Abstract
Background
Despite advances in combined modality therapy, outcomes in head and neck squamous cell cancer (HNSCC) remain dismal with five-year overall survival rates of less than 50%. Prognostic biomarkers are urgently needed to identify patients with a high risk of death after initial curative treatment. Methylation status of the paired-like homeodomain transcription factor 2 (PITX2) has recently emerged as a powerful prognostic biomarker in various cancers. In the present study, the clinical performance of PITX2 methylation was validated in a HNSCC cohort by means of an independent analytical platform (Infinium HumanMethylation450 BeadChip, Illumina, Inc.).
Methods
A total of 528 HNSCC patients from The Cancer Genome Atlas (TCGA) were included in the study. Death was defined as primary endpoint. PITX2 methylation was correlated with overall survival and clinicopathological parameters.
Results
PITX2 methylation was significantly associated with sex, tumor site, p16 status, and grade. In univariate Cox proportional hazards analysis, PITX2 hypermethylation analyzed as continuous and dichotomized variable was significantly associated with prolonged overall survival of HNSCC patients (continuous: hazard ratio (HR) = 0.19 [95%CI: 0.04–0.88], p = 0.034; dichotomized: HR = 0.52 [95%CI: 0.33–0.84], p = 0.007). In multivariate Cox analysis including established clinicopathological parameters, PITX2 promoter methylation was confirmed as prognostic factor (HR = 0.28 [95%CI: 0.09–0.84], p = 0.023).
Conclusion
Using an independent analytical platform, PITX2 methylation was validated as a prognostic biomarker in HNSCC patients, identifying patients that potentially benefit from intensified surveillance and/or administration of adjuvant/neodjuvant treatment, i.e. immunotherapy.
Introduction
With an annual incidence of approximately 61,760 cases and 13,190 estimated deaths, head and neck squamous cell carcinoma (HNSCC) is ranked among the leading causes of cancer-related death in the United States [1]. While tobacco and alcohol abuse traditionally are the primary risk factors for HNSCC, a subset of tumors with oropharyngeal location is strongly associated with high risk human papilloma virus (HPV) infections [2]. HPV-positive (HPV+) cancers are more responsive to chemotherapy and radiation, and patients have shown improved overall survival rates compared to HPV-negative (HPV-) cancer patients [3]. The underlying molecular mechanisms for this clinical observation, however, are not fully understood. In a first comprehensive study of genomic alterations in 279 HNSCC patients, smoking-related tumors frequently harbored loss of function mutations of TP53 and CDKN2A in addition to multiple copy number alterations, whereas HPV-associated tumors were predominantly characterized by activating PIK3CA mutations [4]. Mechanisms explaining the differing treatment response of HPV+ and HPV- HNSCC, however, still need to be elucidated, particularly in the light of emerging alternative HNSCC treatment concepts.
Despite initial treatment with curative intent, recurrence rates in HNSCC patients remain high leading to local or distant recurrences in 30% and 25%, respectively, and five-year overall survival rates of less than 50% [5–6]. Reliable prognostic biomarkers are urgently needed to identify patients at risk of disease recurrence and subsequent death, as these patients might beefit from an intensified first-line treatment and surveillance. Current treatment regimens, including surgery, definite or adjuvant radiochemotherapy, elicit a number of severe side effects leaving little room for treatment intensification [7–8]. Emerging therapeutic strategies, e.g. anti-PD-1 antibodies pembrolizumab and nivolumab, which have recently been approved for the treatment of advanced and metastatic HNSCC, are promising options for the management of high risk patients [9–10]. In 2006, the anti-EGFR monoclonal antibody cetuximab was further approved for the treatment of localized disease in combination with radiotherapy and as a single agent in metastatic and recurrent cancers [11]. Particularly for high-risk patients, employing cetuximab in an adjuvant setting has been shown to be feasible [12]. Identifying high-risk patients, however, remains a challenge for both the surgical pathologist and the clinician. So far, only few prognostic biomarkers have shown promising potential. The detection of HPV in tumor tissue, for instance, has been reported as a strong prognostic biomarker predicting reduced risk of death [13]. In a study of 141 HNSCC patient samples, truncating mutations of TP53 resulting in loss of function of the tumor suppressor gene were further associated with worse overall survival (HR = 2.54, p = 0.008) [14]. The investigation of DNA promoter methylation to identify biomarkers in formalin-fixed paraffin-embedded (FFPE) tissue has shown encouraging results. In brief, epigenetic changes are mediated through binding of a methyl- or hydroxymethyl-group to a cytosine-phosphate-guanin (CpG)-dinucleotide. Clustered CpG-dinucleotides, CpG-islands, are found in promoter regions and their methylation or demethylation modulates gene activity [15].
