Abstract
Five commercial broiler flocks were treated with a fluoroquinolone for a clinically relevant infection. Fresh feces from individual chickens and environmental samples were cultured for campylobacters before, during, and weekly posttreatment until slaughter. Both Campylobacter jejuni and C. coli were isolated during all treatment phases. An increased proportion of quinolone-resistant strains was seen during treatment, and these strains persisted posttreatment. One quinolone-resistant isolate of each species, each serotype, and each phage type from each sample at all treatment phases was examined for its phenotype and mechanism of resistance. Two resistant phenotypes were isolated: Nalr Cipr and Nalr Cips. The majority (269 of 290) of fluoroquinolone-resistant isolates, whether they were C. jejuni or C. coli, had a mutation in gyrA that resulted in the substitution Thr-86→Ile. The other gyrA mutations detected were Thr-86→Ala (n = 17) and Asp-90→Asn (n = 10). The genotypic variation, based on the silent mutations in gyrA identified by the denaturing high-performance liquid chromatography pattern and DNA sequencing, was used to supplement typing data and provided evidence for both the spread of preexisting resistant strains and the selection of spontaneous resistant mutants in treated flocks. Multidrug resistance was significantly (P < 0.01) associated with resistance to ciprofloxacin. Twenty-five percent (73 of 290) of ciprofloxacin-resistant isolates but only 13% (24 of 179) of susceptible isolates were resistant to three or more unrelated antimicrobial agents. In conclusion, quinolone-resistant campylobacters were isolated from commercial chicken flocks in high numbers following therapy with a veterinary fluoroquinolone. Most ciprofloxacin-resistant isolates had the GyrA substitution Thr-86→Ile. Resistant isolates were isolated from the feces of some flocks up to the point of slaughter, which may have consequences for public health.
Campylobacter spp. are a common cause of gastroenteritis in humans, and while most cases of campylobacteriosis do not require antimicrobial therapy, treatment may be essential for vulnerable patients and for the management of invasive disease. Fluoroquinolones have been widely used for the treatment of Campylobacter infections, but the incidence of resistance among Campylobacter jejuni and C. coli strains isolated from humans increased significantly throughout the world during the 1990s (10, 29).
It has been widely postulated that the increase in the numbers of fluoroquinolone-resistant campylobacters isolated from human infections results from the emergence of resistant strains in poultry and their subsequent consumption (9, 15, 37). Recent studies have reported a high frequency of fluoroquinolone-resistant campylobacters among poultry flocks (7, 37). Fluoroquinolone-resistant campylobacters have not been detected in domestically acquired human infections in Australia, and this has been attributed to the fact that fluoroquinolones have not been licensed for use in food animals (36).
The primary target of fluoroquinolones in Campylobacter is DNA gyrase, and resistance arises as a result of mutations in the quinolone resistance-determining region (QRDR) of the gyrA gene, which encodes the A subunit of the target enzyme (17, 38). The majority of highly fluoroquinolone-resistant clinical isolates of C. jejuni have the GyrA substitution Thr-86 to Ile (5, 15, 30, 33, 38, 41), which is sufficient to confer high-level resistance. Other less common substitutions, as well as silent polymorphisms, have been reported in the QRDR (20, 30, 33, 38, 41). Substitutions in the B-subunit gene, gyrB, have yet to be documented in fluoroquinolone-resistant campylobacters. One report (16) has described mutations in parC associated with fluoroquinolone resistance in C. jejuni, but that finding has not been supported by other studies (1, 22, 27, 30). Evidence for an efflux pump with broad specificity was found in two laboratory-derived multiply antibiotic-resistant C. jejuni strains (6). Recently, a gene encoding an efflux pump protein, CmeB, has been described, and inactivation of cmeB by insertional mutagenesis has been shown to increase the susceptibility of C. jejuni to several antibiotics, including ciprofloxacin (21, 31).
To explore the hypothesis that fluoroquinolone-resistant campylobacters arise in poultry during treatment, this study investigated the incidence and mechanism of fluoroquinolone resistance in Campylobacter strains isolated from commercial broiler flocks treated for a clinically relevant infection with a veterinary fluoroquinolone. The aims of the study were to determine (i) the baseline incidence of fluoroquinolone resistance in flocks prior to fluoroquinolone exposure; (ii) the level of resistance in treated flocks; (iii) whether fluoroquinolone-resistant campylobacters spread through a flock as a result of the selection of one or several resistant clones; (iv) whether the types of fluoroquinolone-resistant campylobacters and the mechanisms of resistance that emerge in poultry isolates are similar or identical to those in isolates from humans; and finally, (v) whether the mechanisms of resistance in poultry isolates confer sufficiently high-level resistance to be untreatable in human infections, i.e., whether they confer full clinical resistance.
This large study was performed by three groups in the United Kingdom: the Food Microbiology Collaborating Unit, Bristol; the Campylobacter Reference Unit (CRU), London; and the Antimicrobial Agents Research Group, Birmingham. The prevalence and subtypes of ciprofloxacin-resistant campylobacters isolated from five commercial chicken flocks treated with a therapeutic fluoroquinolone are described in detail in the accompanying article (20a). This report describes the incidence and mechanism of fluoroquinolone resistance in campylobacters isolated from fluoroquinolone-treated flocks.
MATERIALS AND METHODS
Treatment of flocks and sampling procedures.
An alert system was put in place for chicken flocks reared in southwest England and elsewhere, whereby the Food Microbiology Collaborating Unit was informed when a commercial flock was about to be treated with a fluoroquinolone for a clinically relevant infection (20a). Six flocks were treated, although one flock (flock 2) remained campylobacter free throughout the sampling period. Birds from flock 1 (ca. 500 barn-reared birds), flock 3 (a free-range flock of ca. 300 birds), flock 4 (a large broiler flock of ca. 20,000 birds), and flock 5 (ca. 1,250 free-range birds) were all treated with difloxacin (Dicural; Fort Dodge Animal Health); and flock 6 (ca. 5,000 free-range birds) received enrofloxacin (Baytril; Bayer). All flocks were treated as instructed by a veterinarian, in accordance with the procedure recommended by the manufacturer (10 mg per kg per bird for 5 days administered in the drinking water).
