Abstract
The evolution of eusociality is one of the major transitions in evolution, but the underlying genomic changes are unknown. We compared the genomes of 10 bee species that vary in social complexity, representing multiple independent transitions in social evolution, and report three major findings. First, many important genes show evidence of neutral evolution as a consequence of relaxed selection with increasing social complexity. Second, there is no single road map to eusociality; independent evolutionary transitions in sociality have independent genetic underpinnings. Third, though clearly independent in detail, these transitions do have similar general features, including an increase in constrained protein evolution accompanied by increases in the potential for gene regulation and decreases in diversity and abundance of transposable elements. Eusociality may arise through different mechanisms each time, but would likely always involve an increase in the complexity of gene networks.
The evolution of eusociality involves changes in the unit of natural selection, from the individual to a group (1). Bees evolved eusociality multiple times and are extremely socially diverse (2) (Fig. 1), but all pollinate angiosperms, including many crops essential to the human diet (3). Simple eusociality may be facultative or obligate, and both forms are characterized by small colonies with a reproductive queen and one or more workers that, due to social and nutritional cues, forego reproduction to cooperatively care for their siblings (2). Further evolutionary elaborations have led to complex eusociality, “superorganisms” with colonies of several thousand individuals, sophisticated modes of communication, and morphological specializations for division of labor (4).
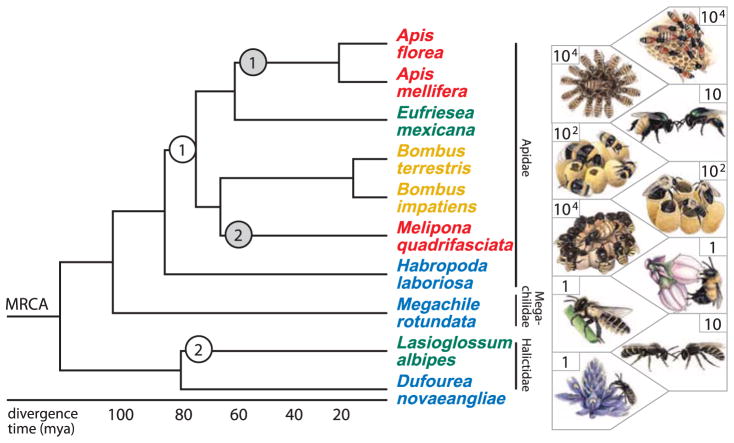
Fig. 1. Phylogeny and divergence times (28) of bees selected for genome analysis.
We analyzed two independent origins of simple eusociality from a solitary ancestor, one each in Apidae (white circle 1) and Halictidae (white circle 2), and two independent elaborations of complex eusociality in honeybees (gray circle 1) and stingless bees (gray circle 2). Most bees mate once, but honeybees mate with multiple males. All bees eat pollen and nectar from flowering plants. Species names are colored according to degree of social complexity: blue: ancestrally solitary; green: facultative simple eusociality; orange: obligate simple eusociality; red: obligate complex eusociality. The social biology of E. mexicana is unknown, but is representative of the facultative simple eusocial life history (29). Numbers in each box are approximate colony size on a log scale. MRCA, most recent common ancestor; mya, millions of years ago.
Theory predicts that the evolution of simple eusociality involves increased regulatory flexibility of ancestral gene networks to create specialized reproductive and nonreproductive individuals, and the evolution of complex eusociality requires genetic novelty to coordinate emergent properties of group dynamics (5). To test these predictions, we analyzed five de novo and five publicly available draft genome sequences of 10 bee species from three families, representing two independent origins of eusociality in Apidae and Halictidae and two independent elaborations of simple to complex eusociality in two apid tribes [Apini (honeybees) and Meliponini (stingless bees); Fig. 1]. The draft genomes were of comparable, high quality (supplementary materials).
We found that the transition from solitary to group life is associated with an increased capacity for gene regulation. We scanned the promoter regions of 5865 single-copy orthologs among the 10 species to calculate a motif score [representing the number and binding strength of experimentally characterized transcription factor binding sites (TFBSs)] for 188 Drosophila melanogaster TFs (6) with at least one ortholog in each of the 10 bees, and correlated motif score with social complexity, using phylogenetically independent contrasts (7). Of 2101 significantly correlated motif-gene pairs, 89% were positive and 11% negative, showing that TFs tend to have increased capacity to regulate genes in eusocial species of bees, relative to solitary species (Fig. 2A, supplementary materials).