Previously, the methylation status of the paired-like homeobox transcription factor 2 (PITX2) and its adjacent long non-coding RNA (PANCR) have been described as promising tissue-based biomarkers in HNSCC, and PITX2 hypermethylation has been associated with better overall survival in HNSCC patients [16]. Mutation of the PITX2 gene, located on chromosome 4q25, causes the developmental disorder Axenfeld-Rieger syndrome type I, which is characterized by ophtalmological and cardiovascular abnormalities and dental hypoplasia [17–18]. In recent years, PITX2 has attracted interest as a potential biomarker in malignant tumors, i.e. non-small cell lung cancer (NSCLC), biliary tract cancers, prostate cancer, and breast cancer [16, 19–27]. In NSCLC, PITX2 hypermethylation was identified as significant predictor of progression-free survival [28]. In hormone receptor-positive, lymph node-negative breast cancer, high levels of PITX2 methylation were, in contrast, associated with a high risk of recurrence [25–27]. In addition, PITX2 methylation has been shown to be predictive of response to adjuvant anthracycline-based chemotherapy in node-positive, hormone receptor-positive breast cancer patients [24]. Therawis GmbH (Munich, Germany) in collaboration with Qiagen N.V. (Venlo, The Netherlands) have recently announced the development of a commercially available assay exploiting the additional predictive potential of PITX2 methylation in breast cancer patients. In prostate cancer, PITX2 hypermethylation has been associated with biochemical recurrence, contributing to individualized risk assessment as a single assay and in combination with PITX3 methylation analysis [20, 22–23, 29]. In addition to its known role in the development of several organs, PITX2 also mediates cell cycle progression by regulating the transcription of cyclin A1, cyclin D2, and p21, therefore providing sufficient evidence to implicate PITX2 in tumorigenesis [30–32]. Moreover, PITX2 is over-expressed on a protein level in a number of solid tumors and has been associated with cancer progression [33]. Several studies have demonstrated that PITX2 methylation can be quantified accurately and robustly in various clinically relevant specimens, i.e. FFPE biopsies, FFPE sections, and microdissected cells from FFPE sections [28, 34–37]. Hence, PITX2 methylation is an ideal candidate for the implementation into a clinical routine setting.
The Cancer Genome Atlas (TCGA) has provided an unprecedented platform for researchers to study genomic, transcriptomic, proteomic, and methylomic data across a multitude of different cancer types [38]. Given the need for biomarkers for treatment stratification in HNSCC patients and the encouraging results regarding PITX2, this study aimed at validating the clinical performance of PITX2 methylation for the outcome prediction of HNSCC patients by means of an independent analytical platform, i.e. Infinium HumanMethylation450 BeadChip, in the TCGA HNSCC patient cohort.
Materials and methods
Ethical approval
The present study is based entirely upon data generated by the TCGA research network (www.cancergenome.nih.gov). All patients included in TCGA have been enrolled following strict human subjects protection guidelines, informed consent and IRB (Institutional Review Board) review of protocols. All patients provided informed consent (written).
Patients
All head and neck squamous cell carcinoma patients (n = 528) from the TCGA cohort (Project Id: TCGA-HNSC) were included into the present study (https://portal.gdc.cancer.gov/projects/TCGA-HNSC). Survival was defined as time to death by any cause (overall survival, OS) and censored after five years (1,825 days).
Statistical analyses
Gene methylation data were publicly available and downloaded from the UCSC Xena browser (www.xena.ucsc.edu). Methylation analysis was performed using the Infinium HumanMethylation450 BeadChip Kit (Illumina, Inc., San Diego, CA, USA). Methylation levels were calculated as previously described [39–41]. For statistical analysis, SPSS version 24 (SPSS Inc., Chicago, IL, USA) was used. Survival analyses were conducted by Kaplan-Meier and Cox proportional hazard regression analyses. P-values refer to Wald and log-rank tests, respectively. The correlation between age and methylation was tested using the Spearman’s rank correlation coefficient (ρ). For comparison between groups, Mann-Whitney U test, one-way analysis of variance (one-way ANOVA), and Fisher’s exact test were employed. P-values lower than 0.05 were considered significant.
Results
PITX2 promoter methylation in HNSCC patients
Gene methylation data from 528 patients with histologically confirmed HNSCC were available, including tumor tissues in all cases and matched normal adjacent tissues in 50 cases (9.5%). Histological tumor subtypes comprised 517 (97.9%) squamous cell cancers of the usual type, ten (1.9%) cases of the basaloid type, and one (0.2%) spindle cell variant.
