Abstract
Tetracycline-resistant Helicobacter pylori strains have been increasingly reported worldwide. However, only a small number of tetracycline-resistant strains have been studied with regard to possible mechanisms of resistance and those studies have focused on mutations in the tetracycline binding sites of 16S rRNA-encoding genes. We here report studies of 41 tetracycline-resistant H. pylori strains (tetracycline MICs, 4 to 32 μg/ml) from North America (n = 12) and from East Asia (n = 29). DNA sequence analyses of 16S rRNA-encoding genes revealed that 22 (54%) of the resistant isolates carried one of five different single-nucleotide substitutions (CGA, GGA, TGA, AGC, or AGT) at the putative tetracycline binding site (AGA965-967). Single-nucleotide substitutions were associated with reduced ribosomal binding and with slightly increased tetracycline MICs (1 to 2 μg/ml). The 19 tetracycline-resistant isolates with no detectable mutations in the tetracycline binding site had normal tetracycline-ribosome binding. All tetracycline-resistant isolates, including those with and those without mutations in the tetracycline binding site, showed decreased accumulation of tetracycline. These results suggest that tetracycline resistance is multifactorial, involving alterations both in ribosomal binding and in membrane permeability.
Helicobacter pylori infection is etiologically associated with chronic gastritis, peptic ulcers, gastric adenocarcinoma, and primary gastric lymphoma (4, 10). The antibiotics commonly used to treat H. pylori infection include amoxicillin, clarithromycin, tetracycline, and metronidazole given as combination therapy with two or more antibiotics plus an antisecretory drug and/or bismuth. Over time, the success rates of eradication therapy have fallen as the prevalence of antibiotic resistant H. pylori has increased (11, 12).
Tetracycline is a major component of quadruple therapy for H. pylori infection, and resistant H. pylori strains are presently uncommon in the United States and Europe (1, 21). However, tetracycline resistance has been increasingly reported in Brazil (22), Korea (14), Japan (15), Lebanon (31), Estonia (20), and India (32). A remarkable 59% of H. pylori strains from China were reported as resistant to tetracycline (37).
Tetracycline is a protein synthesis inhibitor, active against gram-positive and -negative bacteria, chlamydiae, mycoplasmas, rickettsia, and some protozoan parasites (2). Tetracycline inhibits bacterial growth by disrupting codon-anticodon interactions at the ribosome, specifically, by binding to the subunit of 30S, preventing attachment of aminoacyl-tRNA to the acceptor site. Photoaffinity labeling and chemical footprinting studies have shown that ribosomal protein S7 and 16S rRNA bases G693, A892, U1052, C1054, G1300, and G1338 all contribute to tetracycline binding (3, 24, 26, 30).
In H. pylori studies, tetracycline resistance has been attributed to mutations in the 16S rRNA-encoding genes that affect the binding site of tetracycline (8, 35). For example, studies of an isolate from The Netherlands (8) and of two isolates from Australia (35) showed that high-level tetracycline resistance (MIC, 8 to 64 μg/ml) was associated with triple mutations (AGA965-967 to TTC) at the putative tetracycline binding site of the 16S rRNA-encoding genes. Six other tetracycline-resistant isolates with lower-level tetracycline resistance (MIC ≤ 4 μg/ml) also had single- or double-nucleotide substitutions at AGA965-967 of the 16S rRNA-encoding genes (5). Here, we report characterization of tetracycline-resistant clinical H. pylori isolates from North America (United States and Canada) and East Asia (Korea and Japan), only approximately half of which had alterations in the putative tetracycline binding sites of 16S rRNA-encoding genes.
MATERIALS AND METHODS
H. pylori and culture conditions.
Tetracycline-resistant H. pylori strains were identified from 413 isolates obtained from the antrum and corpus of 227 patients from North America (United States, n = 174; Canada, n = 53). In addition, 29 tetracycline-resistant H. pylori strains from East Asia (Korea, n = 22; Japan, n = 7) that have been previously reported (15) were also included in this study. All stock cultures were maintained at −80°C in brain heart infusion (BHI) broth supplemented with 20% glycerol (Sigma Co. St. Louis, Mo.). The frozen cultures were cultured on nonselective BHI agar plates when needed.
Determination of MIC.
Drug MIC measurements for the H. pylori strains were performed by the serial twofold agar dilution method as described previously (16). Briefly, agar dilution plates were prepared using Mueller-Hinton agar as the base medium. Aged sheep blood (2 weeks old) was added to the Mueller-Hinton base medium at a concentration of 5%. The ranges for the antibiotic dilutions were 0.015 to 256 μg for amoxicillin, metronidazole, tetracycline (Sigma), and clarithromycin (Abbott Laboratories, Abbott Park, Ill.). Fresh H. pylori isolates (2- to 3-day cultures) were prepared in sterile saline and adjusted to a no. 2 McFarland standard. Using a Steers-type replicating device, 1 to 5 μl of the adjusted inoculum was delivered to the agar plates. All plates were incubated with CampyPak Plus (Becton Dickinson BBL, Cockeysville, Md.) at 37°C for 3 days. Metronidazole-resistant H. pylori ATCC 43504 was used as a quality control organism. Any test in which the drug MIC for the quality control organism was outside the approved range (64 to 256 μg of metronidazole/ml) was discarded, and the test was repeated. The resistance breakpoint used for tetracycline was a MIC of >2 μg/ml, as we proposed previously (14).