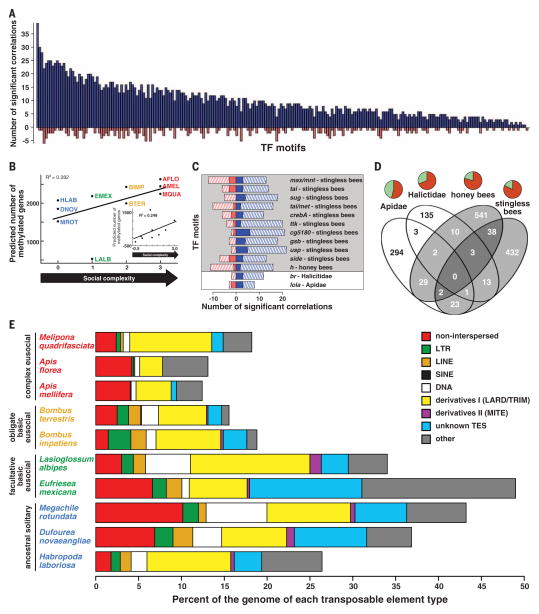
Fig. 2. Genomic signatures of evolutionary transitions from solitary to group life.
(A) Increasing social complexity is associated with increasing presence of cis-regulatory TFBSs in promoter regions. Each bar represents a TFBS for which presence correlates significantly with social complexity (blue: positive; red: negative). (B) Relationship between predicted numberof methylated genes and social complexity before and after (inset) phylogenetic correction (see text for statistics). (C) TFBS motifs showing a relationship between social complexity and evolutionary rate of coding and noncoding sequences in different lineages. Bar length indicates the number of significant correlations (blue: positive; red: negative) between each motif score and social complexity (from Table 1) among genes evolving faster (solid) or slower (hatched) in lineages with different levels of social complexity [from (D)]. Background shading follows circle shading in Fig. 1. (D) Number of genes for which evolutionary rate is faster or slower in lineages with higher compared to lower social complexity. Pie charts represent the proportion of genes evolving slower (light green) or faster (dark orange) with increased social complexity. Venn diagram shading follows circle shading in Fig. 1. (E) Complex eusocial species have a reduced proportion of repetitive DNA compared to other bees (see text for statistics). LTR, long terminal repeat; LINE, long interspersed element; SINE, short interspersed element; DNA, DNA transposon; LARD, large retrotransposon derivative; TRIM, terminal repeat retrotransposon in miniature; MITE, miniature inverted-repeat transposable element; TES, transposable elements.
Further evidence for increased capacity for gene regulation throughout social evolution is a positive ranked correlation between social complexity and the number of genes predicted to be methylated (7) (Spearman’s rho = 0.76, P = 0.01; phylogenetically corrected Spearman’s rho = 0.64, P = 0.06; Fig. 2B; bioinformatics predictions validated with bisulfite sequencing data for three invertebrate species; supplementary materials). DNA methylation affects gene expression in a variety of ways (8). Thus, this result suggests an expansion in regulatory capacity with increasingly sophisticated sociality.
The potential for increased regulatory capacity was further revealed at the protein-coding level. Increased social complexity also is associated with rapid evolution of genes involved in coordinating gene regulation. A Bayesian phylogenetic covariance analysis (9) of 5865 single-copy orthologs identified 162 genes with accelerated evolution in species with increased social complexity (7) (additional data table S3). These rapidly evolving genes were significantly enriched (P < 0.05) for Gene Ontology (GO) terms related to regulation of transcription, RNA splicing, ribosomal structure, and regulation of translation (supplementary text and tables S11 and S12). Similar results have been reported for bee and ant species (10–13); our findings reveal the underlying causes. Approximately two-thirds of these genes are under stronger directional selection in species with increasingly complex eusociality, but we also detected nonadaptive evolution. One-third of the rapidly evolving genes are under relaxed purifying selection in species with complex eusociality, possibly due to reduced effective population sizes (14).
We also found an additional 109 genes, significantly enriched (P < 0.05) for functions related to protein transport and neurogenesis, which evolve slower with increased social complexity (supplementary text, table S13, and additional data table S3). This includes orthologs of derailed 2 and frizzled, which function as Wnt signaling receptors in Drosophila synaptogenesis (15), and rigor mortis, a nuclear receptor involved in hormone signaling (16). A similar pattern of reduced evolutionary rate has been described for genes expressed in human and honey bee brains, potentially due to increasing pleiotropic constraint in complex gene networks (17, 18). Constrained protein evolution of neural and endocrine-related genes seems at odds with the evolution of complexity, but this constraint appears to be compensated for, or perhaps driven by, increased capacity for gene regulation.
