Abstract
The tumor microenvironment imposes physical and functional constraints on the antitumor efficacy of adoptive T cell immunotherapy. Preclinical testing of different T cell preparations can help in the selection of efficient immune therapies, but in vivo models are expensive and cumbersome to develop, while classical in vitro 2D models cannot recapitulate the spatiotemporal dynamics experienced by T cells targeting cancer. Here, we describe an easily customizable 3D model, in which the tumor microenvironment conditions are modulated and the functionality of different T cell preparations is tested. We incorporate human cancer hepatocytes as a single cell or as tumor cell aggregates in a 3D collagen gel region of a microfluidic device. Human T cells engineered to express tumor-specific T cell receptors (TCR–T cells) are then added in adjacent channels. The TCR–T cells’ ability to migrate and kill the tumor target and the profile of soluble factors were investigated under conditions of varying oxygen levels and in the presence of inflammatory cytokines. We show that only the 3D model detects the effect that oxygen levels and the inflammatory environment impose on engineered TCR–T cell function, and we also used the 3D microdevice to analyze the TCR–T cell efficacy in an immunosuppressive scenario. Hence, we show that our microdevice platform enables us to decipher the factors that can alter T cell function in 3D and can serve as a preclinical assay to tailor the most efficient immunotherapy configuration for a specific therapeutic goal.
Keywords: Immunology, Therapeutics
A 3D microfluidic assay was developed to recapitulate key aspects of the spatiotemporal dynamics experienced by T cells targeting cancer cells in a solid tissue.
Introduction
Immunotherapy holds great promise as a novel approach to cancer treatment because of its unique potential to evade genetic and cellular mechanisms of drug resistance and target tumor cells while sparing normal tissues (1, 2). Results of clinical trials with autologous T lymphocytes engineered with chimeric antigen receptors or T cell receptors (TCRs) specific for antigens expressed on cancer cells provide compelling evidence of their clinical potential (3). However, robust clinical results have been definitively achieved only in nonsolid tumors (e.g., hematological malignancies) (4, 5), while adoptive T cell therapy against solid primary tumors or metastasis has only shown limited success (6, 7).
These differences in clinical efficacy are multifactorial and extend beyond the selection of the appropriate tumor antigens and the affinity of the introduced TCR or chimeric antigen receptor for the specific tumor antigen. Other constraints imposed by the tumor cell itself or the microenvironment can also influence the efficacy of T cell therapy. These include physical restrictions, such as the limited accessibility of cancer cells, intrinsic properties of the tumor cells that influence their immunogenicity and sensitivity to immune effectors, and the presence of inhibitory metabolic processes and soluble inhibitors in the parenchyma surrounding cancer cells, all of which can adversely affect T cell immunotherapy efficacy (8). For these reasons, strategies to modify the functional features of adoptive T cells were developed to overcome such barriers, thus enabling T cells to better penetrate the extracellular matrix (9). In addition, increased cytotoxicity in the tumor microenvironment has been achieved through metabolic modifications (10) or by downregulation of checkpoint inhibitors (11).
Preclinical testing to evaluate the efficacy of the different modifications introduced in engineered T cells for cancer immunotherapy, however, still relies primarily on animal models. These models are not only expensive and time consuming to create and study, but also are not fully able to recapitulate human pathology; additionally, tests in animal models lack the flexibility to control individual parameters in distinct anatomical locations (12). In vitro testing offers an alternative, but 2D in vitro models cannot mimic the spatiotemporal dynamics encountered by T cells targeting cancer cells within solid parenchyma. In addition, such testing methodologies have neglected the obvious 3D morphology of solid tumors and the tumor cell clusters necessary for metastasis formation (13–15). Here, we developed an easily reproducible and simple system to test whether TCR-engineered T cells can overcome the physical and metabolic barriers present in the tumor microenvironment. The system is composed of a microdevice-based cellular assay in which cancer cells and T cells interact in a 3D space. Conditions can be precisely regulated and the ability of distinct preparations of engineered T cells to reach and kill cancer cells can be tracked over time. We demonstrate that this 3D microdevice assay can measure the antitumor efficacy of TCR-engineered T cells and that it can be easily customized to study the effect of specific biological conditions and clinical scenarios on their function.
Results
3D microfluidic tumor model can quantify the time-dependent cytotoxicity and migratory features of TCR-engineered T cells.
