Abstract
Acute Myeloid Leukemia (AML) is a hematologic malignancy with a poor prognosis. Recent genome-wide sequencing studies have identified frequent mutations in genes encoding members of the cohesin complex. Mutations in cohesin contribute to myeloid malignancies by conferring enhanced self-renewal of hematopoietic stem and progenitor cells but the mechanisms behind this phenotype have not been fully elucidated. Of note, cohesin mutations are highly prevalent in acute megakaryocytic leukemia associated with Down syndrome (DS-AMKL), where they occur in over half of patients. Evidence suggests that cohesin mutations alter gene expression through changes in chromatin accessibility and/or aberrant targeting of epigenetic complexes. In this review we discuss the pathogenic mechanisms by which cohesin mutations contribute to myeloid malignancies.
Somatic mutations in myeloid malignancies
Myeloid malignancies represent a heterogeneous group of hematopoietic diseases that occur from birth to late in life. The two most common myeloid malignancies are myelodysplastic syndrome (MDS) and AML. In MDS, aberrations in the function of hematopoietic stem cells (HSCs) lead to dysplastic bone marrow with peripheral cytopenias, and in AML to a preponderance of leukemic blasts [1,2]. In both children and adults, MDS can result in death due to profound and prolonged cytopenias and/or transformation to AML. For both MDS and AML, there are distinct clinical sub-types based upon morphology, initial presentation, history of prior therapies, and additional criteria which make both diseases heterogeneous.
What causes AML or MDS in the majority of patients remains unknown, but a host of genomics data reveals that MDS and AML share some overlapping features [3–5]. First, unlike other cancers, MDS and AML tend to have fewer somatic mutations. In both diseases there are typically 5–15 somatic mutations [3,5,6]. MDS and AML appear to have a lower frequency of somatic mutations than most other cancers [3,5,6] such as ALL (12–26; [7]) or breast cancer (33–66; [8]). Importantly, while roughly 25 genes are recurrently mutated within AML, mutations in over 250 additional genes have also been identified [6]. This heterogeneity points to the difficulties in developing targeted therapies to treat these diseases. Thus, while the landscape of mutations present in AML/MDS is well described [3,6,9,10], which combination of somatic mutations are critical to AML development remains unclear.
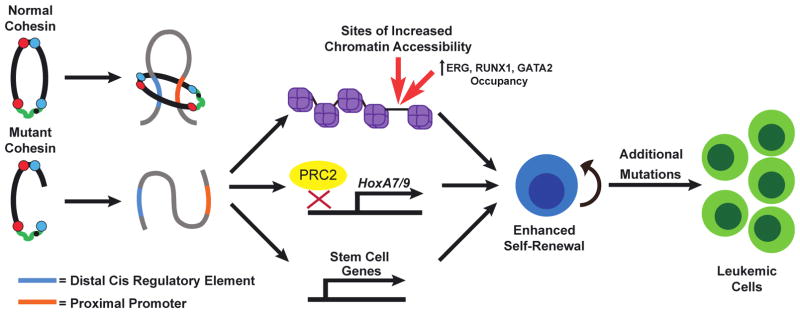
Given the diverse sub-types of AML and MDS, it is difficult to make general statements about prognosis. For example, in patients with AML there is a distinct segregation of prognosis based upon age, with younger patients having an overall survival approaching 50% and older patients having a long-term survival of less than 25% [11]. Even in the face of high-dose chemotherapy and/or allogeneic bone marrow transplantation, treatment of either MDS or AML remains only successful in about half of patients. This illustrates the need for novel, targeted therapies to improve long-term survival for these patients. Given the genetic heterogeneity of MDS/AML, it remains a challenge to develop therapies that are effective against multiple sub-types, so focusing on a set of mutations and how they contribute to the leukemogenic process may allow for the development of novel, targeted treatments. Recently, mutations in the cohesin complex were identified in acute myeloid leukemia (AML) [3,4,6,12–14]. In addition, a unique sub-type of AML, acute megakaryoblastic leukemia associated with Down Syndrome (DS-AMKL) exhibits cohesin mutations in almost 50% of patients [15]. A number of groups have investigated mechanisms by which cohesin mutations may underlie AML (Figure 1, key figure), which will be the focus of this review [16–20].
Figure 1, Key Figure. Cohesin mutations contribute to acute myeloid leukemia.
Wild-type cohesin mediates gene promoter-enhancer interactions to maintain proper gene expression. Cohesin mutations disrupt cohesin ring assembly which results in impaired differentiation and enhanced self-renewal of HSPCs. Recent research shows that disruption of cohesin in hematopoietic cells results in alterations in chromatin accessibility and/or disrupted targeting of epigenetic regulators such as the PRC2 complex, and drives a stem cell gene expression program. In the presence of additional oncogenic mutations, such as FLT3-ITD, cohesin haploinsufficiency leads to leukemia.
Cohesin Mutations in Myeloid Neoplasms
One of the most surprising discoveries following genomic sequencing of 200 de novo AMLs by the Cancer Genome Atlas (TCGA) was the presence of mutations within genes encoding the cohesin complex (STAG2, RAD21, SMC3, SMC1A) in 13% of patients [6]. Mutations in the cohesin complex are mutually exclusive in AML and DS-AMKL, heterozygous, distributed throughout the gene bodies, and they are often frameshift (17.39%) and missense mutations (69.57%; [12]). Overexpression of mutant forms of cohesin complex members in vitro disrupts cohesin complex assembly suggesting these mutations act at least partially as dominant negatives [18]. STAG2 is encoded on the X-chromosome, and therefore mutations would be thought to result in a null-allele. Whether STAG1 or possibly STAG3 can partially compensate for the loss of STAG2 during mitosis remains an important question within the field. In addition, whether women display skewed lyonization in their AML to favor clones with a wild-type or mutant STAG2 allele remains unknown. Evidence suggests that cohesin mutations can occur early or late in both pediatric and adult AML patients and promote leukemogenic transformation [4,12–15]. Importantly, a study of adult AML patients found that the clonal hierarchy of cohesin mutations was comparable with NPM1 mutations [12], which have been proposed to occur early during leukemogenic transformation [21], implying that cohesin mutations can be an early event during leukemia development. Canonically, loss of cohesin function results in premature sister chromatid separation [22,23]. However, AML cells are typically not aneuploid, and almost half of TCGA AML patients had normal karyotypes, implying that gross aneuploidy was unlikely to be a driving factor in their leukemogenesis. In fact, cohesin mutated hematopoietic malignancies are not associated with a statistically significant increase in complex karyotypes or specific chromosomal changes (−7, +8, del 5q) that are often found in myeloid malignancies [3,4,6,12,24]. Subsequent studies have found similar cohesin mutation frequencies not only in AML, but also in other myeloid malignancies such as chronic myelomonocytic leukemia (CMML), chronic myeloid leukemia (CML), myeloproliferative neoplasms (MPNs), and MDS [4,24]. Notably, over half (53%) of patients with DS-AMKL have mutations in cohesin complex members [15]. In AML the most commonly co-occurring mutation is NPM1, which is rarely present in MDS [3,4,6,12,24]. Other common mutations with cohesin in AML include TET2, RUNX1, ASXL1, EZH2, and Ras genes [3,6,12]. In DS-AMKL, mutations in EZH2, JAK2, JAK3, ASXL1, and Ras genes commonly occur with cohesin subunit mutations [15].