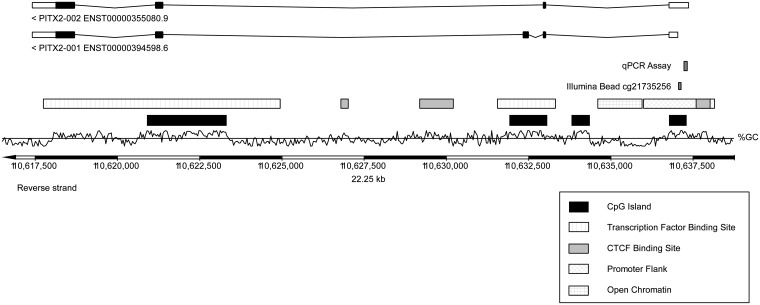
A previous report employing quantitative real-time PCR (qPCR) assays investigated four CpG-dinucleotides within the sequence context CGGGAGCCGGAGCCGGGAGAGCG located on chromosome 4:110637267–110637289 (Ensembl.org genome assembly: GRCh38.p10) [16]. In the current TCGA dataset, the Illumina HumanMethylation450 BeadChip bead cg21735256 was analyzed. This bead is located in the vicinity (approximately 200 base pairs distance) of the previously published CpG-dinucleotides and belongs to the same CpG-island (Fig 1). Accordingly, methylation of the cg21735256 bead was assumed to have the same transcriptional consequences for the PITX2 gene. PITX2 DNA methylation ranged from 2.1% to 67.5% (mean: 13.0%, median: 8.2%) in tumor tissue and from 4.5% to 14.4% (mean: 8%, median: 7.5%) in normal tissue. A significant difference in methylation levels was detected when comparing tumor and normal tissue (p = 0.003).
Fig 1. Organization of the PITX2 gene.
Genomic organization of the PITX2 gene and locations of the PITX2 qPCR assay [16] and the Illumina HumanMethylation450 BeadChip bead cg21735256. The information was taken from Ensembl Homo sapiens version GRCh38.p7.
Association of PITX2 promoter methylation with clinicopathological parameters
Significant differences in PITX2 methylation status were found between male and female patients in the TCGA HNSCC cohort (p = 0.026). This may in part be explained by a difference in smoking habits with female patients being mainly non-smokers (Fisher’s exact test, p < 0.001). No difference in pack years was seen in the patients who reported a history of smoking (p = 0.35). Further, no gender difference was observed regarding HPV-status (p = 0.15). However, HPV-status was only reported for a fifth (19.9%) of all samples, so this result may well be skewed. Nonetheless, there was a difference in PITX2 DNA methylation in HPV- versus HPV+ patient samples with the latter patient group showing significantly higher methylation levels (p < 0.001). This finding was in concordance with previously reported data [16]. Additionally, HPV+ tumors were more frequently located in the oropharynx, and oroparyngeal cancers presented with significantly higher methylation levels compared to tumors from other sites (p < 0.001) [42]. Poorly differentiated and undifferentiated cancers had significantly higher methylation levels compared to well or moderately differentiated tumors (p < 0.001). No difference in PITX2 methylation was found for age, smoking status, and number of pack years (analyzing males and females together), history of alcohol consumption, tumor (T) and nodal (N) categories, race, and surgical margin status (Table 1).
Table 1. Association of PITX2 methylation with clinicopathological characteristics.
Association of PITX2 methylation with clinicopathological characteristics in HNSCC patients of the TCGA cohort (n = 528).
| Characteristic | No. [%] of patients | Mean PITX2 methylation [mean ± standard deviation) | p-value | |
|---|---|---|---|---|
| All patients | 528 (100) | 12.7 (±10.9) | ||
| Sex | 0.026* | |||
| Female | 142 (26.9) | 10.9 (±8.1) | ||
| Male | 386 (73.1) | 13.3 (±11.7) | ||
| Age (years) | 0.65 | |||
| Mean | 61 | |||
| Median | 61 | |||
| n ≤ Median | 282 (53.4) | 13.0 (±11.6) | ||
| n > Median | 245 (46.4) | 12.3 (±10.2) | ||
| Unknown | 1 (0.2) | |||
| Smoking status | 0.61 | |||
