Abstract
We have recently identified a new family of multidomain oxidoreductase (redox) enzymes, the MICALs, that directly regulate the actin cytoskeletal elements necessary for the morphology, motility, and trajectory of cells. Our genetic assays reveal that Mical is both necessary and sufficient for actin organization and cellular effects in vivo and our biochemical assays with purified Mical protein reveal that Mical utilizes its redox activity to directly disassemble actin filaments. These results identify Mical proteins as novel actin disassembly factors and uncover a redox signaling mechanism that directly regulates the actin cytoskeleton. These results have also set the stage for in-depth characterization of the Mical enzyme. However, it has been difficult to obtain sufficient amounts of highly-pure Mical protein to conduct further biochemical, structural, imaging, catalytic, and other high-precision studies. Herein, we describe a means for expressing high levels of soluble recombinant Mical protein in bacteria. Likewise, we have designed a new purification strategy that enables the rapid and efficient purification of milligram quantities of highly-pure and >99% active Mical protein. This new strategy for generating large amounts of highly-pure and active Mical protein will aid research objectives designed to characterize the biochemical, enzymology, and structural biology of Mical and its effects on actin filament dynamics.
Keywords: MICALs, Semaphorin, Plexin, flavoprotein monooxygenase, F-actin disassembly, repulsion, axon guidance, guidance cues
Introduction
Cells require the assembly of actin filaments beneath their plasma membranes in order to produce the forces necessary for motility. Regulating the organization and dynamics of these actin filaments leads to changes in the shape, trajectory, and connectivity of these cells. Multiple extracellular guidance cues and their cell surface receptors have now been identified that regulate actin dynamics in motile cells and their processes [1-4], but we still know little of how these instructive cues present on the cell surface exert their precise effects on the actin cytoskeleton. Semaphorins, for example, are one of the largest families of these guidance cues but use poorly understand mechanisms to directly regulate the movement and navigation of cells and their membranous extensions that are critical for neural function, immunity, angiogenesis, cardiovascular health, and pathologies including cancer [5-8].
We have recently identified a new cytosolic protein, Mical, that provides a direct link through which Semaphorins regulate the actin cytoskeleton [9-12]. Mical directly binds to both the Semaphorin receptor Plexin and actin, and directly disassembles actin filaments (Figure 1A; [9, 10]). Mical is a member of a phylogenetically-conserved family of proteins, the MICALs, that are unusual in that they combine a flavin-binding (flavoprotein) NADPH-dependent oxidoreductase enzymatic moiety with a number of domains found in cytoskeletal-associated proteins (Figure 1B; [9]). Mical utilizes its NADPH-dependent redox enzymatic domain to post-translationally oxidize individual actin filament subunits, simultaneously severing filaments and decreasing polymerization (Figure 1C; [11]). Mical therefore is an important new class of actin disassembly factors that uses a catalytic post-translational redox mechanism to alter F-actin stability. However, this work and the further characterization of the biochemical activity of Mical has been hindered by the inability to purify large amounts of Mical protein [9, 13]. We now present a simple method for expressing and rapidly purifying large amounts of active Mical protein.
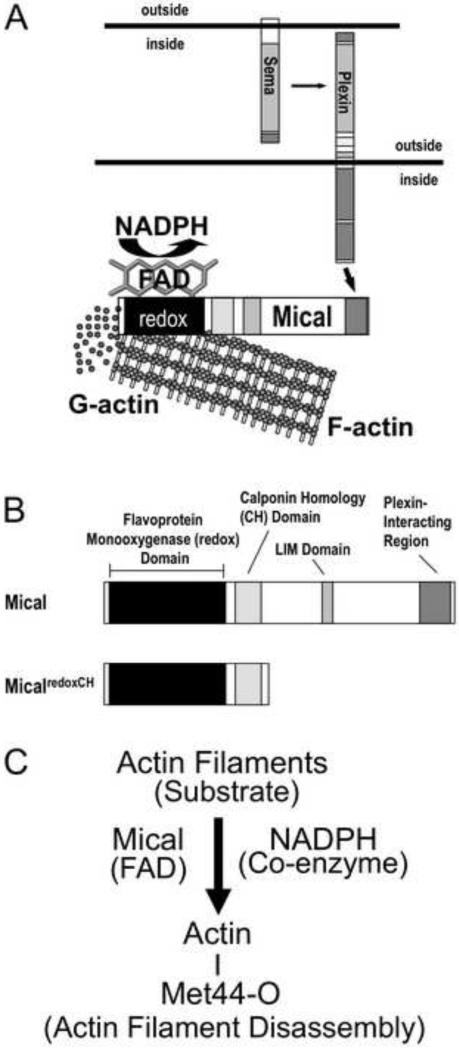
Figure 1. The Semaphorin-Plexin signaling molecule Mical is an actin regulatory enzyme.
((A) The Semaphorins (Semas) belong to one of the largest families of guidance cues and associate with their large, cell surface Plexin receptors to regulate the organization of the actin cytoskeleton. We have recently found that Mical proteins provide a direct physical link between Plexins and the actin cytoskeleton by directly associating with Plexin through their C-terminus and with filamentous actin (F-actin) with their N-terminus [10]. Likewise, our results reveal that Mical directly disassembles F-actin into G-actin using its flavin adenine dinucleotide (FAD) and nicotinamide adenine dinucleotide phosphate (NADPH) – binding redox domain [10-12].
(B) Members of the MICAL family of proteins contain a flavoprotein monooxygenase (redox) domain, a calponin homology (CH) domain, a LIM domain and a Plexin-interacting motif. We have found that the redox and CH domains of Mical (MicalredoxCH) are both necessary and together are sufficient for Mical's effects on the actin cytoskeleton in vivo [10].
(C) F-actin is a specific substrate for the FAD-binding protein (flavoenzyme) Mical, which in the presence of its coenzyme NADPH directly oxidizes (O) the Methionine (Met) 44 amino acid residue of individual actin subunits to induce actin filament disassembly and altered actin polymerization.
Materials and Methods
Chemicals and Chromatography Resins
Basic chemicals were obtained from Sigma Chemical, Inc., St. Louis, MO, USA, while Complete® EDTA-free protease inhibitor cocktail tablets were obtained from Roche Applied Science, Indianapolis, IN, USA and Imidazole was obtained from EMD Biosciences, San Diego, CA, USA. Conical centrifuge tubes were obtained from Corning Life Sciences (Corning, NY, USA), and Amicon Ultra centrifugal filters (Ultracel-50k) were obtained from Millipore, Billerica, MA, USA. Homogenization was performed with a high-pressure homogenizer (EmulsiFlex-C5, Avestin, Inc., Ottawa, Ontario, Canada). HISTRAP FF, HIPREP 26/10 Desalting columns, Superdex™ 200, 10/300GL gel filtration column, and AKTA Purifier UPC 10 were from GE Healthcare Bio-Sciences Corporation (Piscataway, NJ, USA). The Uno S6 column was obtained from Bio-Rad Company (Hercules, CA, USA).
