Abstract
Background
Alignment-free sequence comparison approaches have been garnering increasing interest in various data- and compute-intensive applications such as phylogenetic inference for large-scale sequences. While k-mer based methods are predominantly used in real applications, the average common substring (ACS) approach is emerging as one of the prominent alignment-free approaches. This ACS approach has been further generalized by some recent work, either greedily or exactly, by allowing a bounded number of mismatches in the common substrings.
Results
We present ALFRED-G, a greedy alignment-free distance estimator for phylogenetic tree reconstruction based on the concept of the generalized ACS approach. In this algorithm, we have investigated a new heuristic to efficiently compute the lengths of common strings with mismatches allowed, and have further applied this heuristic to phylogeny reconstruction. Performance evaluation using real sequence datasets shows that our heuristic is able to reconstruct comparable, or even more accurate, phylogenetic tree topologies than the kmacs heuristic algorithm at highly competitive speed.
Conclusions
ALFRED-G is an alignment-free heuristic for evolutionary distance estimation between two biological sequences. This algorithm is implemented in C++ and has been incorporated into our open-source ALFRED software package (http://alurulab.cc.gatech.edu/phylo).
Keywords: Alignment-free methods, Sequence comparison, Phylogeny reconstruction
Background
Accurate estimation of the evolutionary distance between two sequences is fundamental and critical to phylogenetic analysis aiming to reconstruct the correct evolutionary history and estimate the time of divergence between species. One popular approach to evolutionary distance estimation relies on sequence alignment. Typically, the pipeline for alignment-based phylogenetic inference generally works by three steps. Firstly, we perform all-to-all pairwise sequence alignment to gain a pairwise distance matrix for the input sequences. The evolutionary distance between two sequences in the matrix is typically inferred from an optimal alignment, e.g. equal to one minus percent identity in the optimal alignment. Secondly, we construct a guide tree from the pairwise distance matrix and then conduct progressive alignment of multiple sequences following the order determined by the guide tree. Finally, we infer a phylogenetic tree from the resulting multiple alignments using a tree inference program which can be distance-, parsimony-, bayesian, or likelihood-based. Nevertheless, it needs to be stressed that we could also choose to construct a phylogenetic tree directly from the pairwise distance matrix computed in the first step, using some distance-based tree construction algorithm such as unweighted pair group method with arithmetic mean (UPGMA) [1] or neighbor-joining (NJ) [2].
Although they may have high accuracy, alignment-based approaches involve high computational cost, thus resulting in slow speed. This is because pairwise alignment using dynamic programming has a quadratic complexity with respect to sequence length. This is even more challenging when constructing the phylogenetic tree for a large number of sequences, especially long sequences (e.g. eukaryotic genomes). In this case, some research efforts have been devoted to accelerating the tree construction using high performance computing architectures [3–6]. In addition to acceleration, as an alternative to alignment-based approaches, alignment-free approaches emerge and become popular, mainly owing to their speed superiority. For instance, given a collection of d sequences of average length n, the time complexity for pairwise distance matrix computation can be as high as O(d 2 n 2) when using pairwise alignment. In contrast, by using alignment-free exact k-mer (a k-mer is a string of k characters) counting, the whole computation can be done in O(d 2 n) time, significantly reducing the run-time by a factor of n. Moreover, alignment-free approaches are capable of overcoming some difficulties, which challenge alignment-based approaches, such as genetic recombination and shuffling during the evolution process.
A variety of alignment-free approaches have been proposed, most of which are based on the concept of sequence seeding that extracts fixed- or variable- length substrings from a given sequence. Based on fixed-length seeding, there are two kinds of alignment-free approaches: exact k-mer counting [7] and spaced k-mer counting [8]. For the exact k-mer counting approach, it builds a k-mer frequency (or occurrence) vector for each sequence and computes the pairwise distance using some distance measure based on the frequency vectors. Example distance measures include Euclidean distance [9], Kullback-Lebler divergence [10] and the one proposed by Edgar [11]. The Edgar’s distance measure models the similarity between two sequences as the fraction of exact k-mers shared by them, and then computes the pairwise distance by subtracting the similarity value from one. This distance measure has been shown to be highly related to genetic distance and has been used in other applications like metagenomic sequence classification [12]. For the spaced k-mer counting approach, it allows character mismatches between k-mers at some predefined positions and usually employs multiple pattern templates in order to improve accuracy.
