Abstract
Background and Aims
Mucoid structures that coat the epithelium play an essential role in keeping the intestinal microbiota at a safe distance from host cells. Encroachment of bacteria into the normally almost-sterile inner mucus layer has been observed in inflammatory bowel disease and in mouse models of colitis. Moreover, such microbiota encroachment has also been observed in mouse models of metabolic syndrome, which are associated low-grade intestinal inflammation. Hence, we investigated if microbiota encroachment might correlate with indices of metabolic syndrome in humans.
Methods
Confocal microscopy was used to measure bacterial-epithelial distance of the closest bacteria per high-powered field in colonic biopsies of all willing participants undergoing cancer screening colonoscopies.
Results
We observed that, among all subjects, bacterial-epithelial distance was inversely correlated with body mass index, fasting glucose levels, and hemoglobin A1C. However, this correlation was driven by dysglycemic subjects, irrespective of body mass index, whereas the difference in bacterial-epithelial distance between obese and nonobese subjects was eliminated by removal of dysglycemic subjects.
Conclusions
We conclude that microbiota encroachment is a feature of insulin resistance–associated dysglycemia in humans.
Keywords: Metabolic Syndrome, Mucus Layer, Microbiota
Abbreviations used in this paper: BMI, body mass index; HPF, high-powered field; IBD, inflammatory bowel disease; PBS, phosphate-buffered saline; TLR, Toll-like receptor
Graphical abstract
See editorial on page 324.
Summary.
We previously reported that, in multiple murine models of low-grade intestinal inflammation, development of metabolic syndrome correlates with encroachment of bacteria into the normally sterile inner colonic mucus layer. Here, we report that microbiota encroachment is also a feature of metabolic disease, particularly insulin resistance–associated dysglycemia, in humans.
The intestinal tract is inhabited by a large diverse community of bacteria collectively referred to as gut microbiota. When stably maintained, at an appropriately safe distance from epithelial cells, gut microbiota provides a benefit to the host, especially in terms of energy harvest and promotion of immune development.1 However, disturbance of the microbiota-host relationship can drive chronic gut inflammation, including Crohn's disease and ulcerative colitis, collectively referred to as inflammatory bowel disease (IBD).2, 3 Accordingly, patients with IBD, and persons deemed to be at elevated risk for IBD development, exhibit alterations in gut microbiota composition and, moreover, exhibit altered bacteria localization.4, 5 Specifically, IBD-prone individuals frequently display gut bacteria close to, and/or in direct contact with, the epithelium, often accompanied by a thinner or disorganized mucus layer, whereas in control subjects the dense inner layer of mucus rarely exhibits bacteria.6, 7 Such encroaching bacteria are thought to play a role in triggering the activation of the mucosal immune system that characterizes IBD.
Studies in mice suggest that alteration of the host-microbiota can also result in more mild forms of inflammation characterized by modest elevations in proinflammatory gene expression that associate with metabolic syndrome. For example, loss of genes involved in innate immune-mediated recognition of bacteria, such as Toll-like receptor (TLR) 5, TLR2, and NLRP6, resulted in alterations in microbiota composition, low-grade inflammation, and a metabolic syndrome–like phenotype that could be transferred via fecal transplant indicating a central role for the microbiota in these mouse models.8, 9, 10, 11 Such alterations in microbiota result in microbiota encroachment that can be envisaged to play a role in driving elevated proinflammatory gene expression.12 Microbiota encroachment, low-grade inflammation, and metabolic syndrome could be induced, in wild-type mice, by administration of dietary emulsifiers leading to the suggestion that this ubiquitous class of food additives might be a contributor to the post-mid-20th-century increased incidence of metabolic syndrome.13 However, whether microbiota encroachment might be a feature of metabolic syndrome in humans has not been investigated and, hence, was the focus of this study.
Methods
Human Subjects
Subjects were enrolled at the Veteran's Administration Hospital (Atlanta, GA), following their provision of informed consent using procedures approved by the institutional review board, in consecutive fashion from August 1, 2013, to December 31, 2015 (Table 1). Inclusion criteria were subjects undergoing colonoscopy for screening for colon cancer who were at least 21 years of age and had no major health problems besides diabetes. Exclusion criteria were greater than 75 years of age, history of IBD, having a history of systemic neurologic or muscular disorder (eg, Parkinson disease, multiple sclerosis, or Alzheimer disease), or having significant comorbid conditions (eg, chronic liver disease or malignancy) or laboratory abnormalities that preclude colonoscopy. Additionally, patients with history of recent significant gastrointestinal bleeding were excluded from the study. A history, focusing on history of diabetes and gastrointestinal complaints including any prior trials of medications for these complaints, was obtained by review of the medical record database and interview by a study associate. A limited review of the patient medical record was conducted to determine control of diabetes as shown by glycosylated hemoglobin and fasted serum glucose levels. During the colonoscopy procedure 2 mucosal biopsies were taken in the left colon approximately 40 cm from the anus using regular forceps. The biopsies were immediately placed in Carnoy fixative and analyzed by confocal microscopy as described later.
Table 1.
Clinical Metadata of Human Patients Used in the Study
| Patient # | Age | Sex | Race | Weight (kg) | Height (cm) | BMI (kg/m2) | Blood glucose concentration (mg/dL) | HbA1C (%) | DM | Cholesterol (mg/dL) | Triglyceride (mg/dL) | LDL (mg/dL) | Average distance of closest bacteria to intestinal epithelial cells (μm) | PMH | Medication | Antibiotic | Bowel habits |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 68.5 | M | White | 96.6 | 180.3 | 30.00 | 104 | ND | No | 56 | 123 | 60 | 19.00 | Asthma, HLD, HTN | Diazepam, escitalopram, HCTZ/triamterene, losartan, prazosin, ASA | None | ND |
| 2 | 66.5 | M | White | 86.2 | 182.9 | 25.80 | 98 | 5.9 | No | 167 | 90 | 88 | 25.67 | RA, SICCA, HTN | Amlodipine, ASA, atorvastatin, folic acid, gabapentin, methotrexate, prednisone, ranitidine | Topical clindamycin and metronidazole | ND |