| Non-Smoker | 122 (23.1) | 13.2 (±11.6) | ||
| Smoker | 393 (74.4) | 12.6 (±10.8) | ||
| Unknown | 13 (2.5) | |||
| Pack years | 0.18 | |||
| (≤ 40) | 168 (31.8) | 13.5 (±11.5) | ||
| (> 40) | 130 (24.6) | 11.7 (±10.2) | ||
| Unknown | 230 (43.6) | |||
| History of alcohol consumption | 0.99 | |||
| Yes | 352 (66.7) | 12.7 (±11) | ||
| No | 165 (31.3) | 12.7 (±11) | ||
| Unknown | 11 (2.1) | |||
| Tumor site | <0.001* | |||
| Oral cavity | 250 (47.3) | 9.9 (±7.6) | ||
| Oropharynx | 151 (28.6) | 16.5 (±14.4) | ||
| Hypopharynx | 10 (1.9) | 14.4 (±6.6) | ||
| Larynx | 177 (22.2) | 13.7 (±9.5) | ||
| Race | 0.66 | |||
| White | 452 (85.6) | 13 (±11.3) | ||
| Black or African American | 48 (9.1) | 10.9 (±7.9) | ||
| Asian | 11 (2.1) | 11.7 (±9.3) | ||
| American Indian or Alaska Native | 2 (0.4) | 17.8 (±13) | ||
| Unknown | 15 (2.78) | |||
| Pathologic tumor category (pT) | 0.15 | |||
| Tis/T1/T2 | 190 (36) | 12.7 (±10.4) | ||
| T3/T4 | 276 (52.3) | 11.3 (±9.8) | ||
| Unknown | 62 (11.7) | |||
| Pathologic nodal category (pN) | 0.42 | |||
| N0 | 180 (34.1) | 12.1 (±9.7) | ||
| N1 | 68 (12.9) | 10.8 (±8) | ||
| N2 | 172 (32.6) | 11.0 (±9.5) | ||
| Unknown | 108 (20.5) | |||
| p16 | <0.001* | |||
| Negative | 74 (14) | 9.7 (±6.5) | ||
| Positive | 41 (7.8) | 25.7 (±16.2) | ||
| Unknown | 413 (78.2) | |||
| Grade (G) | <0.001* | |||
| G1 | 63 (11.9) | 9.8 (±6.3) | ||
| G2 | 311 (58.9) | 11.3 (±9.2) | ||
| G3 | 125 (23.7) | 14.9 (±13.3) | ||
| G4 | 7 (1.3) | 28.5 (±11.1) | ||
| Unknown | 22 (4.2) | |||
| 0.003* | ||||
| G1 and G2 | 374 (70.8) | 11.1 (±8.8) | ||
| G3/G4 | 132 (25) | 15.6 (±13.5) | ||
| Surgical margin | 0.83 | |||
| Negative | 407 (77.1) | 11.7 (±9.8) | ||
| Positive | 60 (11.4) | 12.0 (±11.1) | ||
| Unknown | 61 (11.6) | |||
Mann-Whitney U test for sex, smoking status, history of alcohol consumption, pT, p16, grade (dichotomized), surgical margin; One-Way ANOVA for tumor site, pN, grade, race; Spearman’s rank correlation for age, pack years.
* significant feature
PITX2 methylation status as independent prognostic factor for overall survival
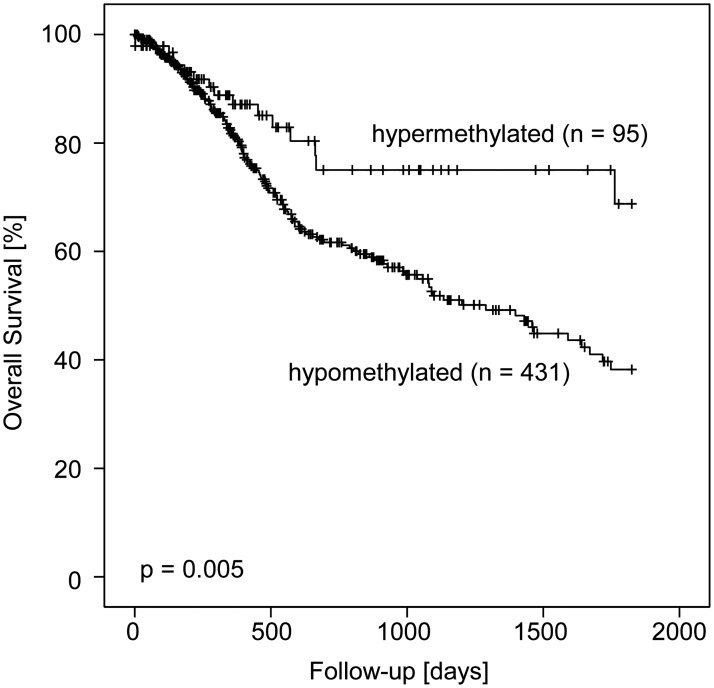
In order to avoid overfitted results, Cox regression analysis was performed using PITX2 methylation as a continuous variable. Univariate analysis revealed a significantly reduced risk of death for patients with hypermethylated tumors (hazard ratio (HR) = 0.19 [95%CI: 0.04–0.88], p = 0.034). For the dichotomization of DNA methylation values, patients were stratified according to an optimized cut-off (20.3%) using a publicly available cut-off calculator [43]. As a result, 433 (82.0%) tumor samples were assigned to the PITX2 hypomethylated group, and 95 (18.0%) specimens were allocated to the PITX2 hypermethylated group. In univariate Cox proportional hazards analysis, patients with hypermethylated tumors had a significantly reduced risk of death compared to patients with hypomethylated tumors (HR = 0.52 [95%CI: 0.33–0.84], p = 0.007). These findings were further confirmed in Kaplan-Meier survival analysis (p = 0.005, Fig 2). In multivariate analysis, PITX2 methylation added significant prognostic information to established clinicopathological parameters, i.e. age, T and N category (Table 2). Although poorly differentiated tumors had significantly higher methylation levels in our study, histological tumor grading did not add further independent information in survival analyses.