Based on variable-length seeding, there are three kinds of approaches: the average common substring (ACS) method [13], the k-mismatch ACS (k-ACS) method [14, 15] and the mutation distances (K r) [16]. The distance based on these methods can be computed using suffix trees/arrays. Given two sequences, the ACS method first calculates the length of the longest substring that starts at each position i in one sequence and matches some substring of the other sequence. Subsequently, it averages and normalizes all of the lengths computed to represent the similarity of the two sequences. Finally, the resulting similarity value is used to compute the pairwise distance. The time complexity of the ACS method is directly proportional to the sum of lengths of the two sequences.
In contrast, the k-ACS method computes the pairwise distance by finding substring pairs with upto k mismatches, given two sequences. Specifically, instead of determining the longest common substrings, the k-ACS method aims to find the longest substring starting at each position in one sequence and matching some substring in the other sequence with upto k mismatches. The mutation distances is closely related to ACS, where the difference is only in the conversion from the similarity value to a pairwise distance.
Unlike the ACS method, the solutions to the k-ACS method involves high computational cost. For example, an algorithm given by Leimeister and Morgenstern [14] takes O(k n 2) time in the worst case, which is certainty not a suitable replacement of alignment based methods. However, they proposed a faster algorithm, namely kmacs, that computes an approximation to k-ACS based distance. Another algorithm by Apostolico et al. runs in O(n 2/ logn) time [17]. This raises an open question, whether the exact k-ACS based distance can be computed in strictly sub-quadratic time. Initial attempts were focused on the special case of k=1 [18, 19]. Later, Aluru et al. [15, 20] positively answered this question by presenting an algorithm with a worst case run time of O(n logk n) for any constant k. The algorithm is much more complicated than the original ACS method and even the k-ACS approximation by [14]. Moreover the practical variant of this algorithm can get quite slow for even moderately large values of k due to its exponential dependency on k [21]. However, this algorithm has its merit as the first sub-quadratic time algorithm for exact k-ACS computation for any positive integer k. A recently proposed algorithm by Pizzi is based on filtering approaches [22]. In summary, on one hand, we have a fast approximation algorithm [14] and on the other hand, we have an exact (theoretical) algorithm [15], that might work well for small values of k in practice. Inspired by both algorithms, we introduce a new greedy heuristic for alignment-free distance estimation, named ALFRED-G. The heuristic is implemented in C++ and has been incorporated into our open-source ALFRED software package (http://alurulab.cc.gatech.edu/phylo).
We use X and Y to denote the two sequences to be compared. The length of sequence X is denoted by |X|, its ith character by X[i], and the substring that starts at position i and ends at position j by X[i…j]. For brevity, we use X i to denote the suffix of X starting at i. The total length of X and Y is denoted by n. A key data structure in our algorithm is the generalized suffix tree (GST). The GST of X and Y is a compact trie of all suffixes of X and Y. It consists of n leaves and at most n−1 internal nodes. Corresponding to each leaf, there is a unique suffix of X or Y. The edges are labeled with a sequence of characters. The string-depth of a node u is the length of the string obtained by concatenating the edge labels on the path from the root of GST to u. The space and the construction time of GST are O(n) [23]. For any (i,j) pair, |LCP(X i,Y j)|, the length of the longest common prefix of X i and Y j is same as the string-depth of the lowest common ancestor node of the leaves corresponding to X i and Y j. Using GST, we can compute it in constant time. Also, we can compute |LCP k(X i,Y j)|, the length of the longest common prefix of X i and Y j with first k mismatches ignored, in O(k) time as follows. Let z=|LCP(X i,Y j)|, then for any k≥1,
| 1 |
Problem definition
The k-mismatch average common substring of X w.r.t. Y, denoted by ACS k(X,Y) is defined as the average of the length of the prefix of a suffix of X, that appears as a substring of Y within hamming distance k. Specifically, let λ k(i)= maxj|LCP k(X i,Y j)|, then
| 2 |
The distance Dist k(X,Y), based on ACS k is given below [13, 14].