Molecular Biology and Protein Expression
In order to obtain high-quality recombinant Mical protein, we have pursued a number of different bacterial expression and purification strategies ([9, 10, 13], including trying to adopt strategies that have been used to express different parts of Mical and specific portions of MICALs from other species (e.g., [13-15]). However, these expression strategies and constructs have not provided a rapid, simple, and efficient means to generate abundant levels of active Mical protein for different biochemical and F-actin assays in vitro and matching with regions of the Mical protein used in vivo ([13], data not shown). Since, our previous work with purified recombinant Mical protein revealed that the oxidoreductase (redox) and calponin homology (CH) domains of Mical (Figure 1B) are together sufficient for Mical's biochemical activity and for its effects on actin filaments in vivo and in vitro [10], we have concentrated our efforts on purifying a portion of the Mical protein containing both the redox and CH domains (MicalredoxCH). We have used the pET43.1bNG plasmid (see Figure 1 in Ref [16]) containing Drosophila MicalredoxCH for this work and it was transformed into ArcticExpress Competent cells (Stratagene, La Jolla, CA, USA) using heat shock at 42°C for one minute in a water bath and bacteria were selected for up-take of the plasmid by their ability to grow on LB/agar plates containing 50 μg/ml carbenicillin (Research Products International [RPI] Corp, Mount Prospect, IL, USA). A single clone was then selected from the culture plate using sterile technique and inoculated into 150ml TB culture medium containing 50 μg/ml carbenicillin, 2mM MgSO4 (Fisher Scientific, Pittsburgh, PA, USA), and 20μg/ml gentamycin (Sigma Chemical, Inc., St. Louis, MO) and shaken at 37°C overnight in an Innova44 shaking incubator (New Brunswick, Edison, New Jersey, USA). The overnight culture was then diluted (1:40) into 6 L of TB medium containing 50 μg/ml carbenicillin, mixed thoroughly, and then divided into 1 L aliquots that were placed into six 2.8L flask after which time 2ml of Antifoam B emulsion (Sigma Chemical, Inc., St. Louis, MO, A6707-500ml) were added into each flask. Bacteria were then cultured at 30°C with shaking at 225rpm for ~8 hrs. Isopropyl-β-D-thiogalactoside (IPTG) was then added to the culture at a final concentration of 0.5mM to induce expression of the Mical protein and the culture was then incubated with shaking at 10°C for 24 hrs. The culture was then poured into 1L centrifuge bottles (Nalge Nunc International Corporation, Rochester, NY, USA), which were spun in a Beckman J-6M Induction Drive Centrifuge at 3500rpm (2623 × g) for 30min to collect the bacteria pellet. The supernatant was then drained from the bottle and the bacteria pellet/bottle was then fast frozen in liquid nitrogen and stored at −80°C.
Homogenization and Clarification of Cell Lysates
Frozen bacteria pellets from 6 L TB media culture was thawed at room temperature for 30min. One tablet of Complete® EDTA-free Protease Inhibitors was dissolved in 100 ml of lysis buffer (10mM Tris-HCl, pH8.0, 500mM NaCl, 3mM β-mercaptoethanol, 20mM imidazole) according to the manufacturer's instructions. This protease inhibitor cocktail was then added to the cell pellets and allowed to stir using a stirrer plate at 4°C and stirrer bars until the pellet had completely dissolved (about 10min). The dissolved sample was then loaded into a high-pressure homogenizer (EmulsiFlex-C5, Avestin, Inc., Ottawa, Ontario, Canada) pre-cooled for 1 hr using a refrigerated circulating bath set at 4°C, and the bacteria were broken by increasing the pressure to 40 psi. In all cases, the output pressure was within 5,000-10,000 psi and the temperature of the sample was controlled by either immersion of the machine/tubing in ice or cooling and applying water using a re-circulating water bath. Initially, the cells were swirled during lysis to prevent cells from getting stuck in the machine and the lysis step was repeated two to three times (up to five times). As an alternative, the bacteria was lysed using a Misonix Ultrasonic Liquid Processor or similar and sonicating for 15 minutes at 50 amplitude, 5s on and 5s off. The cloudy supernatant was then further clarified by transferring the sample into a 250 ml NALGENE centrifuge bottle with a sealing cap and centrifuging the homogenized sample at 22,000g for 2 h with a Beckman J2-MC centrifuge. The resultant supernatant was transferred into another centrifuge bottle and spun for 30min at the same speed. The supernatant was then filtered using Durapore membrane filters (0.45μm, Millipore).
Ni2+-NTA Chromatography and SDS-PAGE
Nickel-nitrilotriacetic acid (Ni2+-NTA) chromatography was done using standard approaches by loading the sample into a 5ml HisTrapFF1-GE affinity column with buffer Ni-A (10mM Tris-HCl, pH8.0, 500mM NaCl, 5% glycerol, 3mM β-mercaptoethanol, 20mM imidazole) and washed with >20 column volumes (CV) of buffer Ni-A at flow rate of 1ml/min (faster flow rates [2-3 ml/min] were also employed if the pressure in the system stayed below 0.35 Mpa). Prior to eluting the MicalredoxCH protein, it is possible to remove much of the chaperonin-associated proteins Cpn60 and Cpn10 from the Ni2+-NTA affinity column by washing with 20 CV of dissociation buffer (20 mM Hepes pH 7.0, 10 mM MgCl2, 5 mM ATP, and 150 mM KCl)) [17]. The dissociation buffer was washed out with 10 CV of buffer Ni-A and MicalredoxCH protein was eluted with elution buffer Ni-B (10mM Tris-HCl, pH8.0, 500mM NaCl, 5% glycerol, 3mM β-mercaptoethanol, 250mM imidazole). The eluates were saved in one mL aliquot samples in microcentrifuge tubes using a FRAC-920 fraction collector (GE Healthcare Bio-Sciences Corporation, Piscataway, NJ, USA). 10 μL of each sample was then mixed with 3.3 μL of 4x Laemmli Sample Buffer (250mM Tris, pH6.8, 8% SDS, 40% glycerol, 0.032% bromophenol blue, 20% β-mercaptoethanol) and loaded onto an 1.5mm-thick SDS-PAGE gel composed of 10% separating gel (10% Acryl and bisacryl [29:1], 375mM Tris-HCl, pH8.8, 0.1% SDS, 0.1% ammonium persulfate and 0.08% TEMED) and 4% stacking gel (4% Acryl and bisacryl [29:1], 125mM Tris-HCl, pH6.8, 0.1% SDS, 0.15% ammonium persulfate and 0.125% TEMED). A Mini Format 1-D Electrophoresis unit (Bio-Rad Company, Hercules, CA, USA) was used for all protein electrophoresis.
Sample Desalting, Thrombin Digestion, and Ion Exchange Chromatography
Samples containing the appropriately sized bands were combined and desalted by loading them onto a HiPrep™ 26/10 column (GE Healthcare Bio-Sciences Corporation, Piscataway, NJ, USA) and eluting them with S-A Buffer (20mM NaPO4, pH7.5, 10mM NaCl, 5% glycerol, 1mM DTT). Samples were then thrombin (Roche Diagnostics GmbH, Germany) digested to remove the Nus solubility tag by incubation with 100 μl of 10 μg/μl thrombin at RT for 4 hrs. A small amount of the digested sample was then run on a 10% SDS-PAGE gel to check the efficiency of the thrombin digestion. After the thrombin digestion was confirmed to be complete, the sample in S-A buffer was loaded onto a MonoS column at a flow rate of 1 ml/min. The column was then washed with 10CV of S-A buffer (until the UV reading was stable, indicating that protein was not continuing to wash off the column). The samples were then collected in 1 ml aliquots following elution with 35 CV of an elution buffer S-B (20mM NaPO4, pH 7.5, 1 M NaCl, 5% glycerol, 1mM DTT). 10 μL samples from each collection tube were then mixed with Laemmli Sample Buffer and electorphoresed on an SDS-PAGE gel.