We next investigated whether these molecular evolution patterns involve similar sets of genes and cis-regulatory elements among the early (facultative and obligate simple eusociality) and advanced (complex eusociality) stages of independent social transitions. We identified lineage-specific differences in coding sequences and promoter regions of 1526 “social genes” for which evolutionary rate (dN/dS) is faster or slower with increased social complexity in two independent origins and two independent elaborations of eusociality (7) (Fig. 1). Among these lineage-specific social genes, we found common patterns of cis-regulatory evolution: gains of TFBSs in the promoters of genes that evolve slower with increasing social complexity (Fig. 2C and supplementary text). This suggests that a shared feature of both independent origins and elaborations of eusociality is increasingly constrained protein evolution with increasing potential for novel gene expression patterns. The TFs responsible for this pattern were different for each social transition, even though our analysis was limited to highly conserved TFs (Table 1). Several function in neurogenesis or neural plasticity, or are prominent regulators of endocrine-mediated brain gene expression in honeybees (19, 20).
Table 1.
Transcription factors (TFs) and corresponding motifs associated with origins and elaborations of eusociality in bees. [Motif names: Fly Factor Survey (6); supplementary text.]
| Motif | D. melanogaster TFs | Hypergeometric test P-value |
|---|---|---|
| Solitary to simple eusociality–Apidae | ||
| lola_PQ_SOLEXA | Lola | 0.0047 |
| Solitary to simple eusociality–Halictidae | ||
| br_PL_SOLEXA_5 | Br | 0.0016 |
| Simple eusociality to complex eusociality–honeybees | ||
| h_SOLEXA_5 | dpn, h | 0.0027 |
| Simple eusociality to complex eusociality–stingless bees | ||
| Side_SOLEXA_5 | E_spl, HLHm3, HLHm5, HLHm7, HLHmbeta, HLHmdelta, HLHmgamma, Side | 0.0008 |
| usp_SOLEXA | EcR, svp, usp | 0.0013 |
| CrebA_SOLEXA | CrebA | 0.0040 |
| CG5180_SOLEXA | CG5180 | 0.0044 |
| tai_Met_SOLEXA_5 | Mio_bigmax, tai_Met | 0.0045 |
| ttk_PA_SOLEXA_5 | Ttk | 0.0078 |
| gsb_SOLEXA | gsb, Poxn, prd | 0.0083 |
| tai_SOLEXA_5 | Tai | 0.0100 |
We found further lineage-specific differences among the rapidly evolving “social genes” themselves. Genes undergoing accelerated evolution at the origins of eusociality were significantly enriched for GO terms related to signal transduction in both Apidae and Halictidae, but they shared only six genes (6 out of 354 and 167 genes, respectively; hypergeometric test, P = 0.82; Fig. 2D and additional data tables S5 and S6). Rapid evolution of signal transduction pathways may be a necessary step in all origins of eusociality to mediate intracellular responses to novel social and environmental stimuli (10), but selection appears to have targeted different parts of these pathways in each independent transition. Caste-specific expression and other analyses of these genes are needed to determine their function in eusociality.
Genes showing signatures of rapid evolution with the elaborations of complex eusociality were also highly disparate between honeybees and stingless bees, with only 43 shared genes and no shared enriched GO terms (43 out of 625 and 512 genes, respectively; hypergeometric test, P = 0.70; Fig. 2D and additional data tables S5 and S6). In addition, only 2 out of 5865 single-copy orthologs showed a signature of convergent evolution by fitting a dendrogram based on social complexity significantly better than the accepted molecular phylogeny (7) (supplementary text and fig. S21). Similarly, families of major royal jelly protein genes, sex-determining genes, odorant receptors, and genes involved in lipid metabolism expanded in some, but not all, lineages of complex eusocial bees (7) (Table 2 and supplementary text). These results suggest that gene family expansion is associated with complex eusociality as predicted (5), but involves different genes in each case. Despite striking convergence of social traits among the superorganisms (4), the final stages of transformation to this level of biological organization do not necessarily involve common molecular pathways.
Table 2.