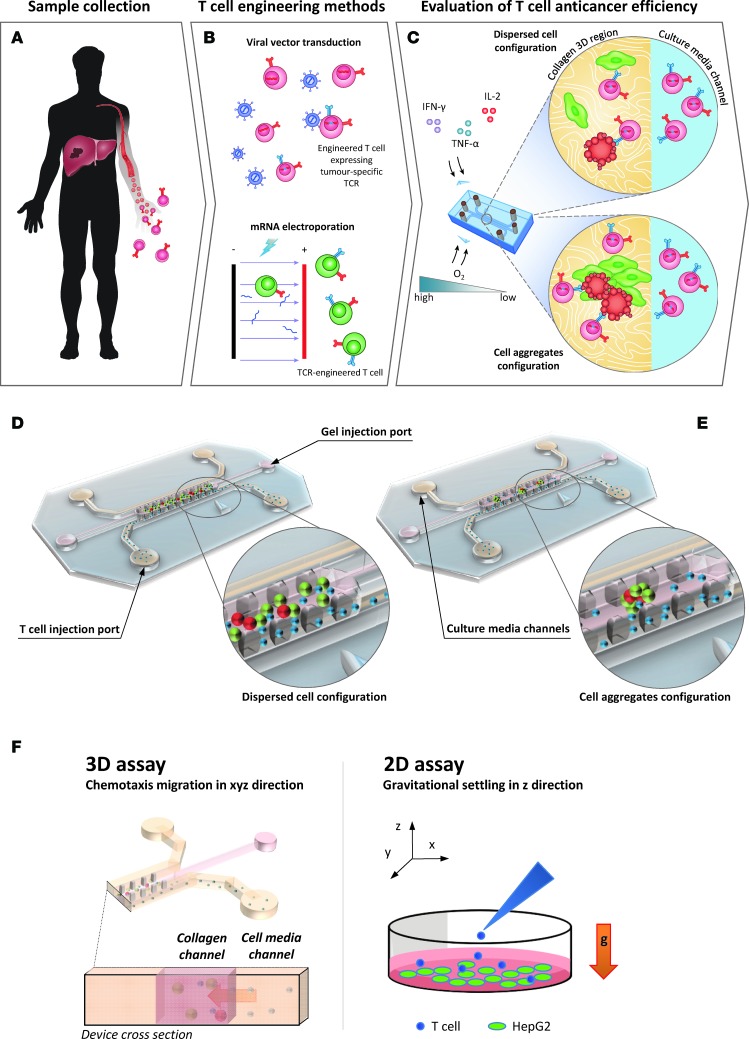
During adoptive T cell therapy against solid tumors, the engineered T cells first have to migrate from blood into tissues to perform their antitumor function. We utilized a previously validated 3D microdevice (16) in which effector T cells must migrate from medium-filled side channels into a central collagen-filled region (the “tissue”) to encounter their target (Figure 1). The central region, defined by two rows of trapezoidal pillars that confine the collagen type I solution prior to gelation, is seeded with hepatocellular carcinoma (HepG2-Env) cells expressing both hepatitis B virus (HBV) envelope protein (HBsAg) and GFP. Once the gel is polymerized, T cells engineered via retroviral transduction to express a specific TCR recognizing the complex HLA-A0201 molecule and hepatitis B Env183-191 epitope (TCRe–T cells) are injected into an adjacent medium channel in contact with the collagen region.
Figure 1. Evaluation of TCR-engineered T cell function using a 3D microfluidic device.
Factors influencing the functionality of TCR-engineered T cells were assessed using the microdevice, and the information obtained will then be used to improve the in vivo efficiency of the engineered T cells. (A–C) The workflow of the presented work is explained. (D and E) 3D rendering of the devices in disperse cell configuration and aggregate configuration. (F) The predominantly gravity-driven migration of engineered T cells along the z axis, as occurs in a 2D well-based assay, compared with the directional chemotaxis in a 3D microdevice.
The TCRe-redirected T cells used here already have been shown in vitro and in vivo to recognize and kill natural hepatocellular carcinoma (HCC) cells that express HBV viral antigen due to HBV-DNA integration in an HLA-A0201–restricted manner (17). The HepG2-Env cells used as target cells express HBV envelope antigen covalently linked to GFP. Thus, the green fluorescence provides visual confirmation of the expression of HBV antigen in the target cells. Note that HepG2 is an HLA-A0201–positive hepatoblastoma-derived cell line and that HEPG2-Env cells are recognized specifically by the TCRe–T cells (18).
By labeling the engineered TCRe–T cells with a fluorescent dye (CellTracker Violet BMQC) we can visualize their location in the device. With the inclusion of a live/dead discrimination dye (DRAQ7) in the culture medium, we can detect cell death events marked by the appearance of DRAQ7 fluorescence when the dye enters cells with compromised membrane integrity and binds to DNA.
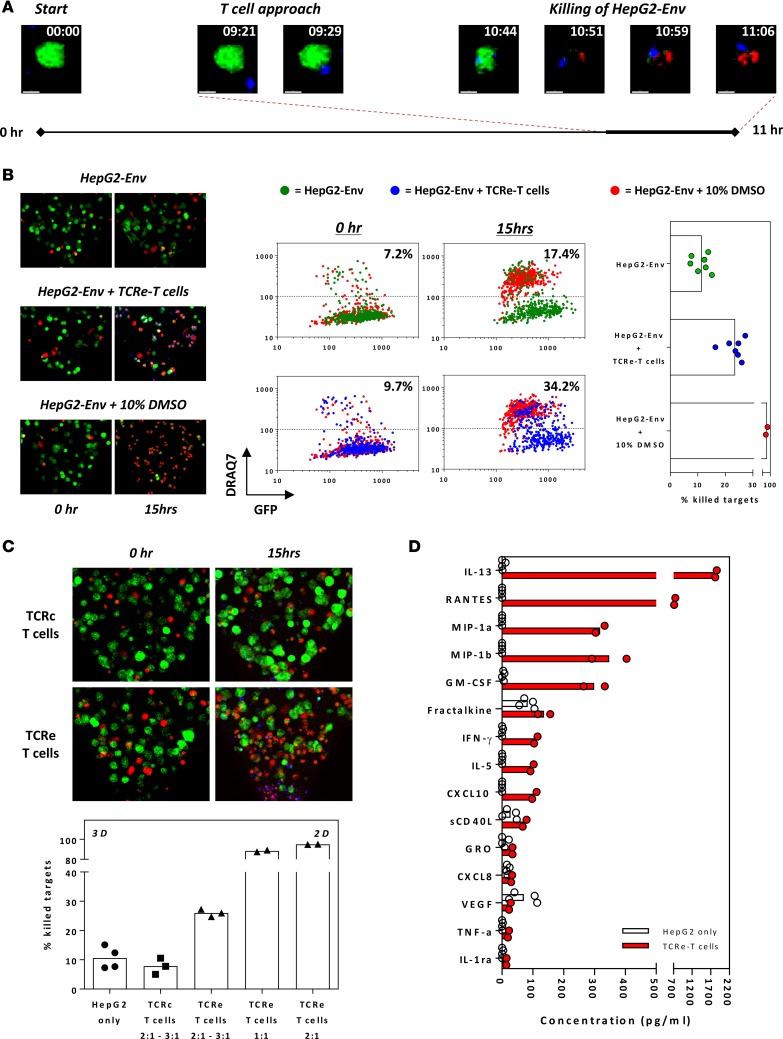
We first performed overnight live-imaging experiments in which we visualized, in real time, the migratory behavior of TCRe–T cells, their interaction with target cells, and eventual target cell death. Time-lapse confocal imaging (Supplemental Video 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.89762DS1) reveals the invasion of T cells from the media channel into the gel and the killing process against HCC. This is also illustrated by a representative sequence of time-lapse images, derived from these experiments, of a single HepG2-Env cell over 11 hours (Figure 2A). At approximately 9.5 hours, a single TCRe–T cell approaches the HepG2-Env target cell, and the two cells begin to interact. This is followed by death of the target cell approximately 1.5 hours later, as shown by the increase in DRAQ7 fluorescence dye.