| 3 | 53.5 | M | Black | 68.0 | 182.9 | 20.39 | 75 | 5.7 | No | 205 | 62 | 80 | 30.50 | COPD | Albuterol, tiotropium, mometasone | None | ND |
| 4 | 65.5 | M | White | 87.4 | 175.3 | 28.50 | 97 | 6.0 | No | 199 | 167 | 122 | 18.60 | CAD, HLD, HTN, kidney stone, ischemic colitis | Lisinopril, metoprolol, pravastatin, | None | ND |
| 5 | 62.5 | M | Black | 105.2 | 175.3 | 34.33 | 148 | 7.7 | Yes | 167 | 129 | 92 | 10.80 | DM, hepatitis C, HTN, prostate cancer, cerebral thrombosis with infarction, OSA | Amlodipine, atorvastatin, gabapentin, glipizide, HCTZ, lisinopril, metformin, oxybutynin | None | Fecal incontinence |
| 6 | 63.5 | M | Black | 85.3 | 185.4 | 24.60 | 106 | 6.8 | Yes | 131 | 82 | 74 | 23.33 | DM, HTN, HLD | ASA, atenolol, diltiazem, losartan, metformin, pravastatin | None | GI ROS neg |
| 7 | 71.5 | M | White | 78.9 | 177.8 | 25.00 | 97 | 4.8 | No | 201 | 27 | 75 | 16.43 | CAD, HTN, Raynaud | Lisinopril, pravastatin | None | ND |
| 8 | 59.5 | M | Hispanic | 92.5 | 170.2 | 32.02 | 191 | 7.0 | Yes | 181 | 254 | 87 | 8.25 | DM, HTN, hepatitis C, asthma | Acarbose, albuterol, atorvastatin, gabapentin, glipizide, insulin, losartan, omeprazole, salsalate | Topical metronidazole | GI ROS neg |
| 9 | 50.5 | M | Black | 85.3 | 180.3 | 26.28 | 95 | 6.1 | No | 155 | 187 | 80 | 27.33 | Polysubstance abuse, BPH | Atorvastatin, tamsulosin | None | GI ROS neg |
| 10 | 61.5 | M | Black | 117.0 | 182.9 | 35.06 | 89 | 5.8 | No | 213 | 103 | 148 | 25.00 | HTN, DJD, HLD | Amlodipine, HCTZ, omeprazole, pravastatin | None | GI ROS neg |
| 11 | 47.5 | M | Black | 156.5 | 182.9 | 46.89 | 113 | 7.0 | Yes | 139 | 93 | 83 | 7.44 | OSA, DM, HTN, alcoholic fatty liver, HLD | Diclofenac, insulin, lisinopril, metformin | None | Constipation |
| 12 | 61.5 | M | White | 95.3 | 188.0 | 27.02 | 174 | 9.1 | Yes | 137 | 163 | 64 | 6.75 | DM, COPD, GERD, CAD, lung CA | Atenolol, gabapentin, hydralazine, HCTZ, insulin, lovastatin, metformin, omeprazole, tamsulosin | None | ND |
| 13 | 57.5 | M | White | 93.4 | 182.9 | 28.00 | 97 | 6.1 | No | 234 | 197 | 166 | 29.20 | Bronchitis | Prednisone | Moxifloxacin | GI ROS neg |
| 14 | 51.0 | M | Black | 93.9 | 170.2 | 32.49 | 95 | 6.0 | No | 180 | 65 | 118 | 28.00 | HTN, CVA | Docusate, HCTZ, potassium, terazosin, ASA, hydralazine, metoprolol, rosuvastatin | None | ND |
| 15 | 65.0 | M | Black | 119.3 | 172.7 | 40.07 | 80 | 6.0 | No | 213 | 307 | 106 | 19.25 | BPH, gout, HTN, HLD | Lisinopril, nifedipine | None | Hematochezia |
| 16 | 52.0 | M | Black | 137.4 | 188.0 | 38.98 | 189 | 10.0 | Yes | 169 | 346 | 69 | 4.75 | DM, OSA, HTN, GERD, gout, HLD | Allopurinol, ASA, atenolol, colchicine, gemfibrozil, HCTZ, Vicodin, insulin, synthroid, losartan, omeprazole, verapamil | Azithromycin | ND |
| 17 | 31.0 | M | Black | 68.5 | 180.3 | 21.10 | 114 | 5.3 | No | 195 | 164 | 121 | 21.67 | Heart transplant, CKD, anemia, GERD, hypothyroid, CHF | Diltiazem, magnesium, cellcept, pantoprazole, pravastatin, ranitidine, tacrolimus, valganciclovir | None | GI ROS neg |
| 18 | 51.0 | M | White | 78.5 | 177.8 | 24.87 | 106 | 5.9 | No | 202 | 121 | 141 | 26.50 | HLD, HTN, hypothyroid | HCTZ, lisinopril, synthroid, naproxen, omeprazole | None | ND |
| 19 | 56.0 | M | Black | 89.4 | 190.5 | 24.67 | 109 | 5.8 | No | 222 | 81 | 161 | 27.25 | Polysubstance abuse | Amoxicillin, Vicodin, Zofran | Amoxicillin | Diarrhea |
| 20 | 60.0 | M | Black | 123.4 | 180.3 | 38.20 | 70 | 8.2 | Yes | 150 | 66 | 92 | 18.50 | HTN, DM | Chlorthalidone, glipizide, lisinopril, metformin | None | GI ROS neg |
| 21 | 57.0 | M | White | 117.9 | 170.2 | 40.81 | 108 | 6.6 | Yes | 155 | 166 | 76 | 13.86 | Fatty liver, DM, HTN, HLD | Atenolol, chlorthalidone, atorvastatin, lisinopril, metformin | None | GI ROS neg |
| 22 | 54.0 | M | Hispanic | 104.3 | 182.9 | 31.26 | 314 | 7.4 | Yes | 196 | 155 | 128 | 5.17 | DM, HTN | B12, diclofenac, glipizide, insulin, losartan, metformin, pravastatin | None | ND |
| 23 | 60.0 | M | Black | 116.1 | 182.9 | 34.79 | 135 | 6.4 | Yes | 127 | 60 | 81 | 15.00 | DM, HTN | Hydrocodone, atorvastatin, lisinopril, metformin, aspirin | None | GI ROS neg |
| 24 | 72.0 | M | White | 102.1 | 182.9 | 30.58 | 102 | ND | No | 147 | 168 | 59 | 32.57 | HTN, hepatitis B | Lisinopril, metoprolol, hydrochlorothiazide, aspirin | None | GI ROS neg |
| 25 | 72.0 | M | Black | 93.0 | 182.9 | 27.96 | 107 | 6.2 | No | 161 | 115 | 97 | 33.00 | HTN | Omeprazole, hydrocholthiazide | None | GI ROS neg |
| 26 | 57.0 | M | White | 78.5 | 174.0 | 25.60 | 101 | 5.8 | No | 184 | 75 | 115 | 28.00 | HTN, seizures | Propranolol, pantoprazole, trazodone | None | GI ROS neg |
| 27 | 70.0 | M | Black | 104.8 | 188.0 | 29.72 | 119 | 5.1 | No | 155 | 65 | 86 | 31.60 | CKD, gout, hyperlipidemia, HTN, hypothyroidism | Ibuprofen, lubiprostone, amlodipine, allopurinol, atorvastatin, levothyroxine, losartan, colchicine | None | Constipation |
| 28 | 46.0 | M | White | 110.7 | 190.5 | 30.56 | 95 | 5.7 | No | 188 | 48 | 114 | 37.50 | GERD, HTN | Tramadol, ibuprofen, omeprazole, hydrochlorothiazide | None | ND |
| 29 | 55.0 | M | White | 118.9 | 190.5 | 33.47 | 108 | 7.5 | Yes | 415 | 351 | 277 | 13.00 | DM, HTN, CKD, hyperlipidemia, psoriasis | Amlodipine, glipizide, hydrochlorothiazide, pravastatin | None | GI ROS neg |
| 30 | 36.0 | M | Black | 79.4 | 188.0 | 21.92 | 88 | ND | No | ND | ND | ND | 28.75 | None | Codeine, ibuprofen, cholecalciferol | None | Diarrhea/some blood in stool |
| 31 | 63.0 | M | Black | 99.2 | 188.0 | 28.13 | 98 | ND | No | 172 | 49 | 95 | 36.00 | Deep vein thrombosis, hepatitis C | Multivitamin, thiamine | None | GI ROS neg |
| 32 | 74.0 | M | White | 105.2 | 182.9 | 32.43 | 232 | 9.3 | Yes | 186 | 239 | 104 | 4.33 | CAD, DM, CKD, HTN, GERD | Calcitriol, docusate, cyanocobalamin, aspirin, metoprolol, carbidopa, amlodipine, furosemide, insulin, divalproex | None | GI ROS neg |
| 33 | 70.0 | M | White | 90.7 | 177.8 | 28.76 | 106 | 6 | No | 215 | 179 | 140 | 25.20 | DJD, CAD, hyperlipidemia | None | None | None |
| 34 | 54.0 | M | Black | 108.4 | 182.9 | 32.48 | 113 | 5.8 | No | 200 | 276 | 105 | 32.25 | Hypertension, hyperlipidemia, gout | Losartan, allopurinol, omeprazole, trazadone, niacin, carvedilol | None | None |
| 35 | 60.0 | F | Black | 90.3 | 172.7 | 30.32 | 90 | 5.7 | No | 189 | 89 | 106 | 40.00 | Goiter, osteoarthritis, migraine headaches, chronic back pain | Cholecalciferol, psyllium, naproxen | None | Constipation |
| 36 | 70.0 | M | Black | 99.8 | 172.7 | 33.52 | 272 | 7.7 | Yes | 223 | 73 | 165 | 5.00 | HTN, hyperlipidemia, GERD, schizophrenia, BPH | Insulin, potassium, tamsulosin, finasteride, atorvastatin, carvedilol, metformin | None | None |
| 37 | 59.0 | M | Black | 92.1 | 177.8 | 29.19 | 99 | ND | No | 171 | 93 | 112 | 31.67 | HTN, spinal stenosis, hyperlipidemia | Methocarbamol, naproxen, hydrochlorothiazide, gabapentin, loratidine | None | None |
