Abstract
Androgen receptor (AR) signaling is fundamental to prostate cancer and is the dominant therapeutic target in metastatic disease. However, stringent androgen deprivation therapy (ADT) regimens decrease quality of life and have been largely unsuccessful in curtailing mortality. Recent clinical and pre-clinical studies have taken advantage of the dichotomous ability of AR signaling to elicit growth-suppressive and differentiating effects by administering hyper-physiological levels of testosterone. In this study, high-throughput drug screening identified a potent synergy between high-androgen therapy and YM155, a transcriptional inhibitor of Survivin (BIRC5). This interaction was mediated by the direct transcriptional upregulation of the YM155 transporter SLC35F2 by the AR. Androgen-mediated YM155-induced cell death was completely blocked by the overexpression of multidrug resistance (MDR) transporter ABCB1. SLC35F2 expression was significantly correlated with intra-tumor androgen levels in four distinct patient-derived xenografts (PDX) models, and with AR-activity score in a large gene expression dataset of castration resistant metastases. A subset of tumors had significantly elevated SLC35F2 expression and therefore, may identify patients who are highly responsive to YM155 treatment.
Implications
The combination of androgen therapy with YM155 represents a novel drug synergy, and SLC35F2 may serve as a clinical biomarker of response to YM155.
Keywords: Androgen Receptor, YM155, SLC35F2, Androgen, Drug Synergy, prostate adenocarcinoma
Introduction
Prostate cancer (PC) is the most frequently diagnosed cancer in men and the second leading cause of male cancer mortality(1). The androgen receptor (AR) is a master regulator of prostate development and the maintenance of prostate epithelial cell viability and secretory activity. The AR-directed transcriptional program also serves to maintain the survival of PC and is the focal point for therapeutic strategies in localized disease where it is combined with radiation treatment, and for advanced disease where AR pathway suppression remains first-line therapy. The physiological role of the AR changes as prostate cells undergo tumorigenesis and progression(2). Notably, as AR signaling promotes cellular differentiation and can suppress proliferation, early oncogenic events including MYC gain, loss of tumor suppressors such as PTEN and TP53, and ERG overexpression allow transforming cells to uncouple the suppressive functions of AR signaling while benefiting from growth and metabolic advantages(3–5).
While suppressing AR activity through ligand reduction in the form of androgen deprivation therapy (ADT) is initially effective in metastatic PC, disease progression, termed castration-resistant prostate cancer (CRPC), inevitably manifests after 2-3 years. This occurs due to re-activation of AR through AR amplification, AR mutations, intra-tumoral synthesis of androgens, and the production of AR splice variants(6). The relationship between AR and PC further evolves upon progression to CRPC through genomic(7), cistromic(8), and transcriptional(9) alterations. Due to the maintenance of AR activity, survival benefits are achieved by re-targeting AR signaling with next-generation AR-directed therapeutics, though responses are generally measured in months rather than years(10,11). While resistance is again often accompanied by persistent AR activity, prolonged and effective AR suppression can induce epithelial-to-mesenchymal-transition (EMT) (12–14), acquisition of stem-like cell characteristics(13,15–17), and trans-differentiation into an AR-null neuroendocrine phenotype. There are also significant complications and quality of life issues that arise with long-term ADT(18,19). For these reasons, there has been a longstanding interest in the discovery of therapeutic modalities that may acutely synergize with AR-directed therapy to improve long-term outcomes in men with advanced PC.
AR activity can be dichotomous in action by promoting PC growth under normal circumstances and retarding PC growth when overstimulated with excessive androgens. AR signaling induces cell-cycle arrest, in part, by upregulating negative regulators of the cell cycle (20,21). AR is also a licensing factor for DNA replication and excessive androgens prevents AR-cycling and re-licensing resulting in cell-cycle block(22,23). Additionally, AR can recruit TOP2B to sites of active transcription resulting in double strand DNA breaks(24).
The observation that PC cells can adjust to too much or too little AR-signaling over time has led to the development of an approach termed bipolar androgen therapy (BAT) that rapidly cycles androgen levels to maximize the suppressive benefits of ADT while attempting to prevent PCs from adapting to a static low androgen environment(25,26). This approach has been evaluated in phase I and II clinical trials(27,28). Furthermore, the use of sustained high-dose androgens or bipolar therapy has also been evaluated to exploit potential synergies with other therapeutics such as ionizing radiation(29). In this study, we sought to determine if the growth suppressive effects of high dose androgen, or other cellular activities regulated through AR, would synergize with anti-cancer drugs to further inhibit proliferation or induce PC cell death.
Materials and Methods
Cell culture and drug screen
The LNCaP cell line (ATCC, CRL-1740) was cultured in RPMI-1640, no phenol red (GIBCO, cat#11835030) with 10% FBS (GIBCO, cat#10437-02). VCaPs (ATCC, CRL-2876) were cultured in DMEM/F-12, no phenol red (GIBCO, cat#21041025) with 10% FBS. Cell lines were lineage and mycoplasma validated by DDC (DNA Diagnostic Center) Medical (Fairfield, OH, USA). Cells were cultured for no more 20 passages from the validated stocks. Drug screening was carried out at Quellos High Throughput Screening Core (University of Washington, Seattle, WA). The epigenetics, apoptosis and stem cell modifier screening libraries were obtained from SelleckChem. LNCaPs were plated in 384 well plates at 2500 cells per well in 50μL of complete media (10% FBS) with either 10uM enzalutamide, 10nM R1881, or DMSO control using a PerkinElmer Wellmate Dispenser. They were incubated at 37°C in 5% CO2 overnight. The next day, the compound libraries were added using a CyBio CyBi-Well Vario outfitted with tip washing stations and a 384-head equipped with a pin tool using 50nL slotted pins (V&P Scientific, San Diego, CA). Fifty nanoliters of compounds dissolved in 100% DMSO (1,000× concentrated) were adsorbed onto the pins and washed off into cell assay wells containing 50μL of complete media (0.1% DMSO final). Plates were then incubated for 72 to 75 hours at 37°C, 5% CO2. Cells were harvested with CellTiter-Glo (Promega, cat#G7572) according to manufacturer's protocol and read with PerkinElmer Envision Multi-label Plate Reader outfitted with a plate stacker and ultra-sensitive luminescence detection. Viability signal was blank subtracted and normalized to DMSO control and plotted using Microsoft Excel and Tibco Spotfire.
Isobolographic analyses and dose response curves
LNCaP or VCaP cells were plated into 96 well plates at 5000 cells/well in complete growth media (10% FBS). The next day a 1/4 dose dilution series of YM155 was added in 25uL complete media following a 1/5 R1881 dose dilution series or a 1/3 enzalutamide dose dilution series also in 25uL in complete growth media. Cells were incubated for five days, 37°C 5% CO2, then harvested with 30uL/well CellTiter-Glo (Promega, cat#G7572). LNCAP and VCAP cells were plates as described for isobolographic analysis except either 10nM R1881 or 2.5 nM Testosterone was added and cultured for 5 days unless otherwise noted. IC50 values were generated with GraphPAD Prism 6 and evaluated with the extra sum of squares F-test applying a p-value of 0.05 (n=4).