Gel Filtration Chromatography, Buffer Exchange, and Sample Concentration
Samples eluded from the ion exchange column containing the appropriately sized bands were loaded into a Millipore Amicon Ultra centrifugal filter (Ultracel-50 kDa cutoff) and centrifuged for 15 min at 2623 × g with a Beckman J-6M Induction Drive Centrifuge in order to decrease the volume. To further polish protein quality, samples were loaded onto a Superdex™ 200, 10/300GL gel filtration column (GE Healthcare Bio-Sciences Corporation, Piscataway, NJ, USA) that had been pre-equilibrated with gel filtration buffer (20 mM NaPO4, 200 mM NaCl, 5% glycerol, 1 mM DTT). Samples were washed with gel filtration buffer and collected in 500 μl aliquots. 5 μl of each aliquot was subjected to SDS-PAGE. Samples that were highly enriched for the appropriately sized band (MicalredoxCH protein) were then concentrated by loading the sample into a Millipore Amicon Ultra centrifugal filter (Ultracel-50kDa cutoff) and centrifuging as described above until the volume was reduced to less than 1 ml. The MicalredoxCH protein was then changed into its storage buffer by adding ~14 ml of ice-cold Mical protein Storage buffer (10mM Tris-HCl, pH8.0 100mM NaCl, 5% glycerol, 1mM DTT). The sample was then mixed evenly and centrifuged to concentrate it to <1 ml. This addition of ~14 ml of ice-cold Mical protein Storage buffer and concentration of the sample to <1 ml was repeated two more times with a final centrifugation step to decrease the volume to less than 500 μl.
Protein Quantification and Western Blotting
Protein concentration was determined by adding 2 μl to the platform of a Nanodrop spectrophotometer and measuring the absorption at 280 nm [18]. For Western blot analysis, 0.5 μg of purified MicalredoxCH protein and 5 μl Precision Plus Protein Standards (Bio-Rad Company, Hercules, CA, USA) were loaded onto a 10% SDS-PAGE gel. After electrophoresis, the protein was transferred onto a PVDF membrane (Immobilon P, Millipore, Billerica, MA). The membrane was blotted with 5% dry milk / PBS for 1 hr at room temperature. A 1:2000 dilution of a Mouse 6 His tag antibody (His tag Monoclonal antibody, Novagen, EMD Biosciences, San Diego, CA, USA)/ 5% dry milk/ PBS was then added to the membrane and incubated overnight at 4°C. The membrane was washed 5 times with PBS for 10 min each and then incubated in a 1:10,000 dilution of HRP-conjugated, sheep anti-mouse IgG antibody (NA931V, Amersham, GE Healthcare Bio-Sciences Corporation, Piscataway, NJ, USA) in 5% dry milk/ PBS for 1 hr at room temperature. The membrane was then washed 5 times with PBS for 10 min each, incubated in SuperSignal™ West Pico Chemiluminescent substrate (Thermo Scientific, Rockford, IL, USA) for 1 minute at RT, and then exposed to BioMax Light Film (Kodak, Rochester, NY, USA).
UV-visible Spectroscopy and Related Analyses
UV-visible spectroscopy was done using standard approaches [19] and either a Nanodrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA) or fluorescence spectrophotometer (Spectra Max M2; Molecular Devices, Sunnyvale, CA, USA). Scanning wavelengths between 250nm and 700nm were used. Data was then loaded into Excel software (Microsoft Corp., Redmond, WA, USA) and presented using GraphPad software (La Jolla, CA, USA). Calculating the percentage of purified MicalredoxCH protein that is bound to FAD (i.e., the percentage of MicalredoxCH protein that will be active) was done using standard approaches [20, 21] such that the concentration of the purified MicalredoxCH protein was determined as described above by adding 2 μl to the platform of a Nanodrop spectrophotometer and measuring the absorption at 280 nm. The concentration of FAD in the purified sample was then determined using standard approaches [22], by denaturing MicalredoxCH protein with 0.2% SDS, pelleting the denatured MicalredoxCH protein, measuring the absorbance of the free FAD in the sample, and then using the Beer-Lambert law (Absorption at 459 nm = ε[extinction coefficient, also known as molar absorptivity] × C [concentration in M] × l [path length in cm of the cuvette in which the sample is contained]) to calculate the concentration of FAD. The concentration of FAD in the purified sample was then divided by the concentration of MicalredoxCH in the purified sample to determine the percentage of purified MicalredoxCH protein that is bound to FAD (see also Figure 3B).
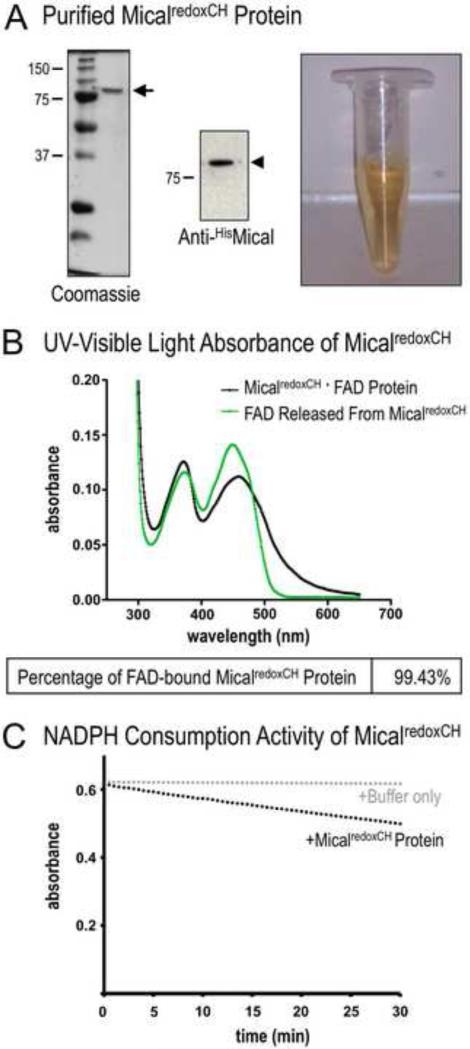
Figure 3. Determining the purity of the MicalredoxCH protein.
(A) Purified Mical protein in the presence of its prosthetic group (co-factor) FAD. SDS-PAGE/Coomassie-staining (left) and anti-His immunoblotting (middle) reveal both the presence of His-tagged MicalredoxCH protein (arrowhead) and the purity of the protein sample (arrow). Note also that this purified Mical protein is yellow in color (right). In particular, active Mical protein requires FAD to be purified along with the Mical protein backbone and FAD generates a yellowish color that can be used to confirm the presence of FAD. MW in kDa.