Up to 14 samples of fresh feces were collected from individual chickens pretreatment (1 to 5 days prior to the start of treatment), during treatment (2 to 5 days after the start of treatment), and after treatment (weekly for up to 4 weeks posttreatment) until the flocks were slaughtered. Samples were also collected from the environment (pooled feed and litter, drinking or puddle water, and swabs of broiler house walls and floors). Campylobacter was isolated by direct or enrichment culture, as described in the accompanying report (20a). Up to six isolates from each fecal sample and three isolates from each environmental sample (a total of 1,630 isolates) were sent to CRU for species identification, serotyping, and phage typing and were screened for fluoroquinolone resistance by using breakpoint testing (20a, 35).
Referral of isolates from CRU.
One quinolone-resistant isolate of each species, phage type, and serotype was investigated from each fecal or environmental sample from each flock during each treatment phase. At least three sensitive isolates from each treatment phase per flock were chosen for comparison with resistant isolates. Bacteria were grown on Mueller-Hinton agar plus 5% horse blood at 37°C in 7.5% CO2 and were stored at −80°C on Protect beads (Technical Service Consultants Ltd., Heywood, United Kingdom).
Determination of antibiotic resistance.
The agar doubling dilution procedure recommended by the NCCLS Campylobacter Working Group (24) was used throughout the study. Mueller-Hinton medium plus 5% defibrinated horse blood and incubation at 37°C in 7.5% CO2 were used to determine the MICs of marker antibiotics of each chemical class (ciprofloxacin, nalidixic acid, erythromycin, tetracycline, chloramphenicol, trimethoprim, kanamycin, and ampicillin), marker dyes (acridine orange and ethidium bromide), marker detergents (sodium deoxycholate and sodium dodecyl sulfate), and marker disinfectants (triclosan and cetrimide). C. jejuni NCTC 11168 and C. coli NCTC 11366 were used as control strains. As no internationally recognized breakpoint concentrations are available for Campylobacter spp., designation of antibiotic susceptible or resistant was made with reference to the guidelines of the British Society for Antimicrobial Chemotherapy and NCCLS for human infections (23, 25) to determine the relevance of any resistance observed to public health. MICs of ≥8 μg/ml for ethidium bromide or ≥16 μg/ml for acridine orange, sodium deoxycholate, and sodium dodecyl sulfate were taken to indicate resistance. All susceptibility data were confirmed on at least two separate occasions.
Detection of mutations in gyrA and gyrB.
Bacteria were grown on Mueller-Hinton agar containing 5% defibrinated horse blood at 37°C in 7.5% CO2 for 48 h. Bacterial colonies were harvested from the agar plate, and a turbid suspension was prepared in sterile distilled water. Genomic DNA was extracted with a DNAace spin cell culture kit (Bioline, London, United Kingdom). The QRDR of C. jejuni gyrA was amplified by PCR with primers 293 (5′-GCCTGACGCAAGAGATGGTT-3′) and 343 (5′-CATCGCAGCGGCACTATCAC-3′) to generate an amplimer of 259 bp covering codons 39 to 123 of gyrA. The QRDR of C. coli gyrA was amplified with primers 344 (5′-TCCTGATGCTAGAGATGGCT-3′) and 345 (5′-CCATCACCATCGATAGAACC-3′) to generate an amplimer of 246 bp covering codons 39 to 118 of gyrA. PCR was performed with a reaction mixture volume of 50 μl by using PCR Master Mix (Abgene, Epsom, United Kingdom), 0.5 μg of DNA, and 250 nM each primer. An initial denaturation was carried out at 94°C for 5 min, followed by 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 10 min. The QRDR of gyrB was amplified as two fragments: amplimer A was amplified with primers 341 (5′-TAGAGGAAGAGAAGCAGCGA-3′) and 342 (5′-CTTCACCTATACCACAGCCA-3′), and amplimer B was amplified with primers 348 (5′-AGCTATACTGCCTTTGCGTG-3′) and 349 (5′-GATCCATCAACATCCGCATC-3′), which generated amplimers of 305 bp (codons 380 to 480) and 186 bp (codons 442 to 502), respectively. PCR of gyrB was essentially the same as that for gyrA, except that a lower annealing temperature of 48°C was used. All primers were synthesized commercially by MWG Biotech (Milton Keynes, United Kingdom).
Mutations in gyrA were detected by denaturing high-performance liquid chromatography (DHPLC) analysis (8). PCR amplimers from each isolate were mixed with an equal quantity of DNA (5 μg) amplified from a wild-type control strain (C. jejuni NCTC 11168 or C. coli NCTC 11366, as appropriate). The DNA mixture was denatured at 95°C for 4 min and then slowly reannealed by cooling to 35°C at 1°C per min to allow the formation of heteroduplexes, as described previously (8). Duplex products were screened for mutations in gyrA by using the WAVE nucleic acid fragment analysis system (Transgenomic, Crewe, United Kingdom), essentially as described previously (8). The column temperature used for analysis of C. jejuni gyrA was 58°C, and that used for analysis of C. coli was 57°C. Elution profiles were analyzed with Navigator software (version 1.4.1 and, later, version 1.5.1). DNA sequencing of novel DHPLC patterns was performed commercially by the Functional Genomics Laboratory, University of Birmingham. The sequences were compared to the published DNA sequences of C. jejuni (38; GenBank accession number L04566) and C. coli (40; GenBank accession number AF092101).
Mutations in C. jejuni gyrB were also detected by DHPLC analysis. Analysis of the gyrB gene with Navigator software showed that mutations could be detected by DHPLC only in the first 200 bp of the PCR amplimer (amplimer A) due to the melting profile of this section of the gene. Therefore, a set of primers was designed to amplify a further 186 bp (amplimer B). Both amplimers were analyzed by DHPLC as described above; amplimer A was analyzed with a column temperature of 57°C, and amplimer B was analyzed with two column temperatures: 55 and 56°C. The data from all analyses were compared to enable the QRDR of gyrB to be screened for mutations. DNA was sequenced as described above for any strains with novel DHPLC patterns.
RESULTS
Prevalence and subtypes of ciprofloxacin-resistant Campylobacter spp. in fluoroquinolone-treated flocks.
Both C. jejuni and C. coli were isolated from samples collected during all treatment phases. Ciprofloxacin-resistant C. jejuni and/or C. coli strains were detected pretreatment in four flocks, but they constituted a very small proportion of the campylobacters present. A rapid increase in the proportion of ciprofloxacin-resistant campylobacters was observed during treatment, and this increase persisted posttreatment. During treatment nearly 100% of campylobacters were resistant, and in some flocks a high proportion of resistant strains persisted for up to 4 weeks after treatment.