Western blots
Western blots were run on NuPAGE 4-12% Bis-tris gels (ThermoFisher, cat#NP0321) with MOPS SDS buffer (ThermoFisher, Cat#NP0001) then transferred to PVDF membranes (ThermoFisher, cat#LC2002) for SLC35F2 or nitrocellulose membranes (ThermoFisher, cat#LC2000) for other proteins with NuPAGE transfer buffer (ThermoFisher, Cat#NP0006). Membranes were blocked in TBS with 0.1% Tween-20, 5% Milk, and 2.5% BSA. The antibodies used were: SLC35F2 (Proteintech, cat#25526-1-AP), GAPDH (Cell Signaling Technology, cat#2118), ATG5 (Cell Signaling Technology, cat#8540), Beclin (Cell Signaling Technology, Cat# 3495), γH2AX (Cell Signaling Technology, Cat# 9718), PARP (Cell Signaling Technology, Cat# 9542), BIRC5 (abcam, cat#ab76424), and Caspase3 (Cell Signaling Technology, Cat#9662).
qRT-PCR and RNA-seq
Gene expression microarray data was analyzed as described previously(30). Expression across benign prostate (n = 24), primary tumors (n = 33), and CRPC (n = 171) was plotted in GraphPad Prism version 7.00 (GraphPad Software, La Jolla, California) using normalized, log2 transformed microarray signal intensities. The AR activity score was determined by the expression of a 20-gene signature(31) and calculated as described previously(32). Briefly, the activity score is defined as the sum of the expression Z-scores converted to a percent. Pearson's correlation coefficient of AR activity and SLC35F2 expression (normalized, log2 signal intensity) in patient CRPC tumors (n = 171) was assessed using the cor.test function in R. qRT-PCR was performed Power Sybr Green (Applied Biosystems, cat#4367659) and run on a BioRad CFX384 real time system according to manufacturer's recommendation. Primers used were: RPL13A_F-CCTGGAGGAGAAGAGGAAAGAGA, Hs_RPL13A_R-TTGAGGACCTCTGTGTATTTGTCAA, Arv567es_F TGCTGGACACGACAACAA, Arv567es_R GCAGCTCTCTCGCAATCA SLC35F2_F-AGGCAAACTCTTCACCTGGAAT, and SLC35F2_R-AAATATTCCAGGTGAAGAGTTTGCC.
ChIP-seq and ChIP-qPCR
Previously published ChIP-seq data for AR was obtained from the following Gene-Expression Omnibus and Sequence Read Archive: LNCaP-1F5 Vehicle reps 1-3 (GSM973815-GSM973817), LNCaP-1F5 DHT reps 1-3 (GSM973818-GSM973820)(33). LNCaP ethanol (GSM353643), LNCaP R1881 (GSM353644)(34). LNCaP Vehicle (GSM696839), LNCaP R1881 stimulated (GSM696840)(35). Raw reads were aligned to hg19 with bowtie(36). Reads that had more than a single alignment were suppressed. Data was visualized with IGV(37).
AR-directed ChIP-qPCR was performed as previously descrived(38). Briefly, LNCaP cells in 5% CSS medium were pretreated with MDV3100 for 2 hours and followed by 10nM of DHT for 4 hours. Chromatin immunoprecipitation was performed using anti-AR antibody (Santa Cruz, 816×). The qPCR analysis was carried out using the SYBR Green method on the QuantStudio 3 Real-Time PCR system (Applied Biosystems). The primers used were SLC35F2_AR1_F:5′-AGAGAATCGTCCTTCAGAACC, SLC35F2_AR1_R:5′-GGACTGAGCACAAACAAACC, SLC35F2_AR2_F:5′-GGTCACTACCAAATGAACTGATCATG, SLC35F2_AR2_R:5′-AGTAGATAAGAAGGCTGACACCTG, SLC35F2_AR3_F:5′-GTTGAACTAACAGAGGTTTCAG, SLC35F2_AR3_R:5′-GATATGAATCAATACGGGCTGGCAC.
Overexpression and Knockdown vectors
The SLC35F2 ORF was obtained from a previously published ORF library(39) and expressed using the pLX304 (Addgene, Plasmid#, 25890) lentivirus backbone. The ABCB1 expression vector was constructed by from pHAMDRwt (Addgene, Plasmid#10957) by PCR amplifying out ABCB1 using the primers ABCB1_F- gccaccATGGATCTTGAAGGGGACCGCAATGG and ABCB1_R-TCACTGGCGCTTTGTTCCAGCCTGGAC and cloning it into PCR8-GW-TOPO (ThermoFisher, cat#K250020) then a gateway reaction was used to clone into the pL6.3/V5 lentivirus (ThermoFisher, V53306). The SLC35F2 shRNAs were the GIPZ Open Biosystems Human shRNAmirs V3LHS_377258 and V2LHS_154477.
LuCaP Human Prostate Cancer Xenografts
Animal studies were carried out in strict accordance with NIH guidelines, and with protocols approved by the Fred Hutchinson Center and the University of Washington Institutional Animal Care Use Committees. All surgeries were performed under isofluorane anesthesia, and all efforts were made to minimize suffering. Five different LuCaP patient derived xenografts (PDX) models established as part of the University of Washington tissue bank (40,41) were used (LuCaP 23, LuCaP 35, LuCaP 96, LuCaP 86.2 and LuCaP 136). All lines express the wild type AR and secrete PSA. Intact 6-8 week old male C.B-17 SCID mice (Charles River Laboratories, San Diego, CA) were implanted subcutaneously with 30mm3 tumor pieces. When tumors reached an average of 100mm3, a subset of mice were castrated (Cx). Tumor volume was determined by the following formula (long and short axis lengths in mm): long × (Short(x002C6)2)/2. Tumors from a subset of mice in each cohort were harvested at day 7 and day 21 after castration, and the rest of the animals were followed and sacrificed until tumors exceeded 1000 mm3 (End of Study, EOS) or sacrificed if animals became compromised. Xenografts were harvested and flash frozen for determination of tissue androgens and extraction of total RNA.
Steroid Measurements
Methods for determination of steroids in tissue samples by mass spectrometry were as previously reported(42). Briefly, frozen tissues were weighed, added to 60C water containing deuterated internal standards, heated to 60C for 10 minutes, and homogenized using a tissue homogenizer (Precellys; Bertin, Rockville, MD); supernatant was extracted twice with hexane (ethyl acetate [80:20 v/v]), and the organic layer was dried (SpeedVac; Thermo Scientific,Waltham, MA), derivatized with 0.025 M hydroxylamine hydrochloride for 24 hours at room temperature to form oximes, and quantified using liquid chromatography electrospray-ionization tandem mass spectrometry. The lower limit of detection for steroids in tissue was 0.49 pg/sample (0.02 pg/mg) for DHT and testosterone.
Results
The survivin/BIRC5 inhibitor YM155 synergizes with androgen therapy to suppress prostate cancer cell proliferation
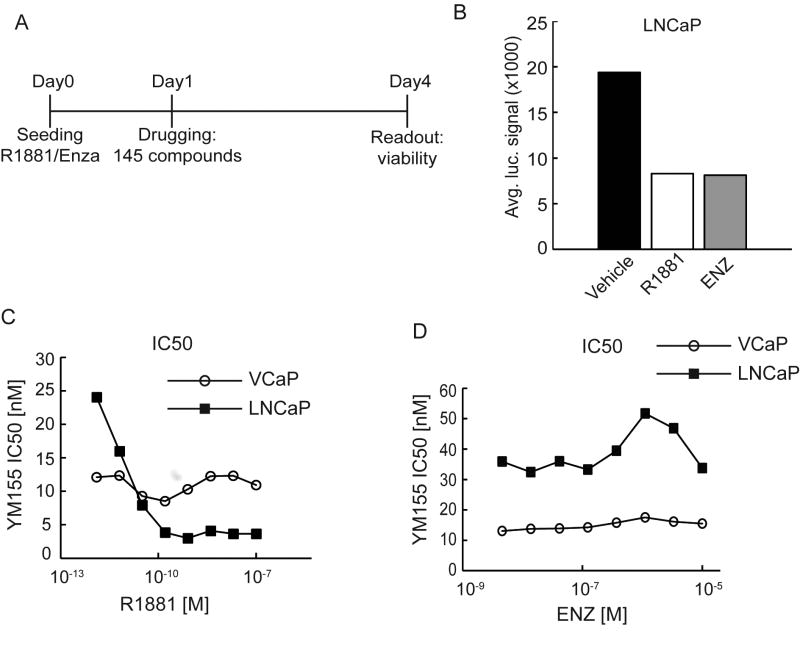
The LNCaP PC cell line grows maximally in 10% FBS containing media and growth is repressed when androgens are added (FIG S1). To identify drugs capable of enhancing the activity of AR inhibition or AR activation, we screened a library of 145 well-characterized pharmacological agents that impair cancer cell proliferation or survival using the androgen-sensitive PC cell line LNCaP (FIG 1A). Briefly, we plated LNCaP cells in standard growth media containing 10% FBS with either the AR antagonist enzalutamide (ENZ), the synthetic androgen R1881, or DMSO vehicle. After 24 hours, a 14-point range of concentrations of each library compound was applied to the plates and incubated for an additional 72 hrs at which point cell viability was quantitated. Strikingly, in control cultures with no other added drugs, R1881 suppressed cell proliferation as potently as the ENZ treatment (FIG 1B). No compounds synergized with ENZ under these conditions. However, YM155, which suppresses transcription of the anti-apoptotic protein survivin/BIRC5, displayed a supra-additive effect with R1881 .