(B) The isoalloxazine ring system of FAD that generates the yellow/orange color of FAD (see Figure 3A) is also responsible for light absorption in the UV and visible spectral range such that the oxidized form of FAD has two peaks at ~360nm and ~450nm [19] (green line). Note, that the UV-visible light absorption spectra of the purified MicalredoxCH protein sample also reveal peaks at ~360 nm and ~450 nm (black line, peaks at 372nm and 459nm). This characteristic absorption spectra is a hallmark of a flavin binding protein. Characterization of the absorption spectra also allows a determination of the amount of the purified MicalredoxCH that is bound to FAD. The concentration of purified Mical in this sample is 4.2 mg/ml (50.16 μM) (see Materials and Methods). The concentration of FAD in this sample is 49.87 μM (0.4487/8998 M−1cm−1) (see Materials and Methods). Thus, 99.43% (49.87 μM/50.16 μM) of MicalredoxCH protein in the sample is bound to FAD.
(C) We confirmed the enzymatic activity of purified MicalredoxCH protein by following the conversion of Mical's co-enzyme NADPH to NADP+. This conversion (consumption) of NADPH can be followed by measuring the change in absorbance at 340 nm (NADPH absorbs light at 340 nm, while NADP+ does not). Note the change (decrease) in absorbance of NADPH over time in the presence of MicalredoxCH protein (black dots) but not with buffer only (the buffer used to store MicalredoxCH; gray dots). Consistent with our previous results [11], adding Mical's substrate, F-actin, into this assay further increases (in a F-actin concentration-dependent manner) the consumption of NADPH by MicalredoxCH protein (not shown).
Catalytic (NADPH-Consumption) Activity of Purified Mical Protein
Examining Mical's ability to consume its co-enzyme NADPH was done using standard approaches and based on the ability of NADPH to absorb light at 340 nm, while NADP+ does not [11]. To do this, 294 μl of DTT/General Actin Buffer (5 mM Tris-HCl pH8.0 and 0.2 mM CaCl2, 1mM DTT) was added into two 1.7 ml microcentrifuge tubes (tubes 1 and 2). As a control, 3 μl of MicalredoxCH protein storage buffer (10mM Tris-HCl, pH8.0 100mM NaCl, 5% glycerol, 1mM DTT) was added into tube 1. For the experimental tube (tube 2), 3 μl of 60μM purified MicalredoxCH protein was added (to generate a final concentration of 600 nM). The temperature of a Microplate reader (Spectra Max M2; Molecular Devices, Sunnyvale, CA, USA) was then set to 25°C and programmed to monitor absorbance intensity every 30 sec at 340 nm. The mixture from tube 1 was then placed into a Quartz and Glass Micro Cell (Fisherbrand, Fisher Scientific, Pittsburgh, PA, USA) and the absorbance was pre-read every 30 sec for 3 min to establish a zero point. 3 μl of 10 mM NADPH (MP Biomedicals, Solon, OH, USA, 100 μM final concentration) was then added into the same Micro Cell. The Micro Cell was then covered with a small piece of parafilm and mixed by inverting 5 times. The absorbance was recorded every 30 sec for 30 min to get results for the buffer only negative control. The mixture from tube 2 was then transferred into a Fisherbrand Quartz and Glass Micro Cell and the absorbance was pre-read every 30 sec for 3 min (the zero point). 3 μl of 10 mM NADPH (100 μM final concentration) was then added into the same Micro Cell and inverted 5 times to mix as described above. The absorbance was then recorded every 30 sec for 30 min to get the experimental result using the purified MicalredoxCH protein. The saved results from the spectrophotometer were then loaded into GraphPad Prism (La Jolla, CA, USA). The zero points of both samples were then normalized according to the pre-read results (zero point results) and the curve was presented using GraphPad Prism.
Actin polymerization Assay
Standard approaches for a pyrene-labeled actin polymerization assay was employed ([10-12, 23]; Cytoskeleton, Inc.) such that ATP (final concentration: 200μM) and 1M DTT (final concentration: 1 mM) were added into 1ml ice-cold General Actin Buffer (5mM Tris-HCl pH8.0 and 0.2 mM CaCl2). This General Actin Buffer was then used to dilute pyrene-labeled muscle actin (Cytoskeleton Inc, Denver, CO, USA) to 0.1mg/ml (2.3 μM). The mixture was pipetted up and down to mix after which time it was incubated on ice for 1 hr to depolymerize actin oligomers that had formed during storage. The actin was then centrifuged at 60,000 rpm (150,000 × g) at 4°C for 1 hr in a TLA-120.1 rotor and an Optima™ TLX Ultracentrifuge (Beckman Coulter, Inc., Fullerton, CA, USA) to remove residual actin nucleating centers after which the supernatant was transferred to a new tube on ice. Actin was then added into wells of a 96-well plate (100μl/well) and NADPH (MP Biomedicals, Solon, OH, USA, final: 200 μM) was added into some of the wells. A 2x actin polymerization buffer (10mM Tris-HCl pH7.5, 100 mM KCl, 4mM MgCl2, 2mM EGTA, 1mM DTT, 0.4mM ATP) containing either MicalredoxCH protein (final concentration: 600 nM) or the buffer used to store Mical (10mM Tris-HCl pH8.0, 100 mM NaCl, 5% glycerol, 1 mM DTT) was then added into each well to induce actin polymerization at 25° C. Actin was used at a final concentration of 1.1 μM for each of the conditions (Storage Buffer + actin, NADPH + actin, MicalredoxCH + actin, MicalredoxCH + NADPH + Actin). Fluorescence intensity was immediately monitored at 407 nm with excitation at 365 nm by a fluorescence spectrophotometer (Spectra Max M2; Molecular Devices, Sunnyvale, CA, USA). After normalization with the blank control, all curves were shifted to the same zero point as G-actin only controls. The XY scatter method was used to draw the dynamic actin polymerization curves with GraphPad Prism software (La Jolla, CA, USA).
Results and Discussion
In order to obtain high-quality recombinant Mical protein for biochemical, catalytic, imaging, and structural biological applications, we have pursued a number of different bacterial expression and purification strategies [9, 10, 13]. While each of these approaches has been successful in generating Mical protein that has been used for characterizing Mical and its effects on actin filaments [9-12], our approaches have also been confounded by problems of both low solubility and yield, and they have not provided a rapid, simple, and efficient means to generate abundant levels of Mical protein. Therefore, the objectives of this study were to: (1) establish a system to express high amounts of soluble Mical protein; (2) develop a simple and efficient chromatography protocol to purify large amounts of Mical protein; (3) characterize the purified Mical protein to establish its authenticity and integrity with respect to its correct mass, flavin/cofactor content, and NADPH/co-enzyme consumption activity; and (4) ascertain that the purity and catalytic efficiency of the purified Mical protein are sufficient for the characterization of Mical and its effects on actin filaments.
A new strategy to generate high levels of soluble MicalredoxCH protein
Our previous analysis of purified recombinant Mical protein revealed that the oxidoreductase (redox) and calponin homology (CH) domains of Mical (Figure 1B) are together sufficient for Mical's biochemical activity and for its effects on actin filaments in vivo and in vitro [10]. Therefore, we have concentrated our efforts on purifying a portion of the Mical protein containing both the redox and CH domains (MicalredoxCH). Our previous work [9, 10, 13] revealed that Mical protein was most soluble in bacteria when we expressed Mical fused to a Nus solubility tag [24-27]. However, the organization of the available pET43.1 vector that contained a Nus tag also contained His purification tags on both sides of a thrombin protease cleavage site, making it difficult to use His/nickel affinity chromatography to separate the Mical target protein from the Nus tag following thrombin-mediated protein cleavage. Therefore, we modified the pET43.1b vector to change the position of the thrombin cleavage site and the N-terminal His tag and inserted the redoxCH portion of Mical (Figure 1B) within this modified vector, which we called pET43.1bNG (see Figure 1 in Ref [16]).