Prior to treatment a variety of campylobacter subtypes were present, predominantly susceptible subtypes of C. jejuni. Considerable changes in both species and subtype prevalence were observed during and after treatment, but no single fluoroquinolone-resistant clone became dominant. Instead, resistant C. coli strains or a mixture of resistant C. coli and C. jejuni strains became dominant. The resistant subtypes which emerged and became dominant were not always the same as those detected pretreatment. A detailed analysis of the prevalence and the subtypes of Campylobacter spp. is reported in the accompanying article (20a).
Antimicrobial susceptibility.
One resistant isolate of each species, serotype, and phage type was selected from each fecal or environmental sample for further study; and three or more susceptible isolates were selected at random from each flock during each treatment phase for comparison. The Antimicrobial Agents Research Group in Birmingham received a total of 469 isolates from CRU, and these represented 29% of all isolates typed by CRU (20a). Of these 469 isolates, 290 isolates were ciprofloxacin resistant (MICs ≥ 2 μg/ml). Fifty-three percent (174 of 326) of the C. jejuni isolates and 82% (115 of 141) of the C. coli isolates were ciprofloxacin resistant. Most ciprofloxacin-resistant isolates (282 of 290) were correctly identified as resistant by breakpoint testing by CRU.
Determination of the nalidixic acid and ciprofloxacin MICs revealed that there were two quinolone-resistant phenotypes: both nalidixic acid and ciprofloxacin resistant (Nalr Cipr; n = 290) and nalidixic acid resistant and ciprofloxacin susceptible (Nalr Cips; n = 13). Eight C. jejuni isolates and five C. coli isolates had the Nalr Cips phenotype and were typically inhibited by 0.25 or 0.5 μg of ciprofloxacin per ml (see Table 3).
TABLE 3.
Relationship between ciprofloxacin and nalidixic acid MICs and GyrA substitution in C. jejuni and C. coli
| Resistance phenotypea | Species | GyrA substitutionb | No. of isolates | Ciprofloxacin MIC (μg/ml)
|
Nalidixic acid MIC (μg/ml)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 50% | 90% | Range | Geometric mean | 50% | 90% | Range | Geometric mean | ||||
| Nalr Cipr | C. jejuni | Ile-86 | 157 | 32 | 64 | 2-128 | 22.7 | 64 | 128 | 8->128 | 73.2 |
| Ala-86 | 4 | —c | — | 4-32 | — | — | — | 16->128 | — | ||
| Asn-90 | 10 | 32 | 128 | 8-128 | 39.4 | 128 | >128 | 64->128 | 97.0 | ||
| C. coli | Ile-86 | 111 | 8 | 16 | 4-64 | 12.0 | 128 | 128 | 32->128 | 97.3 | |
| Ala-86 | 1 | — | — | 8 | — | — | — | 64 | — | ||
| Nalr Cips | C. jejuni | Ile-86 | 1 | — | — | 1 | — | — | — | 128 | — |
| Ala-86 | 7 | — | — | 0.25-0.5 | — | — | — | 64-128 | — | ||
| C. coli | Ala-86 | 5 | — | — | 0.25-0.5 | — | — | — | 64 | — | |
Nalr Cipr, resistant to nalidixic acid (MICs ≥ 32 μg/ml) and ciprofloxacin (MICs ≥ 2 μg/ml); Nalr Cips, nalidixic acid resistant (MICs ≥ 32 μg/ml) and ciprofloxacin sensitive (MICs ≤ 1 μg/ml).
Ile-86, Thr-86→Ile; Ala-86, Thr-86→Ala; Asn-90, Asp-90→Asn.
—, the MIC50, MIC90, and geometric mean were not calculated when there were <10 isolates.
MDR.
As there is increasing evidence that exposure to a fluoroquinolone can select for bacteria that are resistant to multiple agents of unrelated chemical classes as well as fluoroquinolones, all isolates were examined for their susceptibilities to at least one agent representative of each chemical class used in human medicine. Isolates were divided into those that were ciprofloxacin susceptible (MIC ≤ 1 μg/ml) and those that were ciprofloxacin resistant (MIC ≥ 2 μg/ml) to determine whether there was an association between fluoroquinolone resistance and resistance to any other agents (Table 1). All isolates were resistant to trimethoprim (MICs ≥ 2 μg/ml); and most were resistant to acridine orange, sodium dodecyl sulfate, and sodium deoxycholate (MICs ≥ 16 μg/ml), regardless of their fluoroquinolone susceptibilities. Both trimethoprim and sodium deoxycholate were present in the charcoal cefoperazone deoxycholate agar (Oxoid) selective medium used for the isolation of campylobacter (20a). These agents were therefore excluded from the definition of a multiple-drug-resistant (MDR) phenotype. Chloramphenicol resistance was rare (Table 1).
TABLE 1.
Cross-resistance to antimicrobial agents and dyes of Campylobacter spp. isolated from commercial poultry flocksa
| Agent | Concn (≥μg/ml) used to determine resistance | % Resistance among Cips isolates (no. of isolates) in flock:
|
% Resistance among Cipr isolates (no. of isolates) in flock:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 3 | 4 | 5 | 6 | 1 | 3 | 4 | 5 | 6 | ||
| ERY | 4 | 2 (1) | 5 (2) | 24 (6) | 26 (10) | 7 (2) | 29 (23) | 2 (1) | 29 (12) | 3 (2) | 25 (15) |
| CHL | 8 | 2 (1) | 2 (1) | 16 (4) | 0 (0) | 4 (1) | 5 (4) | 0 (0) | 20 (8) | 0 (0) | 2 (1) |
| TET | 8 | 9 (4) | 0 (0) | 36 (9) | 0 (0) | 78 (21) | 51 (40) | 0 (0) | 68 (28) | 19 (13) | 61 (37) |
| KAN | 16 | 13 (6) | 10 (4) | 56 (14) | 10 (4) | 7 (2) | 10 (8) | 19 (8) | 49 (20) | 13 (9) | 3 (2) |
| AMP | 16 | 28 (13) | 71 (30) | 20 (5) | 13 (5) | 59 (16) | 83 (65) | 74 (31) | 80 (33) | 25 (17) | 36 (22) |
| EtBr | 8 | 15 (7) | 17 (7) | 40 (10) | 18 (7) | 59 (16) | 41 (32) | 2 (1) | 20 (8) | 54 (37) | 77 (47) |
| MDR | 4 (2) | 5 (2) | 24 (6) | 5 (2) | 44 (12) | 32 (25) | 0 (0) | 46 (19) | 9 (6) | 38 (23) | |
| Total | 46 | 42 | 25 | 39 | 27 | 78 | 42 | 41 | 68 | 61 | |
Cips, MICs ≤ 1 μg/ml; Cipr, MICs ≥ 2 μg/ml. MDR indicates resistance to three or more of the following agents: erythromycin (ERY), chloramphenicol (CHL), tetracycline (TET), kanamycin (KAN), ampicillin (AMP), or ethidium bromide (EtBr).