| 38 | 32.0 | M | Black | 81.6 | 180.3 | 25.16 | 94 | 5.3 | No | 187 | 286 | 94 | 34.33 | Posttraumatic stress disorder, depression | Omeprazole, bupropion, ferrous sulfate, folic acid, hydroxyzine, melatonin, prazosin | None | None |
| 39 | 59.0 | M | White | 83.9 | 172.7 | 28.19 | 77 | 5.8 | No | 157 | 116 | 95 | 50.00 | Posttraumatic stress disorder, depression, chronic hepatitis C, GERD | Hydrocodone, gabapentin, tamsulosin, ibuprofen, prazosin, ledipasvir/sofobuvir, paroxetine, psyllium, creon, methocarbamol, verapamil | None | Diarrhea |
| 40 | 49.0 | M | Black | 91.2 | 175.3 | 29.74 | 96 | 5.4 | No | 257 | 179 | 175 | 32.67 | BPH, migraines, HTN, asthma, hyperlipidemia, GERD | Mirtazapine, quetiapine, atorvastatin, tamsulosin, topiramate, cyanocobalamin | None | None |
| 41 | 55.0 | M | Black | 99.8 | 167.6 | 35.58 | 95 | 5.9 | No | 224 | 68 | 158 | 35.50 | HTN, hyperlipidemia, sleep apnea, depression | Losartan, amlodipine, atorvastatin | None | None |
| 42 | 60.0 | M | White | 97.1 | 185.4 | 28.39 | 133 | 6.8 | Yes | 256 | 976 | 71 | 7.67 | DM, HTN, GERD, posttraumatic stress disorder, depression | Allopurinol, trazadone, citalopram, amlodipine, atenolol, indomethacin, lisinopril, lovastatin, omeprazole, sildenafil | None | Diarrhea |
ASA, aspirin; BPH, benign prostate hyperplasia; CA, cancer; CAD, coronary artery disease; CHF, congestive heart failure; CKD, chronic kidney disease; COPD, chronic obstructive pulmonary disease; CVA, cerebrovascular accident; DJD, degenerative joint disease; DM, diabetic mellitus; GERD, gastroesophageal reflux disease; GI ROS, gastrointestinal review of systems; HbA1C, hemoglobin A1C; HCTZ, hydrochlorothiazide; HLD, hyperlipidemia; HTN, hypertension; LDL, low-density lipoprotein; ND, not determined; OSA, osteoarthritis; PMH, past medical history; RA, rheumatoid arthritis.
Localization of Bacteria and Quantitation of Bacterial-Epithelial Distance by Fluorescent In Situ Hybridization/Confocal Microscopy
Mucus immunostaining was paired with fluorescent in situ hybridization, as previously described,14 to analyze bacteria localization at the surface of the intestinal mucosa. Briefly, colonic tissues (proximal colon, 2 cm from the cecum) containing fecal material were placed in methanol-Carnoy fixative solution (60% methanol, 30% chloroform, 10% glacial acetic acid) for a minimum of 3 hours at room temperature. Tissue were then washed in methanol 2 × 30 minutes, ethanol 2 × 15 minutes, ethanol/xylene (1:1) 15 minutes, and xylene 2 × 15 minutes, followed by embedding in paraffin with a vertical orientation. Sections of 5 μm were performed and dewaxed by preheating at 60°C for 10 minutes, followed by xylene 60°C for 10 minutes, xylene for 10 minutes, and 99.5% ethanol for 10 minutes. Hybridization step was performed at 50°C overnight with EUB338 probe (5’-GCTGCCTCCCGTAGGAGT-3’, with a 5' labeling using Alexa 647) diluted to a final concentration of 10 μg/mL in hybridization buffer (20 mM Tris–HCl, pH 7.4, 0.9 M NaCl, 0.1% sodium dodecyl sulfate, 20% formamide). After washing 10 minutes in wash buffer (20 mM Tris–HCl, pH 7.4, 0.9 M NaCl) and 3 × 10 minutes in phosphate-buffered saline (PBS), PAP pen (Sigma, St. Louis, MO) was used to mark around the section and block solution (5% fetal bovine serum in PBS) was added for 30 minutes at 4°C. Mucin-2 primary antibody (rabbit H-300, Santa Cruz Biotechnology, Dallas, TX) was diluted 1:1500 in block solution and applied overnight at 4°C. After washing 3 × 10 minutes in PBS, block solution containing antirabbit Alexa 488 secondary antibody diluted 1:1500, phalloidin-tetramethylrhodamine B isothiocyanate (Sigma) at 1 μg/mL and Hoechst 33258 (Sigma) at 10 μg/mL was applied to the section for 2 hours. After washing 3 × 10 minutes in PBS slides were mounted using Prolong antifade mounting media (Life Technologies, Carlsbad, CA). Observations of bacterial localization and quantitation of bacterial-epithelial distance were performed in a blinded manner by the first author (B.C.) via confocal microscopy. Instrument software was used to determine the distance between bacteria and epithelial cell monolayer. For each subject, 5 high-powered fields (HPF) were arbitrarily selected with the following inclusion criteria: (1) the presence of stained bacteria, (2) the presence of a clear and delimitated mucosal layer, and (3) the presence of an intact mucus layer. For each HPF, the distance between the 5 closest bacteria and the epithelium was determined. Thus, each bacterial-epithelial distance indicated by a point in the figures is, in fact, the average distance of 25 bacteria-epithelial distances.
Immunofluorescence Staining of CD19 and CD68 Cells and Quantitation by Confocal Microscopy
Colonic tissues (proximal colon, 2 cm from the cecum) were placed in methanol-Carnoy fixative solution (60% methanol, 30% chloroform, 10% glacial acetic acid) for a minimum of 3 hours at room temperature. Tissue were then washed in methanol 2 × 30 minutes, ethanol 2 × 15 minutes, ethanol/xylene (1:1) 15 minutes, and xylene 2 × 15 minutes, followed by embedding in paraffin with a vertical orientation. Sections of 5 μm were performed and deparaffinized/rehydrated by xylene >100% ethanol >95% ethanol >70% ethanol >50% ethanol >distilled water washes (2 × 10 minutes). Antigen retrieval was performed by placing the section in boiling (microwaves) 10-mM sodium citrate buffer (pH 6.0) and subsequently maintained at a subboiling temperature for 10 minutes. Slides were let to cool down 30 minutes at room temperature and subsequently washed twice in distilled water. Tissue was permeabilized by 2 × 10 minutes treatment with 1% fetal bovine serum in PBS containing 0.4% Triton X-100 (Sigma) and blocked by incubating the tissue sections with 5% fetal bovine serum in PBS for 30 minutes at room temperature. Antihuman CD19 (clone HIB19, ebioscience, Santa Clara, CA) or antihuman CD68 (clone KP1, ebioscience) primary antibody were diluted 1:100 in PBS/5% fetal bovine serum and applied overnight at 4°C. After washing 3 × 10 minutes in PBS, PBS/5% fetal bovine serum solution containing antimouse Alexa 488 secondary antibody (Abcam, Cambridge, United Kingdom) diluted 1:500 and Hoechst 33258 (Sigma) at 10 μg/mL was applied to the section for 2 hours at 4°C. After washing 3 × 10 minutes in PBS, slides were mounted using Prolong antifade mounting media. Observations and quantitation of CD19+ and CD68+ cells were performed in a blinded manner by the first author (B.C.) via confocal microscopy. Instrument software was used to determine the number of positive cells per field. For each subject, 3 HPF were arbitrarily selected. For each HPF, the number of positive cells was determined.
