Zhou Q., Xia L., Zeng L. L., Li Z. P., Wang G., Yang Z. Z. (2012) Human apurinic/apyrimidinic endonuclease 1 translocalizes to mitochondria after photodynamic therapy and protects cells from apoptosis. Cancer Sci. 103, 882–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Y., Guo C., Fishel M. L., Wang Z. Y., Vasko M. R., Kelley M. R. (2009) Role of APE1 in differentiated neuroblastoma SH-SY5Y cells in response to oxidative stress: use of APE1 small molecule inhibitors to delineate APE1 functions. DNA Repair (Amst.) 8, 1273–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Unnikrishnan A., Raffoul J. J., Patel H. V., Prychitko T. M., Anyangwe N., Meira L. B., Friedberg E. C., Cabelof D. C., Heydari A. R. (2009) Oxidative stress alters base excision repair pathway and increases apoptotic response in apurinic/apyrimidinic endonuclease 1/redox factor-1 haploinsufficient mice. Free Radic. Biol. Med. 46, 1488–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim H. W., Cho K. J., Lee B. I., Kim H. J., Kim G. W. (2009) Post-ischemic administration of peptide with apurinic/apyrimidinic endonuclease activity inhibits induction of cell death after focal cerebral ischemia/reperfusion in mice. Neurosci. Lett. 460, 166–169 [DOI] [PubMed] [Google Scholar]
- 52.Kam M. K., Lui V. C. (2015) Roles of Hoxb5 in the development of vagal and trunk neural crest cells. Dev. Growth Differ. 57, 158–168 [DOI] [PubMed] [Google Scholar]
- 53.Kachgal S., Mace K. A., Boudreau N. J. (2012) The dual roles of homeobox genes in vascularization and wound healing. Cell Adhes. Migr. 6, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunn J., Simmons R., Thabet S., Jo H. (2015) The role of epigenetics in the endothelial cell shear stress response and atherosclerosis. Int. J. Biochem. Cell Biol. 67, 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang H. P., Liu W. J., Guo Q. L., Bai Y. Q. (2016) Effect of silencing HOXA5 gene expression using RNA interference on cell cycle and apoptosis in Jurkat cells. Int. J. Mol. Med. 37, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van der Wal A. C., Becker A. E. (1999) Atherosclerotic plaque rupture--pathologic basis of plaque stability and instability. Cardiovasc. Res. 41, 334–344 [DOI] [PubMed] [Google Scholar]
- 57.Freitas A. A., de Magalhães J. P. (2011) A review and appraisal of the DNA damage theory of ageing. Mutat. Res. 728, 12–22 [DOI] [PubMed] [Google Scholar]
- 58.Swain U., Rao K. S. (2012) Age-dependent decline of DNA base excision repair activity in rat cortical neurons. Mech. Ageing Dev. 133, 186–194 [DOI] [PubMed] [Google Scholar]
- 59.Vigelso A., Dybboe R., Hansen C. N., Dela F., Helge J. W., Guadalupe Grau A. (2015) GAPDH and beta-actin protein decreases with aging, making Stain-Free technology a superior loading control in Western blotting of human skeletal muscle. J. Appl. Physiol. (1985) 118, 386–394 [DOI] [PubMed] [Google Scholar]
- 60.Mazzola J. L., Sirover M. A. (2005) Aging of human glyceraldehyde-3-phosphate dehydrogenase is dependent on its subcellular localization. Biochim. Biophys. Acta 1722, 168–174 [DOI] [PubMed] [Google Scholar]
- 61.Torregrosa-Muñumer R., Gómez A., Vara E., Kireev R., Barja G., Tresguerres J. A., Gredilla R. (2016) Reduced apurinic/apyrimidinic endonuclease 1 activity and increased DNA damage in mitochondria are related to enhanced apoptosis and inflammation in the brain of senescence- accelerated P8 mice (SAMP8). Biogerontology 17, 325–335 [DOI] [PubMed] [Google Scholar]