Abstract
Targeted gene disruption by in vitro transposon mutagenesis has been used to identify the genes required for biosynthesis of the Haemophilus influenzae Rd cell wall under standard cultivation conditions. Of the 28 genes known to be associated with the cell wall biosynthetic pathway, 14 were determined to be essential.
Judicious identification and validation of novel protein targets for therapeutic intervention represent the first critical steps in developing new antimicrobials. Key elements in target selection include conservation across a broad range of human pathogens, essentiality of the target for bacterial viability or pathogenesis, and known function of the target for the development of screening assays.
Transposon mutagenesis has become an important tool for determining bacterial gene essentiality (3). Akerley et al. identified essential Haemophilus influenzae genes by in vitro transposition and genetic footprinting, looking for open reading frames in which transposons could not integrate as evidence of essentiality (1). Reich and coworkers used in vitro transposon mutagenesis to create a random insertional library in H. influenzae. The library was then subjected to PCR and Southern analysis to determine the positions of the chromosomal insertions, thereby identifying essential and nonessential genes (4). Recently, Akerley and colleagues applied high-density in vitro transposon mutagenesis to the entire H. influenzae genome to identify genes essential for growth or viability (2). However, of the 1,725 genes examined, the essentiality of 453 could not be determined. We have reported previously on the use of a rapid in vitro transposition method for insertionally inactivating genes in H. influenzae (5). Here we describe the application of this technology to determining the essentiality of a compiled list of the H. influenzae genes currently associated with cell wall biosynthesis (http://www.genome.jp/kegg/pathway.html).
PCR amplification of the target gene with a flanking primer pair, in vitro transposon mutagenesis, and competent cell transformation were performed as previously described (5). PCR screening of surviving clones for inserts within the N terminus of the target gene by internal gene primers was used to identify nonessential genes. If no inserts were detected after an additional round of screening targeting small colonies with reduced growth rates, then the surviving colonies were rescreened with the flanking primer pair for the presence of a transposon inserted within the flanking sequences as a control to ensure the efficiency of the mutagenic reaction. Essential genes were defined by the lack of inserts within the N terminus but present in the flanking regions surrounding the target gene. A list of all of the primer pairs used in this work is available upon request.
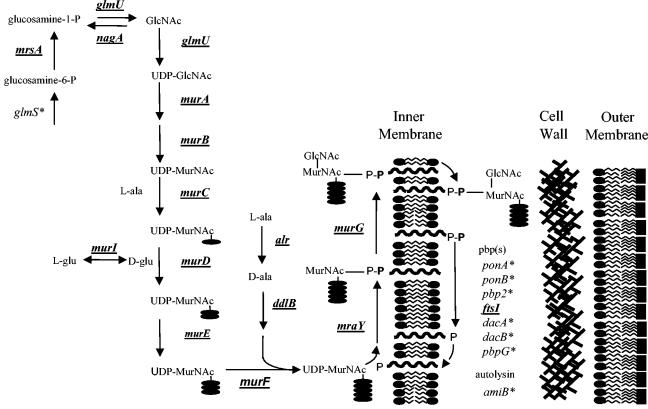
A summary of cell wall biosynthetic gene essentiality is shown in Table 1 and illustrated in Fig. 1. All of the genes directly involved in peptidoglycan biosynthesis, except for mrsA, are essential. Although the two individual mrsA genes can be independently inactivated, a functional MrsA enzyme is essential for bacterial survival. Six of the seven penicillin-binding proteins are not essential; only ftsI (penicillin-binding protein 3) is essential. Of the remaining seven genes associated with cell wall biosynthesis, only nagA is essential.
TABLE 1.
Summary of H influenzae cell wall gene essentiality
| Gene | Locus | No. of Tn inserts/total no. of colonies screened | Essential in H. influenzae Rd |
|---|---|---|---|
| alr | HI1575 | 0/96 | Yes |
| amiB | HI0066 | 3/96a | No |
| dacA | HI0029 | 22/48 | No |
| dacB | HI1330 | 3/48 | No |
| ddlB | HI1140 | 0/96 | Yes |
| ftsI | HI1132 | 0/96 | Yes |
| glmS | HI0429 | 28/48 | No |
| glmU | HI0624 | 0/96 | Yes |
| glnA | HI0865 | 5/96a | No |
| lpp | HI1579 | 22/48 | No |
| mepA | HI0197 | 25/48 | No |
| mraY | HI1135 | 0/96 | Yes |
| mrsAb | HI1337 | No | |
| mrsAb | HI1463 | No | |
| mtgA | HI0831 | 5/96a | No |
| murB | HI0268 | 0/96 | Yes |
| murC | HI1139 | 0/96 | Yes |
| murD | HI1136 | 0/96 | Yes |
| murE | HI1133 | 0/96 | Yes |
| murF | HI1134 | 0/96 | Yes |
| murG | HI1138 | 0/96 | Yes |
| murI | HI1739 | 0/96 | Yes |
| murZ | HI1081 | 0/96 | Yes |
| nagA | HI0140 | 0/96 | Yes |
| pbp2 | HI0032 | 6/48 | No |
| pbpG | HI0364 | 25/48 | No |
| ponA | HI0440 | 36/96a | No |
| ponB | HI1725 | 16/48 | No |
| slt | HI0829 | 14/48 | No |
Exhibits a slow-growth phenotype compared to the wild type.