Figure 1.
YM155 synergizes with high-androgen therapy to suppress prostate cancer viability. A) Design of high-throughput screen in LNCaP grown in 10% FBS media supplemented with either 1nM R1881, 10uM Enzalutamide (ENZ), or DMSO. B) Average CellTiter-Glo luciferase signal (viability) for all control wells on drug screen 96 hours post cell seeding. C) Inhibitory concentration for 50% viability (IC50) plotted for various doses of R1881 and D) enzalutamide (ENZ).
To validate the synergy between R1881 and YM155 we measured YM155 IC50 values in response to varying R1881 concentrations. These data demonstrated a potent synergistic interaction between R1881 and YM155 in LNCaP cells with R1881 increasing LNCaP sensitivity to YM155 from IC50 24 nM without R1881 to 3.85 nM with an R1881 concentration of 160 pM (FIG 1C). Synergy with YM155 was not achieved with ENZ (FIG 1D). Surprisingly, the VCaP PC cell line, which has amplification of AR, was initially more sensitive to YM155 but not further sensitized to YM155 by the addition of androgen (FIG 1C).
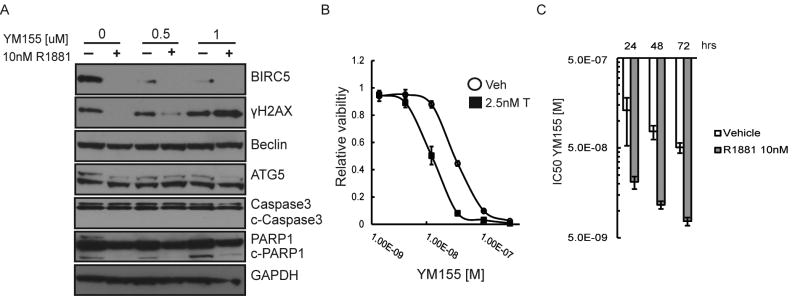
The mechanism by which YM155 is reported to elicit cell death is controversial and varies based on cell line, time, and YM155 concentration(43). However, YM155 is reported to suppress BIRC5 expression, increase DNA-damage, and induce autophagy(43). Given that BIRC5 is involved in DNA repair(44), and DNA-damage can induce autophagy(45), we sought to determine the action of YM155-mediated cellular effects before significant cell loss and secondary effects occurred. To this end we performed Western blots on LNCaP cells pre-incubated for 48 hours with 10 nm R1881 or vehicle control and subsequently dosed with 0.5 uM or 1 uM of YM155 for 24 hours (in contrast to the proliferation studies carried out at 96 hours). R1881 alone suppressed BIRC5 levels, consistent with androgen-induced G1 cell-cycle arrest(21) (FIG 2A). YM155 and R1881 coordinately suppressed BIRC5 levels (FIG 2A). Modest DNA-damage (γH2AX) was observed at the higher dose of YM155 with R1881 (FIG 2A). Reductions in γH2AX and cleaved PARP1 were observed in AR-suppressed cells in the control and low-dose YM155 groups, consistent with reduced replicative stress(46) and DNA-repair supportive effects of AR signaling(47) (FIG 2A). Additionally, there was no change in levels of autophagy (as measured by Beclin and AT5 levels) and negligible changes in apoptosis (as measured by cleaved-Caspase3 and PARP1) between the YM155 and control groups (FIG 2A).
Figure 2.
High-dose androgens suppress BIRC5 levels and increase YM155 mediated DNA-damage. A) Western blots for BIRC5 (survivin), γH2AX, Beclin, ATG5, Caspase3, and PARP1 on LNCaP treated for 24 hrs with 0.5 or 1 uM YM155 with and without a 48 hour pre-incubation with 10nM R1881. B) LNCaP viability (CellTiter-Glo) in response to YM155 with or without 2.5 nM Testosterone (T). (Error = standard deviation) (n=4) C) YM155 IC50 values of LNCaP pre-incubated 24hrs with 10nM R1881 or vehicle control then exposed to a dose range of YM155 for 24, 48, and 72 hrs (Error = 95% confidence interval, n=4).
To discount the possibility that the effects we observed are specific to R1881, a potent synthetic androgen, we performed a dose response curve of LNCaP viability to YM155 with a physiological dose of testosterone (2.5 nM). The addition of testosterone (T) also increased LNCaP sensitivity to YM155 (FIG 2B). To determine the time dependence of androgen and YM155 synergy we pre-incubated LNCaPs with 10 nM R1881 or vehicle for 24hrs and then determined YM155 IC50 values after 24, 48, and 72hrs (FIG 2C). Both the vehicle and R1881 groups increased sensitivity to YM155 with longer incubation times, consistent with reported observations that efficacy of YM155 increases with time(48). Given that R1881 potently suppresses BIRC5 without inducing apoptosis and does not induce DNA-damage under these experimental conditions (FIG 2A), these data suggest the mechanism by which androgen therapy sensitizes cells to YM155 is mediated through AR transcriptional activity rather than through genotoxicity or loss of BIRC5.
The membrane transport protein SLC35F2 is regulated by androgen receptor activation
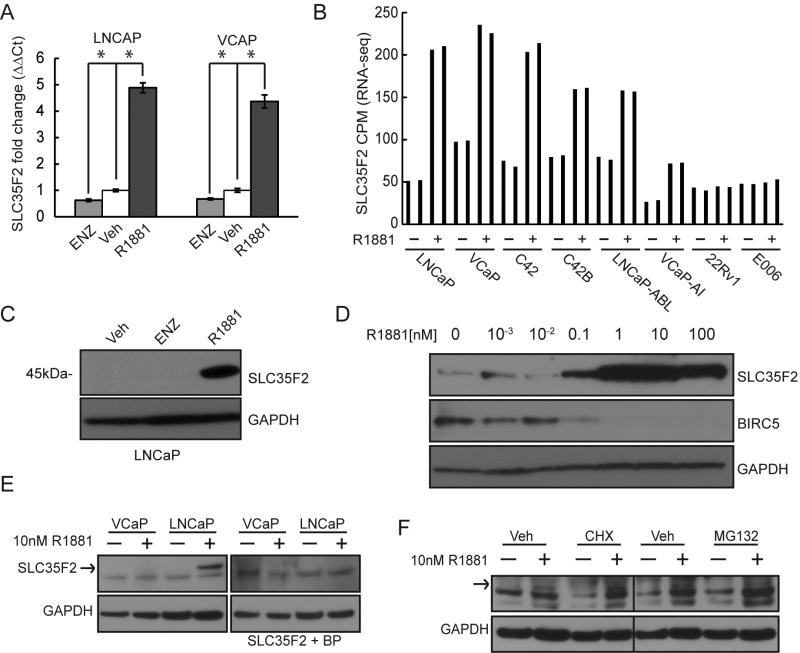
A recent study demonstrated that YM155 is transported by SLC35F2, a member of the solute carrier group of membrane transport proteins(49). Therefore, one possible mechanism of the R1881-YM155 synergy is through transcriptional upregulation of SLC35F2 by AR. To establish a transcriptional relationship we performed qRT-PCR to measure SLC35F2 transcript levels in LNCaP and VCaP cells cultured with 10 nM R1881 or ethanol vehicle for 48 hours. In cells treated with 10nM R1881, transcript levels were approximately fivefold higher than the vehicle control in LNCaP and approximately 4-fold higher in VCaP (FIG 3A). Importantly, a similar increase was not seen with 10 nM ENZ, suggesting transcriptional upregulation of SLC35F2 is not simply associated with cell cycle arrest, which occurs in response to either R1881 or ENZ (FIG 3A). To compare expression levels across multiple PC cell lines we plotted the RNA-seq counts-per-million (CPM) of SLC35F2 after exposure to 1 nM R1881 or vehicle control for 24hrs. The PC cell lines used were: androgen sensitive lines LNCaP and VCaP; castration-resistant lines C42, C42B, LNCAP-ABL, and VCaP-AI; and androgen-independent lines 22Rv1 and E006. As expected, SLC35F2 transcription levels were regulated by androgens with the exception of the androgen-independent lines (FIG 3B).