Figure 2. Engineered HBV-specific T cells invade and specifically kill HBV antigen–expressing HCC cells.
(A) Timeline of notable events during a representative 11-hour live-imaging assay (~7-minute acquisition intervals; experiment performed twice), in which engineered HBV Env183-191–specific T cells (TCRe–T cells) were introduced into the device containing GFP-expressing HepG2-Env cells cultured in a 3D collagen matrix. Engineered T cells were labeled with CellTracker BMQC (blue), while DRAQ7 (red) was added in the culture media. HepG2-Env target cells are shown in green (GFP). The magnified maximum intensity projections of a single HepG2-Env cell are shown at the indicated times. Scale bar: 10 μM. (B) Representative maximum intensity projections of a region of the collagen gel showing HepG2-Env cells at 0 and 15 hours after incubation alone, with the addition of 10% DMSO, or with engineered TCRe–T cells. The mean fluorescence intensity (MFI) of GFP and DRAQ7 of each HepG2-Env target cell identified in Imaris was plotted at 0 and 15 hours after incubation at the respective conditions. Devices in which HepG2-Env cells were cultured with DMSO (red) were plotted in the background for reference, with the percentage of dead target cells quantified in devices with (blue) or without (green) the addition of TCRe–T cells at time points shown. The graph shows the percentage of killing quantified for the respective conditions; each point represents an individual experiment. Original magnification, ×10. (C) Representative maximum intensity projections of a region of the collagen gel showing HepG2-Env cells at 0 and 15 hours after incubation with engineered TCRe–T cells or Core18-27–specific T cells (TCRc–T cells). The mean percentage of killed target cells after overnight incubation without T cells or with engineered TCRe- or TCRc- specific T cells at the antigen-specific E/T ratios are shown as bars; each point represents an individual experiment. All experiments were repeated in at least 3 microfluidic devices. The amount of target cells killed when TCRe–T cells were analyzed using a 2D well-based killing assay is shown for comparison. Original magnification, ×10. (D) 41 secreted factors were quantified using multiplex bead-based assay. Factors with detected concentrations less than 10 pg/ml in all samples were removed from analysis. The mean concentration of secreted factors present in the supernatant retrieved from microdevices in which only HepG2-Env target cells were seeded (n = 4) or cocultured with retroviral-transduced TCRe–T cells (n = 2).
These live observations were further validated in multiple static imaging experiments, in which quantification of HepG2-Env death was performed before and after overnight incubation with engineered TCRe–T cells (Figure 2B). Microdevices with only HepG2-Env target cells seeded in the collagen gel region, without the addition of engineered TCRe–T cells, were used to quantify the basal amount of HepG2-Env death (Figure 2B), while the addition of toxic levels of DMSO (10%) acted as a positive control (Figure 2B). Without the addition of engineered T cells, low levels of basal HepG2-Env death (~10% increase in dead cells) were typically observed (Figure 2B). Conversely, overnight exposure to 10% DMSO killed nearly all the HepG2-Env target cells, which were identified as cells with reduced GFP and increased DRAQ7 fluorescence intensity (Figure 2B). Addition of engineered TCRe–T cells resulted in a robust and quantifiable decrease in the number of live HepG2-Env target cells (Figure 2, B and C), confirming that the engineered T cells were capable of invading the collagen gel and killing the HepG2-Env cells. The specificity of TCR-redirected T cell killing was clearly observed in the 3D microdevice, since addition of TCR-redirected T cells specific for an HLA-A02010/core 18-27 epitope (TCRc–T cells) not presented by HepG2-Env target cells caused only basal amounts of HepG2-Env death (Figure 2, B and C).
Since the cellular dynamics in the microdevice, in which chemotaxis of T cells into the gel region precedes target cell interaction, differs from a classical 2D well-based assay in which gravity-driven settling of T cells is predominant (Figure 1F), we measured the cytotoxicity of engineered TCRe–T cells using both systems to determine whether this could result in an observable difference. Coculturing the target cells and TCRe–T cells for 15 hours in a 3D microdevice resulted in the detection of killing of approximately 25% of the target cells, while the classical 2D well-based cytotoxicity assay detected an almost complete killing of all target cells, even at a lower antigen-specific effector/target (E/T) ratio (Figure 2C). The different results from the two systems shows how the complex spatial and migratory characteristics unique to the 3D assay system can profoundly alter the observable cytotoxic function of TCRe–T cells.
We also noted that recruitment of human T cells engineered to express tumor-specific T cell receptors (TCR–T cells) within the solid-phase collagen region required the presence of the HepG2-Env target cells, as seeding of engineered T cells into microdevices without HepG2-Env cells did not induce the migration of TCR–T cells into the collagen gel using the same experimental setup (Supplemental Figure 1). These data suggest that chemokines produced by target cells can be sensed by TCR–T cells but also that the supernatant in the microdevice can be possibly used to depict the cytokine/chemokine environment present in the system. A complex array of different cytokines/chemokines was present in the supernatants of the 3D microdevice (Figure 2D). Some of these were constitutively detected only in the presence of target cells (fractalkine, CXCL8, VEGF), while antigen-specific recognition (TCRe–T cells and HepG2-Env cells) clearly boosts the production of several cytokines/chemokines (RANTES, MIP-1α, MIP-1β, IFN-γ, CXCL-10).