Individual gene copies are not essential; however, MrsA function is essential.
FIG. 1.
The H. influenzae cell wall biosynthetic pathway. Essential enzymes are underlined and in bold. Nonessential enzymes are indicated by asterisks.
The compiled list of the H. influenzae cell wall biosynthetic genes revealed two genes encoding MrsA (phosphoglucosamine mutase), HI1337 and HI1463. A direct sequence comparison showed that a 5,432-bp segment of the chromosome encompassing mrsA had been duplicated. Determining mrsA essentiality began with attempting to insertionally inactivate each copy of mrsA. A single mrsA PCR amplicon containing sequence solely within the duplicated region was synthesized, mutagenized, and transformed into H. influenzae. Screening with primers unique to sequence upstream of each mrsA gene and within mrsA itself identified colonies containing insertions in either HI1337 or HI1463, but not in both genes within the same cell. Having established that each individual gene was not essential, two H. influenzae strains were then constructed containing an allelic replacement of either HI1337 or HI1463 with a chloramphenicol resistance marker to determine the essentiality of MrsA enzyme function. First, insertional inactivation with the original randomly mutagenized mrsA PCR amplicon was performed in each deletion strain, designated 1337Δ or 1463Δ, to determine the essentiality of the remaining copy of the mrsA gene. No insertions in the remaining mrsA gene were found in either strain. Next, two mrsA gene amplicons containing a transposon insertion in either HI1337 or HI1463 from the initial insertion study were generated. The mrsA::Tn gene amplicon from HI1337 was transformed into HI1463Δ, and the mrsA::Tn amplicon from HI1463 was transformed into HI1337Δ, selecting for kanamycin resistance. Neither transformation produced kanamycin-resistant colonies. Taken together, these results demonstrate that both mrsA genes are functional and that at least one functional copy of the mrsA gene must be present for cell viability.
Determining gene essentiality by insertional inactivation can be complicated by several factors, including protein fusion, operon structure, growth conditions, and gene duplication.
Misidentification of essential genes from enzyme activity that is present because of protein fusion with the gene for kanamycin resistance is all but eliminated because the transposon was engineered to include stop codons in all three reading frames in both orientations. Distinguishing between a null transformation result and true essentiality relied on detecting inserts within the sometimes small intergenic regions of proposed operons or sequence mapping insertions within the C-terminal end of an essential gene (murF). Examples of the lack of polarity of the transposon were evident when colonies containing insertions in small intergenic regions between adjacent essential genes were all viable. We compensated for the possibility of cellular growth rates affecting gene essentiality by extending the incubation period of the recombinants after the initial round of PCR screening another 24 h and then selecting the smallest colonies for additional screening. Four cell wall genes identified with this slow-growth phenotype, amiB, glnA, mtgA, and ponA, although not essential, are required for optimal growth. By individually targeting duplicate gene copies, our technique provides an avenue for addressing their essentiality. For example, we were able to show that although each individual copy of mrsA could be inactivated independently, one functional copy of mrsA must be present, thereby identifying the enzymatic step catalyzed by MrsA as essential.
The results presented here closely mirror those obtained with Escherichia coli, with a few exceptions. The gene ddlB is essential in H. influenzae but not in E. coli because E. coli carries two copies of the d-Ala-d-Ala ligase gene, ddlA and ddlB. The nagA gene is essential in H. influenzae but not in E. coli. This may represent metabolic differences between H. influenzae and E. coli. The pbp2 gene is not essential in H. influenzae, but the E. coli homolog is essential. Since variation in pbp essentiality between species is known, this result is not unexpected.
Nonessential genes were defined in this study as cellular survival with the target gene insertionally inactivated. Nonessential gene function could be compensated for by other cellular proteins, as this study addressed essentiality at the genetic level and not at the enzymatic level (with the exception of MrsA). Our survey also included only the cell wall biosynthetic genes known to date; H. influenzae may yet contain genes of unknown function that are involved in cell wall biosynthesis. Although insertional mutagenesis is a highly reliable way of determining gene essentiality, multiple approaches need to be used to ultimately validate target essentiality in pathogenic bacteria as one step in the process toward the development of novel antimicrobials.
Acknowledgments
We thank the members of the former Pharmacia Discovery Core Sequencing Lab for sequencing contributions.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, A. Camilli, D. J. Lampe, H. M. Robertson, and J. J. Mekalanos. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA 95:8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moir, D. T., K. J. Shaw, R. S. Hare, and G. F. Vovis. 1999. Genomics and antimicrobial drug discovery. Antimicrob. Agents Chemother. 43:439-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich, K. A., L. Chovan, and P. Hessler. 1999. Genome scanning in Haemophilus influenzae for identification of essential genes. J. Bacteriol. 181:4961-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trepod, C. M., and J. E. Mott. 2004. Identification of the Haemophilus influenzae tolC gene by susceptibility profiles of insertionally inactivated efflux pump mutants. Antimicrob. Agents Chemother. 48:1416-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]