| 3 |
Methods
Approximating ACSk(·,·)
It is observed that ACS k(·,·) can be easily computed in O(n 2 k) time via |X|×|Y| number of |LCP k(·,·)| queries, which is clearly not affordable. The first attempt to circumvent this issue was made by Leimeister and Morgenstern [14], who presented a heuristic method, named kmacs, that quickly computes an approximation to ACS k(X,Y). The key idea is to replace λ k(i) with λ k′(i) in the equation for ACS k, where α i=a r g maxj|LCP(X i,Y j)| and . Using GST, we can compute α i for all values of i in O(n) time. Therefore, λ k′(i) for all values of i and the corresponding distance can be easily obtained in O(n k) time. Note that the ratio of λ k(i) to λ k′(i) can be as high as Θ(n). Nonetheless, it has been shown that for most practical cases, the average of the latter can serve as a good approximation to the average of the former.
Our approach
The idea is to follow a simple adaptation of Aluru et al.’s exact algorithm [15] for 1-mismatch case and then use the heuristic approach by Leimeister and Morgenstern [14] to extend the result to k-mismatch. Specifically, our approximation to ACS k is obtained by replacing λ k(i) in the equation for ACS k by λ k″(i), where β i=a r g maxj|LCP 1(X i,Y j)| and . To compute β i for i=1,2,…,|X|, we first construct GST and an array A[1,|X|]. Then for each internal node u in GST, process the set of suffixes corresponding to the leaves in the subtree of u. Let h be the string-depth of u. Then (h+1) is the first position, in which the prefixes of two suffixes in can differ. We sort all suffixes in by treating the (h+1)th character all suffixes to be identical, or equivalently first (h+1) characters to be the same. To do so, we follow the steps below:
Map each to a pair (X i,k e y), where key is the lexicographic rank of the suffix X i+h+1 among all suffixes of X and Y. In other words, key is the lexicographic rank of the suffix obtained by deleting the first (h+1) characters of X i. Using GST, we can compute key in constant time.
Likewise, map each to a pair (Y j,k e y), where key is the lexicographic rank of Y j+h+1 among all suffixes of X and Y.
Sort all pairs in the ascending order of key.
For each pair (X i,·), find the closest pairs, say (Y a,·) and (Y b,·), towards the left and right side (if they exist) that are created from a suffix of Y, and update A[i]←a r g maxj∈{a,b,A[i]}|LCP 1(X i,Y j)|.
After processing all internal nodes as described above, compute the following and report it as our approximation to ACS k(X,Y)
It can be easily verified that A[i] will be correctly updated to β i while processing the lowest common ancestor node of the leaves corresponding to X i and . The overall run time is , where H is the height of GST and its expected value is O(logn) [24].
Implementation
ALFRED-G is implemented in C++ and is incorporated in our open-source ALFRED software package (http://alurulab.cc.gatech.edu/phylo). This algorithm takes a collection of sequences as input and computes an approximation to ACS k(·,·) for all pairs of sequences. For this, we have used the open-source libdivsufsort library [25] to construct the suffix array (SA) and have used the implementations in the SDSL library [26] to build the corresponding LCP table (using the Kasai algorithm [27]) and the range minimum query (RMQ) table (using the Bender-Farach’s algorithm [28]). (Note that the operations on a suffix tree can be simulated using the corresponding SA, inverse SA, LCP array and RMQ table). The SDSL library has support for using bit compression techniques to reduce the size of the tables and arrays in exchange for slower query time. However, we don’t compress these data structures, and instead we have used 32-bit integers for indices as well as prefix lengths.
Results and discussion
Benchmark datasets
We have assessed the performance of ALFRED-G for the reconstruction of phylogenetic trees by using three sequence datasets, which contain prokaryotic DNA sequences, eukaryotic DNA sequences, and protein sequences, respectively. The prokaryotic sequence dataset consists of 27 Primate mitochondrial genomes, which was previously studied by [16] in order to assess the performance of alignment-free approaches for phylogenetic tree reconstruction. In the study, a reference tree was constructed based on a multiple alignment of the sequences.
The eukaryotic sequence dataset is constructed by Newton et al. [29] from 32 Roseobacter genomes, by extracting 70 universal single-copy genes for the 32 genomes with each gene being completely sequenced in all genomes and having no ambiguous start/stop sites. The 70 genes for each genome are, subsequently, concatenated and aligned with ClustalW in Geneious 4.0 (available from http://www.geneious.com) using Escherichia coli K12 substrain MG1655 as the outgroup. The multiple sequence alignment file is available at http://alurulab.cc.gatech.edu/phylo, from which the raw sequences corresponding to the 32 Roseobacter genomes are extracted and then used in our study. In our study, we have used the phylogenetic tree presented in Newton et al. [29] as the reference tree.