Genomic DNA extraction and random amplified polymorphism DNA (RAPD) analysis.
H. pylori cells grown on BHI agar plates (2 plates) for 2 to 3 days were washed with TE buffer (10 mM Tris HCl [pH 8.0], 1 mM EDTA) and centrifuged at 15,000 rpm (12,000 × g) for 2 min. The cells were then suspended in lysis buffer (10 mM NaCl, 20 mM Tris HCl [pH 8.0], 25 mM EDTA, 0.5% sodium dodecyl sulfate, 100 μg of proteinase K/ml) and incubated at 50°C for 3 h. Cell lysates were extracted with phenol and chloroform (1:1). Genomic DNA was precipitated by ethanol and quantified by spectrophotometer at an optical density of 260 nm. The quality of the genomic DNA was assessed by 0.7% agarose gel electrophoresis with 1× TAE buffer (40 mM Tris HCl [pH 8.0], 20 mM sodium acetate, 1 mM EDTA) with ethidium bromide staining.
Each 100-μl reaction mixture of RAPD-PCR contained 50 ng of template DNA, 20 pmol of random amplified primer pMV 17B (17), 10 mM Tris-Cl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, 3 mM MgCl2, 0.2 mM concentrations each of four deoxynucleoside triphosphates (Pharmacia LKB Biotechnology, Piscataway, N.J.), and 1 U of Taq polymerase (Promega, Madison, Wis.). PCR amplifications were performed with an automated thermocycler (MJ Research, New York, N.Y.) with an initial denaturation step (95°C for 5 min) followed by 40 cycles of denaturation (94°C for 30 s), annealing (37°C for 1 min), and extension (72°C for 1 min), with one final extension (72°C for 10 min). Aliquots (5 μl) of the PCR-amplified product were resolved in 1% agarose gels containing 0.5× TAE and stained with ethidium bromide. The RAPD-PCR amplifications were performed twice to confirm reproducibility of the genomic DNA fingerprinting for each H. pylori isolate. The DNA band patterns were visualized by ethidium bromide staining under a short-wavelength UV light source and photographed with Polaroid 667 film.
DNA sequence analysis of 16S rRNA-encoding genes.
A portion of a 16S rRNA-encoding gene containing three putative tetracycline binding sites (8, 36) was amplified from the tetracycline-resistant H. pylori isolate. A 320-bp PCR fragment containing the putative tetracycline binding sites A (GGTGC1052-1056) and C (AGA965-967) was amplified by primer pair 16STCR-1 (5′-GGTAGTCCACGCCCTAAACGA-3′) and 16STCR-2 (5′-GGGTTGCGCTCGTTGCGGGA-3′). A 464-bp PCR fragment containing the putative tetracycline binding site B (GACGT1196-1200) was amplified by primer pair 16STCR-3 (5′-TCGTGTCGTGAGATGTTGGG-3′) and 16STCR-4 (5′-AGGAGGTGATCCAACCGCAG-3′). In addition, rrnA-specific primers (F1 and F2) and rrnB-specific primers (F3 and F4) used in combination with primer R-1 were also employed to determine data for both 16S rRNA-encoding genes as previously described (8). PCR conditions were identical to those previously described (16). Each PCR fragment was purified using a PCR product purification kit (QIAGEN Co., Hilden, Germany) before being used for DNA sequence determinations by use of a system from SeqWright (Houston, Tex.) or Macrogen (Seoul, Korea).
Natural transformation.
Natural transformation was performed using the methods described by Haas et al. (13). Briefly, recipient H. pylori cells (107 per ml) were suspended in 10% horse serum BHI broth in a 50-ml Falcon tube and incubated for 6 h. The cells (200 μl) were dispensed into a 96-well microtiter plate with the addition of 1 μg of genomic DNA and incubated for 18 h under microaerobic conditions. Then, the cells were spread on selective BHI agar plates containing 0.5 to 2 μg of tetracycline per ml and incubated for up to 5 days.
Interruption of a tetA (P) gene in tetracycline-resistant H. pylori.
To interrupt the tetA (P) gene (HP 1165) (33) from H. pylori ATCC 700392, a PCR fragment for the tetA (P) gene (1,161 bp of full length) was amplified by using PCR with a primer pair TetAB [5′-ATGTTAAGGAAAAACATTTTA-3′] and TetBE [5′-TCACTCATCAAACGGCTTA-3′] that covered from 1229436 to 1230597 of the H. pylori 26695 complete genome (33) and PCR conditions described previously (17). The PCR-amplified fragment was inserted into pBluescript SK+ (Stratagene, La Jolla, Calif.), and the fragment was cleaved by a restriction enzyme, SphI, which divided the tetA (P) gene into 320- and 841-bp lengths. A chloramphenicol resistance gene cassette (cat) (16) was inserted into the SphI site after blunt ends were made using a Klenow enzyme. The resulting recombinant plasmid containing tetA (P)::cat was used to transform tetracycline-resistant clinical H. pylori isolates 5255A and KH179A and the transformed resistant H. pylori cells as previously described (16).