Fig 2. Kaplan-Meier survival analysis.
Kaplan-Meier survival analysis of overall survival in 528 HNSCC patients stratified according to PITX2 DNA methylation status. Overall survival in patients with PITX2 hypermethylated HNSCC was significantly improved compared to patients with PITX2 hypomethylated tumors.
Table 2. Univariate and multivariate Cox proportional hazards analyses.
Univariate and multivariate Cox proportional hazards analyses on overall survival in 528 HNSCC patients. Multivariate Cox proportional hazard analysis was conducted including only variables that showed significance in univariate analysis (PITX2 methylation [dichotomized variable; cut off: 20.3%], T category, N category, age).
| Variable | Univariate Cox | Multivariate Cox | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| PITX2 methylation (continuous) | 0.19 (0.04–0.88) | 0.034* | NA | NA |
| PITX2 methylation (dichotomized) | 0.52 (0.33–0.84) | 0.007* | 0.28 (0.09–0.84) | 0.023* |
| pT3/4 vs. pT1/2 | 1.57 (1.14–2.17) | 0.004* | 1.84 (1.20–2.84) | 0.006* |
| pN1/2 vs. pN0 | 1.62 (1.12–2.35) | 0.010* | 1.50 (1.01–2.20) | 0.044* |
| Age (continuous variable) | 1.02 (1.01–1.03) | 0.003* | 1.02 (1.00–1.04) | 0.044* |
| p16 (positive vs. negative) | 0.66 (0.18–2.48) | 0.54 | NA | NA |
| Grade (G3,G4 vs. G1,G2) | 0.87 (0.70–1.07) | 0.18 | NA | NA |
| Surgical margin (positive vs. negative) | 1.45 (0.97–2.16) | 0.17 | NA | NA |
NA: Not applicable, variate not included into multivariate analysis
* significant feature
Discussion
HNSCC is a common cancer type with a dismal prognosis in the event of recurrent or metastatic disease. Consequently, there is a pressing need for reliable prognostic biomarkers which might aid the risk stratification of patients and clinical decision-making process with regard to primary treatment and subsequent surveillance. Methylation analysis of PITX2 has recently been shown to predict overall survival in HNSCC patients [16]. Encouraged by these data, we sought to investigate PITX2 methylation in an independent HNSCC cohort by means of an additional innovative technology.
In the present study, PITX2 DNA hypermethylation was associated with improved overall survival, thereby validating our previous results in the TCGA dataset. Also in line with prior results, PITX2 methylation further added independent prognostic information to established clinicopathological parameters like age, T and N category. Combining both studies, there is now strong evidence for the high analytical performance and prognostic value of PITX2 methylation in almost one thousand HNSCC patients. Implementing PITX2 methylation into prospective clinical trials is therefore highly desirable to further support these findings. The robust prognostic performance of PITX2 as biomarker in HNSCC is further corroborated, since different CpG-sites were analyzed in the present study, hereby demonstrating that the prognostic value is not only limited to the previously published CpG-sites [16]. In earlier publications, PITX2 testing was conducted using qPCR and DNA microarray methodologies [34–35]. The employment of methylation data generated by means of the Illumina HumanMethylation450 BeadChip poses an additional strength of the present study, as the application of different diagnostic platforms not only increases the value of a biomarker but also allows for an area-wide implementation of the biomarker into clinical routine.
The biological function of PITX2 in tumorigenesis has been linked to its role as key player in cell cycle regulation. Hypermethylation is assumed to result in silencing of the PITX2 gene, therefore disrupting cell cycle progression and proliferation. The same phenomenon has been shown for NSCLC [28], a tumor which, like HNSCC, is known for its strong association with a history of smoking. Interestingly, in hormonally dependent tumors like breast and prostate cancer, PITX2 DNA hypermethylation has been associated with adverse overall survival [25–27, 29]. This might point at a role of PITX2 in tumorigenesis which lies beyond cell cycle regulation. However, it remains unclear how methylation actually affects gene transcription. Wang and colleagues could not identify a correlation between low PITX2 protein expression and methylation in pancreatic ductal adenocarcinoma but rather suggested that PITX2 acts as a tumor suppressor by inhibiting the TGFβ1-Smad4-pathway [44]. In embryonic development, isoforms of PITX2 exert various functions in different organs, thus possibly providing an explanation for its diverse and sometimes opposing role in tumorigenesis [45].
In HNSCC, PITX2 DNA-methylation is significantly associated with HPV status and tumor site. This underscores that HPV-associated tumors are molecularly distinct from smoking-associated tumors. It has also been established by now that integration of HPV into the genome might result in altered DNA methylation [46–47]. Thus, it cannot be ruled out that PITX2 DNA methylation acts as a surrogate marker for HPV infection. Neither HPV status nor the tumor site was associated with survival in the present study. Information on HPV status, however, was missing for the majority of the cohort. Thus, further studies are needed to investigate the association of PITX2 methylation and HPV.