Isolates in each flock were designated MDR and were resistant to three or more of the following agents: chloramphenicol, kanamycin, ampicillin, erythromycin, and ethidium bromide. Apart from flock 3, larger numbers of fluoroquinolone-resistant strains from each flock were MDR than sensitive. Only two isolates from flock 3 were MDR (Table 1). Twenty-five percent (73 of 290) of ciprofloxacin-resistant isolates and 13% (24 of 179) of ciprofloxacin-susceptible isolates were MDR. Statistical analysis by the χ2 test showed that MDR was significantly associated with ciprofloxacin resistance (P < 0.01). Only 1 of the 13 Nalr Cips isolates was MDR.
The MICs of the disinfectants triclosan and cetrimide were determined for all MDR isolates (n = 97). The MICs of the agents for C. jejuni NCTC 11168 were 4 and 8 μg/ml, respectively. For the MDR isolates, the triclosan MIC range was 2 to 128 μg/ml, with an MIC at which 50% of isolates are inhibited (MIC50; median), MIC90, and mode MIC of 64 μg/ml and a geometric mean MIC of 55.1 μg/ml. The MIC range for cetrimide was 1 to >128 μg/ml, with an MIC50 (median) of 32 μg/ml, an MIC90 and a mode MIC of 64 μg/ml, and a geometric mean MIC of 35.2 μg/ml. These values did not differ for ciprofloxacin-susceptible and -resistant MDR isolates.
Role of gyrA in fluoroquinolone resistance.
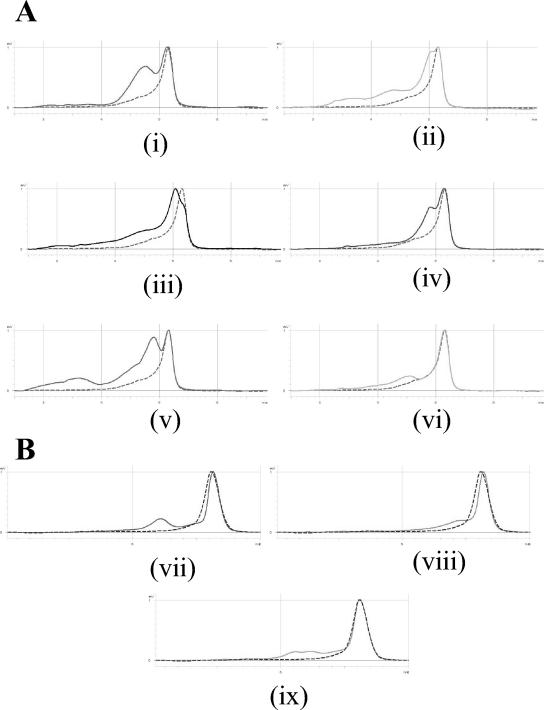
DNA sequencing of each novel DHPLC pattern identified 15 gyrA genotypes among the C. jejuni isolates. Nine patterns were associated with quinolone-resistant (Nalr Cipr and Nalr Cips) isolates, and six patterns were associated with sensitive strains, including wild-type C. jejuni NCTC 11168. Each pattern corresponded to one or more nucleotide changes in the DNA sequence compared with the sequence of the wild type. However, the majority of these changes did not confer an amino acid substitution. DNA sequencing of several representative isolates with each pattern revealed that the majority of resistant isolates possessed an amino acid substitution in gyrA, in which Thr-86 was replaced with Ile (Table 2). The different patterns were due to the presence of different silent mutations, in addition to the substitution mutation (types A to E and L; Fig. 1). For 17 strains, Thr-86 was replaced by Ala (types F and N), and 10 C. jejuni strains had the gyrA mutation Asp-90→Asn (type G; Fig. 1).
TABLE 2.
Substitutions in GyrA detected in flocks 1 and 3 to 6
| GyrA substitutiona | No. of isolatesb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Flock 1, Nalr Cipr | Flock 3
|
Flock 4
|
Flock 5
|
Flock 6, Nalr Cipr | Total | ||||
| Nalr Cipr | Nalr Cips | Nalr Cipr | Nalr Cips | NALr Cipr | Qs | ||||
| Ile-86 | 72 | 40 | 0 | 40 | 1 | 65 | 0 | 51 | 269 |
| Ala-86 | 1 | 1 | 12 | 0 | 0 | 1 | 0 | 2 | 17 |
| Asn-90 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 8 | 10 |
| Ile-60 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
Ile-86, Thr-86→Ile; Ala-86, Thr-86→Ala; Asn-90, Asp-90→Asn; Ile-60, Val-60→Ile.
Nalr Cipr, resistant to nalidixic acid (MICs ≥ 32 μg/ml) and ciprofloxacin (MICs ≥ 2 μg/ml); Nalr Cips, nalidixic acid resistant (MICs ≥ 32 μg/ml) and ciprofloxacin sensitive (MICs ≤ 1 μg/ml); Qs, quinolone sensitive.
FIG. 1.
DHPLC elution traces for C. jejuni and C. coli isolates with mutations in gyrA. (A) C. jejuni (i) type A (Thr-86→Ile, ACA→ATA), (ii) type B (His-81, CAC→CAT; Thr-86→Ile, ACA→ATA), (iii) type C (His-81, CAC→CAT; Thr-86→Ile, ACA→ATA; Ser-119, AGT→AGC; Ala-120, GCC→GCT), (iv) type G (Asp-90→Asn, GAT→AAT), (v) type E (His-81, CAC→CAT; Thr-86→Ile, ACA→ATA; Gly-110, GGC→GGT), and (vi) type F (His-81, CAC→CAT; Thr-86→Ala, ACA→GCA). (B) C. coli (vii) CC/A (Thr-86→Ile, ACT→ATT; Phe-99, TTT→TTC), (viii) CC/B (His-81, CAC→CAT; Thr-86→Ile, ACA→ATA; Gly-113, GGA→GGT; Ile-115, ATA→ATC), and (ix) CC/C (Val-60→Ile, GTA→ATA; Phe-99, TTT→TTC). The wild-type pattern (dotted line) is shown on each elution trace: C. jejuni NCTC 11168 (A) and C. coli NCTC 11366 (B). Isolates with gyrA code CC/B were C. coli but had a gyrA sequence with a closer identity to that of C. jejuni (the nucleotide changes shown are differences from the C. jejuni gyrA sequence). The polymorphisms seen at His-81 (CAT), Gly-113 (GGT), Ile-115 (ATC), and Ala-120 (GCT) in C. jejuni are present in wild-type C. coli (40).