Tetracycline-ribosome binding assay.
Ribosomes from tetracycline-resistant clinical H. pylori isolates were purified using differential centrifugation as described by Doucet-Populaire et al. (6) and Goldman et al. (9) with modifications. Briefly, H. pylori cells grown on BHI agar plates (three plates) were harvested in phosphate buffer containing 137 mM NaCl and 2.7 mM KCl. The harvested cells were washed twice by centrifugation at 5,000 × g for 5 min at 4°C. The cells were resuspended in 4 ml of buffer A (10 mM Tris-HCl containing 4 mM MgCl2, 100 mM KCl, and 10 mM NH4Cl, pH 7.2) and passed through a French pressure cell (Aminco, Urbana, Ill.) at 600 lb/in2. The cell debris was discarded by centrifugation at 30,000 × g for 30 min at 4°C. The supernatant was centrifuged at 100,000 × g for 90 min at 4°C to pellet the ribosomes. Ribosomes were resuspended in buffer A and stored at −80°C until needed.
The tetracycline-ribosome binding assay commenced by the addition of 75 pmol of [7-3H]tetracycline to different amounts of ribosomes (at an optical density at 260 nm of 1, 2, or 3) in 0.5 ml of buffer A as a reaction mixture. After 30 min, the binding reaction was stopped by diluting the reaction mixture with 3 ml of cold buffer B (10 mM Tris hydrochloride containing 5 mM MgCl2 and 150 mM KCl). The ribosomes were collected on a 0.45-μm-pore-size nitrocellulose filter (Millipore). After three washes with buffer B (3 ml), the filters were transferred to scintillation vials containing scintillation fluid (CytoScint; Fisher Biotech) for radioactivity determination in a Beckman LS 6500 scintillation counter (Beckman Instruments, Palo Alto, Calif.).
Antibiotic accumulation assay.
Antibiotic accumulation in H. pylori cells was performed as previously described (18). Briefly, H. pylori cells were grown on BHI agar plates without any antibiotic for 2 to 3 days and harvested in 10 ml of assay buffer containing 50 mM KPO4 and 1 mM MgSO4 (pH 6.6). The cells (5 × 109 per ml) were centrifuged and resuspended in 10 ml of the same assay buffer. Antibiotic accumulation assays commenced by the addition of [7-3H]tetracycline (Dupont/NEN Research Product, Boston, Mass.) (0.6 Ci mmol−; 22.2 GBq mmol−). Aliquots (1 ml) were taken every 10 min. After 20 min, each cell suspension was divided in half and 100 μM of CCCP (carbonyl cyanide m-chlorophenylhydrazone) was added to only one half to de-energize the cells but not the other half. Then, 1 ml aliquots were taken every 10 min from each half. Each aliquot was then immediately centrifuged and washed three times in phosphate-buffered saline. The resulting pellets were then diluted in scintillation fluid (CytoScint; Fisher Biotech) and analyzed for radioactivity in a Beckman LS 6500 scintillation counter (Beckman Instruments).
RESULTS
Tetracycline-resistant H. pylori.
Twelve tetracycline-resistant H. pylori isolates (tetracycline MICs, 4 to 32 μg/ml) were taken from 8 (4%) of the 227 North American patients. MICs of amoxicillin, clarithromycin, metronidazole, and tetracycline were measured for the 12 tetracycline-resistant isolates and compared to the susceptible reference H. pylori ATCC 700392 results (Table 1). In addition, we studied 29 isolates from East Asia that were reported previously (15). Tetracycline resistance levels among these isolates resulted in tetracycline MICs that ranged from 4 to 16 μg/ml (15).
TABLE 1.
MICs of tetracycline-resistant H. pylori
| H. pylori isolate | MIC (μg/ml)a
|
|||
|---|---|---|---|---|
| Amoxicillin | Clarithromycin | Metronidazole | Tetracycline | |
| ATCC 700392 | 0.125 | 0.031 | 2 | 0.25 |
| (0203A)b | 0.25 | 0.062 | 64 | 8 |
| (1208A and -C) | 0.25 | 0.062 | 8 | 4 |
| 2901A and -C | 0.25 | 16 | 128 | 8 |
| 4442A | 0.5 | 0.062 | 16 | 8 |
| (4516C) | 0.25 | 0.061 | 128 | 4 |
| 4923A and -C | 0.25 | 0.061 | 8 | 4 |
| 5220A and -C | 0.25 | 0.061 | 8 | 8 |
| 5255A | 1 | 0.062 | 64 | 32 |
| Transformed H. pylori colonies 700392 (5255A)c | 0.25-0.5 | 0.062 | 4-8 | 4-8 |
MIC measurements were repeated three times with identical results.
Data in parentheses represent H. pylori isolates from Canada. A, H. pylori isolate from antrum; C, H. pylori isolate from corpus.
700392 (5255A) denotes colonies of H. pylori ATCC 700392 transformed using genomic DNA from clinical H. pylori 5255A. Transformed colonies (5 colonies) were used for three repeated MIC measurements with identical results.
Genotype analysis.