Overall, these data show that the 3D microfluidic system provides a complex environment that can be used to quantify and visualize, over time, the cytotoxicity and migratory features of engineered TCR–T cells, the ability of different cancer cells to attract them, and the cytokine/chemokine microenvironment that is shaped by the interaction between effector and target cells, which cannot be fully recapitulated by classical 2D assays.
Different TCR–T cell preparations exhibit differential antitumor activity.
Having established the 3D microdevice assay, we then used it to address important questions along the engineered T cell immunotherapy pipeline.
TCR–T cells can be engineered using different methods. Viral transduction is the most commonly utilized method, but delivery of the TCR gene construct through mRNA electroporation of in vitro–preactivated primary human T cells can confer advantages, despite the transient nature of TCR expression. In addition to a faster and simpler cellular engineering protocol, transient TCR expression can be seen as an opportunity to monitor excessive toxicity and allow patient treatment with escalating T cell doses. We have previously demonstrated, in a murine xenograft model of HCC, a direct comparison of the antitumor activity of retroviral-mediated stably transduced TCR–T cells (retroV-TCRe–T cells) versus the mRNA-electroporated transiently transfected TCR–T cells (mRNA-TCRe–T cells) (19). Despite the transient TCR expression, we demonstrated that mRNA-TCRe–T cells suppress the growth of established tumor in the model, even though their efficacy was lower than that observed after adoptive transfer of retroV-TCRe–T cells.
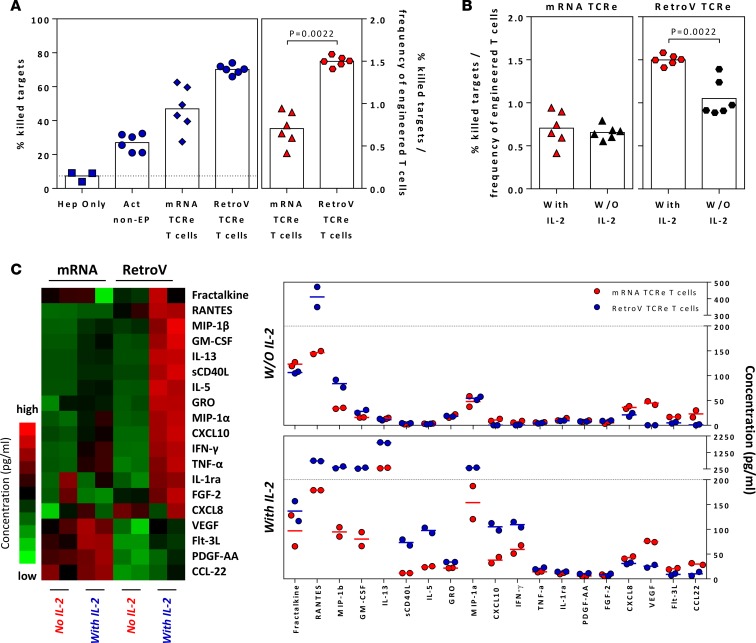
In order to test whether our 3D microdevice can detect the differential antitumor efficacy observed in vivo, we used it for a comparative analysis of the two different TCR–T cell preparations (mRNA-TCR–T cells versus retroV-TCR–T cells) (Figure 3). The ability of activated mRNA-TCR–T cells and retroV-TCR–T cells to lyse HepG2-Env cells embedded in the solid phase of the device was analyzed in the presence or absence of IL-2 in the culture media. These conditions were used as in patients; adoptive T cell transfer is often performed in association with IL-2 treatment (20).
Figure 3. Different preparations of engineered T cells have varying antitumor responses.
(A) The mean percentage of killed target cells when different engineered T cells were seeded into the microdevices in the presence of exogenous IL-2 is shown in blue. The mean percentage of killed target cells normalized to the frequency of successfully engineered T cells is shown in red. Each dot represents a single experiment, and devices without engineered T cells or seeded with nonelectroporated T cells were included as controls. Statistical significance was evaluated with 2-tailed t test. (B) The normalized mean percentage of killed target cells (similar to above) when different engineered T cells were seeded into the microdevices in the presence or absence of exogenous IL-2. Each dot represents a single experiment. Statistical significance was evaluated with 2-tailed t test. (C) Heatmap of the relative concentration of soluble factors detected in the supernatants collected from microdevices, with indicated engineered T cells and IL-2 supplementation, and dot plots showing the summary of the concentration of detected soluble factors. Factors with detected concentrations of less than 10 pg/ml in all samples were removed from analysis. Each column represents a single experiment.
Confirming the antitumor efficacy observed in the murine model, retroV-TCR–T cells displayed greater efficacy than mRNA-TCR–T cells in killing HEPG2-env target cells (Figure 3A). However, the killing ability of mRNA-TCR–T cells was independent of IL-2, since levels of antitumor efficacy in the 3D microdevice were not significantly affected by the absence of IL-2 (Figure 3B).
Analysis of supernatants collected in the 3D microdevice confirmed the strong IL-2 dependency of cytokine/chemokine production in retroV-TCR–T cells (Figure 3C). In contrast, some cytokines/chemokines were produced in greater quantities by only the mRNA-TCR–T cells, even in the absence of IL-2 (VEGF, CCL22, CXCL8). However, when the target cells were cultured in 2D, this chemokine/cytokine profile was no longer observed and instead dominated by CXCL10 (Supplemental Figure 3).
In all, these results show that the two different preparations of TCR–T cells are not only characterized by a different IL-2 dependence, but also by the induction of a distinct functional response that could be influenced by the unique characteristics of a 3D system.
TCR-engineered T cells effectively lyse tumor cell aggregates.