Fewer DHPLC patterns were found among the C. coli isolates. The majority of C. coli isolates produced pattern CC/A, which corresponded to two nucleotide changes, compared to the sequence of the wild type (C. coli NCTC 11366), that conferred the substitution Thr-86 to Ile and a nonsubstitution mutation at codon Phe-99 (Fig. 1). Isolates with the pattern CC/B also had an Ile-86 change, but the gyrA sequence was more similar to that of C. jejuni than to that of C. coli throughout the QRDR. One quinolone-sensitive C. coli isolate had a novel pattern (designated CC/C) that corresponded to the substitution Val-60→Ile (Fig. 1).
To determine whether there was any association between a specific mutation in gyrA and the MICs of nalidixic acid and ciprofloxacin for each species, a detailed analysis of the gyrA DHPLC patterns and MICs was performed (Table 3). Previous studies (30) found that it is difficult to assign an MIC for Campylobacter strains with a specific mutation, whereas previously published data for Escherichia coli (11) and Salmonella (18, 30) have suggested that defined mutations confer specific MICs (within the doubling dilution error of this technique). As had been found previously, no clear MIC was associated with a specific amino acid substitution, and for most patterns a range of ciprofloxacin MICs was obtained (Table 3). The MIC50s (median), MIC90s, and geometric mean MICs for strains with each gyrA substitution are shown in Table 3. For C. jejuni with the Ile-86 substitution, the mode MIC of ciprofloxacin was 32 μg/ml and the mode MIC of nalidixic acid was 64 μg/ml; for C. coli the mode MIC of ciprofloxacin was 8 μg/ml and the mode MIC of nalidixic acid was 128 μg/ml. For C. jejuni strains with the Asn-90 substitution, the mode MICs of ciprofloxacin and nalidixic acid were 128 μg/ml. However, the Ala-86 substitution was typically associated with a Nalr Cips phenotype, with 12 of 17 strains inhibited by 0.25 to 0.5 μg of ciprofloxacin per ml and 64 to 128 μg of nalidixic acid per ml (Table 3).
It was noted that C. coli became the more prevalent species during treatment but that C. jejuni became reestablished posttherapy (20a). It was hypothesized that C. coli not only may be more prevalent but also may be more highly resistant than C. jejuni, enabling survival during fluoroquinolone exposure. However, these data indicate that the same amino acid is replaced in the QRDR of GyrA of resistant C. coli and C. jejuni strains. In fact the MICs for the C. coli strains containing Ile-86 were lower than those for many of the C. jejuni strains with the same substitution (Table 3).
Seventy-three isolates were cultured from the barn environments of four flocks (none were isolated from flock 4). Of these, 36 (49%) were ciprofloxacin resistant and were isolated from drinking water, feed, litter, and surface swabs up to 3 weeks posttreatment. The most common species, serotypes, and phage types of ciprofloxacin-resistant environmental strains were also seen among the fecal isolates (20a), and the same GyrA substitutions were detected among isolates from both sources.
Role of gyrB in fluoroquinolone resistance.
Sixty of the 83 isolates from flock 3 were screened for mutations in gyrB by DHPLC, and representative isolates including all those with novel patterns (n = 20) were sequenced. Twenty-two of the 60 isolates screened were of the wild type. Four patterns in addition to the wild type were found, each of which had silent mutations at two loci in gyrB; these occurred at Leu-407 (TTA→TTG), Pro-415 (CCA→CCG), Phe-440 (TTC→TTT), or Leu-458 (CTA→TTA or CTG). All 38 of these isolates were either quinolone sensitive or resistant with an identified mutation in gyrA. No substitution mutations were found in gyrB in any isolate; therefore, it was decided to screen quinolone-resistant isolates from subsequent flocks for gyrB mutations by DHPLC only if no gyrA mutation was detected. However, no resistant isolates lacking a mutation in gyrA were found in flocks 4 to 6.
Role of gyrA genotype in isolate typing.
It was thought that genotypic variation in gyrA, based on the patterns of silent mutations, could be used to supplement the typing data from CRU for detailed strain fingerprinting. These data were used to investigate whether fluoroquinolone exposure selected a preexisting antibiotic-resistant strain which became dominant or whether a spontaneous mutant was selected from the preexisting susceptible population. In practice, this was difficult to analyze because of the diversity of campylobacter serotypes and phage types within each flock, but there was evidence that both events occurred. In flock 1, both ciprofloxacin-sensitive isolates (gyrA type I; n = 2) and ciprofloxacin-resistant isolates (gyrA type C, n = 1) of C. jejuni serotype HS31 phage type 1 (PT1) were isolated pretherapy. During treatment, only gyrA type C resistant isolates (n = 19) were seen. A mutation at Thr-86 (ACA→ATA) in a gyrA type I strain would give rise to a C genotype, but as resistant isolates of type C were seen pretreatment and no further susceptible type I isolates of this serotype and phage type were isolated, it is suggested that the preexisting antibiotic-resistant strain became dominant. In flock 3, susceptible C. jejuni HS13 PT1 isolates (gyrA type H, gyrB type A) were isolated from feces and the barn environment pretherapy and during treatment. However, resistant isolates of gyrA type F (n = 2) and gyrA type B (n = 2) were also isolated during treatment, and all isolates had the same gyrB genotype and the MICs of all agents for these isolates (Table 4) were identical to those for the susceptible isolates. A single nucleotide change in gyrA type H would give rise to an F or a B genotype. This is evidence that preexisting susceptible strains may have acquired resistance mutations during treatment. Alternatively, type F and type B resistant strains may have been present at low densities in the flock prior to treatment but became dominant as a result of fluoroquinolone exposure. There was no association between the gyrA genotype and any specific serotype or phage type; the different gyrA genotypes were distributed at random throughout the different types identified by CRU.
TABLE 4.