The protein sequence dataset is taken from BAliBASE (v3.0) [30], which is popular benchmark dataset for multiple sequence alignment. We have used 218 sets of protein sequences in BAliBASE, and constructed the reference trees from the corresponding reference alignments using the proml program available in PHYLIP [31], which implements the Maxmimum Likelihood method. For each of the parameter selected for our experiments, we report the average RF-distance of the 218 trees constructed from this set.
Phylogenetic tree construction and comparison
Given a set of d sequences, we first compute the distance between any sequence pair and then construct a pairwise distance matrix of size d×d. Subsequently, the neighbor-joining (NJ) algorithm [2] is applied on the pairwise distance matrix to reconstruct the phylogenetic tree, where the neighbor program in PHYLIP is used. Finally, the topology of the tree is compared with the reference tree using the Robinson-Foulds (RF) distance metric, where the treedist program in PHYLIP is used to compute the RF distance between two trees. Note that the lower the RF distance is, the better the tree topology matches. In particular, if the RF distance equals zero, it means exact topology match between the two trees.
All experiments are preformed in an Apple Macbook Pro (Mid-2012 model) running Mac OS 10.10.4 (OS X Yosemite). The machine features a 2.9 GHz dual-core Intel Core i7-3667U processor with 4MB L3 cache and 8GB RAM.
Performance comparison
As our method is closely related to kmacs, we compared the performance of ALFRED-G with kmacs in terms of speed and accuracy (based on RF-distance) for different values of k, ranging from 0 to 9. Note that for k=0, both kmacs and ALFRED-G are the same as the ACS method.
Figure 1 shows the results for the prokaryotic dataset. It can be observed that for all values of k, ALFRED-G provides either the same or better accuracy (in terms of RF distance). Interestingly, for k=4 and 5, the phylogenetic tree created based on ALFRED-G coincides exactly with the reference tree (see Fig. 2). We notice that the only other alignment-free method, that was able to recreate this exact reference tree is the recently proposed spaced-seed method [8] (but needs careful parameter turning).
Fig. 1.
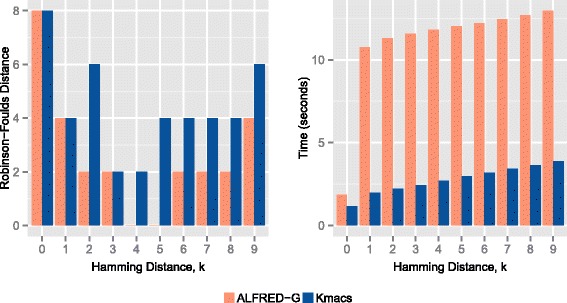
RF distance and run-time plots for the prokaryotic dataset
Fig. 2.

Tree generated by ALFRED-G for the prokaryotic dataset with k=4
Figure 3 shows the results for the eukaryotic dataset. Likewise, our RF distance is never worse than that obtained by kmacs. In particular, when setting k=6,7 and 8, our RF distance is lower, indicating better performance. Figure 4 shows the topological comparison between the tree generated by our approach and the reference tree, which is generated by the Dendroscope software [32].
Fig. 3.

RF distance and run-time plots for the eukaryotic dataset
Fig. 4.

Reference tree and the tree generated by ALFRED-G for the eukaryotic dataset with k=7 (RF distance = 8)
Figure 5 shows the results for the protein dataset. Here both ALFRED-G and kmacs gave almost the same RF score for each value of k. As expected, ALFRED-G is slower than kmacs (by a factor of 2 to 4), however the difference in run-time is independent of k.
Fig. 5.