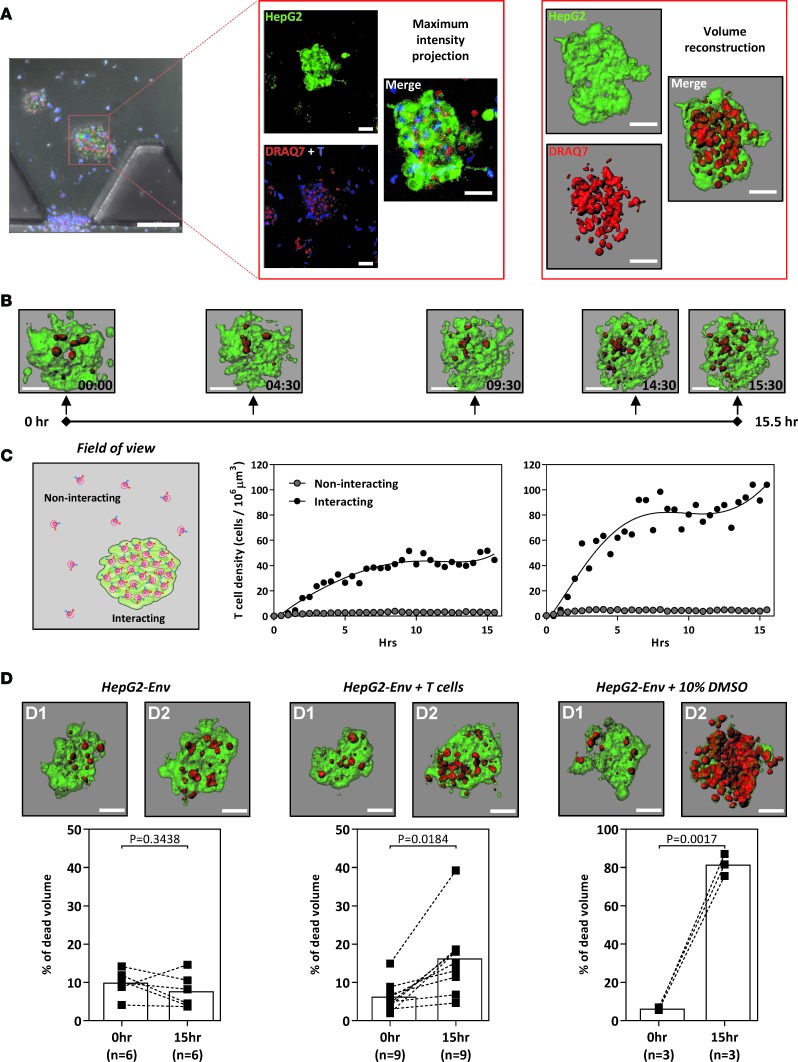
To further develop the 3D microdevice as a platform to analyze the efficacy of TCR–T cell therapy against solid cancer or metastasis, we introduced HepG2-Env target cells as a tumor aggregate into the central collagen region. For this purpose, HepG2-Env target cells were first cultured in a customized Petri dish (21) to obtain tumor aggregates that are 40–100 μm in diameter. These aggregates were then incorporated in the central collagen gel region, while retroV-TCR–T cells were introduced into the flanking media channels in the presence of DRAQ7. Confocal images were acquired as before, and the fluorescent signals were converted into 3D volumes; in Figure 4A, the red volume (DRAQ7 positive) represents the dead portion of the cell cluster, while the green volume (GFP positive) represents the live cells of the aggregate. To quantify the killing, the volume of DRAQ7 fluorescence is expressed as a percentage of the total GFP fluorescence volume.
Figure 4. TCR-engineered T cells lyse hepatocellular carcinoma aggregates.
(A) Schematic illustrating the 3D volume reconstruction of HepG2-Env aggregates embedded in the collagen gel of the microdevice (scale bar: 100 μm [left]; 50 μm [middle and right]). (B) Timeline depicting a representative 15.5-hour live-imaging assay (~30-minute acquisition intervals; experiment performed twice), in which engineered HBV Env183-191–specific T cells were introduced into the device containing GFP-expressing HepG2-Env aggregates cultured in a 3D collagen matrix. DRAQ7 was added in the culture media labeling dead cells red. The reconstructed aggregates were shown at the indicated times (scale bar: 50 μm). (C) The density of engineered T cells randomly moving in empty collagen gel regions (noninteracting) and within HepG2-Env aggregates (interacting) was quantified in 2 live-imaging experiments depicted. (D) Representative reconstructed aggregates at 0 hours and after overnight incubation alone (left), with the addition of 10% DMSO or with engineered HBV-specific T cells (scale bar: 50 μm). The summary of the percentage of dead aggregates was quantified for the respective conditions. Each dot represents a single experiment. Scale bar: 50 μM. Statistical significance was evaluated with 2-tailed t test.
Live-imaging experiments revealed a time-dependent increase in the percentage of dead HepG2-Env target cells, as evidenced by the increase in DRAQ7 volume in the tumor aggregates (Figure 4B). To evaluate the interaction between engineered T cells and the tumor aggregates, we quantified the density of engineered T cells in an empty gel region and within the tumor aggregate volume in the same field of view over a 15-hour time period. This analysis revealed a preferential accumulation of engineered TCR–T cells within the tumor aggregates, as evidenced by the dramatically higher density of engineered T cells in close proximity to the tumor aggregates (~70 cell/106 μm3) compared to that in the surrounding empty collagen gel space (~2 cell/106 μm3) (Figure 4C). Images of tumor aggregates acquired before and approximately 15 hours after the addition of engineered T cells also showed a significant increase in the proportion of killed target cells (from ~5% to ~15%), further corroborating our time-lapse live-imaging findings (Figure 4D). Thus, our 3D microassay can be used to visualize and evaluate the function of engineered TCR–T cells, either against isolated cancer cells or against tumor aggregates, providing what we believe to be a novel dimension to in vitro assays that more accurately represents the in vivo scenario.