Generation of Experimental Mice
All animals used in this study were wild-type animals, on a C57BL/6J genetic background. All mice were bred and housed at Georgia State University (Atlanta, GA) under institutionally approved protocols (Institutional Animal Care and Use Committee No. A14033). Mice were fed with the standard Purina rodent chow LabDiets 5001 (St. Louis, MO) used in this facility.
Streptozotocin-Induced Diabetes
Diabetes was induced in 10-week-old female C57/Bl6 mice by streptozotocin injection, as previously described.15 Briefly, streptozotocin (Sigma) was resuspended in 50 mM sodium citrate buffer and intraperitoneally injected for 5 consecutive days at a dose of 40 mg/kg. During those 5 days, animals were administered water containing 10% sucrose. Ten days after the last injection, animals were fasted for 5 hours and blood glucose and feces were collected. Following euthanasia, colons were collected and placed in Carnoy solution. Localization of bacteria and quantitation of bacterial-epithelial distance by fluorescent in situ hybridization/confocal microscopy was performed, as described previously.
Fasting Blood Glucose Measurement
Mice were placed in a clean cage and fasted for 5 hours. Blood glucose concentration was then determined using a Nova Max Plus Glucose Meter (Billerica, MA) and expressed in mg/dL.
Fecal Glucose Measurement
Feces were resuspended in distilled water at a final concentration of 100 mg/mL. Following heating at 55°C, glucose concentration was determined using the glucose assay kit (GO, Sigma) using a standard curve according to the manufacturer’s protocol.
Statistical Analysis
Linear regression and associated P values were generated using GraphPad Prism software version 6.01 (La Jolla, CA). Significance was determined using Student t test (2-sided). Differences were noted as significant P ≤ .05.
Results
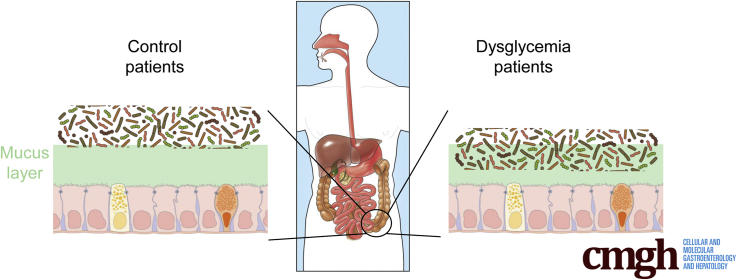
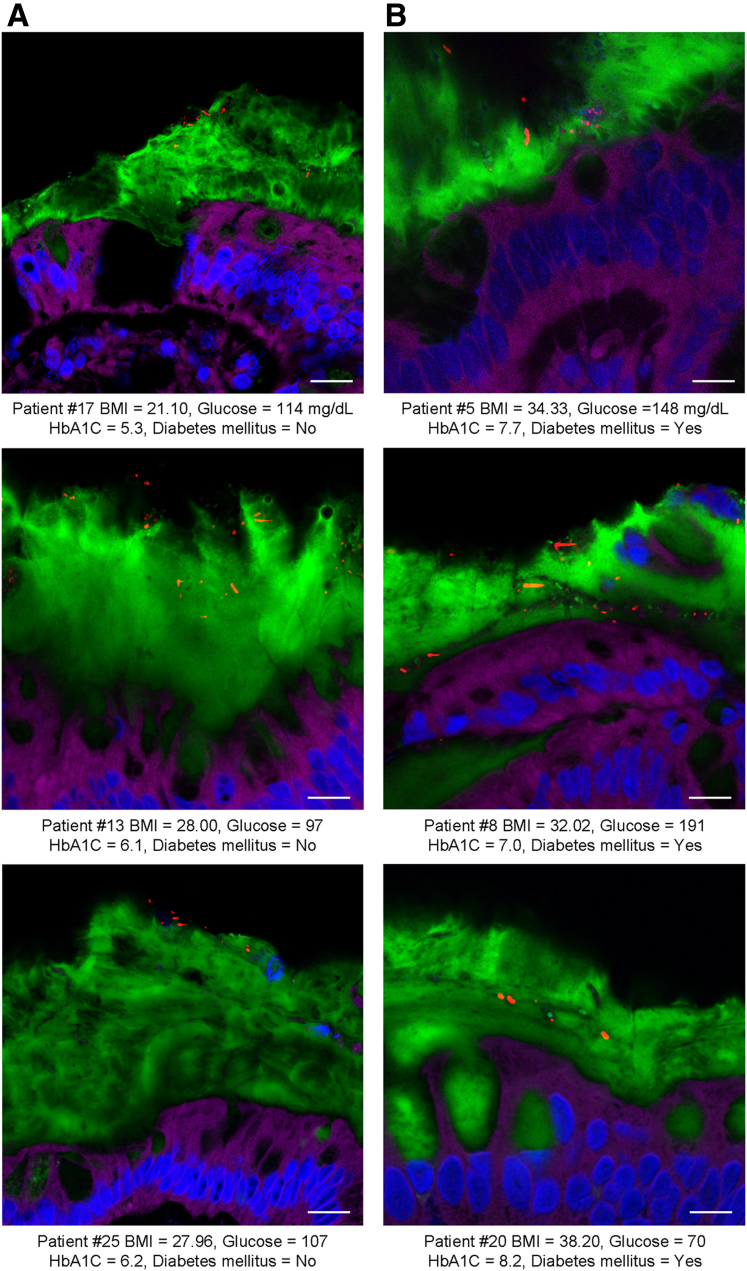
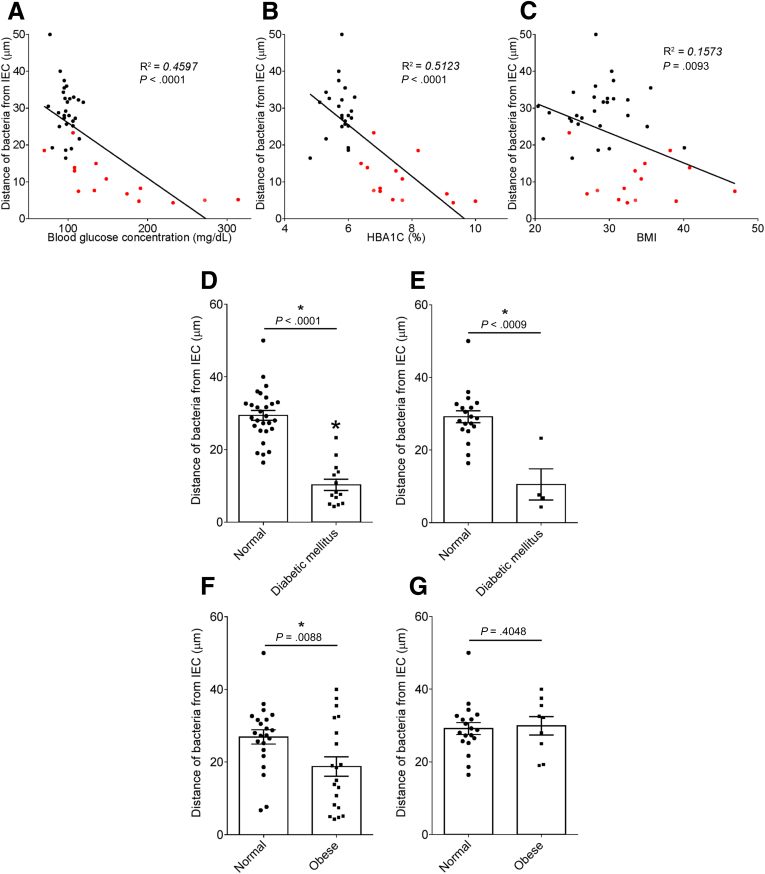
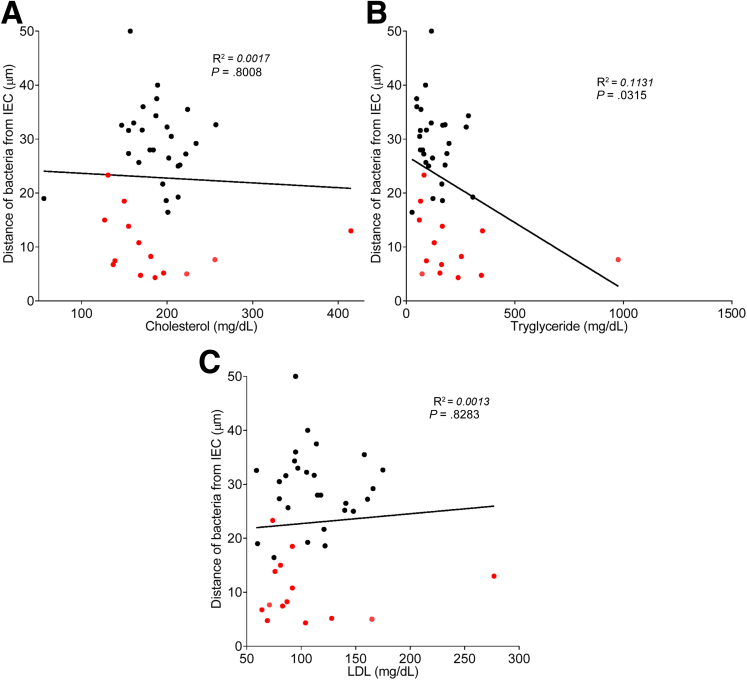
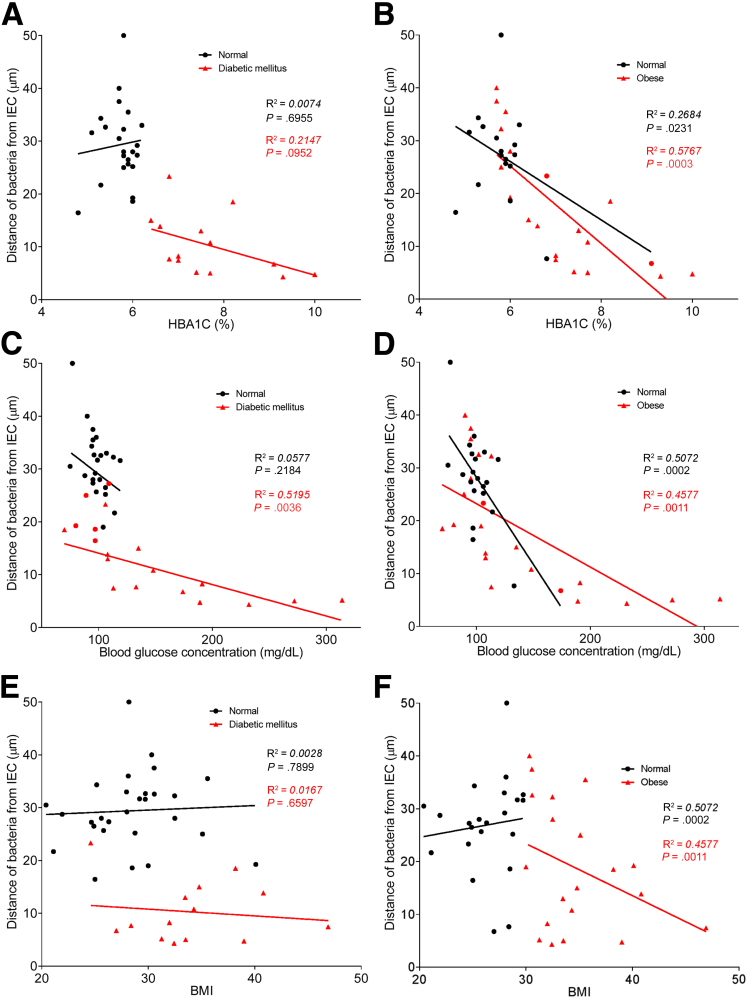
To explore the concept that a perturbed host-microbiota relationship might be a feature of metabolic syndrome, we analyzed microbiota-mucus-epithelial juxtaposition in a cohort of middle-aged Americans (58.1 ± 10.1 years old) undergoing routine cancer-screening colonoscopies (major diseases excluded, as outlined in Methods). As one would expect in such a cohort, most (86%) were overweight, many (45%) were obese, and a third (14 out of 42) had diabetes (Table 1). We obtained 2–3 biopsies per subject from the left colon of each subject and subsequently analyzed