Interestingly, specific translocations have variable co-occurrence rates with cohesin mutations. Core binding factor (CBF) mutations are relatively common in AML, with the two most common translocations being RUNX1-RUNX1T1; t(8;21) or CBFB-MYH11; inv(16). Given that both translocations are associated with a favorable prognosis, one would predict that the co-occurring mutations that ultimately result in AML would be similar. Surprisingly, two different groups have determined mutational differences between these two clinically similar diseases. In one publication, 20% (17 of 85) of patients with RUNX1-RUNX1T1 also had cohesin mutations, whereas no patients (0 of 80) with CBFB-MYH11 had cohesin mutations. Interestingly, patients with RUNX1-RUNX1T1 had a significantly higher number of mutations than CBFB-MYH11 patients (12 versus 8, p<0.0001; [25]), and STAG2 was not mutated in any patient, which is unusual because other myeloid malignancies show a bias for STAG2 mutations compared with other subunits (RAD21, SMC3, SMC1A). Similar results were reported in a second publication [26], but this study also found that the combination of cohesin mutations with alterations in tyrosine kinases conferred a greater risk of relapse in patients with an otherwise “favorable” prognosis [26]. Lastly, recent studies have found that 26% of patients with MLL-PTD leukemia harbor cohesin mutations [27], but additional translocations have not been definitively studied thus far. Whether cohesin mutations cooperate or antagonize other common AML translocations remains unknown. Collectively, these data indicate that cohesin mutations likely play a role in leukemia development but in the context of specific cooperating mutations [25,26].
Whether mutations in cohesin are prognostically significant remains an open question. In AML, it appears that cohesin mutations do not significantly alter prognosis, likely because they co-occur predominantly with NPM1 mutations, making it difficult to evaluate whether cohesin mutations alone confer a worse prognosis [12]. In MDS it appears that mutations in cohesin are associated with lower overall survival, especially in patients surviving >1 year after diagnosis [4]. One of the difficulties in determining cooperativity is the heterogeneity in both AML and MDS, and the presence of additional mutations, which may confound whether cohesin mutations are driving prognosis. Parsing of these important issues will require genomic studies with larger numbers of patients in addition to basic science studies to firmly establish cooperativity of mutations to leukemia development.
Disruption of the cohesin complex
Self-renewal and Differentiation of HSPCs
The identification of cohesin mutations in AML patients raised the intriguing question about whether or not these mutations are causative of the disease, and if so, how they may contribute to leukemogenesis. Our lab and others have investigated this question using in vivo mouse models and cultured cells[16–20]. The common finding is that cohesin mutations are capable of conferring enhanced self-renewal on hematopoietic stem and progenitor cells (HSPCs). Enhanced self-renewal of HSPCs is a common finding of known driver mutations of AML, including TET2 [28,29], NPM1 [30,31] and DNMT3A [32]. The mechanism(s) causing this enhanced self-renewal phenotype following reduction of cohesin function is likely multifactorial. The first studies published by the Levine and Aifantis laboratories were congruent in their conclusions that disruption of cohesin function by shRNA-mediated knockdown or heterozygous conditional deletion confers enhanced serial replating capacity on HSPCs in methylcellulose culture in vitro and in competitive repopulation assays in vivo [16,17]. Using a genetic model, Viny et al., demonstrated that HSPC-specific heterozygous ablation of cohesin (Smc3) causes AML with a latency of about 6 months when combined with the FLT3-ITD mutation [16]. The enhanced self-renewal was later confirmed by other studies using similar approaches in both human and mouse hematopoietic cells [18–20]. Importantly, studies using heterozygous mouse conditional knockout models or RNAi-mediated cohesin knockdown observed enhanced self-renewal that is also observed when mutant forms of cohesin are overexpressed [16–20] suggesting that mutations in the cohesin complex result in a loss of function or act as partial dominant negatives. None of the published studies uncovered evidence that disrupted cohesin function alone is sufficient to drive AML, suggesting that cohesin mutations cooperate with other genetic alterations during AML development.
The mechanism by which cohesin haploinsufficency induces enhanced self-renewal in HSPCs remains an active area of investigation, but one common observation is altered differentiation of higher order HSPCs [16–20]. Although the effects of cohesin depletion on HSPC differentiation varied slightly between the different groups, many similarities were observed (Table 1). The cause of the differences in lineage skewing between the published reports remains unclear but may reflect differences in experimental approaches (shRNA versus forced expression of mutant forms of cohesin versus conditional ablation). Given the differentiation defects observed in cohesin haploinsufficient HSPCs [16–19], it is plausible that at least part of the phenotype arises from partially defective differentiation, and maintenance of higher order progenitors.
Table 1.