Figure 3.
SLC35F2 is regulated by androgen receptor signaling. A) qRT-PCR showing relative fold change of SLC35F2 in LNCaP and VCaP exposed to 10uM Enzalutamide (ENZ), vehicle (veh), 10nM R1881 (error bars = standard deviation, n=6). B) RNA-seq counts-per-million (CPM) of prostate cancer cell lines cultured with 1nM R1881 or vehicle for 24 hrs, each in duplicate C) Western blot of SLC35F2 on LNCaP exposed to 10uM enzalutamide (ENZ), 10nM R1881, or ethanol vehicle. D) Western blots of SLC35F2 and BIRC5 in response to a dose range of R1881 in LNCaP. E) Western blot of SLC35F2 with or without a blocking peptide of SLC35F2 and GAPDH on VCaP and LNCaP cultured with 10nM R1881 for 48hrs. F) Western blot of SLC35F2 on VCaP cultured with 10nM R1881 or vehicle for 48hrs and 5ug/mL cyclohexaminde (CHX) or 5uM MG132 for 24hrs. (* = p <0.05).
The androgen-induced increase in SLC35F2 transcript levels was accompanied by increases in SLC35F2 protein levels in LNCaP cells (FIG 3C). We then compared levels of SLC35F2 protein across a dose range of R1881 in LNCaP cells. In concordance with the 160 pM of R1881 at which the maximum response to YM155 was achieved (FIG 1C), SLC35F2 levels were dramatically increased at 100 pM R1881 (FIG 3D). Of note, SLC35F2 upregulation corresponded with the downregulation of BIRC5 (FIG 3D).
Despite the androgen-mediated increase of SLC35F2 transcript levels in VCaP, SLC35F2 protein levels were barely detectable in VCaP cells cultured for 48 hours with 10 nM R1881 (FIG 3E). A band close to the correct size of 45kD for SLC35F2 was observed by Western blot in VCaP but was not masked by a SLC35F2 blocking peptide, in contrast with AR-regulated product observed in LNCaP cells which was masked by the blocking peptide (FIG 3E). However, a faint band corresponding to SLC35F2 was detected in VCaP cells. Notably, SLC35F2 did not appreciably increase or decrease in VCaP cells incubated with the translational inhibitor cyclohexamide or the proteasome inhibitor MG132, respectively (FIG 3F). Taken together, we hypothesize that the lack of synergy between R1881 and YM155 in VCaP is due to the inefficient translation of additional SLC35F2 transcripts.
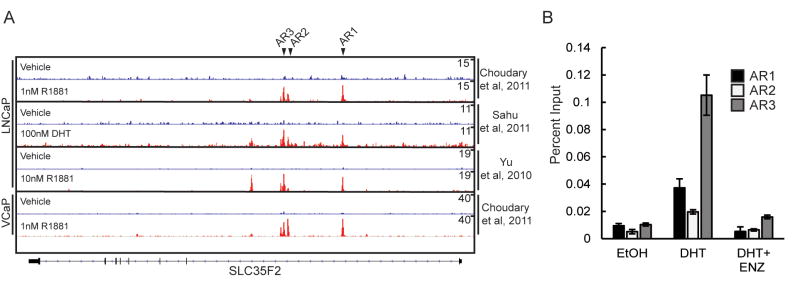
To provide further evidence that SLC35F2 transcription is directly regulated by the AR, we evaluated AR ChIP-seq data from three published studies of LNCaP cells and one study of VCaP cells evaluating AR binding sites throughout the genome. Three peaks, termed AR1-3, were observed in intron 1 of SLC35F2 under androgen exposure but not the androgen depleted control condition (FIG 4A). We designed PCR primers recognizing these regions and performed qPCR on LNCaP lysates following AR crosslinking and immunoprecipitation. Each peak demonstrated a substantial and significant increase in product following androgen treatment relative to either no androgen control or exposure to ENZ (FIG 4B).
Figure 4.
The membrane transport protein SLC35F2 is regulated by androgen receptor activation A) Chromatin immunoprecipitation sequencing (ChIP-seq) histograms of AR binding to the SLC35F2 locus from published datasets. Peaks selected for validation marked with black arrows B) ChIP-qPCR of peaks “AR1-3” in LNCaPs cultured in CSS with DHT, enzalutamide (ENZ), or ethanol vehicle control (EtOH)
SLC35F2 and ABCB1 expression determines cellular response to YM155 exposure
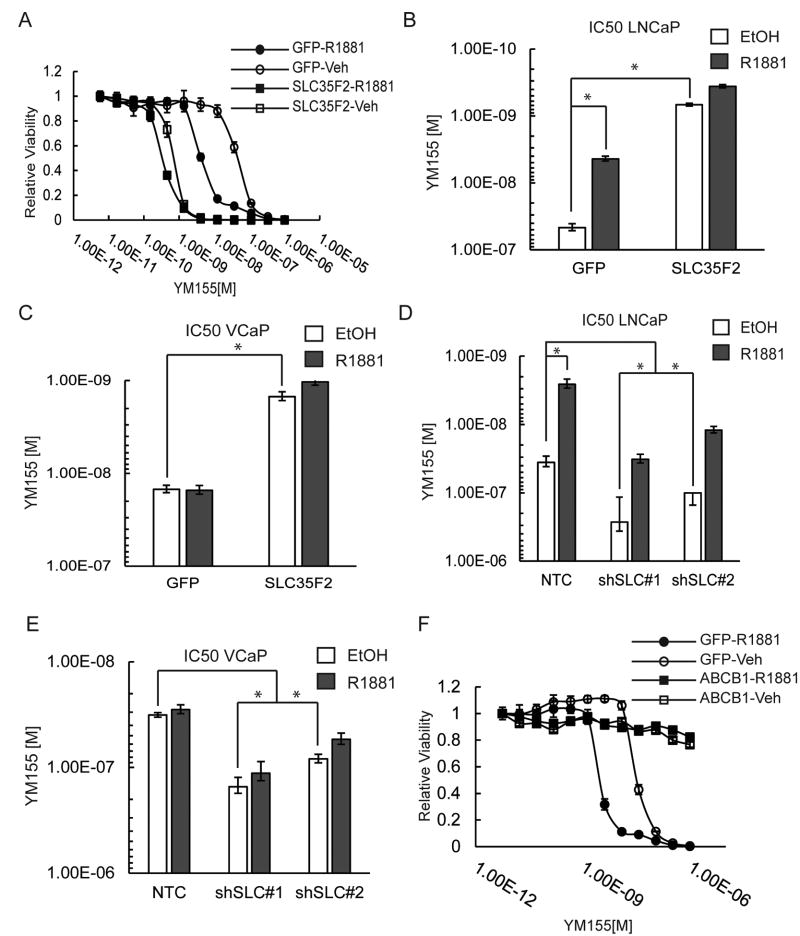
To further examine SLC35F2-mediated YM155 sensitivity we generated SCL35F2 knockdown and overexpression cell lines and measured viability in response to a range of YM155 doses (FIG 5A). IC50 values of YM155 shifted from 46.5 nM in GFP-control LNCaP cells treated with vehicle to 679 pM in LNCaP SCL35F2-overexpressing cells treated with vehicle (∼68 fold change) (FIG 5B). The YM155 IC50 only shifted from 679 pm to 363 pM (∼2 fold) when 10 nM R1881 was added to SLC35F2 overexpressing cells whereas the IC50 values shifted from 46.5 nM to 4.35 nM when 10nM R1881 was added to GFP control cells. In agreement, VCaP sensitivity to YM155 increased from 14.8 nM in the GFP cells with vehicle to 1.49 nM when SLC35F2 was overexpressed. Consistent with the lack of androgen induced changes in SLC35F2 protein levels in VCaP cells, IC50 values did not shift in VCaP cells dosed with 10 nM R1881 or vehicle (FIG 5C, FIG S2A).
Figure 5.