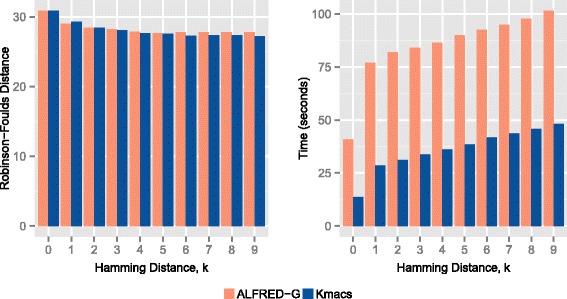
RF distance and run-time plots for the BAliBASE protein dataset
In the earlier work by Leimeister and Morgenstern [14], it has been show that kmacs and spaced-seed [8] are superior to other alignment-free methods, when applied to the aforementioned three datasets. Our experiments show that ALFRED-G is comparable and often more accurate than kmacs, albeit involving higher computational cost. It needs to be mentioned that the comparison with spaced-seed is not as straightforward as with kmacs, because spaced-seed has different input parameters and requires tedious pattern templates tuning. Nevertheless, we have carefully evaluated spaced-seed based on the suggestions from [8]. Our evaluation shows that spaced-seed is able to recover the entire reference tree (i.e. RF distance = 0) for the prokaryotic dataset, in just 4 seconds. However, for the rest, the performance of spaced-seed is roughly comparable to both ours and kmacs.
Conclusions
In this paper, we have introduced a greedy alignment-free approach to estimating the evolutionary distance between two sequences. The core of the heuristic is to identify a 1-mismatch longest substring in sequence Y that appears as a prefix of any given suffix in sequence X, and vice versa. This heuristic has been further applied to reconstruct the phylogenetic tree, given a collection of sequences that are believed to be close enough and have some evolutionary relationship between them. The performance of our heuristic has been evaluated using three real datasets: one prokaryotic dataset, one eukaryotic dataset and one protein dataset, in terms of tree-topology RF score and speed. Our experimental results show that our heuristic can exactly reconstruct the same phylogenetic tree topology with the reference tree for the prokaryotic dataset, whereas kmacs cannot. On the remaining two datasets, our heuristic also demonstrates comparable or even better performance than kmacs. As for speed, our heuristic is slightly slower than kmacs.
Although our heuristic has been shown effective for phylogenetic inference, there are still some limitations that could be improved in the future. Firstly, our heuristic assumes an evolution model having only mismatches, not involving insertions or deletions, for simplicity. This model may not exactly fit the real evolutionary process given a collection of sequences. Nevertheless, our performance evaluation has shown that even though there are some insertions or deletions between sequences (observed from multiple sequence alignment), their evolutionary distances can still be estimated with reasonable accuracy using our heuristic. However, it should be noted that the existence of insertions or deletions may cause our heuristic to underestimate the similarity values, i.e. ACS k(·,·), between sequences, thus overestimating their distances, i.e. Dist k(X,Y).
Secondly, our heuristic assumes that the homologous regions between two sequences are on the same strand. Actually, this is not always the case. Given a homologous region, the substring in sequence X may have an opposite strand to the corresponding homology in sequence Y. In this case, directly applying our heuristic to such sequences may overestimate the distance, since these homologies with opposite strands are not counted in the computation of similarity values. In some sense, we would expect that the estimation accuracy of alignment-free approaches could be further improved by incorporating support for strand differences in homologies.
Thirdly, our heuristic has only used Eq. (3) to estimate the distance from the similarity values computed from Eq. (2). Actually, we usually need to tune distance equations for different similarity computation approaches and even for similarity values in different ranges. For example, Edgar [11] used percent identity D (0≤D≤1) between two sequences as a similarity measure, but proposed to use two different distance computations depending on the value of D. In this case, Edgar computed the distance as − ln(1−D−D 2/5) if D>0.25, and retrieved the distance value from a pre-computed lookup table, otherwise. Hence, it may be beneficial to design some new distance computation equations that better match our approach. Finally, considering the generality and fast speed of our heuristic, we would expect that related research in bioinformatics and computational biology could benefit from our algorithm.
Acknowledgements
This research is supported in part by the U.S. National Science Foundation grant IIS-1416259. We thank the reviewers of this article and its preliminary version [33]. We also thank the authors of [29] for sharing the multiple sequence alignment file for the 32 Roseobacter genomes.
Funding
The funding for publication of the article was by the U.S. National Science Foundation grant IIS-1416259.
Availability of data and material
Both dataset and code are available at http://alurulab.cc.gatech.edu/phylo.