Effects of disrupted cohesin function
| Citation | Model System | Method for Cohesin Depletion | System | Lineage Skewing Phenotype | Chromatin Accessibility Changes | Key Conclusions |
|---|---|---|---|---|---|---|
| [16] | Mouse HSPCs and Bone Marrow Cells | Smc3 Conditional Knockout | In vivo | Decreased LT-HSCs and increased ST-HSCs, and MPPs in animals. | Decreased chromatin accessibility within 30kb and 10kb, in the 5′ and 3′ direction respectively, surrounding TSSs of genes that exhibit decreased expression in cohesin haploinsufficient cells. | Smc3 haploinsufficiency alone fails to impart complete malignant transformation but causes AML when combined with FLT3-ITD mutation. Cohesin haploinsufficiency alters chromatin accessibility |
| Competitive Repopulation Assay | Increased LT-HSCs, ST-HSCs, MPPs, GMPs CMPs, and MEPs in chimeric bone marrow studies. | |||||
| [17] | Mouse HSPCs and Bone Marrow Cells | shRNA-mediated depletion of cohesin complex members | In vivo | Decreased LT-HSCs, ST-HSCs, and B-cells and increased MPPs and GMPs, and myelomonocytic and erythroid cells. | Disrupted cohesin function caused changes in chromatin accessibility that correlated well with observed changes in gene expression. Cohesin-depleted LSKs exhibited increased accessibility of myeloid and erythroid genes. | Cohesin-depletion causes a myeloproliferative neoplasm- like state. Cohesin depletion alters chromatin accessibility. |
| [18] | Human CD34+ HSCs | Overexpression of mutant forms of cohesin subunits and shRNA- mediated depletion | In Vitro | Increased HSCs, and decreased myeloid and erythroid progenitors | Disrupted cohesin function causes a global decrease in chromatin accessibility however key sites, enriched for ERG, GATA2, and RUNX1 consensus binding sites exhibit increased accessibility. | Disrupted cohesin function enforces a stem cell program impairing differentiation of HSCs concomitant with altered chromatin accessibility. |
| In Vivo | Increased myeloid progenitors and decreased erythroid progenitors | |||||
| [19] | Human CD34+ HSCs | RNAi-mediated depletion of cohesin complex members | In Vitro | Increased myeloid and CD34+ cells. | Not Applicable | Cohesin-depletion drives an HSC-specific transcriptional program, but does not impart complete malignant transformation. |
| Competitive Repopulation Assay | Increased myeloid and CD34+ cells. | |||||
| [20] | Mouse HSPCs | shRNA-mediated depletion of Rad21 | In Vitro | No alterations in the numbers of c-kit+ cells or lineage+ cells was detected. | Not Applicable | Rad21 interacts with PRC2; Rad21-depletion alters PRC2 targeting to the Hoxa locus leading to elevated Hoxa7/9 expression. The enhanced self-renewal phenotype requires Hoxa7/9. |
Epigenetic alterations
While the published reports have agreed that disruption of cohesin function impairs differentiation and confers enhanced self-renewal on HSPCs, the mechanism behind this phenotype remains an important area of investigation [16–18,20]. There are several proposed mechanisms which are not mutually exclusive and focus on alterations in gene expression. For example, disruption of cohesin function in HSPCs causes altered expression of a large number of genes but notably causes sustained expression of the HSC factors Hoxa7 and Hoxa9 due to decreased recruitment of polycomb repressive complex 2 (PRC2) to these genes [16,17,20]. These data reveal an interaction between the cohesin and PRC2 complexes and demonstrate reduced levels of the PRC2 mark trimethylated lysine 27 of histone H3 (H3K27me3) at the Hoxa7/9 genes. Combined shRNA-mediated knockdown of cohesin and Hoxa7 or Hoxa9 abrogated this enhanced self-renewal phenotype. This mechanism is consistent with the known role of Hoxa9 in driving hematopoietic stem cell transcriptional programs [33–35].
Another mechanism that has been proposed involves alterations in chromatin accessibility caused by disrupted cohesin function. Three studies recently used the assay for transposase-accessible chromatin using sequencing (ATAC-seq) to identify alterations in chromatin compaction in response to cohesin loss (summarized in Table 1; [16–18]). These reports found that altered chromatin accessibility correlated well with the altered gene expression caused by disruption of cohesin function. There was a global decrease in chromatin accessibility [16–18] including at the transcriptional start sites (TSSs) of genes that exhibit decreased expression in cells with disrupted cohesin function[16]. Additionally, an increase in chromatin accessibility was observed at genes that play a role in myeloid and erythroid differentiation [17]. It remains unclear whether the alterations in chromatin accessibility are a direct effect of disrupted cohesin function, since it is unknown whether cohesin directly binds to the sites with altered chromatin accessibility. Additionally, given that different lineages of hematopoietic cells exhibit different chromatin accessibility profiles [36] the altered chromatin accessibility could be secondary to defects and/or skewing in differentiation. A more thorough characterization of how cohesin-deficiency alters hematopoietic differentiation is necessary to fully address this possibility. It is worth noting that the length of the domain with altered chromatin accessibility is consistent with local chromatin loops involved in transcription factor transactivation [37]. This correlation is intriguing given the known collaboration between cohesin and CCCTC-binding factor (CTCF) in modulating chromatin architecture [38] and establishment of sub-megabase topologically associating domains (sub-TADs) [39]. CTCF and cohesin have been shown to regulate higher order chromatin structure at the HOXA locus [40,41]. More broadly CTCF and cohesin cooperate to establish higher order chromatin structure genome-wide [42]. Disruption of cohesin function doesn’t affect topologically associate domains but instead disrupts local chromatin interactions. Thus, cohesin haploinsufficiency may disrupt sub-TAD architecture, which then influences expression of a large number of genes contained within cohesin-dependent sub-TADs, conferring a clonal advantage to certain sub-populations of cells. This is supported by a study in Importantly, CTCF and cohesin cooperate at sites throughout the genome, but there are CTCF-independent cohesin binding sites [42]. Clearly, mutations that disrupt cohesin function result in altered gene expression, due to improper PRC2 targeting, altering chromatin accessibility, or disrupting sub-TADs, driving a stem cell gene expression program. The consequence of this gene expression program is impaired differentiation and enhanced HSPC self-renewal capacity. Given the diverse roles of cohesin, and that cohesin mutations alone are insufficient to drive leukemogenesis, it is likely that the mechanisms by which cohesin mutations contribute to myeloid malignancies are dependent on the cooperating mutations present in patients. Similar to the alterations in chromatin accessibility, the alterations in gene expression could be secondary to defects in differentiation between cohesin wild-type and cohesin-deficient HSPCs, a possibility that warrants further investigation.