Relative size of select gene families as related to social complexity in bees.
| Family | Function | Eusocial bees compared to solitary bees |
|---|---|---|
| Differences among bees | ||
| Major royal jelly | Brood feeding | Expanded only in Apis |
| Sex determination pathway genes | Sex-specific development | Expanded in some eusocial lineages |
| Odorant receptors | Olfaction | Expanded in complex eusocial lineages |
| Lipid metabolism genes | Metabolic processing of lipids | Expanded in complex eusocial lineages |
| Similarities across bees | ||
| Biogenic amines receptors, neuropeptides, GPCRs* | Neural plasticity | Similar |
| Insulin-signaling and ecdysone pathway genes | Insect development, caste determination in honeybees, behavioral plasticity as adults | Similar |
| Immunity | Infectious disease protection | Similar |
| Cytochrome P450 monooxygenase genes | Detoxification | Similar |
GPCRs, G protein–coupled receptors.
The major transitions in evolution involve a reduction in conflict as the level of natural selection rises from the individual to the group (1). Extending this to intragenomic conflict may explain our finding of decreased diversity and abundance of transposable elements (TEs) with increasing social complexity (7) (regression after phylogenetic correction, F = 8.99, adjusted R2 = 0.47, P = 0.017; Fig. 2E, figs. S42 to S44, and supplementary text). This may be a consequence of increased recombination rates among highly eusocial insects (21, 22) or because key features of complex eusociality lead to decreased exposure to parasites and pathogens that horizontally transmit TEs (4, 23). Eusociality in bees may thus provide natural immunity against certain types of intragenomic conflict.
Our results and those in (10–13) support the prediction that changes in gene regulation are key features of evolutionary transitions in biological organization (5). Our results further reveal the convergent adaptive and nonadaptive evolutionary processes common to both the early and advanced stages of multiple independent transitions from solitary to group living. It is now clear that there are lineage-specific genetic changes associated with independent origins of eusociality in bees, and independent elaborations of eusociality in both bees and ants. This includes different sets of genes showing caste-biased expression across species (24–26) and, as we have shown, evolutionary modifications of TEs, gene methylation, and cis-regulatory patterns associated with the suite of life-history traits that define eusociality. This suggests that if it were possible to “replay life’s tape” (27), eusociality may arise through different mechanisms each time, but would likely always involve an increase in the complexity of gene networks.
Acknowledgments
Data deposition at National Center for Biotechnology Information: H. laboriosa, D. novaeangliae, E. mexicana, M. quadrifasciata, and M. rotundata genome assemblies accession numbers PRJNA279436, PRJNA279825, PRJNA279814, PRJNA279820, and PRJNA66515. Funding for genome sequencing and analysis: BGI, a U.S. National Institutes of Health Pioneer Award (DP1 OD006416) to G.E.R., and European Union Marie Curie International Incoming Fellowship (300837) to G.Z. Additional funding: U.S. National Science Foundation grant DEB-0640690 (M.A.D.G.) and DEB-0743154 (G.E.R. and M.E.H); Danish Council for Independent Research grants 10-081390 (C.L.) and 0602-01170B (C.J.P.G.); Lundbeck Foundation (C.J.P.G); Georgia Tech–Elizabeth Smithgall Watts endowment (M.A.D.G.); Marie Curie International Outgoing Fellowship PIOF-GA-2011-303312 (R.M.W.); Swiss National Science Foundation award 31003A-125350, Commission Informatique of the University of Geneva, Schmidheiny Foundation, and Swiss Institute of Bioinformatics (E.M.Z.); and Natural Sciences and Engineering Research Council of Canada Discovery Grant and Early Research Award (A.Z.). This project was conducted under the auspices of the i5K Initiative. We thank the Roy J. Carver Biotechnology Center (sequencing services); N. Lartillot (advice on Coevol); T. Newman (assistance with DNA extractions); and E. Hadley (assistance with figures). Computational support: D. Davidson, N. Band, D. Slater (University of Illinois), J. H. Kidner and H. Scharpenberg (Martin-Luther-University Halle-Wittenberg); Compute Canada; and Center for High Performance Computing at the University of Utah. J. Himes created the illustrations in Fig. 1. We are grateful to T. Pitts-Singer, H. G. Hall, B. N. Danforth, J. Gibbs, and S. Cardinal for providing bee specimens; R. Ayala for identification of E. mexicana; J. Vega and the staff at Estación de Biología Chamela, Universidad Nacional Autónoma de México (UNAM), for support during collecting trips; and S. A. Cameron for insightful discussion.