Authors’ contributions
ST conceived the algorithm and wrote the initial manuscript; SC implemented the code and performed some experiments; YL wrote the manuscript; AK performed the experiments; SA conceptualized the study. All authors have read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
About this supplement
This article has been published as part of BMC Bioinformatics Volume 18 Supplement 8, 2017: Selected articles from the Fifth IEEE International Conference on Computational Advances in Bio and Medical Sciences (ICCABS 2015): Bioinformatics. The full contents of the supplement are available online at https://bmcbioinformatics.biomedcentral.com/articles/supplements/volume-18-supplement-8.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
From Fifth IEEE International Conference on Computational Advances in Bio and Medical Sciences(ICCABS 2015) Miami, FL, USA.15-17 October 2015
Contributor Information
Sharma V. Thankachan, Email: sharma.thankachan@ucf.edu
Sriram P. Chockalingam, Email: sriram.pc@iitb.ac.in
Yongchao Liu, Email: yliu860@gatech.edu.
Ambujam Krishnan, Email: krishnambujam@gmail.com.
Srinivas Aluru, Email: aluru@cc.gatech.edu.
References
- 1.Sokal RR. A statistical method for evaluating systematic relationships. Univ Kans Sci Bull. 1958;38:1409–38. [Google Scholar]
- 2.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 3.Stewart CA, Hart D, Berry DK, Olsen GJ, Wernert EA, Fischer W. Parallel implementation and performance of fastdnaml-a program for maximum likelihood phylogenetic inference. In: Supercomputing, ACM/IEEE 2001 Conference. IEEE: 2001. p. 32–2.
- 4.Ott M, Zola J, Stamatakis A, Aluru S. Large-scale maximum likelihood-based phylogenetic analysis on the ibm bluegene/l. In: Proceedings of the 2007 ACM/IEEE Conference on Supercomputing. ACM: 2007. p. 4.
- 5.Liu Y, Schmidt B, Maskell DL. Parallel reconstruction of neighbor-joining trees for large multiple sequence alignments using cuda. In: Parallel & Distributed Processing, 2009. IPDPS 2009. IEEE International Symposium On. IEEE: 2009. p. 1–8.
- 6.Zhou J, Liu X, Stones DS, Xie Q, Wang G. Mrbayes on a graphics processing unit. Bioinformatics. 2011;27(9):1255–61. doi: 10.1093/bioinformatics/btr140. [DOI] [PubMed] [Google Scholar]
- 7.Vinga S, Almeida J. Alignment-free sequence comparison-a review. Bioinformatics. 2003;19(4):513–23. doi: 10.1093/bioinformatics/btg005. [DOI] [PubMed] [Google Scholar]
- 8.Leimeister CA, Boden M, Horwege S, Lindner S, Morgenstern B. Fast alignment-free sequence comparison using spaced-word frequencies. Bioinformatics. 2014; 30(14):1991. doi:http://dx.doi.org/10.1093/bioinformatics/btu177,http://dx.doi.org/10.1093/bioinformatics/btu177. [DOI] [PMC free article] [PubMed]
- 9.Blaisdell BE. Effectiveness of measures requiring and not requiring prior sequence alignment for estimating the dissimilarity of natural sequences. J Mol Evol. 1989;29(6):526–37. doi: 10.1007/BF02602924. [DOI] [PubMed] [Google Scholar]
- 10.Wu TJ, Hsieh YC, Li LA. Statistical measures of dna sequence dissimilarity under markov chain models of base composition. Biometrics. 2001;57(2):441–8. doi: 10.1111/j.0006-341X.2001.00441.x. [DOI] [PubMed] [Google Scholar]
- 11.Edgar RC. Muscle: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma. 2004;5(1):1. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun Y, Cai Y, Liu L, Yu F, Farrell ML, McKendree W, Farmerie W. Esprit: estimating species richness using large collections of 16s rrna pyrosequences. Nucleic Acids Res. 2009;37(10):76–6. doi: 10.1093/nar/gkp285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulitsky I, Burstein D, Tuller T, Chor B. The average common substring approach to phylogenomic reconstruction. J Comput Biol. 2006;13(2):336–50. doi: 10.1089/cmb.2006.13.336. [DOI] [PubMed] [Google Scholar]
- 14.Leimeister CA, Morgenstern B. kmacs: the k-mismatch average common substring approach to alignment-free sequence comparison. Bioinformatics. 2014;30(14):2000–8. doi: 10.1093/bioinformatics/btu331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aluru S, Apostolico A, Thankachan SV. Efficient alignment free sequence comparison with bounded mismatches. In: International Conference on Research in Computational Molecular Biology. Springer: 2015. p. 1–12.