Cohesin mutations in DS-AMKL
Acute megakaryoblastic leukemia in children with Down Syndrome
Down syndrome (DS) is the result of trisomy 21 (TS21) and comes with a range of hematologic abnormalities. Up to 10% of children with DS will present at birth with transient abnormal myelopoiesis (TAM), a pre-leukemia that is morphologically very similar to AMKL, but typically resolves spontaneously over a period of weeks to months. Up to 30% of children with a history of TAM will develop frank AMKL during childhood. Of note, DS-AMKL has a more favorable prognosis than non-DS AMKL, with 5 year event free survival rates over 80% [43]. Virtually all DS-AMKL patients have an N-terminal truncation within the hematopoietic transcription factor GATA1, which results in exclusive expression of a shorter isoform named GATA1s. GATA1s has been shown to cause hyperproliferation of an abnormal megakaryocyte progenitor population during early fetal liver hematopoiesis, which subsides during postnatal bone marrow hematopoiesis mirroring the TAM observed in DS patients [44]. Variable allele frequency studies of TAM and DS-AMKL patients show that GATA1s, and mutations in cohesin complex members, CTCF and EZH2 were similar indicating they occur early during DS-AMKL development [15]. The presence of trisomy 21 and GATA1s is necessary but not sufficient for the development of DS-AMKL [45], and progression from TAM to DS-AMKL is associated with multiple other genetic mutations including cohesin [15]. Interestingly, mutations in CTCF occur frequently in DS-AMKL (20%), but are exceedingly rare in other myeloid malignancies. CTCF and cohesin have been found to directly interact [46,47] to regulate a higher order of genomic structure, thus mutations in either could promote global changes in the nuclear architecture, including sub-TAD interactions and/or TAD boundaries themselves. Importantly, CTCF and cohesin mutations were not mutually exclusive, suggesting that disrupting CTCF or cohesin function may have non-overlapping effects on chromatin architecture, both in terms of TAD but also sub-TAD organization [39,42]. How mutations in CTCF may promote AMKL remains unknown and represents an important unanswered question.
Cohesin Subunits are Frequently Mutated in DS-AMKL
Cohesin mutations are especially prominent in DS-AMKL, but the reason for this remains unknown. Similar to AML, cohesin mutations in DS-AMKL are predicted to be heterozygous, loss-of-function, and early events during leukemia development [15]. That cohesin mutations occur in DS-AMKL, but not the pre-leukemic TAM, suggests that cohesin haploinsufficiency may drive oncogenic transformation and progression to megakaryocytic leukemia. Furthermore, mutations in cohesin likely cooperate with chromosome 21 genes and/or GATA1s to promote leukemia and alter megakaryopoiesis. Cohesin may play an important role in differentiation along the megakaryocyte-erythroid lineage, as genes downregulated by SMC3 attenuation are typically enriched in mouse and human megakaryocyte-erythroid progenitors (MEPs) as well as human megakaryocytes [16]. This implies that megakaryocyte differentiation may be blocked by SMC3 deficiency, although this has not yet been described.
Cohesin mutations alter gene expression in DS-AMKL
The cohesin complex regulates expression of several genes located on chromosome 21. For example, cohesin abrogation leads to increased expression of the transcription factor Runx1 [48,49] an important regulator of HSCs and megakaryocytes. Whether this upregulation in Runx1 reflects changes across all HSPC lineages, or reflects altered differentiation is an important unanswered question. Regardless, in the context of trisomy 21, cohesin may regulate transcription from all three alleles, resulting in further alterations in RUNX1 levels in TS21 cells compared to disomic cells. RUNX1 also binds chromatin more frequently in cohesin-mutant hematopoietic cells [18]. Other DS genes that may participate in cohesin-mutant DS-AMKL include ERG and ETS2, which also regulate hematopoietic stem cells and megakaryocytes and have been linked to AMKL [50,51]. Furthermore, in cohesin-mutant human HSPCs, ERG binding motifs were found to be located in areas of more open, accessible chromatin, and knockdown of ERG reversed the block in differentiation caused by disrupted cohesin function[18].
Cohesin haploinsufficiency may also indirectly affect gene transcription through regulation of other transcription factors, including GATA family members. A reduction in cohesin induces expression of GATA1 and GATA2 [9,17,18,48] and regulates their association with chromatin. ATAC-seq peaks unique to cohesin-haploinsufficient cells are either strongly enriched [17,18] or depleted [16] for the GATA consensus binding site, indicating that cohesin loss may alter GATA1 and/or GATA2 binding. It is unknown whether cohesin binding sites are located proximal to these altered transcription factor binding sites, and if cohesin loss directly drives relocalization of transcription regulators. Whether GATA1 binding to chromosomes is in fact increased or decreased when cohesin function is disrupted remains to be determined, as is the binding activity of the GATA1s isoform in this context. The GATA1s protein, which lacks the N-terminal activation domain, retains its DNA binding domains and is able to bind its FOG-1 cofactor [52], but many of its genomic binding sites differ from the wild-type GATA1 [53]. Therefore, the ability of GATA1s to bind to chromatin may be differentially affected by cohesin loss compared to wild-type GATA1. All of these genetic events likely cooperate to alter HSPC differentiation. For example, ETS2 and ERG overexpression produce immature megakaryocyte expansion in GATA1s mice [54]. Whether cohesin mutations affect gene expression in the context of trisomy 21, and alter GATA1 wild-type or GATA1s binding, remains an important unanswered question.
Concluding Remarks
Cohesin mutations are commonly found in AML and DS-AMKL, and evidence suggests that these mutations likely occur early during leukemogenesis [3,4,6,12–15]. Cohesin mutations alone are insufficient to impart complete malignant transformation suggesting that cooperating mutations, such as FLT3-ITD or mutations in NPM1, most likely drive the leukemogenic process [16]. In AML and DS-AMKL many cohesin mutations act as null alleles creating a haploinsufficient state. Additionally, cohesin mutations that produce a functional protein can prevent proper cohesin subunit interactions [17,18]. The consequence of this is enhanced HSPC self-renewal and altered HSPC differentiation [16–20]. The mechanism(s) underlying these phenotypic changes are altered gene expression, disrupted chromatin accessibility [16–18] and altered targeting of epigenetic modulators [20].
The mechanisms that lead to altered chromatin accessibility and disrupted targeting of epigenetic modifiers remain unclear. A likely candidate would be alterations in chromatin architecture and DNA looping. A previous study showed that in non-cycling thymocytes, cohesin cooperates with CTCF to facilitate long-range interactions within architectural domains that contain cohesin-regulated genes [38,55]. In this context cohesin is thought to modulate DNA looping bringing together distal cis-regulatory elements with proximal promoters. Disruption of cohesin function in this setting alters gene expression within pre-existing chromatin compartments. Interestingly, cohesin has been previously shown to regulate Hoxa gene expression by regulating higher order chromatin structure [40,56,57]. Whether disruption of cohesin function in HSPCs alters chromatin architecture surrounding the Hoxa locus remains unclear, but given the maintained expression of Hoxa7 and Hoxa9 in Rad21-depleted HSPCs [20] it is reasonable to speculate that disrupted cohesin function alters chromatin reorganization during HSPC differentiation. Additionally, multiple groups have shown altered chromatin accessibility in HSPCs with disrupted cohesin function [16–18]. How chromatin architecture relates to chromatin accessibility remains unclear, because there is no clear evidence that cohesin directly occupies the regions of altered chromatin accessibility. Computational biology approaches comparing chromosomal capture data with ATAC-seq data would help answer this intriguing question.