Our initial observations with MicalredoxCH protein expressed using the pET43.1bNG vector revealed that 1) MicalredoxCH expressed in this vector was even more soluble in this configuration with the Nus tag than it had been using the pET43.1 vector and 2) the use of the pET43.1bNG vector increased the amount of MicalredoxCH protein that bound to the Ni2+ beads, suggesting that the N-terminal His tag might be better exposed in its new position. However, the on-going use of a number of conventional expression strategies and induction temperatures as low as 14°C still failed to provide high levels of soluble MicalredoxCH protein (data not shown). Our experimental analysis of Mical protein expression using bacteria revealed, however, that lowering the temperature of growth resulted in more soluble Mical protein (data not shown). We therefore turned to bacterial cells engineered to express cold-adapted chaperonin proteins Cpn60 and co-chaperonin Cpn10 from the cryophilic bacterium, Oleispira antarctica [28]. Chaperonins have often been employed to help bacteria process and fold recombinant proteins and Cpn60 and Cpn10 facilitate protein folding at temperatures as low as 4–12°C. Thus, these chaperonins enable both low-temperature expression and assisted processing of proteins, which has been found to increase the abundance of soluble protein [29, 30]. In comparison to more traditional induction temperatures [13], our pilot studies using both standard microbiological and induction procedures [9, 13, 31] and these cold-adapted Arctic Express bacteria revealed an increase in the levels of soluble MicalredoxCH (Figure 2A). These findings suggested that our pET43.1bNG expression vector coupled with expression in cold-adapted Arctic Express bacteria provided a means to produce large amounts of soluble MicalredoxCH protein in bacteria.
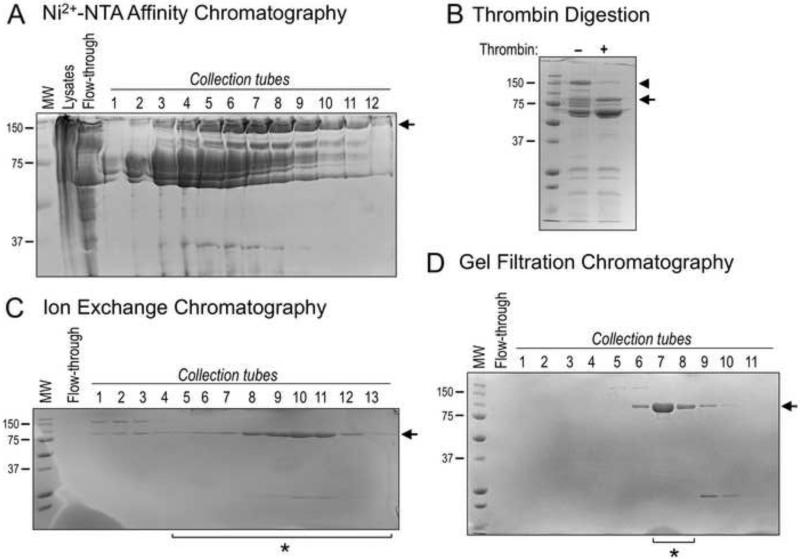
Figure 2. Purification of Recombinant MicalredoxCH protein.
(A) Coomassie-stained bands (arrow) corresponding to the Nus-tagged MicalredoxCH protein were observed in multiple collection tubes following IPTG-induced bacterial expression, lysis, Ni2+-NTA affinity chromatography, and elution with 250 mM imidazole. MW in kDa (also for B-D).
(B) Samples from (A) containing Ni2+-NTA affinity purified Nus-tagged MicalredoxCH protein were combined and subjected to thrombin digestion (+) to remove the Nus tag. A Coomassie-stained band corresponding in size to the cleaved MicalredoxCH protein was observed following thrombin digestion (arrow). Note, that uncleaved Nus-tagged MicalredoxCH protein is readily seen in the absence of thrombin (arrowhead).
(C) As seen following Coomassie-staining, Cation exchange chromatography was used to separate MicalredoxCH protein (arrow) from contaminating proteins, including chaperonins and the Nus tag. Proteins were eluted with a gradient of NaCl by increasing the percentage of S-B buffer. Sample tubes 5 through 13 (*) were utilized for (D).
(D) Gel filtration chromatography allowed the further purification of the MicalredoxCH protein as seen following Coomassie-staining such that the protein collected in tubes 7 and 8 (*) was combined, concentrated to a working volume, and utilized for the results presented in Figures 3 and 4. We find an overall yield of ~2 mg of highly-pure active MicalredoxCH protein can be routinely generated from 6 L of bacterial culture using this purification strategy and the starting culture volume can be scaled-up in order to purify larger amounts of MicalredoxCH protein.
Purification
The step-by-step purification process is described in detail below and summarized in Scheme 1. A representative MicalredoxCH purification process that monitors enzyme purity by SDS-PAGE and Coomassie blue staining is shown in Figure 2.
Scheme 1.
Expression and Purification of Recombinant MicalredoxCH
Homogenization and clarification of bacterial cell lysates
Homogenization of 45 g of cell paste, that was obtained from 6 L cultures (5 × 1010 cells), was performed in 100 ml of lysis buffer and resulted in a cloudy solution that was subjected to high-pressure homogenization and centrifugation at 22,000 × g to remove the larger particular matter. The supernatant was then centrifuged again at 22,000 × g after which time the clarified solution between the pellet and the film at the top of the centrifuge tube was removed, filtered through a 0.45μm filter, and the filtered solution was used for the next purification step (Ni2+− NTA chromatography).
Ni2+ -NTA chromatography
The sample preparation was first loaded into a column containing the Ni2+-NTA and the column was washed repeatedly with a buffer containing 20mM imidazole (buffer Ni-A) to elute contaminating proteins. The washing step was repeated with greater than 20 CV of this 20mM imidazole washing solution and was completed when the chromatogram of the FPLC was returned to baseline at 280nm indicating that no more protein was being eluted from the column. It should also be noted that MicalredoxCH protein is orange/yellow in color on a column (due to its flavin binding). Thus, we also followed the enrichment of the Mical protein by the yellow appearance of the column. To elute the bound MicalredoxCH from the Ni2+-NTA resin, elution buffer containing 250mM imidazole (elution buffer Ni-B) was added to the column and the MicalredoxCH protein was rapidly and completely eluted (Figure 2A). As was expected from similar MicalredoxCH purification work [10, 13], the Ni2+-NTA column provided a major purification step and greatly enriched the MicalredoxCH protein (Figure 2A). However, as can be observed in Figure 2A, several other prominent bands, including the chaperonin-associated proteins Cpn60 and Cpn10, also bound with tight affinity to the Ni2+-NTA column.