Three monoclonal antibodies targeting either EGFR (cetuximab) or PD-1 (pembrolizumab, nivolumab) are currently available for the treatment of HNSCC patients. Only very recently, the immune checkpoint inhibitors pembrolizumab and nivolumab have been approved for patients in a recurrent or metastatic disease setting. However, they might potentially be a therapeutic option for high-risk patients with tumors harboring PITX2 hypomethylation as part of a neoadjuvant or adjuvant therapeutic regimen. With regard to the findings in the present study, evidence is mounting that PITX2 DNA methylation might serve as highly informative prognostic biomarker in HNSCC patients. A prospective randomized trial stratifying patients according to classical and molecular risk factors, however, would need to confirm the predictive value of PITX2 DNA methylation status in HNSCC patients.
Data Availability
All relevant data are within the paper or can be obtained from The Cancer Genome Atlas (TCGA) Research Network (www.cancergenome.nih.gov): https://portal.gdc.cancer.gov/projects/TCGA-HNSC.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Cohen EE, LaMonte SJ, Erb NL, Beckman KL, Sadeghi N, Hutcheson KA, et al. American Cancer Society Head and Neck Cancer Survivorship Care Guideline. CA Cancer J Clin. 2016;66(3):203–39. doi: 10.3322/caac.21343 [DOI] [PubMed] [Google Scholar]
- 2.Sturgis EM, Cinciripini PM. Trends in head and neck cancer incidence in relation to smoking prevalence: an emerging epidemic of human papillomavirus-associated cancers? Cancer. 2007;110(7):1429–35. doi: 10.1002/cncr.22963 [DOI] [PubMed] [Google Scholar]
- 3.Lechner M, Fenton TR. The Genomics, Epigenomics, and Transcriptomics of HPV-Associated Oropharyngeal Cancer—Understanding the Basis of a Rapidly Evolving Disease. Adv Genet. 2016;93:1–56. doi: 10.1016/bs.adgen.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 4.Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi: 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper JS, Pajak TF, Forastiere AA, Jacobs J, Campbell BH, Saxman SB, et al. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N Engl J Med. 2004;350(19):1937–44. doi: 10.1056/NEJMoa032646 [DOI] [PubMed] [Google Scholar]
- 6.Laramore GE, Scott CB, al-Sarraf M, Haselow RE, Ervin TJ, Wheeler R, et al. Adjuvant chemotherapy for resectable squamous cell carcinomas of the head and neck: report on Intergroup Study 0034. Int J Radiat Oncol Biol Phys. 1992;23(4):705–13. doi: 10.1016/0360-3016(92)90664-4 [DOI] [PubMed] [Google Scholar]
- 7.Bernier J, Domenge C, Ozsahin M, Matuszewska K, Lefebvre JL, Greiner RH, et al. Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N Engl J Med. 2004;350(19):1945–52. doi: 10.1056/NEJMoa032641 [DOI] [PubMed] [Google Scholar]
- 8.Mallen-St Clair J, Alani M, Wang MB, Srivatsan ES. Human papillomavirus in oropharyngeal cancer: The changing face of a disease. Biochim Biophys Acta. 2016;1866(2):141–50. doi: 10.1016/j.bbcan.2016.07.005 [DOI] [PubMed] [Google Scholar]
- 9.Busch CJ, Laban S, Knecht R, Hoffmann TK. [Immunotherapeutic studies of head and neck tumors: Highlights of the 2016 ASCO Annual Meeting]. HNO. 2016;64(10):708–16. doi: 10.1007/s00106-016-0238-3 [DOI] [PubMed] [Google Scholar]
- 10.Chow LQ, Haddad R, Gupta S, Mahipal A, Mehra R, Tahara M, et al. Antitumor Activity of Pembrolizumab in Biomarker-Unselected Patients With Recurrent and/or Metastatic Head and Neck Squamous Cell Carcinoma: Results From the Phase Ib KEYNOTE-012 Expansion Cohort. J Clin Oncol. 2016. doi: 10.1200/JCO.2016.68.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rancoule C, Vallard A, Espenel S, Guy JB, Xia Y, El Meddeb Hamrouni A, et al. Immunotherapy in head and neck cancer: Harnessing profit on a system disruption. Oral Oncol. 2016;62:153–62. doi: 10.1016/j.oraloncology.2016.09.002 [DOI] [PubMed] [Google Scholar]
- 12.Gliese A, Busch CJ, Knecht R. [Study results of primary therapy for head and neck tumors: Highlights of the 2016 ASCO Annual Meeting]. HNO. 2016;64(10):717–22. doi: 10.1007/s00106-016-0243-6 [DOI] [PubMed] [Google Scholar]
- 13.Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol. 2015;12(1):11–26. doi: 10.1038/nrclinonc.2014.192 [DOI] [PubMed] [Google Scholar]