One final remaining question is whether cohesin mutations drive DS-AMKL in the same way as in non-DS AML. Gene expression changes imparted by disrupted cohesin function need to be determined in the context of trisomy 21 and GATA1s to determine whether the oncogenic pathways are the same. Notably, human HSCs expressing mutant forms of cohesin exhibit increased binding of RUNX1 to chromatin, and the observed cohesin-dependent block in differentiation is dependent on presence of RUNX1 and ERG [18]. This may be amplified in DS-AMKL where three RUNX1 and ERG copies are present. Additionally, it is not clear whether cohesin-mutant DS-AMKL clones arise from the same cell of origin as cohesin-mutant AML. RAD21 knockdown or overexpression of a RAD21 mutant produced differentiation defects only in the most immature stem and progenitor populations, such as HSCs and MPPs [18], but it is unknown whether cohesin-mutant DS-AMKL cells also have a similar cell of origin, or if they arise from another cell type such as the megakaryocyte-erythroid progenitor. Cohesin mutations, which are present in various myeloid malignancies including AML and DS-AMKL, alter differentiation and enhance self-renewal capacity of HSPCs, however many questions remain (see outstanding questions). Future studies aimed at further elucidating the mechanisms by which cohesin mutations contribute to leukemogenesis will allow for the development of novel targeted therapies for patients with cohesin mutant myeloid malignancies.
Outstanding Questions Box.
Cohesin mutations fall short of imparting complete malignant transformation. What mutations cooperate with cohesin in AML and DS-AMKL?
How do cohesin mutations alter chromatin accessibility?
Does cohesin occupy genomic regions where changes in accessibility are observed?
How do cohesin and CTCF mutations affect TAD boundaries and sub-TAD interactions?
Do cohesin mutations alter targeting of epigenetic modulators other than PRC2? If so, is this mediated by global changes in nuclear architecture?
Can drugs that target epigenetic modulators ameliorate the effects of cohesin mutations in AML and DS-AMKL patients?
Mutations in epigenetic regulators, such as cohesin, are thought to occur early in disease development [4,12–14], therefore a number of therapeutic interventions targeting epigenetic pathways have been proposed aimed at inducing differentiation of or selectively killing leukemic blast cells [58]. For example, Hoxa9 is a key gene that is upregulated in MDS and AML [16,17,20], is known to be upregulated by disruption of cohesin function [20], and is regulated by Dot1l methyltransferase [59]. Dot1l inhibitors are currently in clinical trials for treating AML patients and preclinical studies have shown some efficacy in selectively killing leukemic blast cells [60]. Additionally, HOXA9 expression is regulated by the opposing actions of the PRC2 and COMPASS complexes. PRC2 and the compass complexes repress and activate HOXA9 expression by depositing the histone marks H3K27me3, and H3K4me3 respectively. COMPASS complex inhibitors may prove clinically useful in a similar manner as Dot1l inhibitors. It is reasonable to speculate that treating AML patients with therapeutics targeting epigenetic pathways will selectively kill not only the leukemic blast cells, but also the tumor stem cells. More studies are required to address this intriguing possibility.
Text Box 1. The cohesin complex.
The cohesin complex was first discovered in S. cerevisiae and Drosophila melanogaster during forward genetic screens seeking mutations resulting in precocious sister chromatid segregation during mitosis [61,62]. Later it was discovered in Xenopus and S. Pombe that several of these genes were part of a multiprotein complex called cohesin, which is required for proper sister chromatid cohesion [63,64]. Members of the cohesin complex are remarkably well conserved across species, and in humans the core subunits are SMC1A, SMC3, RAD21, and either STAG1 or STAG2 [23]. These subunits form a heterotrimeric ring that encircles DNA and functions to maintain sister chromatid cohesion, facilitate DNA looping, and stabilize sites of double stranded DNA breaks [22,39,46,65]. Paralogs of SMC1A and STAG1 or STAG2, SMC1B and STAG3 respectively, were originally identified as being essential for meiotic cell division; however, recently these have also been identified in non-canonical cohesin complexes in somatic cells undergoing mitotic cell division [66,67]. Accessory subunits such as WAPAL, NIPBL, PDS5, CDC5A (sororin), ESCO1, and ESCO2 are required for different aspects of cohesin function [23].
Germline mutations in cohesin complex members result in disorders termed cohesinopathies, and include Cornelia de Lange syndrome and Robert’s syndrome [68–71]. The most commonly mutated cohesin complex member that is mutated in cohesinopathies is Nipbl, although other components (Rad21, Smc1a, Smc3) and HDAC8 are mutated [68,72,73] Although precocious sister chromosome segregation has been observed in a subset of patients possessing cohesinopathies [74], many do not exhibit aneuploidy suggesting that non-mitotic functions of cohesin contribute to cohesinopathies. The role of cohesin in regulating sister chromatid cohesion during mitosis has been extensively studied (well reviewed in [23], but the role it plays in regulating gene expression and DNA repair is less well understood. Interestingly, germline mutations in cohesin complex genes lead to chromosomal instability[75], but do not seem to predispose patients to cancer [76]. The study of how germline cohesin mutations contribute to cohesinopathies may provide some insight into possible mechanisms of how somatic cohesin mutations underlie oncogenic processes, including myeloid malignancies.
Figure I. Cohesin structure and functions.
A) The core cohesin subunits SMC3, SMC1A, RAD21, and either STAG1 or STAG2 form the core subunits to generate a cohesin “ring”. In myeloid malignancies, all four core subunits are recurrently mutated with the exception of STAG1. Red circles and blue circles represent the hinge and ATPase head domains respectively of the SMC1 and SMC3 proteins. B) The canonical function of the cohesin complex is during mitosis where it maintains sister chromatin cohesion. C) Cohesin stabilizes DNA loops during interphase, facilitating interactions. Through a direct interaction with CTCF, higher order chromatin structures are stabilized, organizing the genome into topologically associated domains and sub-megabase topologically associated domains. D) Cohesin permits distal cis-regulatory elements such as enhancers to interact with genes and regulate gene expression. CTCF and cohesin co-regulate DNA looping; CTCF can mediate DNA loops independent of cohesin, and vice-versa.
Text Box 2. Cohesin mutations in solid tumors.