Thrombin digestion to remove the Nus solubility tag
Our previous work aimed at purifying the redox portion of the Mical enzyme [9, 13] revealed that the Nus solubility tag [24-27], in contrast to other hydrophilic solubility tags such as GST and MBP, greatly increased the solubility of MicalredoxCH. Therefore, we employed a Nus solubility tag that could be separated from MicalredoxCH by a consensus site for the serine protease thrombin (see Figure 1 in Ref [16]). To remove the Nus tag, we prepared our Ni2+-NTA pooled protein sample by first desalting and exchanging the buffer using a HiPrep 26/10 column. Little to no precipitate formed in our pooled fraction upon desalting and buffer exchange so we proceeded to digest our sample with thrombin. The full-length Nus-tagged MicalredoxCH migrated on an SDS-PAGE at ~150 kDa which was significantly reduced following digestion with thrombin (Figure 2B). Likewise, a prominent band at ~84kDa appeared in our thrombin digested sample that corresponded to the MicalredoxCH protein (Figure 2B). We also noticed the presence of bands corresponding to the chaperonins and also the appearance of a band corresponding in size to the ~56kDa Nus tag (Figure 2B). This indicated at least one more purification step was necessary to separate out the MicalredoxCH protein.
Ion exchange chromatography
We turned to ion exchange chromatography to remove the Nus tag and additional residual proteins including the chaperonins and enrich for our MicalredoxCH protein. Based on its amino acid sequence, the isoelectric point (pI) of our MicalredoxCH protein was predicted to be 7.07 (DNA Star Software, Madison, WI, USA) while the pI of the Nus tag was calculated at 4.43. We therefore selected an anion exchange column (Uno Q6) with a buffer pH above the pI for the MicalredoxCH protein (Tris Buffer, pH8.0). We found however a less than expected ability of the MicalredoxCH protein to bind and be purified to a high level from the anion exchange column at this pH (data not shown). This suggested that the MicalredoxCH protein might be more positively charged then would be predicted. We therefore examined the ability of MicalredoxCH to bind to a cation exchange column (such as a Uno S6 column or a Mono S 5/50 GL Column). Our results revealed that MicalredoxCH protein bound very tightly to the cation exchange column at pH7.5 and could not be eluted when the conductivity was lower than 18 mS/cm. We have also noticed similar ionic properties of different forms of the Mical protein and Mical proteins from other species (data not shown). Moreover, since proteins which bind the Ni-agarose column non-discriminately (i.e., without a His(6)-tag) are likely to carry a strongly negative charge on their surface and therefore would be predicted not to bind with a cation exchange column, this allowed us to further remove residual proteins and enrich for MicalredoxCH. Therefore, we loaded our sample through a cation exchange column at pH7.5 and following washing with 5% S-B buffer (in which the final conductivity was around 10 mS/cm), protein was eluted from the column using a gradient from 5-100% S-B buffer (Figure 2C). Our results revealed that the purity of the MicalredoxCH protein is very high after cation column chromatography (Figure 2C). However, in order to obtain enzyme with even more homogeneity and purity for precision spectroscopic, kinetic studies, and biochemical assays we added an additional purification step based on gel filtration chromatography.
Gel filtration chromatography and Buffer Exchange
We employed a gel filtration column to eliminate the residual lower molecular weight proteins that were still present in our sample (Figure 2C). To prepare for gel filtration chromatography, samples enriched for MicalredoxCH were loaded into an Amicon Ultracel-50 in order to decrease the volume. Samples were then loaded onto a Superdex 200, 10/300GL gel filtration column and washed with gel filtration buffer to enable the different size proteins to separate. During this time, samples were collected in 500 μl aliquots and the separation and purity of the samples were confirmed on an SDS-PAGE gel (Figure 2D). The samples that were highly enriched for MicalredoxCH protein were once again loaded into an Amicon Ultracel-50, after which the MicalredoxCH protein was exchanged into a buffer that we had previously found to be useful for storing active Mical protein [10, 13].
Characterization of the purity, light absorption properties, and catalytic activity of the Mical flavoprotein
The purified MicalredoxCH in its storage buffer was concentrated to a minimum volume using an Amicon Ultra centrifugal filter. A concentration of protein at 4.6 mg/ml was determined by absorption at 280 nm. An overall yield of >2 mg of highly pure MicalredoxCH was generated from 6 L of cell culture using this purification strategy (Figure 3A). Purified MicalredoxCH was confirmed by immunoblotting of the MicalredoxCH protein following SDS-PAGE and membrane transfer (Figure 3A). Likewise, since the Mical redox domain is a flavin adenine dinucleotide (FAD) binding module [9, 10, 13], we examined the color of the protein and found that the purified Mical protein was yellow/orange in color (Figure 3A), a distinguishing feature of proteins that bind flavins (flavoproteins). In particular, it is the isoalloxazine ring system that is the structural component of the flavin cofactor that is responsible for the yellow appearance of FAD, FMN, and flavoproteins. This isoalloxazine ring system is also responsible for light absorption in the UV and visible spectral range such that the oxidized form of the flavin has two peaks at ~360nm and ~450nm. We therefore examined the UV-Visible absorption spectra of our MicalredoxCH purified protein and found that it showed peaks at ~360nm and ~450nm (Figure 3B). To further analyze the purity of our MicalredoxCH protein and the percentage of purified MicalredoxCH protein that is bound to FAD (i.e., the percentage of MicalredoxCH protein that will be active) we took absorption readings of the MicalredoxCH protein at both 280nm (to determine the protein concentration of the apoprotein in the sample) and at 450nm (to determine the amount of flavin in the sample). In particular, the amount of FAD in a sample cannot typically be determined by simply measuring the absorbance of the sample, since when FAD is bound to a protein like MicalredoxCH, the protein backbone shields the FAD from absorbing light (e.g., compare the black and green lines in Figure 3B). Instead, the amount of FAD in the protein sample is determined by denaturing MicalredoxCH (thereby releasing FAD), pelleting the denatured MicalredoxCH protein, measuring the absorbance of the free FAD in the sample (as in the green line in Figure 3B), and then using the Beer-Lambert law to determine the concentration of FAD in the sample. However, it is not necessary to perform these steps each time Mical protein is purified if one knows the extinction coefficient (absorption coefficient) of the FAD when it is bound to the MicalredoxCH protein. For example, we have used standard approaches [20, 21] to determine that the extinction coefficient at 459nm of the MicalredoxCH . FAD protein is 8998 M−1 cm−1. The concentration of FAD in a sample can then be determined by using the Beer-Lambert law (concentration [M] = Absorption459/Extinction Coefficient MicalredoxCH . FAD). In Figure 3B, for example, using a spectrophotometer and a cuvette with a path length of 1 cm, the Absorption459 was determined to be 0.4487 for this sample. Thus as shown in Figure 3B, we were able to calculate the concentration of FAD in this sample and that the amount of active (FAD-bound) purified MicalredoxCH protein that we obtained from this purification strategy is >99%. Finally, since Mical uses the pyridine nucleotide NADPH as a co-enzyme in its redox reactions [10-12], we confirmed the activity of the purified MicalredoxCH protein by measuring the NADPH consumption activity of Mical (Figure 3C).
Biochemical analyses of purified Mical
We have recently found that the redox portion of the Mical protein regulates the biochemical properties of actin filaments [10-12]. In particular, we find using standard actin biochemical assays including pyrene-labeled actin polymerization and depolymerization assays, high-speed actin sedimentation assays, negative-staining electron microscopy (EM) approaches, and total internal reflection fluorescence (TIRF) microscopy that Mical catalytically induces actin depolymerization and suppresses actin polymerization [10-12]. Our previous results, therefore, uncover for the first time a biochemical role for Mical proteins and identify the Micals as important new actin modifying proteins with widespread implications for the control of the actin cytoskeleton. These previous results [10-12] also identify a simple and efficient biochemical assay in which to characterize the activity of our newly purified MicalredoxCH protein.