microbiota localization by confocal microscopy using nondehydrating fixation that preserves mucus structures.6 The standard precolonoscopy consumption of polyethylene glycol used to clean the colon, thus aiding the procedure's diagnostic capabilities, also removes most intestinal bacterial. The remaining bacteria were, in healthy (ie, nonobese, nondiabetic) subjects, almost exclusively observed in outer regions of the mucus layer, whereas in obese persons with diabetes, bacteria could be found in the dense inner mucus and in close proximity to the epithelium (Figure 1A and B). To quantitate this observation, we examined 5 HPF per subject and determined the average distance of the 5 closest bacteria to the epithelium in each field. Among all subjects, we observed an inverse correlation between such microbiota-epithelial distance and parameters that mark metabolic syndrome, namely body mass index (BMI), fasting blood glucose levels, and hemoglobin A1C concentrations (Figure 2A–C), wherein the latter parameters reflecting dysglycemia correlated more closely with encroachment than did BMI (R2 = 0.16, 0.46, and 0.51 for BMI, fasting blood glucose, and hemoglobin A1C vs microbiota-epithelial distance, respectively). A modest correlation was also observed between bacterial-epithelial distance and triglyceride levels, whereas there was not an apparent relationship between microbiota encroachment and cholesterol (Figure 3).
Figure 1.
Microbiota localization in human with or without metabolic syndrome. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution. Representative images of confocal microscopy analysis of microbiota localization; Muc2 (green), actin (purple), bacteria (red), and DNA (blue). (A) Patients without diabetes mellitus. (B) Patients with diabetes mellitus. Bar = 10 μm; n = 42. HbA1C, hemoglobin A1C.
Figure 2.
Metabolic syndrome correlates with microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus fasting blood glucose concentration. (B) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus hemoglobin A1C level. (C) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus body mass index. (D–G) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered field according to the diabetes mellitus (D, E) or the obese (F, G) status. In E, obese patients were removed from the analysis. In G, patients with diabetes mellitus were removed from the analysis. Linear regression line was plotted and R2 and P values were determined. Significance was determined by Student t test. *P < .05); n = 42; red dots represent subjects with diabetes. HbA1C, hemoglobin A1C; IEC, intestinal epithelial cells.