The first report in which cohesin mutations were found in cancer patients observed missense mutations in SMC1A, SMC3, STAG3, and NIPBL in aneuploid colon cancers [77]. Following this initial study a number of other solid tumors were identified that harbor cohesin mutations. These included glioblastoma multiforme, Ewing sarcoma, melanoma, and cervical carcinoma[78]. In this study STAG2 mutations were found to lead to chromosome instability resulting in aneuploidy. STAG2 is the most commonly mutated cohesin subunit in non-hematopoietic malignancies, and depending on context, may or may not be associated with aneuploidy[79]. Interestingly, a recent study found that missense mutations in STAG2 do not affect sister chromatid cohesion whereas nonsense mutations cause defects in sister chromatid cohesin, but rarely do these mutations cause aneuploidy [80]. STAG2 functions as a scaffold protein to facilitate interactions between the cohesin complex and the cohesin loading and unloading proteins WAPL, PDS5A, and PDS5B. Many mutations in STAG2 do not affect its interaction with the core cohesin complex, but instead affects it interactions with accessory proteins[80]. Much work has focused on understanding how STAG2 mutations contribute to oncogenic processes; however, it is less clear how mutations in other cohesin genes underlie non-hematopoietic malignancies. Altered expression of cohesin genes has been linked to various carcinomas including prostate cancer [81], breast cancer [82,83], colorectal cancer [84,85], and cervical cancer [86]. In all these cases increased levels of cohesin proteins were correlated with disease state. Whether the elevated levels of cohesin complex members were contributing to the oncogenic process, and the cause of the elevated cohesin levels was unclear.
Trends Box.
Cohesin mutations have been identified in various myeloid malignancies.
Mutations occur within genes encoding four core subunits of the cohesin complex (SMC3, SMC1A, RAD21, and STAG2) and are heterozygous, ultimately resulting in haploinsufficiency or acting as dominant negatives.
Cohesin mutations confer both enhanced self-renewal and altered differentiation of hematopoietic stem and progenitor cells.
Cohesin mutations cause an overall decrease in chromatin accessibility, but increased accessibility of binding sequences for master hematopoietic transcription factors such as GATA2, RUNX1, and ERG.
The epigenetic writer complex PRC2 displays altered targeting following loss of cohesin, providing a possible therapeutic target for treating patients with cohesin-mutant myeloid malignancies.
Acknowledgments
This review was supported in part by a gift from the Heartland Blood Centers, a part of Versiti (to JDC and SR). S. Rao is supported by grants from the Midwest Athletes Against Childhood Cancer (MACC Fund), Hyundai Hope on Wheels, and an American Society of Hematology Bridge Grant. Additional research support to S. Rao provided by charitable gifts from Ms. Nanette Gardetto, and Mr. Doug Ziegler. MJB is supported by grants from the MACC Fund. Support for this review also was provided by the NCI (R01 CA101774) and the Samuel Waxman Cancer Research Foundation (to JDC). MN is supported by an NCI training grant (T32 CA009560).
Glossary Box
- Acute Myeloid Leukemia (AML)
a myeloid disease characterized by uncontrolled self-renewal and proliferation of myeloid progenitor cells.
- Assay for Transpose Accessible Chromatin with high throughput sequencing (ATAC-Seq)
This assay uses high throughput sequencing to determine the incorporation rate of transposons into chromatin as a measure of chromatin accessibility.
- Chromatin accessibility
The degree of chromatin compaction present in genomic regions. This can be altered by histone modifications, DNA methylation state, and other chromatin modifying enzymes. Closed chromatin is occupied by nucleosomes, while open and accessible chromatin has displaced nucleosomes.
- CCCTC-Binding Factor (CTCF)
A chromatin insulator protein that binds to DNA to establish topologically-associated domains and regulate gene expression.
- DNA looping
A mechanism that modulates transcriptional regulation and establishment of chromatin architectural states whereby proteins form a complex with DNA bringing distal cis-regulatory elements in close proximity to proximal promoters.
- Down syndrome
A germline karyotypic abnormality whereby individuals possess three copies of chromosome 21.
- Down syndrome associated Acute Megakaryocytic Leukemia (DS-AMKL)
a disease present in children with down syndrome characterized by dysplastic, hyperproliferative megakaryoblasts.
- GATA1s
A truncated form of the GATA1 transcription factor that contributes to DS-AMKL disease progression.
- Genomic translocations
The aberrant transfer of DNA segments to other parts of the genome. Often caused by non-homologous end joining (NHEJ) during repair of DNA double strand breaks. These often result in altered transcriptional activity or cause fusion genes that contain peptides from two different genes. Examples include MLL-rearrangements and CBFB-Myh11.
- Hematopoietic Stem Cells (HSCs)
The cells of the hematopoietic system that give rise to all the formed elements of blood. Two types are present, long term (LT) and short term (ST) HSCs.
- Hematopoietic Stem and Progenitor Cells (HSPCs)
The cells of the hematopoietic system comprised of LT- and ST-HSCs, multipotent progenitors (MPPs), common myeloid progenitors (CMPs), granulocyte-macrophage progenitors (GMPs), and megakaryocyte erythroid progenitors (MEPs).
- HoxA cluster
Homeobox A cluster of genes are located on chromosome 7 in humans and are key regulators of various developmental processes including regulation of hematopoietic stem cell self-renewal and differentiation. The HoxA cluster consists of the Hox genes A13, A11, A10, A9, A7, A6, A5, A4, A3, A2, and A1 arranged in 5′–3′ order.
- Myelodysplastic Syndrome (MDS)
A group of myeloproliferative disorders marked by an expansion and failure to differentiate of various myeloid progenitors cells. Some MDS cases progress to AML depending on the underlying genetic causes. Classifications include refractory anemia (RA), refractory anemia with ring sideroblasts (RARS), refractory anemia with excess blasts (RAEB), refractory anemia with excess blasts in transformation (RAEB-T), and chronic myelomonocytic leukemia (CMML).
- Myeloid Neoplasms (MPNs)
A myeloproliferative condition where patients possess excess myeloid progenitors. 4 classifications exist depending on the underlying genetic cause. MPNs include primary myelofibrosis (PMF), chronic myelogenous leukemia (CML), polycythemia vera (PV), and essential thrombocythemia (ET).
- Polycomb Repressive Complex 2 (PRC2)
A histone modifying complex that modulates gene repression by methylating histone H3 on lysine residue 23. The core complex consists of the subunits EED, Suz12, and EZH2 (catalytic subunit).
- Self-renewal
A characteristic of HSPCs whereby they retain their stem cell state and proliferative potential. Self-renewal is often enhanced in HSPC clones that harbor mutations that contribute to various hematopoietic malignancies. HSPCs that possess enhanced self-renewal capabilities will continue to propagate and form colonies beyond 3 passages in methylcellulose in vitro, and will exhibit increased contribution to the hematopoietic compartment in competitive repopulation assays in vivo.