SLC35F2 and ABCB1 expression determine response to YM155. A) YM155 dose response curve for LNCaPs transduced with SLC35F2 or GFP overexpression vectors. B) IC50 values plotted for curves in “A.” C) Same as “B” for VCaPs. D) YM155 IC50 values of LNCaPS transduced with shRNA vectors to SLC35F2 (shSLC) compared to a non-targeting control (NTC) vector. E) Same as “D” for VCaP. F) YM155 dose response curves are plotted for LNCaPs overexpressing ABCB1 with and without 10nM R1881. (error bars: A = standard deviation, B-E 95% confidence interval; * = p <0.05, n=4)
Conversely, the reduction of SLC35F2 by shRNAs decreased the sensitivity of LNCaP cells to YM155, with IC50 values going from 35.8 nM with the non-targeting shRNA to 267nM for SLC35F2-directed shRNA#7 and 100 nM for shRNA#8 (FIG 5D, FIG S2B-C). VCAPs displayed a similar shift in IC50 values following SLC35F2 suppression (FIG 5E, FIG S2D). YM155 has also been reported to be effluxed by the ABCB1 transporter with a consequent resistance to drug treatment(50,51). Strikingly, ABCB1 overexpression rendered LNCaP cells completely resistant to YM155 regardless of androgen levels, suggesting the ratio of SLC35F2 to ABCB1 expression in tumor cells is an important determinant of sensitivity to YM155 (FIG 5F).
SLC35F2 expression correlates with AR activity in metastatic prostate cancer
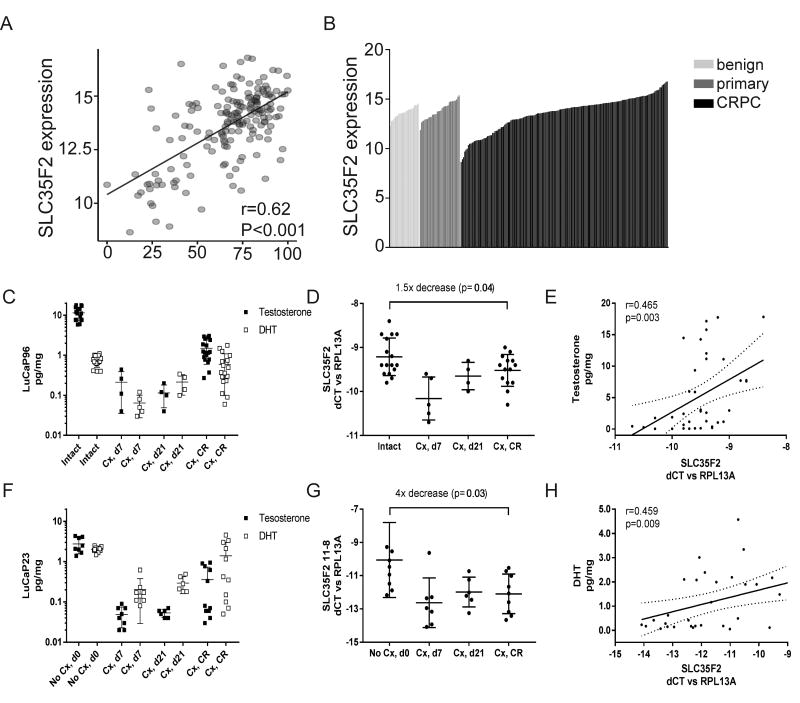
Identifying the patient population most likely to respond to YM155 is crucial for studies designed to establish YM155 as a targeted therapy for PC. To this end, we compared the expression of SLC35F2 with a metric of in vivo AR-activity, which is based on a panel of AR-regulated genes not including SLC35F2(30). We analyzed SLC35F2 transcript levels and calculated an AR activity score for 171 CRPC metastases by gene-expression microarrays(30). SLC35F2 expression was significantly correlated with AR-activity score (r = 0.62, p < 0.001) (FIG 6A). To examine the distribution of SLC35F2 expression across various states of PC progression, we compared microarray signal intensities for SLC35F2 benign prostate (n = 24), primary tumors (n = 33), and CRPC (n = 171) (FIG 6B). Notably, SLC35F2 was broadly expressed in all progression states and spanned a particularly large range of expression in CPRC. A subset of tumors had very high levels of SLC35F2 expression, suggesting that a subset of patients harbor tumors that may be highly responsive to YM155 treatment.
Figure 6.
SLC35F2 expression correlates with AR activity and androgen levels in castration resistant prostate cancer. A) Pearson correlation plot comparing AR activity and SLC35F2 expression in 171 CRPC samples. B) Waterfall plot of microarray signal intensities for SLC35F2 in benign prostate (n = 24), primary tumors (n = 33), and CRPC (n = 171). C) Intratumoral testosterone and dihydrotestosterone (DHT) levels in the LuCaP 96 PDX model in intact mice, at 7 and 21 days post castration (Cx), and after castration resistant (CR) regrowth. D) qRT-PCR for tumoral SLC35F2 expression in LuCaP 96 xenograft samples shown in panel C. E) Spearman correlation comparing intratumoral testosterone levels with normalized SLC35F2 mRNA expression in LuCaP 96 tumors. F) Intratumoral androgen levels in LuCaP 23. G) qRT-PCR for tumoral SLC35F2 expression in LuCaP 23. H) Spearman correlation comparing normalized SLC35F2 levels and intumoral DHT in LuCaP 23. T tests comparing SLC35F2 levels in intact and castrate mice by student's T test.
SLC35F2 expression correlates with androgen levels in xenografts models of prostate cancer
Since SLC35F2 is regulated by androgens, and previous clinical trials of YM155 in PC were performed in men with castrate levels of testosterone (52), we evaluated the relationship between SLC35F2 expression and intra-tumor androgen levels in AR positive patient-derived xenograft (PDX) models before and after castration. Levels of testosterone and dihydrotestosterone (DHT) in LuCaP96 and LuCap23 prostate adenocarcinoma PDX models were substantially decreased at days 7 and 21 after castration (FIG 6C,F) but partially recovered upon progression to castration resistant disease, consistent with previous reports of increased tumoral androgen synthesis leading to elevated levels of androgens in castration resistant tumors (53). Concordantly, tumor SLC35F2 mRNA levels decreased upon castration and increased in castration resistant tumors (FIG 6D), although not necessarily to levels present in tumors from intact mice (FIG 6D,G). Importantly, we observed strong correlations between SLC35F2 expression and tumor testosterone and/or DHT levels in both LuCaP96 (testosterone r=0.456, p=0.003; Fig 6E) and LuCaP23 (DHT r=0.459, p=0.009; Fig 6H), as well as in 2 additional PDX models (LuCaP35 DHT r=0.483, p=0042; LuCap136 DHT r=0.327, p=0.08, FIG S3; data for all models summarized in Table S1), suggesting that exogenous administration of high dose androgens has the potential for further regulating SLC35F2 expression. In contrast, SLC35Fexpression was not associated with tumor androgen levels in the LuCaP86.2 PDX model. This model is known to harbor a genomic AR rearrangement resulting in high level expression of the exon-skipping androgen independent AR splice variant ARV567 (54). Notably, we found that SLC35F2 expression was strongly correlated with ARV567 expression (r=0.745, p=0.0004), consistent with the ability of ligand independent AR variants to stimulate expression of AR target genes(55).
Discussion
BIRC5/survivin is a critical mediator of cancer cell growth, survival, DNA-repair, and therapy resistance(56). Though preclinical studies have repeatedly demonstrated the relevance of BIRC5 as a therapeutic target, clinical efforts to exploit these findings have largely been unsuccessful(57,58). YM155 was originally developed to suppress the transcription of BIRC5(48). Subsequent studies measuring the efficacy of YM155 in over 100 human cancer cell lines and xenografts found YM155 to potently cause cell death across a broad spectrum of cancers with low systemic toxicity in xenografts(59–61). YM155 inhibits BIRC5 by perturbing transcription factor-DNA interactions of Sp-1(62), ILF3(63), p50(64), and NONO(65). However, the anticancer effects of YM155 are unlikely to be due to BIRC5 suppression alone(66). Other targets related to cell survival such as Mcl1, Bcl-2, and Bcl-xl have also been reported(64).