Actin filaments are necessary components of the contractile apparatus of muscle, and also control the shape and movement of non-muscle cells. These filaments are helical polymers composed of multiple individual 42 kD actin proteins (monomers) that connect (polymerize) together with non-covalent bonds and dynamically join and leave the filament at its two ends. The polymerization properties of actin can be reconstituted in vitro by using purified actin and specific constituents and conditions [23]. For example, fluorescently-labeled actins like pyrene actin have long been employed to monitor both the polymerization and depolymerization of actin using standard approaches, where the fluorescence intensity of the pyrene-labeled actin polymer is substantially higher than the pyrene-labeled actin monomer (Figure 4; [23]). Our previous work has revealed that purified MicalredoxCH has no observable effects on its own on actin polymerization or depolymerization [10, 11], and likewise, we find that purified MicalredoxCH prepared using our new expression and purification procedure also has no observable effects in an actin polymerization assay (Figure 4). In particular, as can be observed by following the characteristic increase in fluorescence intensity over time, the addition of MicalredoxCH to actin does not alter its rate or extent of polymerization (Figure 4, black line). However, Mical uses the pyridine nucleotide NADPH as a required coenzyme in its in vitro reactions regulating F-actin dynamics (Figure 1C; e.g., [10-12]), and we find that similar to our previously published results [10-12] that activating our newly purified MicalredoxCH with NADPH results in dramatic alterations to the ability of actin to polymerize (Figure 4). In particular, as measured in changes in fluorescence intensity, we observed a decrease in actin polymerization over time, which was followed by a substantial decrease in the extent of polymerization, the rapid depolymerization of F-actin to G-actin, and the inability of G-actin to reinitiate detectable polymer formation (Figure 4, green line). NADPH on its own has no appreciable effect on actin polymerization. Thus, our results reveal that our newly purified MicalredoxCH exhibits similar effects on actin dynamics to that of our previously purified MicalredoxCH [10-12]. Indeed, we noted a similar but increased activity and effect on F-actin dynamics with our new MicalredoxCH protein (not shown) that is likely the result of our new purification procedure generating recombinant MicalredoxCH protein that is of higher purity than our previously purified recombinant MicalredoxCH protein [13]. Thus, this expression and purification strategy generates highly-active Mical enzyme.
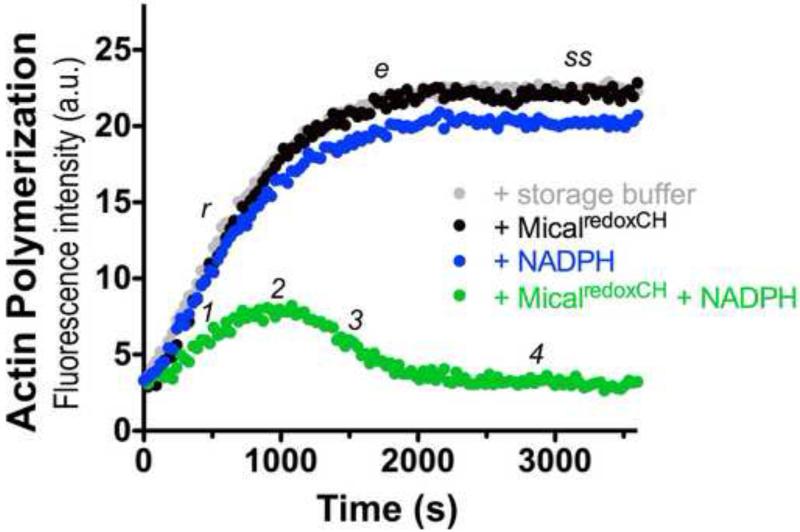
Figure 4. Analysis of the activity of purified MicalredoxCH protein.
Pyrene-labeled actin was used to monitor both the polymerization and depolymerization of actin, where the fluorescence intensity (a.u. [arbitrary units]) of the pyrene-labeled actin polymer is substantially higher than the pyrene-labeled actin monomer. As judged by the increase in fluorescence intensity over time which reveals actin polymerization, the addition of 600 nM of purified MicalredoxCH protein alone to actin (black dots) does not alter the rate (r), extent (e), or steady-state level (ss) of actin polymerization (as compared to an actin only control incubated with the buffer used to store MicalredoxCH; gray dots). In the presence of 100 μM of its NADPH coenzyme, however, purified MicalredoxCH protein (MicalredoxCH + NADPH; green dots) specifically alters the rate of actin polymerization (1) such that over time activated MicalredoxCH induces a decrease in the extent of polymerization (2), the rapid depolymerization of F-actin (3), and the inability of actin to reinitiate polymer formation (4). These results are similar to what we observed with our previously purified active MicalredoxCH enzyme [10-12].
Conclusions
We provide herein a simple, efficient, and detailed approach that utilizes a modified bacterial expression vector, a low temperature expression system, and a simple chromatography set-up to express and purify large amounts of an active form of the redox enzyme Mical. In particular, the purification procedure reported here is unique, yet it is simple, efficient, and highly reproducible, and allows one to go from cell harvesting to >95% pure protein in ~5 days. Moreover, our three-step purification process consisting of consecutive Ni2+−NTA chromatography, ion exchange chromatography, and gel filtration chromatography is capable of yielding over 2 mg of MicalredoxCH. This procedure could also easily be scaled-up in culture volume in order to purify even larger amounts of MicalredoxCH protein. We also find that this MicalredoxCH enzyme can be aliquoted and stored frozen (it is stable in this manner for >18 months). It is also stable upon freeze-thaw and enables one to carry out a large number of experiments with a single preparation of Mical enzyme (e.g., we have used this MicalredoxCH protein in F-actin assays at concentrations as low as 1nM). The quality of the enzyme and its specific activity are also higher than our previously published efforts, and will provide a source of purified protein for further characterizing the biochemical properties and actin regulatory effects of MICAL-family actin disassembly factors. Furthermore, this purification procedure yields a homogenous sample of highly pure enzyme as well as reproducibility from batch to batch that will be useful for crystallographic and other high-precision spectroscopic and kinetic studies, various enzymology and catalytic determinations, additional assay development, high throughput screening, and characterization of inhibitors and activators of Mical. Our work described herein identifying a simple purification strategy for Mical should also prove useful for examining the role of other domains of Mical and their effects on Mical-mediated F-actin alterations. Finally, our preliminary results indicate that this purification strategy also yields high amounts of pure protein for the three human MICAL family members. This will allow the comparison of the different MICALs from invertebrates to humans.
Highlights.