- Topologically Associated Domains (TADs)
A higher order chromatin structure in which different genomic regions preferentially interact to influence gene expression across multiple genes. TAD boundaries tend to be bound by DNA binding proteins such as CTCF and cohesin. TAD compartments also contain sub-megabase genomic interactions termed sub-TADs, which influence expression of genes contained within TADs. Importantly, cohesin and CTCF also bind within TADs, and therefore play a role in both TAD and sub-TAD interactions.
- Trimethylated Lysine 23 on Histone H3 (H3K27me3)
A histone modification that arises from methylation of dimethylated lysine 27 on histone H3. This histone modification is deposited by PRC2 and has a repressive effect on gene transcription.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sperling AS, et al. The genetics of myelodysplastic syndrome: from clonal haematopoiesis to secondary leukaemia. Nat Rev Cancer. 2017;17:5–19. doi: 10.1038/nrc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Döhner H, et al. Acute Myeloid Leukemia. N Engl J Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374:2209–2221. doi: 10.1056/NEJMoa1516192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thota S, et al. Genetic alterations of the cohesin complex genes in myeloid malignancies. Blood. 2014;124:1790–1798. doi: 10.1182/blood-2014-04-567057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haferlach T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2013;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunz JB, et al. Pediatric T-cell lymphoblastic leukemia evolves into relapse by clonal selection, acquisition of mutations and promoter hypomethylation. Haematologica. 2015;100:1442–1450. doi: 10.3324/haematol.2015.129692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leeke B, et al. Cohesin mutations in myeloid malignancies: underlying mechanisms. Experimental Hematology & Oncology. 2014;3:13. doi: 10.1186/2162-3619-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ganguly BB, Kadam NN. Mutations of myelodysplastic syndromes (MDS): An update. Mutat Res Rev Mutat Res. 2016;769:47–62. doi: 10.1016/j.mrrev.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Burnett A, et al. Therapeutic Advances in Acute Myeloid Leukemia. Journal of Clinical Oncology. 2011;29:487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 12.Thol F, et al. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123:914–920. doi: 10.1182/blood-2013-07-518746. [DOI] [PubMed] [Google Scholar]
- 13.Shiba N, et al. Whole-exome sequencing reveals the spectrum of gene mutations and the clonal evolution patterns in paediatric acute myeloid leukaemia. British Journal of Haematology. 2016;175:476–489. doi: 10.1111/bjh.14247. [DOI] [PubMed] [Google Scholar]
- 14.Farrar JE, et al. Genomic Profiling of Pediatric Acute Myeloid Leukemia Reveals a Changing Mutational Landscape from Disease Diagnosis to Relapse. Cancer Research. 2016;76:2197–2205. doi: 10.1158/0008-5472.CAN-15-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshida K, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet. 2013;45:1293–1299. doi: 10.1038/ng.2759. [DOI] [PubMed] [Google Scholar]
- 16.Viny AD, et al. Dose-dependent role of the cohesin complex in normal and malignant hematopoiesis. Journal of Experimental Medicine. 2015;212:1819–1832. doi: 10.1084/jem.20151317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mullenders J, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. Journal of Experimental Medicine. 2015;212:1833–1850. doi: 10.1084/jem.20151323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazumdar C, et al. Leukemia-Associated Cohesin Mutants Dominantly Enforce Stem Cell Programs and Impair Human Hematopoietic Progenitor Differentiation. Cell Stem Cell. 2015;17:675–688. doi: 10.1016/j.stem.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galeev R, et al. Genome-wide RNAi Screen Identifies Cohesin Genes as Modifiers of Renewal and Differentiation in Human HSCs. Cell Reports. 2016;14:2988–3000. doi: 10.1016/j.celrep.2016.02.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fisher JB, et al. The cohesin subunit Rad21 is a negative regulator of hematopoietic self-renewal through epigenetic repression of Hoxa7 and Hoxa9. Leukemia. 2016 doi: 10.1038/leu.2016.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krönke J, et al. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. 2013;122:100–108. doi: 10.1182/blood-2013-01-479188. [DOI] [PubMed] [Google Scholar]
- 22.Peters JM, Nishiyama T. Sister chromatid cohesion. Cold Spring Harbor Perspectives in Biology. 2012;4:a011130–a011130. doi: 10.1101/cshperspect.a011130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peters J-M, et al. The cohesin complex and its roles in chromosome biology. Genes Dev. 2008;22:3089–3114. doi: 10.1101/gad.1724308. [DOI] [PubMed] [Google Scholar]
- 24.Kon A, et al. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 25.Faber ZJ, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48:1551–1556. doi: 10.1038/ng.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duployez N, et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127:2451–2459. doi: 10.1182/blood-2015-12-688705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun QY, et al. Ordering of mutations in acute myeloid leukemia with partial tandem duplication of MLL (MLL-PTD) Leukemia. 2017;31:1–10. doi: 10.1038/leu.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 30.Grisendi S, et al. Role of nucleophosmin in embryonic development and tumorigenesis. Nature. 2005;437:147–153. doi: 10.1038/nature03915. [DOI] [PubMed] [Google Scholar]
- 31.Vassiliou GS, et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat Genet. 2011;43:470–475. doi: 10.1038/ng.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Challen GA, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauvageau G, et al. Differential expression of homeobox genes in functionally distinct CD34+ subpopulations of human bone marrow cells. Proc Natl Acad Sci USA. 1994;91:12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kroon E, et al. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorsteinsdottir U, et al. Overexpression of the myeloid leukemia-associated Hoxa9 gene in bone marrow cells induces stem cell expansion. Blood. 2002;99:121–129. doi: 10.1182/blood.v99.1.121. [DOI] [PubMed] [Google Scholar]
- 36.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seitan VC, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Research. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu M, et al. CTCF controls HOXA cluster silencing and mediates PRC2-repressive higher-order chromatin structure in NT2/D1 cells. Molecular and Cellular Biology. 2014;34:3867–3879. doi: 10.1128/MCB.00567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim YJ, et al. Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proceedings of the National Academy of Sciences. 2011;108:7391–7396. doi: 10.1073/pnas.1018279108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuin J, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proceedings of the National Academy of Sciences. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khan I, et al. Myeloid Leukemia in Down Syndrome. Crit Rev Oncog. 2011;16:25–36. doi: 10.1615/critrevoncog.v16.i1-2.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woo AJ, et al. Developmental differences in IFN signaling affect GATA1s-induced megakaryocyte hyperproliferation. J Clin Invest. 2013 doi: 10.1172/JCI40609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Malinge S, et al. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J Clin Invest. 2012;122:948–962. doi: 10.1172/JCI60455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wendt KS, et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- 47.Ong CT, Corces VG. CTCF: an architectural protein bridginggenome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horsfield JA, et al. Cohesin-dependent regulation of Runx genes. Development. 2007;134:2639–2649. doi: 10.1242/dev.002485. [DOI] [PubMed] [Google Scholar]