Footnotes
SUPPLEMENTARY MATERIALS
www.sciencemag.org/content/348/6239/1139/suppl/DC1
Materials and Methods
Supplementary Text
Figs. S1 to S44
Tables S1 to S32
Additional Data Tables S1 to S12
References (30–151)
Author Contributions
REFERENCES AND NOTES
- 1.Maynard Smith J, Szathmáry E. The Major Transitions in Evolution. Oxford Univ. Press; Oxford, UK: 1995. [Google Scholar]
- 2.Michener CD. The Social Behavior of the Bees. Harvard Univ. Press; Cambridge, MA: 1974. [Google Scholar]
- 3.Klein AM, et al. Proc Biol Sci B. 2007;274:303–313. doi: 10.1098/rspb.2006.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hölldobler H, Wilson EO. The Superorganism: The Beauty, Elegance and Strangeness of Insect Societies. Norton; New York: 2009. [Google Scholar]
- 5.Johnson BR, Linksvayer TA. Q Rev Biol. 2010;85:57–79. doi: 10.1086/650290. [DOI] [PubMed] [Google Scholar]
- 6.Zhu LJ, et al. Nucleic Acids Res. 2011;39:D111–D117. [Google Scholar]
- 7.Materials and methods are available as supplementary materials on Science Online.
- 8.Yan H, et al. Annu Rev Entomol. 2015;60:435–452. doi: 10.1146/annurev-ento-010814-020803. [DOI] [PubMed] [Google Scholar]
- 9.Lartillot N, Poujol R. Mol Biol Evol. 2011;28:729–744. doi: 10.1093/molbev/msq244. [DOI] [PubMed] [Google Scholar]
- 10.Woodard SH, et al. Proc Natl Acad Sci USA. 2011;108:7472–7477. doi: 10.1073/pnas.1103457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harpur BA, et al. Proc Natl Acad Sci USA. 2014;111:2614–2619. doi: 10.1073/pnas.1315506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roux J, et al. Mol Biol Evol. 2014;31:1661–1685. doi: 10.1093/molbev/msu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simola DF, et al. Genome Res. 2013;23:1235–1247. doi: 10.1101/gr.155408.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romiguier J, et al. J Evol Biol. 2014;27:593–603. doi: 10.1111/jeb.12331. [DOI] [PubMed] [Google Scholar]
- 15.Park M, Shen K. EMBO J. 2012;31:2697–2704. doi: 10.1038/emboj.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gates J, Lam G, Ortiz JA, Losson R, Thummel CS. Development. 2004;131:25–36. doi: 10.1242/dev.00920. [DOI] [PubMed] [Google Scholar]
- 17.Brawand D, et al. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 18.Molodtsova D, Harpur BA, Kent CF, Seevananthan K, Zayed A. Front Genet. 2014;5:431. doi: 10.3389/fgene.2014.00431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaff DW, Arnold AP, Etgen AM, Rubin RT, Fahrbach SE, editors. Hormones, Brain and Behavior. Elsevier; New York: 2009. [Google Scholar]
- 20.Chandrasekaran S, et al. Proc Natl Acad Sci USA. 2011;108:18020–18025. doi: 10.1073/pnas.1114093108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilfert L, Gadau J, Schmid-Hempel P. Heredity. 2007;98:189–197. doi: 10.1038/sj.hdy.6800950. [DOI] [PubMed] [Google Scholar]
- 22.Dolgin ES, Charlesworth B. Genetics. 2008;178:2169–2177. doi: 10.1534/genetics.107.082743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaack S, Gilbert C, Feschotte C. Trends Ecol Evol. 2010;25:537–546. doi: 10.1016/j.tree.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldmeyer B, Elsner D, Foitzik S. Mol Ecol. 2014;23:151–161. doi: 10.1111/mec.12490. [DOI] [PubMed] [Google Scholar]
- 25.Ferreira PG, et al. Genome Biol. 2013;14:R20. doi: 10.1186/gb-2013-14-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunt BG, et al. Proc Natl Acad Sci USA. 2011;108:15936–15941. doi: 10.1073/pnas.1104825108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould SJ. Wonderful Life: The Burgess Shale and the Nature of History. Norton; New York: 1989. [Google Scholar]
- 28.Cardinal S, Danforth BN. Proc Biol Sci. 2013;280:20122686. doi: 10.1098/rspb.2012.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardinal S, Danforth BN. PLOS ONE. 2011;6:e21086. doi: 10.1371/journal.pone.0021086. [DOI] [PMC free article] [PubMed] [Google Scholar]