Tetracycline-resistant isolates from the eight North American patients were used for RAPD-PCR fingerprinting genotype analyses. Each isolate had a unique fingerprint. In contrast, fingerprints of paired antrum-corpus isolates were identical except for those for one patient (patient 2901), who was apparently infected by two different tetracycline-resistant H. pylori strains.
DNA sequence analysis of 16S rRNA-encoding genes from tetracycline-resistant H. pylori cells.
Three putative tetracycline binding sites of 16S rRNA-encoding genes (16S rrnAB) were examined by DNA sequence determination of the 41 tetracycline-resistant clinical isolates. DNA sequence alterations for the North American isolates were found only in the putative tetracycline binding site C (AGA965-967) of 16S rrnAB and consisted of single-nucleotide substitutions (A965 to G or T; A967 to C or T) (Table 2). The nucleotide substitutions in paired antrum-corpus isolates (patients 1208, 4923, and 5220) were identical with the exception of the nucleotide substitutions in site C from isolates 2901A and 2901C, which were different (i.e., 2901A, A967 to T; 2901C, A965 to G). Two resistant isolates (4442A and 5255A) did not show nucleotide substitutions in any of the three putative tetracycline binding sites (Table 2).
TABLE 2.
Nucleotide sequence analysis of putative tetracycline binding sites of 16S rRNA-encoding genes from tetracycline-resistant H. pylori isolates
| Type of nucleotide substitution at site C (AGA965-967)a | Tetracycline resistant H. pylori isolate(s)b
|
|
|---|---|---|
| North American isolate(s)d | East Asian isolate(s)c | |
| gGA | (0203A), 2901C, (4516C), 4923A, 4923C | KH84B, KH87B, KH95B, KH164B, KH179A, KH185A, KH259A, KH330A |
| AGc | (1208A), (1208C) | KH69A, KH400A |
| AGt | 2901A | |
| cGA | KH100A | |
| tGA | 5220A, 5220C | KH430B |
| No nucleotide change | 4442A, 5255A | KH55A, KH161A, KH222A, KH292B, KH294A, KH299A, KH422B, KH439A, KH453A, KH461A, JH34A, JH86A, JH86B, JH98A, JH223A, JH244B, JH269B |
Altered nucleotides were shown as a lowercase.
A, H. pylori isolates from antrum; C or B, H. pylori isolates from corpus or body.
East Asian isolates data were from a previous report (15).
Data in parentheses represent H. pylori isolates from Canada.
DNA sequence alterations for the East Asia were also found only in putative tetracycline binding site C (AGA965-967) of 16S rrnAB and consisted of single-nucleotide substitutions (A965 to G, C, or T; A967 to C). However, 17 isolates had no detectable mutations in any of putative tetracycline binding sites of 16S rrnAB. Overall, five different types of single-nucleotide substitutions (AGA965-967 to GGA, AGC, AGT, CGA, and TGA) were identified from 22 tetracycline-resistant isolates, and no mutation in any of the putative tetracycline binding sites of 16S rrnAB was found in the other isolates (19 of 41; 46%). The most common alteration was a single-nucleotide substitution of AGA965-967 to GGA at putative tetracycline binding site C, which was present in both North American and East Asian isolates (Table 2).
Role of single-nucleotide substitutions of the 16S rRNA-encoding genes in tetracycline resistance.
Each of the five different single-nucleotide substitutions was assessed regarding its potential role in tetracycline resistance. PCR-amplified DNA fragments carrying the different nucleotide substitutions were used to transform susceptible H. pylori ATCC 700392. Transformants were selected on agar plates containing only 0.5 to 1 μg of tetracycline per ml. The same nucleotide substitutions were found in the tetracycline-resistant transformants. For example, nucleotide substitution of AGA965-967 to GGA in the PCR fragment was always found in the tetracycline-resistant transformants used for the same PCR fragment. These results confirm that the single-nucleotide substitution at site C appears to be involved in low-level resistance to tetracycline, as reported previously (7).
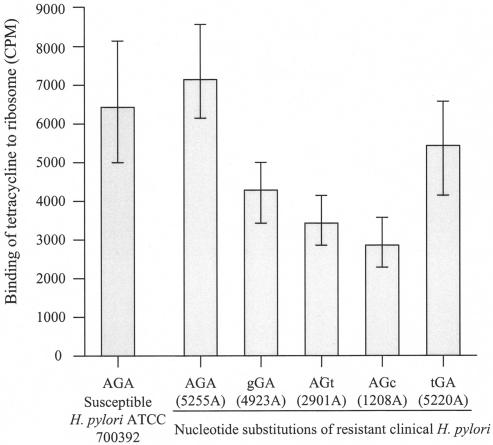
To further analyze the role of single-nucleotide substitutions, tetracycline-ribosome binding assays were performed using the clinical isolates carrying nucleotide substitutions in ribosome binding sites. Ribosomes were isolated from the tetracycline-resistant clinical isolates carrying the different types of nucleotide substitutions (isolates 1208A, 2901A, 4923A, and 5220A), a resistant isolate without a nucleotide substitution in 16S rRNA-encoding genes (isolate 5255A), and the susceptible H. pylori ATCC 700392 as a control. Tetracycline binding to the ribosomes with nucleotide substitutions was decreased 24 to 52% compared to the susceptible H. pylori results (Fig. 1). However, tetracycline binding to the ribosomes from the tetracycline-resistant isolate, 5255A, which had no nucleotide substitution in 16S rRNA-encoding genes, was similar to that of the susceptible H. pylori (Fig. 1).