Evidence for emergence of spontaneous quinolone-resistant mutants from preexisting susceptible C. jejuni HS13 PT1 strains from flock 3
| Treatment phase | Source | Isolate | MIC (μg/ml)a
|
Genotypeb
|
Resistance phenotypec | ||
|---|---|---|---|---|---|---|---|
| CIP | NAL | gyrA | gyrB | ||||
| Pretreatment | Puddle | P751 | 0.12 | 8 | H | A | QS |
| Feces | P765 | 0.12 | 8 | H | A | QS | |
| Treatment | Litter | P762 | 0.12 | 8 | H | A | QS |
| Feces | P769 | 0.12 | 8 | H | A | QS | |
| Feces | P757 | 0.25 | 64 | F | ND | Nalr Cips | |
| Feces | P753 | 0.5 | 64 | F | A | Nalr Cips | |
| Drinking water | P764 | 8 | 128 | B | A | Nalr Cipr | |
| Feces | P756 | 16 | 64 | B | A | Nalr Cipr | |
CIP, ciprofloxacin; NAL, nalidixic acid. The MICs of sodium dodecyl sulfate, ethidium bromide, tetracycline, chloramphenicol, kanamycin, ampicillin, trimethyprim acridine orange, and sodium deoxycholate were identical (±1 dilution) for all eight isolates.
gyrA genotypes were determined by DHPLC analysis and are designated as follows: H, His-81 (CAC→CAT); F and B, as shown in Fig. 1; a single nucleotide change in gyrA type H would give rise to an F or B genotype; gyrB genotype A, Phe-440 (TTC→TTT) and Leu-458 (CTA→TTA); ND, not done.
Nalr Cipr, resistant to nalidixic acid (MICs ≥ 32 μg/ml) and ciprofloxacin (MICs ≥ 2 μg/ml); Nalr Cips, nalidixic acid resistant (MICs ≥ 32 μg/ml) and ciprofloxacin sensitive (MICs ≤ 1 μg/ml); QS, quinolone sensitive.
DISCUSSION
The study described here and in the accompanying article (20a) investigated the incidence and mechanism of fluoroquinolone resistance among campylobacters isolated from five commercial chicken flocks treated with a fluoroquinolone for a clinically relevant infection. The hypothesis explored was that fluoroquinolone use in poultry gives rise to fluoroquinolone-resistant campylobacters that can enter the food chain. This is the first comprehensive, detailed study of commercial flocks rather than models. The proportion of ciprofloxacin-resistant campylobacters increased rapidly during treatment and then declined posttherapy (20a). Changes of the subtypes present in the campylobacter population were observed during and after treatment. Exposure to fluoroquinolones appeared to transiently select for C. coli over C. jejuni (20a). This report focuses on the mechanism of resistance in the quinolone-resistant campylobacters from the five farms.
Previous studies have used various techniques to screen campylobacters for mutations in the QRDR of gyrA. Some, such as single-strand conformation polymorphism analysis (5, 30), are able to distinguish mutations at several locations in the QRDR, while other techniques (restriction fragment length polymorphism analysis [30], real-time PCR with the LightCycler instrument [4], fluorogenic PCR [39], and mismatch amplification mutation assay PCR [40, 41]) screen only for changes at Thr-86. This is the first report to describe the application of DHPLC (8) to the detection of mutations in a campylobacter gene. Fifteen variants of the gyrA gene in C. jejuni were distinguished by use of this technology, and nine of these were associated with a substitution in GyrA. Most commonly, Thr-86 was replaced by Ile, which has previously been shown to be sufficient to render campylobacters resistant to fluoroquinolones, including enrofloxacin and ciprofloxacin. This has also been the predominant substitution in quinolone-resistant isolates from humans in the United Kingdom (15, 30).
Four gyrA patterns were detected among the C. coli isolates, including the wild-type strain. The most common pattern corresponded to a Thr-86→Ile substitution. DNA sequencing of one pattern (pattern CC/B) showed that it was quite different from the published sequence for C. coli (40; GenBank accession number AF092101) and was more similar to that of C. jejuni (38; GenBank accession number L04566). The species of all isolates were determined at CRU, and all isolates were typed at CRU by using established procedures (2, 3, 13, 14). The seven isolates with this pattern were confirmed to be C. coli by the PCR identification method of Fermer and Engvall (12) (data not shown). This phenomenon has previously been observed by the Antimicrobial Agents Research Group (30). The genome of C. jejuni is known to contain hypervariable sequences (26), and the high frequency of local repeats within the genome suggests that there is an increased likelihood of recombination events (28), which may account for this apparent anomaly.
No multiple GyrA substitutions were found in the campylobacters evaluated in this study, and they have been described only rarely in other studies (20, 30). Therefore, it seems unlikely that campylobacters with a Nalr Cips phenotype would become Nalr Cipr by acquiring a second mutation in gyrA, whereas double mutations in gyrA are common in other species, such as E. coli and Salmonella (8, 11).
The presence of additional mechanisms of resistance could contribute to the wide range of fluoroquinolone MICs observed for isolates with the same mutation in gyrA. No mutations in gyrB have been described in quinolone-resistant strains, and campylobacters appear to lack the parC gene; mutations in both these genes have been shown to contribute to high-level resistance in other species (11). The strains screened for changes in gyrB in the present study contained no substitutions that resulted in mutations. There was a clear association between ciprofloxacin resistance and resistance to other agents, with a quarter of the resistant isolates being MDR. Hakanen et al. (19) recently reported a strong association between MDR and resistance to ciprofloxacin in C. jejuni strains isolated from clinical specimens. In a recent study, C. jejuni strains that had a mutation in gyrA and that overexpressed the efflux pump gene cmeB were less susceptible to ciprofloxacin than isolates in which either a gyrA mutation or overexpression of cmeB occurred alone (32). There is evidence that efflux systems other than cmeB and cmeF that may contribute to MDR are present in campylobacters (32). The additive effect of efflux and mutations in gyrA may account, in part, for the range of ciprofloxacin MICs observed for strains with identical mutations in the QRDR. The contributions of efflux and multiple mechanisms of resistance to MDR in these isolates are being investigated.
For all resistant C. coli isolates (except six isolates from flock 3) the mutation in gyrA resulted in the replacement of Thr-86 with Ile. While most resistant C. jejuni strains also had this substitution, other mutations were observed in a few strains from four of the five farms. These data suggest that there is more variation within gyrA of C. jejuni than in gyrA of C. coli and that C. coli may be more clonal than C. jejuni. The increased numbers of C. coli isolates recovered during fluoroquinolone treatment (20a) cannot be explained by the level of resistance (i.e., the MIC) or by the mechanism of resistance. The factor that caused the surge in the numbers of isolates of this species and the subsequent decrease posttreatment remains to be established.