A bacterial expression system for generating high amounts of Mical enzyme
A purification strategy that yields highly-pure and active Mical enzyme
This enzyme is suitable for biochemical, structural, imaging, and catalytic studies
Acknowledgments
We thank Taehong Yang and Hunkar Gizem Yesilyurt for helpful comments on this work and Willem van Berkel and members of the Terman Lab for experimental insights, discussions, and assistance. We also thank Huawei He and Xuewu Zhang for suggestions on the use of the cold-adapted chaperonin proteins and the low temperature expression system. This work was supported by grants from the NINDS (NS073968) and Welch Foundation (I-1749) to J.R.T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Stossel TP. On the Crawling of Animal-Cells. Science. 1993;260:1086–1094. doi: 10.1126/science.8493552. [DOI] [PubMed] [Google Scholar]
- 2.Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 3.Hung R-J, Terman JR. Extracellular inhibitors, repellents, and Semaphorin/Plexin/MICAL-mediated actin filament disassembly. Cytoskeleton. 2011;68:415–433. doi: 10.1002/cm.20527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolodkin AL, Tessier-Lavigne M. Mechanisms and molecules of neuronal wiring: a primer. Cold Spring Harb Perspect Biol. 2011;3:a001727. doi: 10.1101/cshperspect.a001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pasterkamp RJ. Getting neural circuits into shape with semaphorins. Nat Rev Neurosci. 2012;13:605–618. doi: 10.1038/nrn3302. [DOI] [PubMed] [Google Scholar]
- 6.Rehman M, Tamagnone L. Semaphorins in cancer: biological mechanisms and therapeutic approaches. Semin Cell Dev Biol. 2013;24:179–189. doi: 10.1016/j.semcdb.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Kumanogoh A, Kikutani H. Immunological functions of the neuropilins and plexins as receptors for semaphorins. Nat Rev Immunol. 2013;13:802–814. doi: 10.1038/nri3545. [DOI] [PubMed] [Google Scholar]
- 8.Worzfeld T, Offermanns S. Semaphorins and plexins as therapeutic targets. Nat Rev Drug Discov. 2014;13:603–621. doi: 10.1038/nrd4337. [DOI] [PubMed] [Google Scholar]
- 9.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 10.Hung RJ, Yazdani U, Yoon J, Wu H, Yang T, Gupta N, Huang Z, van Berkel WJ, Terman JR. Mical links semaphorins to F-actin disassembly. Nature. 2010;463:823–827. doi: 10.1038/nature08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung RJ, Pak CW, Terman JR. Direct redox regulation of F-actin assembly and disassembly by Mical. Science. 2011;334:1710–1713. doi: 10.1126/science.1211956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung RJ, Spaeth CS, Yesilyurt HG, Terman JR. SelR reverses Mical-mediated oxidation of actin to regulate F-actin dynamics. Nat Cell Biol. 2013;15:1445–1454. doi: 10.1038/ncb2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta N, Terman JR. Characterization of MICAL flavoprotein oxidoreductases: Expression and solubility of different truncated forms of MICAL. In: Frago S, Gomez-Moreno C, Medina M, editors. Flavins and Flavoproteins 2008; Proceedings 16th International Symposium on Flavins and Flavoproteins; Zaragoza. Prensas Universitarias De Zaragoza; 2008. pp. 345–350. [Google Scholar]
- 14.Siebold C, Berrow N, Walter TS, Harlos K, Owens RJ, Stuart DI, Terman JR, Kolodkin AL, Pasterkamp RJ, Jones EY. High-resolution structure of the catalytic region of MICAL (molecule interacting with CasL), a multidomain flavoenzyme-signaling molecule. Proc Natl Acad Sci U S A. 2005;102:16836–16841. doi: 10.1073/pnas.0504997102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadella M, Bianchet MA, Gabelli SB, Barrila J, Amzel LM. Structure and activity of the axon guidance protein MICAL. Proc Natl Acad Sci U S A. 2005;102:16830–16835. doi: 10.1073/pnas.0504838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta N, Wu H, Terman JR. A modified bacterial expression vector for expressing and purifying Nus solubility-tagged proteins. Data in Brief. doi: 10.1016/j.dib.2016.07.032. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joseph RE, Andreotti AH. Bacterial expression and purification of interleukin-2 tyrosine kinase: single step separation of the chaperonin impurity. Protein Expr Purif. 2008;60:194–197. doi: 10.1016/j.pep.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoscheck CM. Quantitation of Protein. Method Enzymol. 1990;182:50–68. doi: 10.1016/0076-6879(90)82008-p. [DOI] [PubMed] [Google Scholar]
- 19.Chapman SK, Reid GA, editors. Flavoprotein Protocols. Humana Press; Totowa, NJ: 1999. [Google Scholar]
- 20.Macheroux P. UV-visible spectroscopy as a tool to study flavoproteins. Methods Mol Biol. 1999;131:1–7. doi: 10.1385/1-59259-266-X:1. [DOI] [PubMed] [Google Scholar]
- 21.Leferink NG, van den Berg WA, van Berkel WJ. l-Galactono-gamma-lactone dehydrogenase from Arabidopsis thaliana, a flavoprotein involved in vitamin C biosynthesis. FEBS J. 2008;275:713–726. doi: 10.1111/j.1742-4658.2007.06233.x. [DOI] [PubMed] [Google Scholar]
- 22.Aliverti A, Curti B, Vanoni MA. Identifying and quantitating FAD and FMN in simple and in iron-sulfur-containing flavoproteins. Methods Mol Biol. 1999;131:9–23. doi: 10.1385/1-59259-266-X:9. [DOI] [PubMed] [Google Scholar]
- 23.Cooper JA. Actin filament assembly and organization in vitro. In: Carraway KL, Carraway CAC, editors. The Cytoskeleton: A Practical Approach. Oxford University Press; New York: 1992. pp. 47–71. [Google Scholar]
- 24.Terpe K. Overview of tag protein fusions: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2003;60:523–533. doi: 10.1007/s00253-002-1158-6. [DOI] [PubMed] [Google Scholar]
- 25.Davis GD, Elisee C, Newham DM, Harrison RG. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol Bioeng. 1999;65:382–388. [PubMed] [Google Scholar]
- 26.Harrison RG. Expression of soluble heterologous proteins via fusion with NusA protein. InNovation. 2000;11:4–7. [Google Scholar]
- 27.Kohl T, Schmidt C, Wiemann S, Poustka A, Korf U. Automated production of recombinant human proteins as resource for proteome research. Proteome Sci. 2008;6:4. doi: 10.1186/1477-5956-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferrer M, Chernikova TN, Yakimov MM, Golyshin PN, Timmis KN. Chaperonins govern growth of Escherichia coli at low temperatures. Nat Biotechnol. 2003;21:1266–1267. doi: 10.1038/nbt1103-1266. [DOI] [PubMed] [Google Scholar]
- 29.Ferrer M, Lunsdorf H, Chernikova TN, Yakimov M, Timmis KN, Golyshin PN. Functional consequences of single:double ring transitions in chaperonins: life in the cold. Mol Microbiol. 2004;53:167–182. doi: 10.1111/j.1365-2958.2004.04077.x. [DOI] [PubMed] [Google Scholar]
- 30.Vera A, Gonzalez-Montalban N, Aris A, Villaverde A. The conformational quality of insoluble recombinant proteins is enhanced at low growth temperatures. Biotechnol Bioeng. 2007;96:1101–1106. doi: 10.1002/bit.21218. [DOI] [PubMed] [Google Scholar]
- 31.Wu H, Gao J, Sharif WD, Davidson MK, Wahls WP. Purification, folding, and characterization of Rec12 (Spo11) meiotic recombinase of fission yeast. Protein Expr Purif. 2004;38:136–144. doi: 10.1016/j.pep.2004.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]