Functionality of TCR-engineered T cells can be assessed under hypoxic and inflammatory conditions in the 3D microdevice.
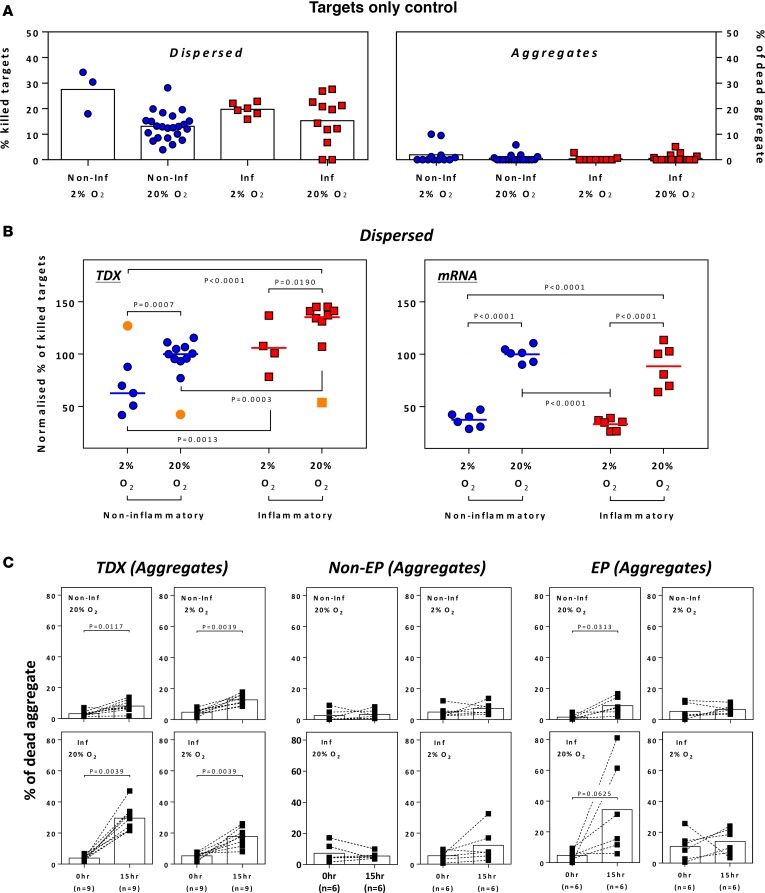
Metabolic changes or inflammatory conditions in the tumor microenvironment can also modulate the efficacy of immunotherapy. We therefore used the 3D microassay to directly investigate the effect of hypoxia and inflammatory cytokines (IFN-γ and TNF-α), prominent features in solid tumor (14), on the function of TCRe–T cells. Through the addition of IFN-γ and TNF-α to the assay media, we mimicked certain aspects of the tumor inflammatory environment, whereas experiments conducted in 2% or 20% O2 examined the effect of hypoxia on TCRe–T cell function (Figure 5). mRNA-TCRe– and retroV-TCRe–T cells were tested to determine their ability to lyse the dispersed (Figure 5B) or aggregated (Figure 5C) target cells.
Figure 5. Cytotoxicity of TCR-engineered T cells under hypoxic and inflammatory conditions.
(A) Spontaneous death of HepG2-Env target cells cultured under specified conditions. (B) Normalized percentage of killed target cells when retroviral-transduced (left) or activated mRNA-electroporated (right) TCR-engineered T cells were cocultured with dispersed HepG2-Env target cells under specified conditions. Results were normalized to data obtained under typical in vitro assay conditions (20% oxygen and no inflammatory cytokines). Each point represents a single experiment. Outliers are indicated in orange. Statistical significance was evaluated with 2-way ANOVA with Tukey’s multiple comparison test. (C) The percentage of dead HepG2-Env aggregates before and after overnight coculture with retroviral-transduced (left), activated nonelectroporated (middle), or mRNA-electroporated (right) TCR-engineered T cells under specified conditions. Each dot represents a single experiment. Statistical significance was evaluated with 2-tailed t test.
In the presence of IFN-γ/TNF-α (inflammatory condition) or the different oxygen concentrations (2% and 20%), we observed that these factors had no significant effect on the spontaneous death of the target cells in either dispersed or aggregate forms (Figure 5A). Similarly, introduction of non-TCR-engineered T cells in the 3D microassay did not effect the spontaneous death of dispersed (Figure 5B) or aggregate (Figure 5C) HepG2-Env target cells. This allowed us to isolate and investigate the effect of O2 levels and inflammatory cytokines on the ability of TCR–T cells to kill dispersed or aggregated hepatoma cells.
We first observed reduced killing of dispersed HepG2-Env cells by both TCR–T cell preparations in 2% O2 in comparison to 20% O2 (Figure 5B). However, while the ability of retroV-TCRe–T cells to lyse dispersed targets was augmented by the presence of inflammatory cytokines, mRNA-TCRe–T cells were not affected (Figure 5B). The presence or absence of inflammatory cytokines, however, clearly affected the ability of both TCR–T cell preparations to lyse tumor aggregates (Figure 5C). In the absence of IFN-γ/TNF-α, the lysis of tumor aggregates was minimal irrespective of O2 levels or different TCR preparations (Figure 5C). Robust killing ability was instead clearly observed with the addition of IFN-γ and TNF-α, in particular with high O2 conditions (20% O2 levels, Figure 5C).
These data demonstrate that different cellular and biochemical variables can be introduced in our 3D microdevice, allowing the specific determination of their role in modulating the antitumor ability of TCR–T cell preparations.
2D well-based cytotoxicity assay neglects the effect of hypoxia on engineered T cell function.