Figure 3.
Circulating lipid profile does not correlate with microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus cholesterol concentration. Linear regression line was plotted and R2 and P values were determined. (B) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus triglyceride concentration. Linear regression line was plotted and R2 and P values were determined. (C) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus low-density lipoprotein concentration. Linear regression line was plotted and R2 and P values were determined. n = 42; red dots represent subjects with diabetes. IEC, intestinal epithelial cells; LDL, low-density lipoprotein.
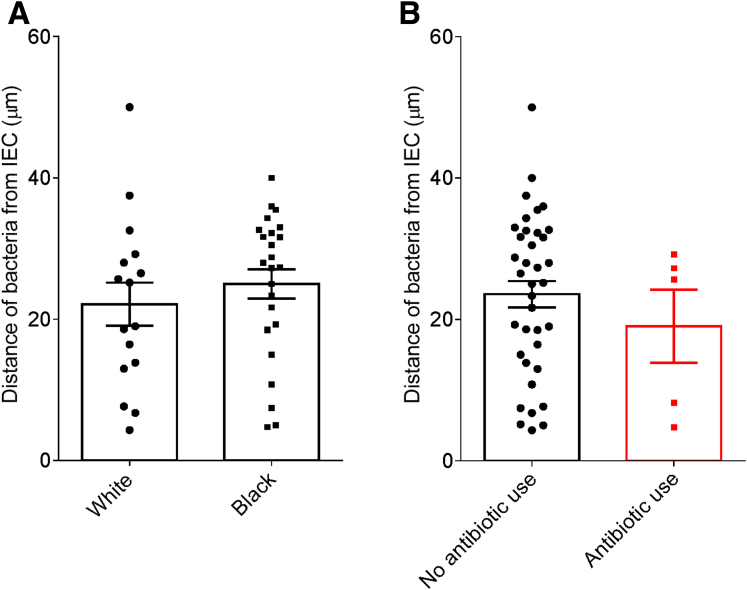
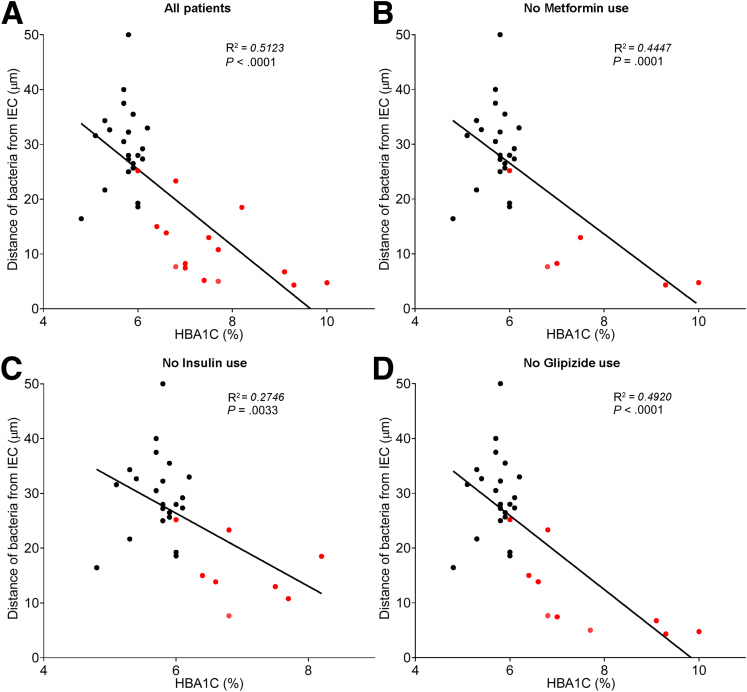
In accord with dysglycemia correlating with microbiota encroachment, stratifying subjects with and without diabetes indicated that microbiota-epithelial distance was reduced by almost 3-fold in patients with type 2 diabetes (Figure 2D). This pattern held true, and remained statistically significant, even if all obese subjects were removed from the analysis (Figure 2E), although only a few nonobese subjects had diabetes. Moreover, within the subjects with type 2 diabetes, disease severity, as reflected by fasting glucose and hemoglobin A1C, also correlated inversely with microbiota-epithelial distance (Figure 4). Stratifying the entire cohort as obese (BMI >30) or nonobese (BMI <30) also found a significant albeit lesser reduction in bacterial-epithelial distance in obese subjects (Figure 2F). However, this difference was completely eliminated by removal of subjects with diabetes (Figure 2G). Accordingly, BMI was not proportional to bacterial epithelial distance among subjects without diabetes (Figure 4). Ethnicity or antibiotic use did not significantly correlate with microbiota-epithelial distance (Figure 5). The inverse correlation observed between dysglycemia and bacterial-epithelial distance among subjects with diabetes was maintained if subjects prescribed metformin, insulin, or glipizide were excluded from the analysis (Figure 6). Together, these results indicate that microbiota encroachment is a feature of type 2 diabetes, wherein it correlates with disease severity, whereas the correlation between BMI and encroachment largely reflected the high rate of type 2 diabetes among middle-aged obese subjects.
Figure 4.
Dysglycemia correlates with microbiota encroachment in human with diabetes. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus hemoglobin A1C level (A, B), fasting blood glucose concentration (C, D), or body mass index (E, F). Linear regression line was plotted and R2 and P values were determined. n = 42; red dots represent subjects with diabetes (A, C, and E) or obese subjects (B, D, and F). HbA1C, hemoglobin A1C; IEC, intestinal epithelial cells.
Figure 5.
Ethnicity or antibiotic use do not correlate with microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered fields according to the ethnicity. (B) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered fields according to the antibiotic use status. IEC, intestinal epithelial cells.
Figure 6.
Antidiabetic drug use does not impact microbiota encroachment in human. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution, followed by confocal microscopy analysis of microbiota localization. (A) Distances of the closest bacteria to intestinal epithelial cells was measured in 5 high-powered fields per sample and plotted versus hemoglobin A1C level. Linear regression line was plotted and R2 and P values were determined. (B) Patients using metformin drug were removed from the analysis. (C) Patients using insulin drug were removed from the analysis. (D) Patients using glipizide drug were removed from the analysis. Linear regression line was plotted and R2 and P values were determined. n = 42; red dots represent subjects with diabetes. HbA1C, hemoglobin A1C; IEC, intestinal epithelial cells.