Not all resistant isolates were of the same type, demonstrating that resistance was not a result of the spread of a single resistant clone but that numerous clones were selected by fluoroquinolone treatment (20a). The persistence of highly quinolone-resistant campylobacters up to 4 weeks posttreatment has possible consequences for human health. The gyrA mutations identified among poultry isolates are identical to those described in human clinical isolates, and the majority conferred high-level ciprofloxacin resistance. Although it has been presumed that a human infection caused by a food-borne ciprofloxacin-resistant campylobacter results in treatment failure, there is little evidence to confirm that this occurs. The proportion of campylobacter infections in humans that are treated with an antibiotic is also thought to be low, but no published data are yet available. However, it has been shown that the duration of diarrhea is prolonged in patients infected with quinolone-resistant C. jejuni (34).
In conclusion, quinolone-resistant campylobacters were isolated from commercial chicken flocks in high numbers following therapy with a veterinary fluoroquinolone. Most ciprofloxacin-resistant isolates had a gyrA mutation, with the replacement of Thr-86 by Ile. However, the high incidence was not due to the spread of a single resistant clone throughout each flock, as resistant isolates of different species, serotypes, and phage types were identified within each flock. Resistant isolates were isolated from the feces of some flocks up to the point of slaughter, which may have consequences for public health.
Acknowledgments
This study was funded by Defra (project code OZ0501). We thank Transgenomic for support. The Functional Genomics Laboratory is funded by a BBSRC grant (grant 6/JIF13209). L.J.V.P. is a recipient of the Bristol-Myers Squibb unrestricted grant in infectious diseases.
We are grateful to Haddy Wadda, Lilian Pumbwe, Gil Domingue, Karen Martin, Marco Siccardi, and Lisa Williams for technical and scientific support to this project.
REFERENCES
- 1.Bachoual, R., S. Ouabdesselam, F. Mory, C. Lascols, C. J. Soussy, and J. Tankovic. 2001. Single or double mutational alterations of GyrA associated with fluoroquinolone resistance in Campylobacter jejuni and Campylobacter coli. Microb. Drug Resist. Mechanisms Epidemiol. Dis. 7:257-261. [DOI] [PubMed] [Google Scholar]
- 2.Best, E. L., E. J. Powell, C. Swift, K. A. Grant, and J. A. Frost. 2003. Applicability of a rapid duplex real-time PCR assay for speciation of Campylobacter jejuni and Campylobacter coli directly from culture plates. FEMS Microbiol. Lett. 229:237-241. [DOI] [PubMed] [Google Scholar]
- 3.Bolton, F. J., D. R. A. Wareing, M. B. Skirrow, and D. N. Hutchinson. 2001. Identification and biotyping of campylobacters, p. 151-161. In G. R. Board, D. Jones, and F. A. Skinner (ed.), Identification methods in applied and environmental microbiology. Blackwell Scientific Publications, Oxford, United Kingdom.
- 4.Carattoli, A., A. Dionisi, and I. Luzzi. 2002. Use of a LightCycler gyrA mutation assay for identification of ciprofloxacin-resistant Campylobacter coli. FEMS Microbiol. Lett. 214:87-93. [DOI] [PubMed] [Google Scholar]
- 5.Charvalos, E., E. Peteinaki, I. Spyridaki, S. Manetas, and Y. Tselentis. 1996. Detection of ciprofloxacin resistance mutations in Campylobacter jejuni gyrA by nonradioisotopic single-strand conformation polymorphism and direct DNA sequencing. J. Clin. Lab. Anal. 10:129-133. [DOI] [PubMed] [Google Scholar]
- 6.Charvalos, E., Y. Tselentis, M. M. Hamzehpour, T. Kohler, and J. C. Pechere. 1995. Evidence for an efflux pump in multidrug-resistant Campylobacter jejuni. Antimicrob. Agents Chemother. 39:2019-2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuma, T., T. Ikeda, T. Maeda, H. Niwa, and K. Okamoto. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from broilers in the southern part of Japan from 1995 to 1999. J. Vet. Med. Sci. 63:1027-1029. [DOI] [PubMed] [Google Scholar]
- 8.Eaves, D. J., E. Liebana, M. J. Woodward, and L. J. V. Piddock. 2002. Detection of gyrA mutations in quinolone-resistant Salmonella enterica by denaturing high-performance liquid chromatography. J. Clin. Microbiol. 40:4121-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endtz, H. P., G. J. Ruijs, B. van Klingeren, W. H. Jansen, R. T. van der, and R. P. Mouton. 1991. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J. Antimicrob. Chemother. 27:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, M. J., Y. F. Jin, V. Ricci, and L. J. V. Piddock. 1996. Contributions of individual mechanisms to fluoroquinolone resistance in 36 Escherichia coli strains isolated from humans and animals. Antimicrob. Agents Chemother. 40:2380-2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fermer, C., and E. O. Engvall. 1999. Specific PCR identification and differentiation of the thermophilic campylobacters, Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J. Clin. Microbiol. 37:3370-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frost, J. A., A. N. Oza, R. T. Thwaites, and B. Rowe. 1998. Serotyping scheme for Campylobacter jejuni and Campylobacter coli based on direct agglutination of heat-stable antigens. J. Clin. Microbiol. 36:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frost, J. A., J. M. Kramer, and S. A. Gillanders. 1999. Phage typing of C. jejuni and C. coli. Epidemiol. Infect. 123:47-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaunt, P. N., and L. J. V. Piddock. 1996. Ciprofloxacin resistant Campylobacter spp in humans: an epidemiological and laboratory study. J. Antimicrob. Chemother. 37:747-757. [DOI] [PubMed] [Google Scholar]