FIG. 1.
Patterns of ribosome binding to tetracycline in tetracycline-susceptible and -resistant H. pylori. Tetracycline-resistant H. pylori 5255A has no nucleotide substitution, and other tetracycline-resistant H. pylori isolates have nucleotide substitutions in the 16S rRNA-encoding genes. H. pylori ATCC 700392, a tetracycline-susceptible strain, was used as a positive control. The results represent the means of three independent experiments; the vertical bars represent standard deviations.
Role of putative tetracycline resistance protein [tetA (P)] in tetracycline resistance.
The putative tetracycline resistance protein [tetA (P)] was identified from the complete H. pylori genome (strain 2669) (33). However, the tetA (P) gene may not be functional for production of tetracycline resistance, because strain 26695 is susceptible to tetracycline. To test whether the tetA (P) gene was involved in tetracycline resistance among the tetracycline-resistant clinical isolates, we interrupted the tetA (P) gene in resistant clinical isolates 5255A and KH179A and in the transformed resistant strains by use of a chloramphenicol resistance gene cassette as described in Materials and Methods. All of the tetA (P)-interrupted resistant strains remained resistant to tetracycline at levels identical to those seen with the parental resistant strains, suggesting that the tetA (P) gene was not be involved in the tetracycline resistance in these strains.
The role of genomic DNA extracted from tetracycline-resistant H. pylori in tetracycline resistance.
Genomic DNA extracted from the tetracycline-resistant clinical isolate without nucleotide substitutions among the three putative tetracycline binding sites of the 16S rRNA-encoding genes (isolate 5255A) was used for natural transformation of susceptible H. pylori ATCC 700392. Several transformed H. pylori colonies grew on BHI agar plates containing 2 μg of tetracycline per ml after 3 to 4 days of incubation whereas there was no growth of the negative control (i.e., without a DNA source) on the same agar plates. DNA sequence analyses of five transformed tetracycline-resistant H. pylori colonies confirmed the absence of nucleotide substitutions in the 16S rRNA-encoding genes. Tetracycline MICs for the five transformed resistant H. pylori colonies ranged from 4 to 8 μg/ml (Table 1).
The role of genomic DNA extracted from the tetracycline-resistant clinical isolate with a nucleotide substitution among the tetracycline binding sites of the 16S rRNA-encoding genes was also examined. These experiments used the transformants reported previously (15), which were derived from susceptible H. pylori strains ATCC 700392 and ATCC 43629 by use of genomic DNA from the clinical resistant H. pylori strains KH84B and KH179A. DNA sequences of the 16S rRNA-encoding genes were examined among three transformants from the two susceptible recipient H. pylori strains (a total 12 transformants: 3 from H. pylori ATCC 700392/KH84B, 3 from H. pylori ATCC 700392/KH179A, 3 from H. pylori ATCC 43629/KH84B, and 3 from H. pylori ATCC 43629/KH179A). All transformants, except ATCC 700392/KH84B and ATCC 700392/KH179A, contained the nucleotide substitution at putative tetracycline binding site C (AGA965-967 to GGA). The transformants ATCC 700392/KH84B and ATCC 700392/KH179A, however, did not have detectable mutations at any of the putative tetracycline binding sites. These results suggest that although a single-nucleotide substitution in the 16S rRNA-encoding genes is associated with low levels of tetracycline resistance, additional determinants transferred from the donor genomic DNA to the susceptible H. pylori strains are needed to produce tetracycline MICs in the range of 4 to 8 μg/ml.
Role of tetracycline accumulation in resistance.
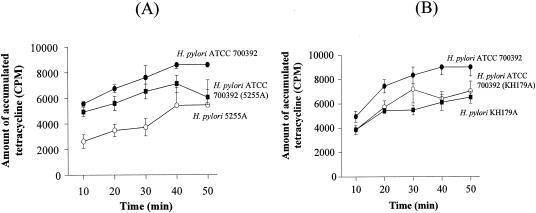
To further elucidate possible tetracycline resistance mechanisms, we examined tetracycline accumulation and active efflux of tetracycline as previously described (18). We used the clinical resistant H. pylori 5255A and the transformed resistant H. pylori ATCC 700392 (5255A) isolates and also included the susceptible parental H. pylori ATCC 700392 strain as a control. Tetracycline accumulations were decreased in both the transformed resistant H. pylori and the clinical resistant H. pylori 5255A strains. However, the degree of the reduction in the level of the resistant transformant was less than that of the resistant clinical isolate (20 to 80% reduction of the resistant clinical isolate) (Fig. 2A). This change may reflect the fact that MIC levels for the transformants (tetracycline MIC, 4 to 8 μg/ml) were less than that for the clinical isolate (tetracycline MIC, 32 μg/ml) (Table 1). Decreased accumulation of tetracycline was also observed with the clinical resistant H. pylori KH179A isolate and the transformed resistant H. pylori ATCC 700392 (KH179A) isolate reported previously (15) (Fig. 2B). We also examined whether the resistant isolates were associated with an energy-coupled active efflux mechanism by using CCCP as a de-energizing agent. Addition of CCCP at 20 min in the accumulation assays did not increase tetracycline accumulation in any of the strains, and identical results were repeatedly obtained (data not shown). Overall, these observations suggest that the decreased accumulation of tetracycline may be one of important tetracycline resistance mechanisms responsible for the development of high-level tetracycline resistance.