- 14.Lindenbergh-van der Plas M, Brakenhoff RH, Kuik DJ, Buijze M, Bloemena E, Snijders PJ, et al. Prognostic significance of truncating TP53 mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2011;17(11):3733–41. doi: 10.1158/1078-0432.CCR-11-0183 [DOI] [PubMed] [Google Scholar]
- 15.Leenen FA, Muller CP, Turner JD. DNA methylation: conducting the orchestra from exposure to phenotype? Clin Epigenetics. 2016;8:92 doi: 10.1186/s13148-016-0256-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sailer V, Holmes EE, Gevensleben H, Goltz D, Droge F, de Vos L, et al. PITX2 and PANCR DNA methylation predicts overall survival in patients with head and neck squamous cell carcinoma. Oncotarget. 2016. doi: 10.18632/oncotarget.12417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang TC, Summers CG, Schimmenti LA, Grajewski AL. Axenfeld-Rieger syndrome: new perspectives. Br J Ophthalmol. 2012;96(3):318–22. doi: 10.1136/bjophthalmol-2011-300801 [DOI] [PubMed] [Google Scholar]
- 18.Semina EV, Reiter R, Leysens NJ, Alward WL, Small KW, Datson NA, et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14(4):392–9. doi: 10.1038/ng1296-392 [DOI] [PubMed] [Google Scholar]
- 19.Uhl B, Dietrich D, Branchi V, Semaan A, Schaefer P, Gevensleben H, et al. DNA Methylation of PITX2 and PANCR Is Prognostic for Overall Survival in Patients with Resected Adenocarcinomas of the Biliary Tract. PloS one. 2016;11(10):e0165769 doi: 10.1371/journal.pone.0165769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes EE, Goltz D, Sailer V, Jung M, Meller S, Uhl B, et al. PITX3 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients after radical prostatectomy. Clin Epigenetics. 2016;8:104 doi: 10.1186/s13148-016-0270-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinarskaja A, Schulz WA, Ingenwerth M, Hader C, Arsov C. Association of PITX2 mRNA down-regulation in prostate cancer with promoter hypermethylation and poor prognosis. Urol Oncol. 2013;31(5):622–7. doi: 10.1016/j.urolonc.2011.04.010 [DOI] [PubMed] [Google Scholar]
- 22.Banez LL, Sun L, van Leenders GJ, Wheeler TM, Bangma CH, Freedland SJ, et al. Multicenter clinical validation of PITX2 methylation as a prostate specific antigen recurrence predictor in patients with post-radical prostatectomy prostate cancer. J Urol. 2010;184(1):149–56. doi: 10.1016/j.juro.2010.03.012 [DOI] [PubMed] [Google Scholar]
- 23.Weiss G, Cottrell S, Distler J, Schatz P, Kristiansen G, Ittmann M, et al. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol. 2009;181(4):1678–85. doi: 10.1016/j.juro.2008.11.120 [DOI] [PubMed] [Google Scholar]
- 24.Hartmann O, Spyratos F, Harbeck N, Dietrich D, Fassbender A, Schmitt M, et al. DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res. 2009;15(1):315–23. doi: 10.1158/1078-0432.CCR-08-0166 [DOI] [PubMed] [Google Scholar]
- 25.Harbeck N, Nimmrich I, Hartmann A, Ross JS, Cufer T, Grutzmann R, et al. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008;26(31):5036–42. doi: 10.1200/JCO.2007.14.1697 [DOI] [PubMed] [Google Scholar]
- 26.Nimmrich I, Sieuwerts AM, Meijer-van Gelder ME, Schwope I, Bolt-de Vries J, Harbeck N, et al. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast cancer research and treatment. 2008;111(3):429–37. doi: 10.1007/s10549-007-9800-8 [DOI] [PubMed] [Google Scholar]
- 27.Maier S, Nimmrich I, Koenig T, Eppenberger-Castori S, Bohlmann I, Paradiso A, et al. DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients—Technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer. 2007;43(11):1679–86. doi: 10.1016/j.ejca.2007.04.025 [DOI] [PubMed] [Google Scholar]
- 28.Dietrich D, Hasinger O, Liebenberg V, Field JK, Kristiansen G, Soltermann A. DNA methylation of the homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell lung cancer patients. Diagn Mol Pathol. 2012;21(2):93–104. doi: 10.1097/PDM.0b013e318240503b [DOI] [PubMed] [Google Scholar]
- 29.Vasiljevic N, Ahmad AS, Carter PD, Fisher G, Berney DM, Foster CS, et al. DNA methylation of PITX2 predicts poor survival in men with prostate cancer. Biomark Med. 2014;8(9):1143–50. doi: 10.2217/bmm.14.41 [DOI] [PubMed] [Google Scholar]
- 30.Gallastegui E, Bicer A, Orlando S, Besson A, Pujol MJ, Bachs O. p27Kip1 represses the Pitx2-mediated expression of p21Cip1 and regulates DNA replication during cell cycle progression. Oncogene. 2016. doi: 10.1038/onc.2016.200 [DOI] [PubMed] [Google Scholar]