A phase I pharmacokinetic study of YM155 on 41 patients, including 9 hormone-and docetaxel-refractory patients, determined YM155 was very well tolerated with very few grade 3 and 4 toxicities. Two CPRC patients had PSA responses, in addition to one complete and two partial responses in three patients with non-Hodgkin's lymphoma. A subsequent phase II study in 35 progressing, hormone- and docetaxel-refractory, PC patients found modest single-agent activity of YM155 with 25% of patients achieving prolonged stable disease of ≥18wks(52). Importantly, eligibility requirements for the trial required castrate levels of serum testosterone (≤50ng/mL). It is possible that the drug was inefficiently absorbed by tumor cells due to a loss of AR-mediated SLC35F2 expression. If so, our data suggests YM155 and related compounds may be more effective in the context of intermittent ADT or high-dose T cycles. Intriguingly, our data showing a correlation between tumor androgen levels and SLC35F2 expression also suggests YM155 may be effective in a subset of patients that progress on ADT with elevated levels of intratumoral androgens caused by aberrant expression of steroidogenic enzymes (67).
SLC transporters are among the least studied protein families and many receptors such as SLC35F2 are considered “orphan” with unknown substrates and physiological roles(68,69). However, there is an evolving conception of SLC transporters as mediators of inter-tissue communication or “remote sensing” of metabolites, signaling molecules, and morphogenic compounds(68). A growing body of literature on model-organism knockouts and human genetic diseases reveal complexity in how transporters are regulated and function in the development of an organism(70,71). For example, both developing and mature tissues have selective SLC expression profiles, sensitizing or insulating cells from signals in their environment(70,72). It is tempting to speculate that SLC transport profiles change with developmental and progression states in cancer, potentially presenting targets of opportunity. In support, it has been reported that SLC35F2 is more highly expressed in non-small cell lung cancers than in surrounding tissue(73).
Though controversial, it is increasingly appreciated that drug transport by passive diffusion across a membrane is often negligible and instead mediated by transmembrane proteins that normally transport metabolites(74). Furthermore, drugs designed with “metabolite-likeness”(75) have a greater chance of achieving standards of efficacy such as Lipinski's rule of five(76), suggesting an important role for SLC transporters in drug uptake. Several important cancer drugs have known transporters. SLC22A1/SLC22A2 transports daunorubicin(77) , imatinib(78), cisplatin, and oxaliplatin (79,80); and SLC22A16 transports doxorubicin(81). Further research on SLC transporter regulation and their substrates may lead to precision-medicine guided therapies with high antitumor-subtype efficacies and low systemic toxicities.
In addition to the potential PCa growth-suppressive effects and quality-of-life benefits of high-T therapy, androgens may selectively sensitize PC to YM155. Future studies are needed to evaluate the use of YM155 with high-T therapies. For example, it may be optimal to treat patients with cycles of synergistic therapeutic combinations by alternating testosterone/YM155 with ADT and docetaxel(82) or PI3K/AKT inhibitiors(83). Structure-activity relationship analysis on YM155 examined the determinants of its selective lethality to transformed cells and derived several dioxonaphthoimidazolium analogs with similar potency(66). Future work is required to determine whether molecules derived from YM155 have similar efficacy and transport mechanisms(64). Finally, it is unclear what other factors regulate of SLC35F2 gene expression or how protein levels are being regulated. The VCaP cell line doesn't upregulate SLC35F2 protein levels to the same extent as LNCaPs with androgens, despite similarly upregulating the transcript. One of the great challenges to the study of SLC transporters is their apparent functional redundancy and observed compensatory regulation(68). The mechanism by which cells and tissues coordinately regulate SLC transport profiles is largely unknown. Moreover, the regulation and functional significance of SLC transporters in cancers as well as their relevance to chemotherapeutics is increasingly appreciated yet still poorly understood(84).
In summary, we demonstrated that YM155 synergizes with endogenously achievable levels of androgens to eliminate PC cells. This is due to the upregulation of YM155 transporter SLC35F2 by AR transcriptional activity. However, YM155-mediated cell death can be counteracted by high ABCB1 levels. Furthermore, SLC35F2 expression correlates with AR-activity in CRPC tumors. Interestingly, some castration-resistant PCs express high levels of SLC35F2 despite lower AR-activity scores (Fig. 6A), suggesting SLC35F2 could be used as a biomarker for YM155 susceptibility. Given that YM155 is well-tolerated and synergizes with various therapies(43), YM155 may be an effective and well tolerated co-therapy for PC.
Supplementary Material
Acknowledgments
We thank Steven Plymate MD for discussions.
Financial Support: This work was supported by US National Institutes of Health: Pacific Northwest Prostate Cancer SPORE grant P50 CA097186 (EAM, EC, PSN), P01 CA163227 (EAM, EC, PSN), US Department of Defense awards W81XWH-15-1-0319 (EAM), W81XWH-16-1-0206 (MDN) and the Lucas Foundation.
Footnotes
The authors declare there are no competing financial interests in relation to the work described.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Mills IG. Nat Rev Cancer. Vol. 14. Nature Publishing Group; 2014. Maintaining and reprogramming genomic androgen receptor activity in prostate cancer; pp. 187–98. [DOI] [PubMed] [Google Scholar]
- 3.Antony L, Van Der Schoor F, Dalrymple SL, Isaacs JT. Androgen receptor (AR) suppresses normal human prostate epithelial cell proliferation via AR/??-catenin/TCF-4 complex inhibition of c-MYC transcription. Prostate. 2014;74:1118–31. doi: 10.1002/pros.22828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, et al. Nat Med. Vol. 19. Nature Publishing Group; 2013. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss; pp. 1023–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abeshouse A, Ahn J, Akbani R, Ally A, Amin S, Andry CD, et al. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watson PA, Arora VK, Sawyers CL. Nat Rev Cancer. Vol. 15. Nature Publishing Group; 2015. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer; pp. 701–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson D, Van Allen EM, Wu YM, Schultz N, Lonigro RJ, Mosquera JM, et al. Integrative Clinical Genomics of Advanced Prostate Cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. Nat Genet. Nature Publishing Group; 2015. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis; pp. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen Receptor Regulates a Distinct Transcription Program in Androgen-Independent Prostate Cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bono JSDe, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, et al. new england journalAbiraterone and Increased Survival in Metastatic Prostate Cancer. 2011:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 12.Liu X, Chen X, Rycaj K, Chao H, Deng Q, Jeter C, et al. Systematic dissection of phenotypic , functional , and tumorigenic heterogeneity of human prostate cancer cells. 2015;6 doi: 10.18632/oncotarget.4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rycaj K, Cho EJ, Liu X, Chao HP, Liu B, Li Q, et al. Longitudinal tracking of subpopulation dynamics and molecular changes during LNCaP cell castration and identification of inhibitors that could target the PSA-/lo castration-resistant cells. Oncotarget. 2016;7 doi: 10.18632/oncotarget.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Y, Wang BE, Leong KG, Yue P, Li L, Jhunjhunwala S, et al. Androgen deprivation causes epithelial-mesenchymal transition in the prostate: Implications for androgen- deprivation therapy. Cancer Res. 2012;72:527–36. doi: 10.1158/0008-5472.CAN-11-3004. [DOI] [PubMed] [Google Scholar]