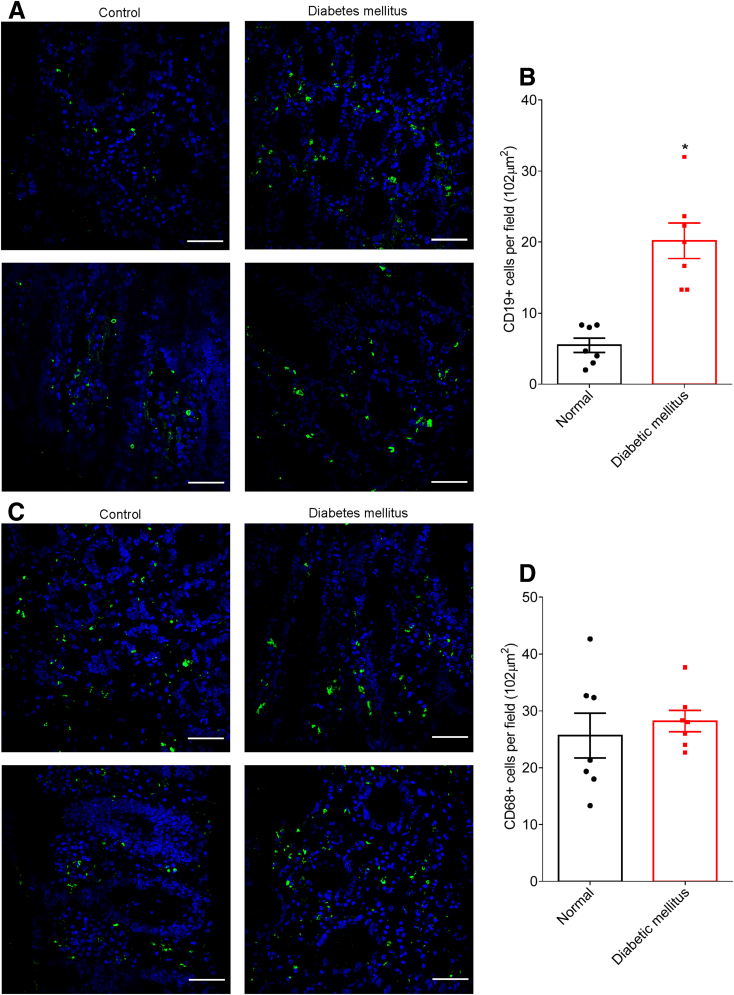
To generate hypotheses as to any possible roles microbiota encroachment might play influencing inflammation, insulin resistance, and/or other aspects of dysglycemia, we next probed colonic tissue sections for levels of various populations of mucosal immune cells in normal subjects and subjects with diabetes. A panel of markers for various types of immune cells was used, with many of those not providing clear cellular staining on our Carnoy-fixed tissues. Yet, we observed a marked increase in cells that stained positive for CD19 (Figure 7A and B), which is considered a fairly specific marker of B lymphocytes in human colon. In contrast, levels of cells expressing CD68, which is preferentially expressed by mucosal phagocytes, especially dendritic cells and macrophages, did not differ between subjects with and without diabetes (Figure 7C and D), thus suggesting that activation of mucosal B cells may result from microbiota encroachment.
Figure 7.
Metabolic syndrome correlates with increased CD19+cell population in the intestinal mucosa. Colonic biopsies were collected during colonoscopy procedure and placed in methanol-Carnoy fixative solution. Representative images of confocal microscopy analysis of CD19 (A) and CD68 (C) staining (green) and DNA (blue). Number of CD19+ (B) and CD68+ (D) cells per field (0.102 mm2). Significance was determined by Student t test. *P < .05). Bar = 50 μm; n = 7.
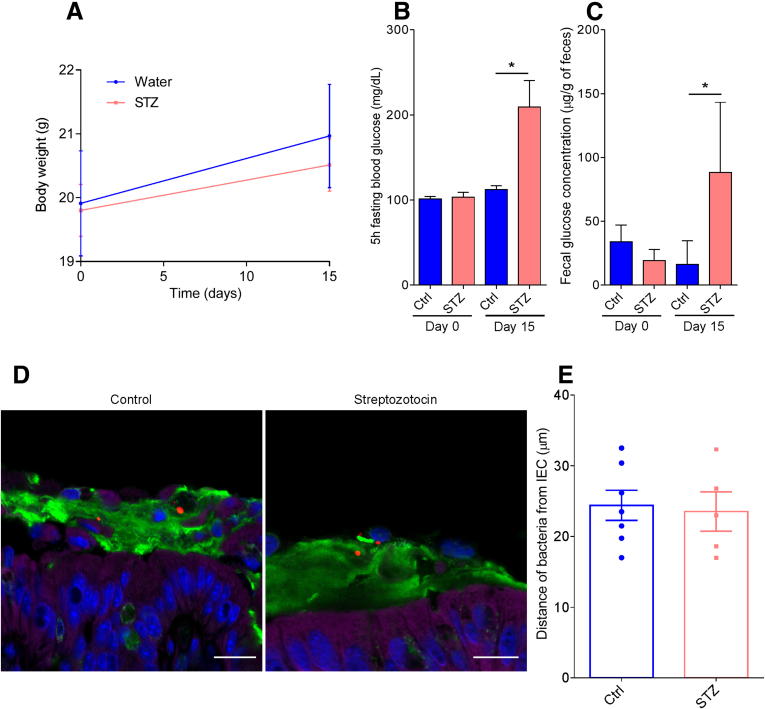
Lastly, considering that microbiota encroachment correlated with dysglycemia rather than obesity per se, we sought to consider the possibility that microbiota encroachment might have resulted from elevations in blood glucose. Specifically, we envisioned that elevated glucose levels might result in a transcolonic gradient that drove bacteria chemotaxis in the mucus layer. To investigate this possibility, we directly induced dysglycemia in mice by repeated injection of streptozotocin, which destroys insulin-producing beta cells.15 Streptozotocin administration resulted in marked obesity-independent elevations in blood glucose (Figure 8A and B) that correlated with elevations in fecal glucose levels (Figure 8C). However, streptozotocin treatment was not sufficient to induce microbiota encroachment (Figure 8D and E) in this short-term (2-week) type 1 diabetes model. This result indicates that, at least in the short term, an increase in fecal glucose concentration may not be sufficient to induce microbiota encroachment into the mucus and, rather, suggests that microbiota encroachment might be related to chronic low-grade inflammatory process that drives the insulin resistance that characterizes type 2 diabetes.
Figure 8.
Streptozotocin-induced type 1 diabetes and associated increased fecal glucose is not sufficient to induce microbiota encroachment in mice. Type 1 diabetes was induced in mice by streptozotocin injection for 5 consecutive days. (A) Body weight over time. (B) Five-hour fasting blood glucose concentration on Day 0 and Day 15. (C) Day 0 and Day 15 fecal glucose concentration. (D) Representative images of confocal microscopy analysis of microbiota localization; Muc2 (green), actin (purple), bacteria (red), and DNA (blue). (E) Distances of the closest bacteria to intestinal epithelial cells per condition over 5 high-powered fields according to the diabetes mellitus status. Significance was determined by Student t test (*P < .05). Bar = 50 μm; n = 5–9. IEC, intestinal epithelial cells; STZ, streptozotocin.
Discussion
The dramatic increase in incidence of metabolic syndrome, and its downstream consequences, compels better understanding of its pathophysiology. Numerous studies have, based on DNA sequencing, associated alterations in gut microbiota composition with various features of this disorder.16, 17, 18, 19, 20 However, how these changes in the species or genomic composition of the microbiota impacts how it interacts the host is far from clear. We subscribe to the central hypothesis that alterations in the microbiota are an underlying cause of low-grade inflammation that desensitizes metabolic signaling, including but not limited to insulin receptor signaling, that promotes hyperphagia and other aspects of metabolic syndrome.21, 22, 23 Our work in mice suggests that 1 means by which the altered microbiota might promote low-grade inflammation might be via attaining greater proximity to the cells and receptors, hence mediating proinflammatory gene expression on detection of bacteria and their metabolites. Specifically, we have previously observed bacterial infiltration into the mucus layer, herein referred to as microbiota encroachment, in mice developing metabolic syndrome as a result of loss of the flagellin receptor TLR5, feeding of synthetic dietary emulsifiers, or from feeding obesogenic diets.12, 13, 24 Herein, we report that microbiota encroachment is also a prominent feature of a human metabolic disease, namely type 2 diabetes, thus underscoring the usefulness of these models in investigating pathophysiology underlying human metabolic disease. Furthermore, we observed that, in human subjects, adiposity per se did not correlate with encroachment, thus providing an insight that had not been gleaned from our mouse models of metabolic syndrome.