- 49.Marsman J, et al. Cohesin and CTCF differentially regulate spatiotemporal runx1 expression during zebrafish development. Biochim Biophys Acta. 2014;1839:50–61. doi: 10.1016/j.bbagrm.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Rainis L, et al. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Research. 2005;65:7596–7602. doi: 10.1158/0008-5472.CAN-05-0147. [DOI] [PubMed] [Google Scholar]
- 51.Salek-Ardakani S, et al. ERG is a megakaryocytic oncogene. Cancer Research. 2009;69:4665–4673. doi: 10.1158/0008-5472.CAN-09-0075. [DOI] [PubMed] [Google Scholar]
- 52.Wechsler J, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet. 2002;32:148–152. doi: 10.1038/ng955. [DOI] [PubMed] [Google Scholar]
- 53.Chlon TM, et al. Global transcriptome and chromatin occupancy analysis reveal the short isoform of GATA1 is deficient for erythroid specification and gene expression. Haematologica. 2015;100:575–584. doi: 10.3324/haematol.2014.112714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood. 2009;113:3337–3347. doi: 10.1182/blood-2008-08-174813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seitan VC, et al. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 2011;476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noordermeer D, et al. The dynamic architecture of Hox gene clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 57.Soshnikova N, Duboule D. Epigenetic Temporal Control of Mouse Hox Genes in Vivo. Science. 2009;324:1320–1323. doi: 10.1126/science.1171468. [DOI] [PubMed] [Google Scholar]
- 58.Wouters BJ, Delwel R. Epigenetics and approaches to targeted epigenetic therapy in acute myeloid leukemia. Blood. 2016;127:42–52. doi: 10.1182/blood-2015-07-604512. [DOI] [PubMed] [Google Scholar]
- 59.Bernt KM, et al. MLL-Rearranged Leukemia Is Dependent on Aberrant H3K79 Methylation by DOT1L. Cancer Cell. 2011;20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daigle SR, et al. Selective Killing of Mixed Lineage Leukemia Cells by a Potent Small-Molecule DOT1L Inhibitor. Cancer Cell. 2011;20:53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michaelis C, et al. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 62.Guacci V, et al. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Losada A, et al. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tóth A, et al. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Merkenschlager M, Nora EP. CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annu Rev Genom Human Genet. 2016;17:17–43. doi: 10.1146/annurev-genom-083115-022339. [DOI] [PubMed] [Google Scholar]
- 66.Revenkova E, et al. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat Cell Biol. 2004;6:555–562. doi: 10.1038/ncb1135. [DOI] [PubMed] [Google Scholar]
- 67.Mannini L, et al. SMC1B is present in mammalian somatic cells and interacts with mitotic cohesin proteins. Sci Rep. 2015;5:18472. doi: 10.1038/srep18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krantz ID, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36:631–635. doi: 10.1038/ng1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tonkin ET, et al. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004;36:636–641. doi: 10.1038/ng1363. [DOI] [PubMed] [Google Scholar]
- 70.Vega H, et al. Roberts syndrome is caused by mutations in ESCO2, a human homolog of yeast ECO1 that is essential for the establishment of sister chromatid cohesion. Nat Genet. 2005;37:468–470. doi: 10.1038/ng1548. [DOI] [PubMed] [Google Scholar]
- 71.Stedman W, et al. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deardorff MA, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 2012;489:313–317. doi: 10.1038/nature11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boyle MI, et al. Cornelia de Lange syndrome. Clinical Genetics. 2015;88:1–12. doi: 10.1111/cge.12499. [DOI] [PubMed] [Google Scholar]
- 74.Kaur M, et al. Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet. 2005;138:27–31. doi: 10.1002/ajmg.a.30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cucco F, Musio A. Genome stability: What we have learned from cohesinopathies. Am J Med Genet. 2016;172:171–178. doi: 10.1002/ajmg.c.31492. [DOI] [PubMed] [Google Scholar]
- 76.Schrier SA, et al. Causes of death and autopsy findings in a large study cohort of individuals with Cornelia de Lange syndrome and review of the literature. Am J Med Genet. 2011;155:3007–3024. doi: 10.1002/ajmg.a.34329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barber TD, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proceedings of the National Academy of Sciences. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Solomon DA, et al. Mutational Inactivation of STAG2 Causes Aneuploidy in Human Cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Balbás-Martínez C, et al. Recurrent inactivation of STAG2 in bladder cancer is not associated with aneuploidy. Nat Genet. 2013;45:1464–1469. doi: 10.1038/ng.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JS, et al. Intact Cohesion, Anaphase, and Chromosome Segregation in Human Cells Harboring Tumor-Derived Mutations in STAG2. PLoS Genet. 2016;12:e1005865–18. doi: 10.1371/journal.pgen.1005865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Porkka KP, et al. RAD21 and KIAA0196 at 8q24 are amplified and overexpressed in prostate cancer. Genes Chromosomes Cancer. 2004;39:1–10. doi: 10.1002/gcc.10289. [DOI] [PubMed] [Google Scholar]
- 82.Xu H, et al. Enhanced RAD21 cohesin expression confers poor prognosis and resistance to chemotherapy in high grade luminal, basal and HER2 breast cancers. Breast Cancer Res. 2011;13:R9. doi: 10.1186/bcr2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan M, et al. Enhanced RAD21 cohesin expression confers poor prognosis in BRCA2 and BRCAX, but not BRCA1 familial breast cancers. Breast Cancer Res. 2012;14:R69. doi: 10.1186/bcr3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deb S, et al. RAD21 cohesin overexpression is a prognostic and predictive marker exacerbating poor prognosis in KRAS mutant colorectal carcinomas. Br J Cancer. 2014;110:1606–1613. doi: 10.1038/bjc.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang J, et al. Role of SMC1A overexpression as a predictor of poor prognosis in late stage colorectal cancer. BMC Cancer. 2015;15:90. doi: 10.1186/s12885-015-1085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oikawa K, et al. Expression of a novel human gene, human wings apart-like (hWAPL), is associated with cervical carcinogenesis and tumor progression. Cancer Research. 2004;64:3545–3549. doi: 10.1158/0008-5472.CAN-03-3822. [DOI] [PubMed] [Google Scholar]