- 15.Bishop JL, Davies A, Ketola K, Zoubeidi A. Regulation of tumor cell plasticity by the androgen receptor in prostate cancer. Endocr Relat Cancer. 2015;22:R165–82. doi: 10.1530/ERC-15-0137. [DOI] [PubMed] [Google Scholar]
- 16.Bishop JL, Sio A, Angeles A, Roberts ME, Azad a, Chi KN, et al. PD-L1 is highly expressed in Enzalutamide resistant prostate cancer. 2014;6 doi: 10.18632/oncotarget.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seiler D, Zheng J, Liu G, Wang S, Yamashiro J, Reiter RE, et al. Enrichment of putative prostate cancer stem cells after androgen deprivation: upregulation of pluripotency transactivators concurs with resistance to androgen deprivation in LNCaP cell lines. Prostate. 2013;73:1378–90. doi: 10.1002/pros.22685. [DOI] [PubMed] [Google Scholar]
- 18.Cary KC, Singla N, Cowan JE, Carroll PR, Cooperberg MR. Impact of androgen deprivation therapy on mental and emotional well-being in men with prostate cancer: analysis from the CaPSURE™ registry. J Urol Elsevier Ltd. 2014;191:964–70. doi: 10.1016/j.juro.2013.10.098. [DOI] [PubMed] [Google Scholar]
- 19.Zejnullahu K, Arevalo MG, Ryan CJ, Aggarwal R. Urol Oncol Semin Orig Investig. Elsevier; 2015. Approaches to minimize castration in the treatment of advanced prostate cancer; pp. 1–7. [DOI] [PubMed] [Google Scholar]
- 20.Kim YC, Chen C, Bolton EC. Androgen receptor-mediated growth suppression of HPr-1AR and PC3-Lenti-AR prostate epithelial cells. PLoS One. 2015;10:1–31. doi: 10.1371/journal.pone.0138286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roediger J, Hessenkemper W, Bartsch S, Manvelyan M, Huettner SS, Liehr T, et al. Supraphysiological androgen levels induce cellular senescence in human prostate cancer cells through the Src-Akt pathway. Mol Cancer. 2014;13:214. doi: 10.1186/1476-4598-13-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Antonio JM, Vander Griend DJ, Isaacs JT. DNA licensing as a novel androgen receptor mediated therapeutic target for prostate cancer. Endocr Relat Cancer. 2009;16:325–32. doi: 10.1677/ERC-08-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vander Griend DJ, Litvinov IV, Isaacs JT. Stabilizing androgen receptor in mitosis inhibits prostate cancer proliferation. Cell Cycle. 2007;6:647–51. doi: 10.4161/cc.6.6.4028. [DOI] [PubMed] [Google Scholar]
- 24.Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, et al. Nat Genet. Vol. 42. Nature Publishing Group; 2010. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements; pp. 668–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denmeade SR, Isaacs JT. Bipolar androgen therapy: The rationale for rapid cycling of supraphysiologic androgen/ablation in men with castration resistant prostate cancer. Prostate. 2010;70:1600–7. doi: 10.1002/pros.21196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isaacs JT, D'Antonio JM, Chen S, Antony L, Dalrymple SP, Ndikuyeze GH, et al. Adaptive auto-regulation of androgen receptor provides a paradigm shifting rationale for bipolar androgen therapy (BAT) for castrate resistant human prostate cancer. Prostate. 2012;72:1491–505. doi: 10.1002/pros.22504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schweizer MT, Antonarakis ES, Wang H, Ajiboye AS, Cao H, Luo J, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: Results from a pilot clinical study. 2015;7 doi: 10.1126/scitranslmed.3010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schweizer MT, Wang H, Luber B, Nadal R, Spitz A, Rosen DM, et al. Bipolar androgen therapy for men with androgen ablation naïve prostate cancer: Results from the phase II BATMAN study. Prostate. 2016 doi: 10.1002/pros.23209. [DOI] [PubMed] [Google Scholar]
- 29.Hedayati M, Haffner MC, Coulter J, Raval RR, Zhang Y, Zhou H, et al. Androgen deprivation followed by acute androgen stimulation selectively sensitizes AR-positive prostate cancer cells to ionizing radiation. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-15-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar A, Coleman I, Morrissey C, Zhang X, True LD, Gulati R, et al. Nat Med. Nature Publishing Group; 2016. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer; pp. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell. 2006;10:321–30. doi: 10.1016/j.ccr.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Cancer Cell. Vol. 19. Elsevier Inc; 2011. Reciprocal Feedback Regulation of PI3K and Androgen Receptor Signaling in PTEN-Deficient Prostate Cancer; pp. 575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sahu B, Laakso M, Pihlajamaa P, Ovaska K, Sinielnikov I, Hautaniemi S, et al. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer Res. 2013;73:1570–80. doi: 10.1158/0008-5472.CAN-12-2350. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, et al. Cancer Cell. Vol. 17. Elsevier Ltd; 2010. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression; pp. 443–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choudhary V, Kaddour-Djebbar I, Lakshmikanthan V, Ghazaly T, Thangjam GS, Sreekumar A, et al. Novel role of androgens in mitochondrial fission and apoptosis. Mol Cancer Res. 2011;9:1067–77. doi: 10.1158/1541-7786.MCR-10-0445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Trapnell C, Pop M, Salzberg S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell. 2005;19:631–42. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Boehm JS, Yang X, Salehi-Ashtiani K, Hao T, Shen Y, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–61. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morrissey C, Roudier MP, Dowell A, True LD, Ketchanji M, Welty C, et al. Effects of androgen deprivation therapy and bisphosphonate treatment on bone in patients with metastatic castration-resistant prostate cancer: results from the University of Washington Rapid Autopsy Series. J Bone Miner Res. 2013;28:333–40. doi: 10.1002/jbmr.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corey E, Quinn JE, Buhler KR, Nelson PS, Macoska JA, True LD, et al. LuCaP 35: A new model of prostate cancer progression to androgen independence. Prostate. 2003;55:239–46. doi: 10.1002/pros.10198. [DOI] [PubMed] [Google Scholar]
- 42.Kalhorn TF, Page ST, Howald WN, Mostaghel EA, Nelson PS. Analysis of testosterone and dihydrotestosterone from biological fluids as the oxime derivatives using high-performance liquid chromatography/tandem mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:3200–6. doi: 10.1002/rcm.3205. [DOI] [PubMed] [Google Scholar]
- 43.Rauch A, Hennig D, Schäfer C, Wirth M, Marx C, Heinzel T, et al. Biochim Biophys Acta - Rev Cancer. Vol. 1845. Elsevier B.V; 2014. Survivin and YM155: How faithful is the liaison? pp. 202–20. [DOI] [PubMed] [Google Scholar]
- 44.Véquaud E, Desplanques G, Jézéquel P, Juin P, Barillé-Nion S. Survivin contributes to DNA repair by homologous recombination in breast cancer cells. Breast Cancer Res Treat. 2016;155:53–63. doi: 10.1007/s10549-015-3657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez-Rocha H, Garcia-Garcia A, Panayiotidis MI, Franco R. Mutat Res - Fundam Mol Mech Mutagen. Vol. 711. Elsevier B.V; 2011. DNA damage and autophagy; pp. 158–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova Oa, Solier S, et al. γH2AX and cancer. Nat Rev Cancer. 2008;8:957–67. doi: 10.1038/nrc2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schiewer MJ, Knudsen KE. Trends Endocrinol Metab. Vol. 27. Elsevier Ltd; 2016. Linking DNA Damage and Hormone Signaling Pathways in Cancer; pp. 216–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakahara T, Takeuchi M, Kinoyama I, Minematsu T, Shirasuna K, Matsuhisa A, et al. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67:8014–21. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 49.Winter GE, Radic B, Mayor-Ruiz C, Blomen Va, Trefzer C, Kandasamy RK, et al. The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat Chem Biol. 2014;10:768–73. doi: 10.1038/nchembio.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lamers F, Schild L, Koster J, Versteeg R, Caron HN, Molenaar JJ. Targeted BIRC5 silencing using YM155 causes cell death in neuroblastoma cells with low ABCB1 expression. Eur J Cancer Elsevier Ltd. 2012;48:763–71. doi: 10.1016/j.ejca.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 51.Iwai M, Minematsu T, Li Q, Iwatsubo T, Usui T. Utility of P-Glycoprotein and Organic Cation Transporter 1 Double-Transfected LLC-PK1 Cells for Studying the Interaction of Suppressant , with P-Glycoprotein. Drug Metab Dispos. 2011;39:2314–20. doi: 10.1124/dmd.111.040733. [DOI] [PubMed] [Google Scholar]