FIG. 2.
Tetracycline accumulation assays for susceptible H. pylori ATCC 700392, transformed tetracycline-resistant H. pylori ATCC 700392 (5255A) and H. pylori ATCC 700392 (KH179A), and tetracycline-resistant clinical H. pylori 5255A and H. pylori KH179A. The results represent the means of three independent experiments; the vertical bars represent standard deviations.
DISCUSSION
Tetracycline-resistant H. pylori strains have increasingly been reported worldwide (1, 14, 22, 23, 28, 37). In this study, 4% of a large group of patients from North America had tetracycline-resistant H. pylori strain infections. RAPD-PCR fingerprinting showed that the resistant isolates from different patients were all of different genotypes, suggesting that the resistance likely does not reflect clonal dissemination of a specific resistant determinant. The tetracycline MICs for the tetracycline-resistant isolates initially ranged from 4 to 32 μg/ml. However, extended growth or step-wise increases in tetracycline concentrations resulted in an increased MIC level up to 64 μg/ml, suggesting that high-level tetracycline resistance results from the cumulative effects of multiple factors.
DNA sequence analyses of putative tetracycline binding sites of 16S rRNA-encoding genes among tetracycline-resistant isolates showed that 54% of tetracycline-resistant isolates contained a single-nucleotide substitution at tetracycline binding site C. Although we confirmed that it was possible to make transformants by use of PCR fragments carrying the single-nucleotide substitutions (7, 8, 35), transformation of susceptible H. pylori strains with single-nucleotide substitutions was associated with lower-level tetracycline MICs than those seen with the parental resistant strain (e.g., MICs of 1 or 2 μg/ml versus ≥4 μg/ml). We also extend prior observations by confirming that single-nucleotide substitutions (A965 to G or T substitutions and A967 to C or T substitutions) were associated with a reduction in tetracycline binding to ribosomes.
Importantly, approximately half (46%) of tetracycline-resistant clinical isolates did not contain nucleotide substitutions at any putative tetracycline binding sites of 16S rRNA-encoding genes. These isolates also expressed higher levels of MICs than those associated with single-nucleotide substitutions in binding site C, suggesting that additional determinants were responsible for the higher levels of resistance. Our data showing the presence of additional resistant determinants in the genomic DNA include the following. (i) Many tetracycline-resistant isolates had no nucleotide substitutions in any of the three putative tetracycline binding sites of the 16S rRNA-encoding genes. (ii) A tetracycline-resistant isolate without a nucleotide substitution at any of the three putative tetracycline binding sites showed tetracycline-ribosome binding similar to that seen with the tetracycline-susceptible H. pylori isolate. (iii) Genomic DNA from the resistant isolate containing a single-nucleotide substitution in the 16S rRNA-encoding genes resulted in higher tetracycline MICs than that seen with the single-nucleotide substitutions. (iv) Genomic DNA from the resistant isolate without any nucleotide substitution at any of the three putative tetracycline binding sites could increase the tetracycline MIC by >2 μg/ml. (v) All transformed tetracycline-resistant H. pylori isolates with a single-nucleotide substitution exhibited lower MIC levels than those of clinical resistant isolates. (vi) Interruption of tetA (P) did not change the level of tetracycline resistance.
Several tetracycline resistance mechanisms have been reported in studies of bacteria (2). In most cases, tetracycline efflux pumps that are specific for the active transport of tetracycline are responsible for tetracycline resistance (19). A second mechanism of tetracycline resistance is mediated through ribosomal protection proteins that reduce the affinity of ribosomes for tetracycline or release the bound tetracycline from the ribosome (34). The third mechanism is an as-yet-unknown chemical modification of tetracycline by an oxidoreductase that requires NADP (2). The fourth mechanism of resistance is a mutation in the 16S rRNA-encoding genes that affects the binding site of tetracycline (7, 29, 35). Tetracycline resistance can be also acquired by decreased membrane permeability, and this is often coupled with additional resistance mechanisms (e.g., active drug efflux, enzymatic degradation of antibiotics, or target alterations) producing high levels of tetracycline resistance (18, 25, 27). We hypothesize that one tetracycline resistance mechanism in H. pylori is decreased membrane permeability and/or active efflux of tetracycline. This idea is based on the fact that the resistant isolates, including the resistant transformant, showed at least slightly increased MIC levels of the tested antibiotics. Our data clearly showed that the tetracycline-resistant clinical isolates had decreased membrane permeability but did not show clear evidence for energy-coupled active efflux when the de-energizing agent CCCP was used. However, these results do not completely exclude the possibility of the active efflux in the resistant strains, because we did not test other de-energizing agents such as arsenate or 2,4-dinitrophenol. Studies with the other de-energizing agents will be needed to confirm whether the tetracycline-resistant isolates are associated with energy-coupled active efflux.