- 16.Haubold B, Pfaffelhuber P, Domazet-Loso M, Wiehe T. Estimating mutation distances from unaligned genomes. J Comput Biol. 2009;16(10):1487–500. doi: 10.1089/cmb.2009.0106. [DOI] [PubMed] [Google Scholar]
- 17.Apostolico A, Guerra C, Landau GM, Pizzi C. Sequence similarity measures based on bounded hamming distance. Theor Comput Sci. 2016;638:76–90. doi: 10.1016/j.tcs.2016.01.023. [DOI] [Google Scholar]
- 18.Flouri T, Giaquinta E, Kobert K, Ukkonen E. Longest common substrings with k mismatches. Inf Process Lett. 2015;115(6):643–7. doi: 10.1016/j.ipl.2015.03.006. [DOI] [Google Scholar]
- 19.Manzini G. Longest common prefix with mismatches. In: International Symposium on String Processing and Information Retrieval. Springer: 2015. p. 299–310.
- 20.Thankachan SV, Apostolico A, Aluru S. A provably efficient algorithm for the k-mismatch average common substring problem. J Comput Biol. 2016;23(6):472–82. doi: 10.1089/cmb.2015.0235. [DOI] [PubMed] [Google Scholar]
- 21.Thankachan SV, Chockalingam SP, Liu Y, Apostolico A, Aluru S. Alfred: a practical method for alignment-free distance computation. J Comput Biol. 2016;23(6):452–60. doi: 10.1089/cmb.2015.0217. [DOI] [PubMed] [Google Scholar]
- 22.Pizzi C. Missmax: alignment-free sequence comparison with mismatches through filtering and heuristics. Algorithm Mol Biol. 2016;11(1):1. doi: 10.1186/s13015-016-0072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiner P. Linear pattern matching algorithms. In: Switching and Automata Theory, 1973. SWAT’08. IEEE Conference Record of 14th Annual Symposium On. IEEE: 1973. p. 1–11.
- 24.Devroye L, Szpankowski W, Rais B. A note on the height of suffix trees. SIAM J Comput. 1992;21(1):48–53. doi: 10.1137/0221005. [DOI] [Google Scholar]
- 25.Mori Y. Libdivsufsort: a lightweight suffix array construction library. 2003.
- 26.Gog S, Beller T, Moffat A, Petri M. From theory to practice: Plug and play with succinct data structures. In: International Symposium on Experimental Algorithms. Springer: 2014. p. 326–37.
- 27.Kasai T, Lee G, Arimura H, Arikawa S, Park K. Linear-time longest-common-prefix computation in suffix arrays and its applications. In: Annual Symposium on Combinatorial Pattern Matching. Springer: 2001. p. 181–92.
- 28.Bender MA, Farach-Colton M. The lca problem revisited. In: Latin American Symposium on Theoretical Informatics. Springer: 2000. p. 88–94.
- 29.Newton RJ, Griffin LE, Bowles KM, Meile C, Gifford S, Givens CE, Howard EC, King E, Oakley CA, Reisch CR, et al. Genome characteristics of a generalist marine bacterial lineage. ISME J. 2010;4(6):784–98. doi: 10.1038/ismej.2009.150. [DOI] [PubMed] [Google Scholar]
- 30.Thompson JD, Koehl P, Ripp R, Poch O. Balibase 3.0: latest developments of the multiple sequence alignment benchmark. Proteins Struct Funct Bioinforma. 2005;61(1):127–36. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- 31.Felsenstein J. {PHYLIP}: phylogenetic inference package, version 3.5 c. 1993.
- 32.Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, Rupp R. Dendroscope: an interactive viewer for large phylogenetic trees. BMC Bioinforma. 2007;8(1):1. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thankachan SV, Chockalingam SP, Liu Y, Krishnan A, Aluru S. A greedy alignment-free distance estimator for phylogenetic inference. In: International Conference on Computational Advances in Bio and Medical Sciences (ICCABS). IEEE: 2015. p. 1–1. [DOI] [PMC free article] [PubMed]