- 16.Gibreel, A., E. Sjogren, B. Kaijser, B. Wretlind, and O. Skold. 1998. Rapid emergence of high-level resistance to quinolones in Campylobacter jejuni associated with mutational changes in gyrA and parC. Antimicrob. Agents Chemother. 42:3276-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gootz, T. D., and B. A. Martin. 1991. Characterization of high-level quinolone resistance in Campylobacter jejuni. Antimicrob. Agents Chemother. 35:840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griggs, D. J., K. Gensberg, and L. J. V. Piddock. 1996. Mutations in gyrA gene of quinolone-resistant salmonella serotypes isolated from humans and animals. Antimicrob. Agents Chemother. 40:1009-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakanen, A., M. Lehtopolku, A. Siitonen, P. Huovinen, and P. Kotilainen. 2003. Multidrug resistance in Campylobacter jejuni strains collected from Finnish patients during 1995-2000. Antimicrob. Agents Chemother. 52:1035-1039. [DOI] [PubMed] [Google Scholar]
- 20.Hakanen, A., J. Jalava, P. Kotilainen, H. Jousimies-Somer, A. Siitonen, and P. Huovinen. 2002. gyrA polymorphism in Campylobacter jejuni: detection of gyrA mutations in 162 C. jejuni isolates by single-strand conformation polymorphism and DNA sequencing. Antimicrob. Agents Chemother. 46:2644-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Humphrey, T. J., F. Jørgensen, J. A. Frost, H. Wadda, G. Domingue, N. C. Elviss, D. J. Griggs, and L. J. V. Piddock. 2005. Prevalence and subtypes of ciprofloxacin-resistant Campylobacter spp. in commercial poultry flocks before, during, and after treatment with fluoroquinolones. Antimicrob. Agents Chemother. 49:690-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, J., L. O. Michel, and Q. Zhang. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob. Agents Chemother. 46:2124-2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo, N., O. Sahin, J. Lin, L. O. Michel, and Q. J. Zhang. 2003. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 47:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacGowan, A., P., and R. Wise. 2001. Establishing MIC breakpoints and the interpretation of in vitro susceptibility tests. J. Antimicrob. Chemother. 48:17-28. [DOI] [PubMed] [Google Scholar]
- 24.McDermott, P. F., S. M. Bodeis, F. M. Aarestrup, S. Brown, M. Traczewski, P. Fedorka-Cray, M. Wallace, I. A. Critchley, C. Thornsberry, S. Graff, R. Flamm, J. Beyer, D. Shortridge, L. J. Piddock, V. Ricci, M. M. Johnson, R. N. Jones, B. Reller, S. Mirrett, J. Aldrobi, R. Rennie, C. Brosnikoff, L. Turnbull, G. Stein, S. Schooley, R. A. Hanson, and R. D. Walker. 2004. Development of a standardized susceptibility test for campylobacter with quality-control ranges for ciprofloxacin, doxycycline, erythromycin, gentamicin, and meropenem. Microb. Drug Resist. 10:124-131. [DOI] [PubMed] [Google Scholar]
- 25.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial disk susceptibility tests; approved standard M2-A8, vol. 23, no. 1. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 27.Payot, S., A. Cloeckaert, and E. Chaslus-Dancla. 2002. Selection and characterization of fluoroquinolone-resistant mutants of Campylobacter jejuni using enrofloxacin. Microb. Drug Resist. Mechanisms Epidemiol. Dis. 8:335-343. [DOI] [PubMed] [Google Scholar]
- 28.Petersen, L., S. L. W. On, and D. W. Ussery. 2002. Visualization and significance of DNA structural motifs in the Campylobacter jejuni genome. Genome Lett. 1:16-25. [Google Scholar]
- 29.Piddock, L. J. V. 1999. Quinolone resistance and campylobacter. Clin. Microbiol. Infect. 5:239-243. [PubMed] [Google Scholar]
- 30.Piddock, L. J. V., V. Ricci, L. Pumbwe, M. J. Everett, and D. J. Griggs. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J. Antimicrob. Chemother. 51:19-26. [DOI] [PubMed] [Google Scholar]
- 31.Pumbwe, L., and L. J. V. Piddock. 2002. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol. Lett. 206:185-189. [DOI] [PubMed] [Google Scholar]
- 32.Pumbwe, L., L. P. Randall, M. J. Woodward, and L. J. Piddock. 2004. Expression of the efflux pump genes cmeB, cmeF and the porin gene porA in multiply antibiotic-resistant Campylobacer jejuni J. Antimicrob. Chemother. 54:341-347. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz, J., P. Goni, F. Marco, F. Gallardo, B. Mirelis, T. J. De Anta, and J. Vila. 1998. Increased resistance to quinolones in Campylobacter jejuni: a genetic analysis of gyrA gene mutations in quinolone-resistant clinical isolates. Microbiol. Immunol. 42:223-226. [DOI] [PubMed] [Google Scholar]
- 34.Smith, K. E., J. M. Besser, C. W. Hedberg, F. T. Leano, J. B. Bender, J. H. Wicklund, B. P. Johnson, K. A. Moore, and M. T. Osterholm. 1999. Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992-1998. N. Engl. J. Med. 340:1525-1532. [DOI] [PubMed] [Google Scholar]
- 35.Thwaites, R. T., and J. A. Frost. 1999. Drug resistance in Campylobacter jejuni, C. coli, and C. lari isolated from humans in north west England and Wales, 1997. J. Clin. Pathol. 52:812-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unicomb, L., J. Ferguson, T. V. Riley, and P. Collignon. 2003. Fluoroquinolone resistance in campylobacter absent from isolates, Australia. Emerg. Infect. Dis. 9:1482-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Looveren, M., G. Daube, L. De Zutter, J. M. Dumont, C. Lammens, M. Wijdooghe, P. Vandamme, M. Jouret, M. Cornelis, and H. Goossens. 2001. Antimicrobial susceptibilities of Campylobacter strains isolated from food animals in Belgium. J. Antimicrob. Chemother. 48:235-240. [DOI] [PubMed] [Google Scholar]
- 38.Wang, Y., W. M. Huang, and D. E. Taylor. 1993. Cloning and nucleotide-sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, D. L., S. R. Abner, T. C. Newman, L. S. Mansfield, and J. E. Linz. 2000. Identification of ciprofloxacin-resistant Campylobacter jejuni by use of a fluorogenic PCR assay. J. Clin. Microbiol. 38:3971-3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zirnstein, G., L. Helsel, Y. Li, B. Swaminathan, and J. Besser. 2000. Characterization of gyrA mutations associated with fluoroquinolone resistance in Campylobacter coli by DNA sequence analysis and MAMA PCR. FEMS Microbiol. Lett. 190:1-7. [DOI] [PubMed] [Google Scholar]
- 41.Zirnstein, G., Y. Li, B. Swaminathan, and F. Angulo. 1999. Ciprofloxacin resistance in Campylobacter jejuni isolates: detection of gyrA resistance mutations by mismatch amplification mutation assay PCR and DNA sequence analysis. J. Clin. Microbiol. 37:3276-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]