The decreased membrane permeability associated with tetracycline resistance is similar to a result presented in our previous report, namely, that amoxicillin-resistant clinical H. pylori was also cross-resistant to tetracycline and that this effect was also mediated by decreased membrane permeability (18). However, total outer membrane protein profiles of the resistant clinical and transformed strains in this study (data not shown) were not similar to those seen with the previously investigated strains (18). It is not clear whether mechanisms of decreased accumulation reported for the previous study (18) and the present study are identical or whether the outer membrane protein profiles and MICs are different between the tetracycline-resistant isolates examined in the previous study and those examined in this study. In summary, our results suggest that tetracycline resistance among clinical H. pylori isolates may be associated with nucleotide substitutions in the tetracycline binding site C of the 16S rRNA-encoding genes as well as with decreased accumulation of tetracycline.
Acknowledgments
This material is based upon work supported in part by the Office of Research and Development Medical Research Service Department of Veterans Affairs and by Public Health Service grant DK56338, which funds the Texas Gulf Coast Digestive Diseases Center. D.H.K. is also supported in part by an award from the Caroline Weiss Law Fund.
REFERENCES
- 1.Boyanova, L., A. Mentis, M. Gubina, E. Rozynek, G. Gosciniak, S. Kalenic, V. Goral, L. Kupcinskas, B. Kantarceken, A. Aydin, A. Archimandritis, D. Dzierzanowska, A. Vcev, K. Ivanova, M. Marina, I. Mitov, P. Petrov, A. Ozden, and M. Popova. 2000. The status of antimicrobial resistance of Helicobacter pylori in Eastern Europe. Clin. Microbiol. Infect. 8:388-396. [DOI] [PubMed] [Google Scholar]
- 2.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chopra, I., P. M. Hawkey, and M. Hinton. 1992. Tetracyclines, molecular and clinical aspects. J. Antimicrob. Chemother. 29:245-277. [DOI] [PubMed] [Google Scholar]
- 4.Crump, M., M. Gospodarowicz, and F. A. Shepherd. 1999. Lymphoma of the gastrointestinal tract. Semin. Oncol. 26:324-337. [PubMed] [Google Scholar]
- 5.Dailidiene, D., M. T. Bertoli, J. Miciuleviciene, A. K. Mukhopadhyay, G. Dailide, M. A. Pascasio, L. Kupcinskas, and D. E. Berg. 2002. Emergence of tetracycline resistance in Helicobacter pylori: multiple mutational changes in 16S ribosomal DNA and other genetic loci. Antimicrob. Agents Chemother. 46:3940-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doucet-Populaire, F., C. Truffot-Pernot, J. Grosset, and V. Jarlier. 1995. Acquired resistance in Mycobacterium avium complex strains isolated from AIDS patients and beige mice during treatment with clarithromycin. J. Antimicrob. Chemother. 36:129-136. [DOI] [PubMed] [Google Scholar]
- 7.Gerrits, M. M., M. Berning, A. H. van Vliet, E. J. Kuipers, and J. G. Kusters. 2003. Effects of 16S rRNA gene mutations on tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 47:2984-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerrits, M. M., M. R. De Zoete, N. L. Arents, E. J. Kuipers, and J. G. Kusters. 2002. 16S rRNA mutation-mediated tetracycline resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 46:2996-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman, R. C., D. Zakula, R. Flamm, J. Beyer, and J. Capobianco. 1994. Tight binding of clarithromycin, its 14-(R)-hydroxy metabolite, and erythromycin to Helicobacter pylori ribosomes. Antimicrob. Agents Chemother. 38:1496-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, D. Y. 1997. Helicobacter pylori infection in the pathogenesis of duodenal ulcer and gastric cancer: a model. Gastroenterology 113:1983-1991. [DOI] [PubMed] [Google Scholar]
- 11.Graham, D. Y. 1998. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology 115:1272-1277. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. Y. 2000. Therapy of Helicobacter pylori: current status and issues. Gastroenterology 118:S2-S8. [DOI] [PubMed] [Google Scholar]
- 13.Haas, R., T. F. Meyer, and J. P. van Putten. 1993. Aflagellated mutants of Helicobacter pylori generated by genetic transformation of naturally competent strains using transposon shuttle mutagenesis. Mol. Microbiol. 8:753-760. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J. J., R. Reddy, M. Lee, J. G. Kim, F. A. El-Zaatari, M. S. Osato, D. Y. Graham, and D. H. Kwon. 2001. Analysis of metronidazole, clarithromycin and tetracycline resistance of Helicobacter pylori isolates from Korea. J. Antimicrob. Chemother. 47:459-461. [DOI] [PubMed] [Google Scholar]