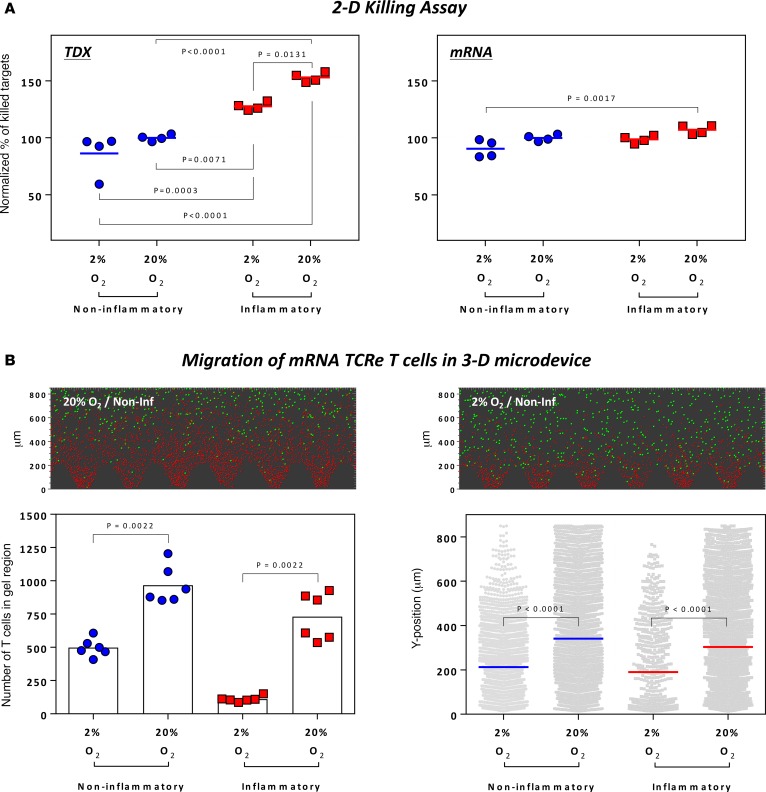
The effect of oxygen levels on the lytic ability of TCR–T cells in the experiments performed in our 3D microdevice is in line with in vivo results obtained in a murine model of EBV-related tumors, in which the adoptive transfer of EBV-specific CTLs accumulated only in the normoxic areas of the tumor, while only a few CTLs were found in the hypoxic areas (22). We performed similar experiments in a 2D model, in which gravitational settling of T cells mediates target cell–T cell interaction (Figure 1F), to determine if TCR–T cell killing of tumor cells was modulated by introducing different variables (inflammatory cytokines and oxygen levels) in this experimental setting.
First, we cocultured retroV-TCRe– or mRNA-TCRe–T cells with luciferase-expressing HepG2-Env target cells at different E/T ratios in a classical 2D well-based assay and quantified the luminance at the end of the experiment to determine the amount of target killing. E/T ratios of 1:4 and 1:8 did not result in complete target death and hence were used to assess the effect of hypoxia and inflammatory cytokines (Supplemental Figure 2). Similar to that observed in our 3D microdevice, in which HepG2-Env target cells were seeded in a dispersed configuration, inflammatory cytokines enhanced the killing of the targets by retroV-TCRe– but not by mRNA-TCRe–T cells (Figure 6A). However, in sharp contrast to the results obtained in our 3D microdevice (Figure 5B), hypoxia did not significantly reduce the killing of HepG2-Env cells by retroV-TCRe– or mRNA-TCRe–T cells (Figure 6A) in the 2D setting.
Figure 6. 2D well-based assays cannot detect the negative effect of hypoxia on the cytotoxicity of engineered T cells.
(A) Normalized percentage of killed target cells when retroV-TCRe (left) or mRNA-TCRe (right) T cells were cocultured with luciferase+ HepG2-Env target cells under specified conditions. Results were normalized to data obtained under typical in vitro assay conditions. Data shown were compiled from experiments with 2 different E/T ratios performed in duplicates, as denoted in Supplemental Figure 2. Statistical significance was evaluated with 2-way ANOVA with Tukey’s multiple comparison test. (B) Representative images of 3D microdevices in which mRNA TCRe–T cells were cultured overnight with HepG2-Env target cells in the respective conditions. Green spheres denote the living HepG2-Env target cells, while red spheres denotes the mRNA TCRe–T cells. The number of T cells present in the gel at the end of the experiment (each dot represents the number of T cells in each microdevice) and the position of all gel-invading T cells in each condition (each dot represent the position of a T cell) are shown. Statistical significance was evaluated with 2-tailed t test.
In order to understand the mechanisms responsible for the discrepancy between the results obtained in the 2D and 3D model, we analyzed whether different oxygen levels effect the migratory behavior of the engineered T cells. We quantified the mRNA-TCRe–T cells that had infiltrated into the gel region and their spatial location (Figure 6B) under various conditions and demonstrated that hypoxia negatively affects the motility of engineered T cells. T cell numbers in the 3D gel matrix volume were significantly lower in hypoxia (Figure 6B), and the invading T cells have a reduced migratory capacity (Figure 6B).
Since the presentation of the appropriate epitope by HLA-I molecules on the cell surface is a determinant of T cell recognition and function, we also analyzed and compared the expression of HLA-I molecules on target cells cultured in 2D and 3D in the presence of inflammatory cytokines or hypoxia (Supplemental Figure 4). Note that the increased HLA-I expression in response to inflammatory cytokines was only observed when target cells were cultured in 2D (this was not recapitulated in the 3D microdevice), while the effect of hypoxia on HLA-I expression remains similar in both systems.
Thus, these data show that our 3D microdevice assay is capable of providing a more holistic analysis of engineered T cell function against a tumor target, which cannot be achieved with the classical 2D well-based assay, with its overtly simplistic nature, which disregards both the migratory behavior of the engineered T cells and intrinsic changes associated with growing cells in 3D space.