- 52.Tolcher AW, Quinn DI, Ferrari A, Ahmann F, Giaccone G, Drake T, et al. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann Oncol. 2012;23:968–73. doi: 10.1093/annonc/mdr353. [DOI] [PubMed] [Google Scholar]
- 53.Montgomery RB, Mostaghel Ea, Vessella R, Hess DL, Kalhorn TF, Higano CS, et al. Maintenance of intratumoral androgens in metastatic prostate cancer: a mechanism for castration-resistant tumor growth. Cancer Res. 2008;68:4447–54. doi: 10.1158/0008-5472.CAN-08-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Y, Hwang TH, Oseth La, Hauge a, Vessella RL, Schmechel SC, et al. AR intragenic deletions linked to androgen receptor splice variant expression and activity in models of prostate cancer progression. Oncogene. 2012;31:4759–67. doi: 10.1038/onc.2011.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KaT, et al. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110:17492–7. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antonio Cheung CH, Huang CC, Tsai FY, Lee JYC, Cheng SM, Chang YC, et al. Survivin - biology and potential as a therapeutic target in oncology. Onco Targets Ther. 2013;6:1453–62. doi: 10.2147/OTT.S33374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Groner B, Weiss A. Targeting Survivin in cancer: Novel drug development approaches. BioDrugs. 2014;28:27–39. doi: 10.1007/s40259-013-0058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Altieri DC. Cancer Lett. Vol. 332. Elsevier Ireland Ltd; 2013. Targeting survivin in cancer; pp. 225–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mehta A, Zhang L, Boufraqech M, Liu-Chittenden Y, Zhang Y, Patel D, et al. Inhibition of Survivin with YM155 Induces Durable Tumor Response in Anaplastic Thyroid Cancer. Clin Cancer Res. 2015;21:4123–32. doi: 10.1158/1078-0432.CCR-14-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, et al. Broad spectrum and potent antitumor activities of YM155, a novel small-molecule survivin suppressant, in a wide variety of human cancer cell lines and xenograft models. Cancer Sci. 2011;102:614–21. doi: 10.1111/j.1349-7006.2010.01834.x. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q, Chen Z, Diao X, Huang S. Cancer Lett. Vol. 302. Elsevier Ireland Ltd; 2011. Induction of autophagy-dependent apoptosis by the survivin suppressant YM155 in prostate cancer cells; pp. 29–36. [DOI] [PubMed] [Google Scholar]
- 62.Cheng Q, Ling X, Haller A, Nakahara T, Yamanaka K, Kita A, et al. Suppression of survivin promoter activity by YM155 involves disruption of Sp1-DNA interaction in the survivin core promoter. Int J Biochem Mol Biol. 2012;3:179–97. [PMC free article] [PubMed] [Google Scholar]
- 63.Nakamura N, Yamauchi T, Hiramoto M, Yuri M, Naito M, Takeuchi M, et al. Interleukin enhancer-binding factor 3/NF110 is a target of YM155, a suppressant of survivin. Mol Cell Proteomics. 2012;11:M111.013243. doi: 10.1074/mcp.M111.013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han S, Ho S, Ali A, Chin TM, Go ML. Dioxonaphthoimidazoliums and phosphorylation of p50 in the NF-κB pathway. 2016 doi: 10.18632/oncotarget.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yamauchi T, Nakamura N, Hiramoto M, Yuri M, Yokota H, Naitou M, et al. Biochem Biophys Res Commun. Vol. 425. Elsevier Inc; 2012. Sepantronium bromide (YM155) induces disruption of the ILF3/p54(nrb) complex, which is required for survivin expression; pp. 711–6. [DOI] [PubMed] [Google Scholar]
- 66.Ho SHS, Sim MY, Yee WLS, Yang T, Yuen SPJ, Go ML. Eur J Med Chem. Vol. 104. Elsevier Masson SAS; 2015. Antiproliferative, DNA intercalation and redox cycling activities of dioxonaphtho[2,3-d]imidazolium analogs of YM155: A structure–activity relationship study; pp. 42–56. [DOI] [PubMed] [Google Scholar]
- 67.Mostaghel Ea, Marck BT, Plymate SR, Vessella RL, Balk S, Matsumoto AM, et al. Resistance to CYP17A1 inhibition with abiraterone in castration-resistant prostate cancer: induction of steroidogenesis and androgen receptor splice variants. Clin Cancer Res. 2011;17:5913–25. doi: 10.1158/1078-0432.CCR-11-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nigam SK. Nat Rev Drug Discov. Vol. 14. Nature Publishing Group; 2014. What do drug transporters really do? pp. 29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, et al. A Call for Systematic Research on Solute Carriers. Cell. 2015;162:478–87. doi: 10.1016/j.cell.2015.07.022. [DOI] [PubMed] [Google Scholar]
- 70.Song Z. Roles of the nucleotide sugar transporters (SLC35 family) in health and disease. Mol Aspects Med. 2013;34:590–600. doi: 10.1016/j.mam.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 71.Hediger MA, Cl??men??on B, Burrier RE, Bruford EA. The ABCs of membrane transporters in health and disease (SLC series): Introduction. Mol Aspects Med. 2013;34:95–107. doi: 10.1016/j.mam.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nishimura M, Suzuki S, Satoh T, Naito S. Tissue-specific mRNA expression profiles of human solute carrier 35 transporters. Drug Metab Pharmacokinet. 2009;24:91–9. doi: 10.2133/dmpk.24.91. [DOI] [PubMed] [Google Scholar]
- 73.Wang J. Highly expressed SLC35F2 in non-small cell lung cancer is associated with pathological staging. Mol Med Rep. 2011:1289–93. doi: 10.3892/mmr.2011.572. [DOI] [PubMed] [Google Scholar]
- 74.Kell DB, Oliver SG. How drugs get into cells: tested and testable predictions to help discriminate between transporter-mediated uptake and lipoidal bilayer diffusion. Front Pharmacol. 2014;5:231. doi: 10.3389/fphar.2014.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kell DB. Nat Rev Drug Discov. Vol. 15. Nature Publishing Group; 2016. Implications of endogenous roles of transporters for drug discovery: hitchhiking and metabolite-likeness; pp. 143–143. [DOI] [PubMed] [Google Scholar]
- 76.Leeson P. Drug discovery: Chemical beauty contest. Nature. 2012;481:455–6. doi: 10.1038/481455a. [DOI] [PubMed] [Google Scholar]
- 77.Andreev E, Brosseau N, Carmona E, Mes-Masson AM, Ramotar D. Sci Rep. Vol. 6. Nature Publishing Group; 2016. The human organic cation transporter OCT1 mediates high affinity uptake of the anticancer drug daunorubicin; p. 20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thomas J, Wang L, Clark RE, Pirmohamed M. Active transport of imatinib into and out of cells : implications for drug resistance. Transport. 2004;104:3739–45. doi: 10.1182/blood-2003-12-4276. [DOI] [PubMed] [Google Scholar]
- 79.Yonezawa A, Masuda S, Yokoo S, Katsura T, Inui Ki. Cisplatin and Oxaliplatin, but Not Carboplatin and Nedaplatin, Are Substrates for Human Organic Cation Transporters (SLC22A1-3 and Multidrug and Toxin Extrusion Family) J Pharmacol Exp Ther. 2006;319:879–86. doi: 10.1124/jpet.106.110346. [DOI] [PubMed] [Google Scholar]
- 80.Zhang S, Lovejoy KS, Shima JE, Lagpacan LL, Shu Y, Lapuk A, et al. Organic cation transporters are determinants of oxaliplatin cytotoxicity. Cancer Res. 2006;66:8847–57. doi: 10.1158/0008-5472.CAN-06-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Okabe M, Unno M, Harigae H, Kaku M, Okitsu Y, Sasaki T, et al. Characterization of the organic cation transporter SLC22A16: A doxorubicin importer. Biochem Biophys Res Commun. 2005;333:754–62. doi: 10.1016/j.bbrc.2005.05.174. [DOI] [PubMed] [Google Scholar]
- 82.Sweeney CJ, Chen YH, Carducci M, Liu G, Jarrard DF, Eisenberger M, et al. Chemohormonal Therapy in Metastatic Hormone-Sensitive Prostate Cancer. N Engl J Med. 2015;373:737–46. doi: 10.1056/NEJMoa1503747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas C, Lamoureux F, Crafter C, Davies BR, Beraldi E, Fazli L, et al. Synergistic targeting of PI3K/AKT pathway and androgen receptor axis significantly delays castration-resistant prostate cancer progression in vivo. Mol Cancer Ther. 2013;12:2342–55. doi: 10.1158/1535-7163.MCT-13-0032. [DOI] [PubMed] [Google Scholar]
- 84.Li Q, Shu Y. Role of solute carriers in response to anticancer drugs. Mol Cell Ther. 2014;2:15. doi: 10.1186/2052-8426-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.