- 31.Liu Y, Huang Y, Zhu GZ. Cyclin A1 is a transcriptional target of PITX2 and overexpressed in papillary thyroid carcinoma. Molecular and cellular biochemistry. 2013;384(1–2):221–7. doi: 10.1007/s11010-013-1801-9 [DOI] [PubMed] [Google Scholar]
- 32.Huang Y, Guigon CJ, Fan J, Cheng SY, Zhu GZ. Pituitary homeobox 2 (PITX2) promotes thyroid carcinogenesis by activation of cyclin D2. Cell Cycle. 2010;9(7):1333–41. doi: 10.4161/cc.9.7.11126 [DOI] [PubMed] [Google Scholar]
- 33.Fung FK, Chan DW, Liu VW, Leung TH, Cheung AN, Ngan HY. Increased expression of PITX2 transcription factor contributes to ovarian cancer progression. PloS one. 2012;7(5):e37076 doi: 10.1371/journal.pone.0037076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dietrich D, Hasinger O, Banez LL, Sun L, van Leenders GJ, Wheeler TM, et al. Development and clinical validation of a real-time PCR assay for PITX2 DNA methylation to predict prostate-specific antigen recurrence in prostate cancer patients following radical prostatectomy. J Mol Diagn. 2013;15(2):270–9. doi: 10.1016/j.jmoldx.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schatz P, Dietrich D, Koenig T, Burger M, Lukas A, Fuhrmann I, et al. Development of a diagnostic microarray assay to assess the risk of recurrence of prostate cancer based on PITX2 DNA methylation. J Mol Diagn. 2010;12(3):345–53. doi: 10.2353/jmoldx.2010.090088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dietrich D, Lesche R, Tetzner R, Krispin M, Dietrich J, Haedicke W, et al. Analysis of DNA methylation of multiple genes in microdissected cells from formalin-fixed and paraffin-embedded tissues. J Histochem Cytochem. 2009;57(5):477–89. doi: 10.1369/jhc.2009.953026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhl B, Gevensleben H, Tolkach Y, Sailer V, Majores M, Jung M, et al. PITX2 DNA Methylation as Biomarker for Individualized Risk Assessment of Prostate Cancer in Core Biopsies. J Mol Diagn. 2017;19(1):107–114. doi: 10.1016/j.jmoldx.2016.08.008 [DOI] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113–20. doi: 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goltz D, Gevensleben H, Dietrich J, Ellinger J, Landsberg J, Kristiansen G, et al. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncoimmunology. 2016;5(10):e1221555 doi: 10.1080/2162402X.2016.1221555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meller S, Zipfel L, Gevensleben H, Dietrich J, Ellinger J, Majores M, et al. CDO1 promoter methylation is associated with gene silencing and is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients. Epigenetics. 2016:1–10. doi: 10.1080/15592294.2016.1241931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goltz D, Gevensleben H, Grunen S, Dietrich J, Kristiansen G, Landsberg J, et al. PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia. 2016. doi: 10.1038/leu.2016.328 [DOI] [PubMed] [Google Scholar]
- 42.Spence T, Bruce J, Yip KW, Liu FF. HPV Associated Head and Neck Cancer. Cancers. 2016;8(8). doi: 10.3390/cancers8080075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Budczies J, Klauschen F, Sinn BV, Gyorffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PloS one. 2012;7(12):e51862 doi: 10.1371/journal.pone.0051862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Q, Li J, Wu W, Shen R, Jiang H, Qian Y, et al. Smad4-dependent suppressor pituitary homeobox 2 promotes PPP2R2A-mediated inhibition of Akt pathway in pancreatic cancer. Oncotarget. 2016;7(10):11208–22. doi: 10.18632/oncotarget.7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kitamura K, Miura H, Miyagawa-Tomita S, Yanazawa M, Katoh-Fukui Y, Suzuki R, et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development. 1999;126(24):5749–58. [DOI] [PubMed] [Google Scholar]
- 46.Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc Natl Acad Sci U S A. 2014;111(43):15544–9. doi: 10.1073/pnas.1416074111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anayannis NV, Schlecht NF, Belbin TJ. Epigenetic Mechanisms of Human Papillomavirus-Associated Head and Neck Cancer. Arch Pathol Lab Med. 2015;139(11):1373–8. doi: 10.5858/arpa.2014-0554-RA [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper or can be obtained from The Cancer Genome Atlas (TCGA) Research Network (www.cancergenome.nih.gov): https://portal.gdc.cancer.gov/projects/TCGA-HNSC.