The consistent correlation of microbiota encroachment with both adiposity and dysglycemia in our mouse models likely reflects that, in these particular models, adiposity and dysglycemia are very consistently correlated, whereas in humans these parameters are generally correlated but yet some individuals have high BMI but maintain good glycemic control. One possible explanation is that our models all involve manipulations that impact host-microbiota interactions in a manner that induces low-grade inflammation, which we hypothesize impairs insulin/leptin signaling in a manner that promotes adiposity and dysglycemia. In contrast, although we hypothesize that altered microbiota/low-grade inflammation may be 1 factor that could promote obesity and its associated disorders including insulin resistance, we presume that humans can become obese for other reasons not involving the microbiota. We anticipate future studies using other mouse models, perhaps involving out bred mice in which we might disentangle obesity, dysglycemia, and perhaps microbiota encroachment.
Another unanticipated observation made herein is that microbiota encroachment in patients with type 2 diabetes was associated with an increase in CD19+ cells, highly likely mucosal B cells. The role of this B-cell response in metabolic syndrome, and it’s interrelationship with microbiota encroachment, merits follow-up studies. We are currently designing such studies in mice, which we anticipate might produce hypotheses to be subsequently tested in humans. At present, we can imagine that a colonic B-cell response might be either detrimental or beneficial but would lean toward the latter, based simply on the philosophy that far more immune responses are beneficial rather than disease-exacerbating. Interestingly, in accord with this possibility, we note that a B-cell response was recently observed to strongly correlate with lack of pathology in patients with celiac disease who were challenged with wheat consumption,25 thus suggesting such B-cell responses may protect and/or restore mucosal homeostasis. We envision that defining the interrelationship between microbiota encroachment, B-cell responses, and metabolic disease may elucidate the pathophysiology of metabolic syndrome and perhaps eventuate in novel strategies to treat and/or prevent this condition.
Footnotes
Author contributions Benoit Chassaing and Andrew T. Gewirtz conceived the project, designed the experiments, interpreted the results, and wrote the manuscript. Shanthi Srinivasan and Shreya M. Raja collected biopsies and critically revised the manuscript. Benoit Chassaing performed experiments and analysis. James D. Lewis performed statistical analysis and helped interpret data.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants DK099071 and DK083890 (A.T.G.), National Institutes of Health grant DK080684 (S.S.), and VA-MERIT (S.S.). B.C. is a recipient of the Career Development Award from the Crohn’s and Colitis Foundation of America.
References
- 1.Hooper L.V., Macpherson A.J. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169. doi: 10.1038/nri2710. [DOI] [PubMed] [Google Scholar]
- 2.Chassaing B., Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 3.Xavier R.J., Podolsky D.K. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 4.Frank D.N., St Amand A.L., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gevers D., Kugathasan S., Denson L.A., Vazquez-Baeza Y., Van Treuren W., Ren B. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johansson M.E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G.C. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swidsinski A., Loening-Baucke V., Herber A. Mucosal flora in Crohn's disease and ulcerative colitis: an overview. J Physiol Pharmacol. 2009;60(Suppl 6):61–71. [PubMed] [Google Scholar]
- 8.Vijay-Kumar M., Aitken J.D., Carvalho F.A., Cullender T.C., Mwangi S., Srinivasan S., Knight R., Ley R.E., Gewirtz A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caricilli A.M., Picardi P.K., de Abreu L.L., Ueno M., Prada P.O., Ropelle E.R., Hirabara S.M., Castoldi Â., Vieira P., Camara N.O., Curi R., Carvalheira J.B., Saad M.J. Gut microbiota is a key modulator of insulin resistance in TLR 2 knockout mice. PLoS Biol. 2011;9:e1001212. doi: 10.1371/journal.pbio.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W.Z., Strowig T., Thaiss C.A., Kau A.L., Eisenbarth S.C., Jurczak M.J., Camporez J.P., Shulman G.I., Gordon J.I., Hoffman H.M., Flavell R.A. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 2012;482:179–185. doi: 10.1038/nature10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Etienne-Mesmin L., Vijay-Kumar M., Gewirtz A.T., Chassaing B. Hepatocyte toll-like receptor 5 promotes bacterial clearance and protects mice against high-fat diet–induced liver disease. Cell Mol Gastroenterol Hepatol. 2016;2:584–604. doi: 10.1016/j.jcmgh.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chassaing B., Ley R.E., Gewirtz A.T. Intestinal epithelial cell toll-like receptor 5 regulates the intestinal microbiota to prevent low-grade inflammation and metabolic syndrome in mice. Gastroenterology. 2014;147:1363–1377. doi: 10.1053/j.gastro.2014.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chassaing B., Koren O., Goodrich J.K., Poole A.C., Srinivasan S., Ley R.E., Gewirtz A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. 2015;519:92–96. doi: 10.1038/nature14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson M.E., Hansson G.C. Preservation of mucus in histological sections, immunostaining of mucins in fixed tissue, and localization of bacteria with FISH. Methods Mol Biol. 2012;842:229–235. doi: 10.1007/978-1-61779-513-8_13. [DOI] [PubMed] [Google Scholar]
- 15.Furman B.L. Streptozotocin-induced diabetic models in mice and rats. Curr Protoc Pharmacol. 2015;70:547. doi: 10.1002/0471141755.ph0547s70. [DOI] [PubMed] [Google Scholar]
- 16.Cavalcante-Silva L.H., Galvao J.G., da Silva J.S., de Sales-Neto J.M., Rodrigues-Mascarenhas S. Obesity-driven gut microbiota inflammatory pathways to metabolic syndrome. Front Physiol. 2015;6:341. doi: 10.3389/fphys.2015.00341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenhill C. Obesity: gut microbiota, host genetics and diet interact to affect the risk of developing obesity and the metabolic syndrome. Nat Rev Endocrinol. 2015;11:630. doi: 10.1038/nrendo.2015.152. [DOI] [PubMed] [Google Scholar]
- 18.Ussar S., Griffin N.W., Bezy O., Fujisaka S., Vienberg S., Softic S., Deng L., Bry L., Gordon J.I., Kahn C.R. Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome. Cell Metab. 2015;22:516–530. doi: 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Festi D., Schiumerini R., Eusebi L.H., Marasco G., Taddia M., Colecchia A. Gut microbiota and metabolic syndrome. World J Gastroenterol. 2014;20:16079–16094. doi: 10.3748/wjg.v20.i43.16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chassaing B., Gewirtz A.T. Gut microbiota, low-grade inflammation, and metabolic syndrome. Toxicol Pathol. 2014;42:49–53. doi: 10.1177/0192623313508481. [DOI] [PubMed] [Google Scholar]
- 21.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 22.Gregor M.F., Hotamisligil G.S. Inflammatory mechanisms in obesity. Annu Rev Immunol. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 23.Chassaing B., Gewirtz A.T. Has provoking microbiota aggression driven the obesity epidemic? Bioessays. 2016;38:122–128. doi: 10.1002/bies.201500116. [DOI] [PubMed] [Google Scholar]
- 24.Zou J., Chassaing B., Gewirtz A.T. Dietary fiber protect against high-fat diet-induced metabolic syndrome through MyD88-mediated IL-22 production that reduces microbiota encroachment. Presented at the DDW meeting. 2017 [Google Scholar]
- 25.Garber M.E., Saldanha A., Parker J.S., Jones W.D., Kaukinen K., Laurila K., Lähdeaho M.L., Khatri P., Khosla C., Adelman D.C., Mäki M. B cells correlate with the extent of gluten-induced intestinal injury in celiac disease. Cell Mol Gastroenterol Hepatol. 2017 doi: 10.1016/j.jcmgh.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]