- 15.Kwon, D. H., J. J. Kim, M. Lee, Y. Yamaoka, M. Kato, M. S. Osato, F. A. El-Zaatari, and D. Y. Graham. 2000. Isolation and characterization of tetracycline-resistant clinical isolates of Helicobacter pylori. Antimicrob. Agents Chemother. 44:3203-3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon, D.-H., F. A. K. El-Zaatari, M. Kato, M. S. Osato, R. Reddy, Y. Yamaoka, and D. Y. Graham. 2000. Analysis of rdxA and involvement of additional genes encoding NAD(P)H flavin oxidoreductase (FrxA) and ferredoxin-like protein (FdxB) in metronidazole resistance of Helicobacter pylori. Antimicrob. Agents Chemother. 44:2133-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon, D. H., K. Hulten, M. Kato, J. J. Kim, M. Lee, F. A. K. El-Zaatari, M. S. Osato, and D. Y. Graham. 2001. DNA sequence analysis of rdxA and frxA from 12 pairs of metronidazole-sensitive and -resistant clinical Helicobacter pylori isolates. Antimicrob. Agents Chemother. 45:2609-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon, D. H., M. P. Dore, J. J. Kim, M. Kato, M. Lee, J. Y. Wu, and D. Y. Graham. 2003. High-level β-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 47:2169-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levy, S. B. 1992. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 36:695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loivukene, K., H. I. Maaroos, H. Kolk, I. Kull, K. Labotkin, and M. Mikelsaar. 2002. Prevalence of antibiotic resistance of Helicobacter pylori isolates in Estonia during 1995-2000 in comparison to the consumption of antibiotics used in treatment regimens. Clin. Microbiol. Infect. 8:598-603. [DOI] [PubMed] [Google Scholar]
- 21.Mégraud, F. 1997. Resistance of Helicobacter pylori to antibiotics. Aliment. Pharmacol. Ther. 11(Suppl. 1):43-53. [DOI] [PubMed] [Google Scholar]
- 22.Mendonca, S., C. Ecclissato, M. S. Sartori, A. P. Godoy, R. A. Guerzoni, M. Degger, and J. Pedrazzoli, Jr. 2000. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxicillin, tetracycline, and furazolidone in Brazil. Helicobacter 5:79-83. [DOI] [PubMed] [Google Scholar]
- 23.Midolo, P. D., M. G. Korman, J. D. Turnidge, and J. R. Lambert. 1996. Helicobacter pylori resistance to tetracycline. Lancet 347:1194-1195. [PubMed] [Google Scholar]
- 24.Moazed, D., and H. F. Noller. 1987. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature 327:389-394. [DOI] [PubMed] [Google Scholar]
- 25.Nikaido, H. 1994. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science 264:382-387. [DOI] [PubMed] [Google Scholar]
- 26.Oehler, R., N. Polacek, G. Steiner, and A. Barta. 1997. Interaction of tetracycline with RNA: photoincorporation into ribosomal RNA of Escherichia coli. Nucleic Acids Res. 25:1219-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Putman, M., H. W. van Veen, and W. N. Konings. 2000. Molecular properties of bacterial multidrug transporters. Microb. Mol. Biol. Rev. 64:672-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Realdi, G., M. P. Dore, A. Piana, A. Atzei, M. Carta, L. Cugia, A. Manca, B. M. Are, G. Massarelli, I. Mura, A. Maida, and D. Y. Graham. 1999. Pretreatment antibiotic resistance in Helicobacter pylori infection: results of three randomized controlled studies. Helicobacter 4:106-112. [DOI] [PubMed] [Google Scholar]
- 29.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnappinger, D., and W. Hillen. 1996. Tetracyclines: antibiotic action, uptake, and resistance mechanisms. Arch. Microbiol. 165:359-369. [DOI] [PubMed] [Google Scholar]
- 31.Sharara, A. I., M. Chedid, G. F. Araj, K. A. Barada, and F. H. Mourad. 2002. Prevalence of Helicobacter pylori resistance to metronidazole, clarithromycin, amoxycillin and tetracycline in Lebanon. Int. J. Antimicrob. Agents 19:155-158. [DOI] [PubMed] [Google Scholar]
- 32.Thyagarajan, S., P. Ray, B. K. Das, A. Ayyagari, A. A. Khan, S. Dharmalingam, U. A. Rao, P. Rajasambandam, B. Ramathilagam, D. Bhasin, M. Sharma, S. Naik, and C. Habibullah. 2003. Geographical difference in antimicrobial resistance pattern of Helicobacter pylori clinical isolates from Indian patients: Multicentric study. J. Gastroenterol. Hepatol. 18:1373-1378. [DOI] [PubMed] [Google Scholar]
- 33.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenny, L. M. Fitzegerald, N. M. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hays, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 34.Trieber, C. A., N. Burkhardt, K. H. Nierhaus, and D. E. Taylor. 1998. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol. Chem. 379:847-855. [DOI] [PubMed] [Google Scholar]
- 35.Trieber, C. A., and D. E. Taylor. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wimberly, B. T., D. E. Brodersen, W. M. Clemons, Jr., R. J. Morgan-Warren, A. P. Carter, C. Vonrhein, T. Hartsch, and V. Ramakrishnan. 2000. Structure of the 30S ribosomal subunit. Nature 407:327-339. [DOI] [PubMed] [Google Scholar]
- 37.Wu, H., X. D. Shi, H. T. Wang, and J. X. Liu. 2000. Resistance of Helicobacter pylori to metronidazole, tetracycline and amoxycillin. J. Antimicrob. Chemother. 46:121-123. [DOI] [PubMed] [Google Scholar]