Mimicking specific clinical scenarios in the 3D microdevice.
In addition to the complex inhibitory features of the tumor microenvironment, the antitumor efficacy of TCR–T cells can also be affected by the systemic administration of therapeutic drugs. This is especially evident in the context of adoptive immunotherapy in patients who undergo allogeneic solid organ transplantation, in which long-term immunosuppressive regimes (required to maintain the transplanted organ) can also suppress the efficacy of adoptively transferred TCR–T cells (23). Understanding the functionality of TCR–T cells in this situation will be of clinical importance.
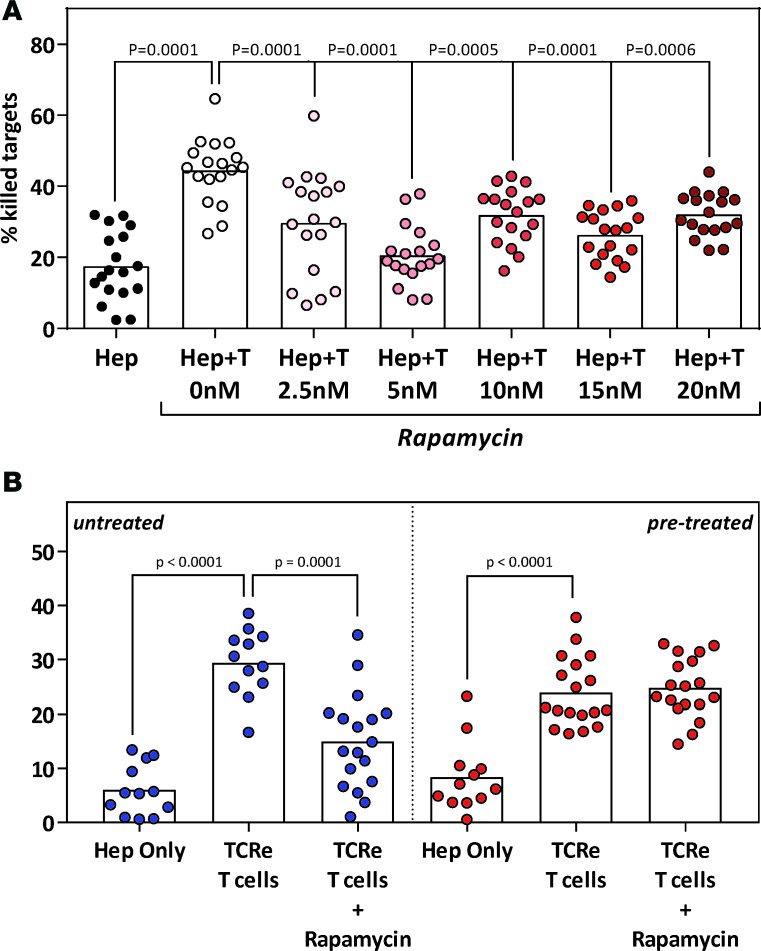
Utilizing the microdevice assay, we first analyzed the function of retroV-TCRe–T cells in the presence of a mTOR inhibitor, a class of immunosuppressant indicated for use in transplantation of various organs, including the liver. Concentrations of rapamycin, spanning the serum trough levels reference range used for clinical monitoring (3.3–22.0 nM) (24), significantly inhibited the cytotoxic function of retroV-TCRe–T cells (Figure 7A). This is not surprising given the immunosuppressive nature of mTOR inhibitors.
Figure 7. TCR–T cells retain their cytotoxicity in a clinical scenario with immunosuppressive mTOR inhibitors.
(A) Cytotoxicity of retroV-TCR–T cells was analyzed in the presence of increasing concentrations of immunosuppressive mTOR inhibitor (rapamycin). HepG2-Env target cells were seeded as before, and rapamycin was introduced with the TCR–T cells. Each dot represents a single experiment. Statistical significance was evaluated with 2-way ANOVA with Tukey’s multiple comparison test. (B) Cytotoxicity of retroV-TCR–T cells against HepG2-Env target cells pretreated for 2 weeks with 5 nM rapamycin was analyzed. Untreated targets were used as controls. Rapamycin concentrations were maintained throughout the experiment to mimic the in vivo condition of a patient under stable immunosuppression. Each dot represents a single experiment. Statistical significance was evaluated with 2-tailed t test.
We next mimicked a scenario in which TCR–T cells were adoptively transferred into a patient under stable immunosuppression. In such cases, immunosuppressant levels are kept near the lower limit of the serum trough levels reference range. As such, HepG2-Env target cells were pretreated with 5 nM rapamycin for 2 weeks before being seeded in a dispersed configuration in the microdevice without removal of rapamycin. RetroV-TCRe–T cells were then introduced into the microdevice and analyzed as before.
Pretreatment with rapamycin did not induce a significant increase in spontaneous HepG2-Env cell death (Figure 7B). Interestingly, unlike what was observed when target cells were not pretreated (Figure 7, blue), rapamycin did not inhibit the cytotoxic efficiency of the retroV-TCRe–T cells against pretreated HepG2-Env cells (Figure 7B, red). This suggests that engineered TCR–T cells could maintain their in vivo antitumor efficacy in the presence of physiological quantities of immunosuppressive mTOR inhibitors. More importantly, it clearly demonstrates how the 3D microdevice could be used to address clinically relevant scenarios.
Discussion
In this work, we demonstrated that a microfluidic device can measure the antitumor efficacy of engineered TCR–T cells under different tumor-like microenvironments. Most studies that evaluate antitumor efficacy of T cells have previously been performed in 2D classic tissue culture plates in which the interaction of T cells and tumor cells is mediated in part by gravity (Figure 1F). In contrast, in our system, we were able to observe and analyze the antitumor efficacy of T cells moving freely from a medium channel into a solid 3D matrix, in which they recognize and subsequently kill tumor cells over a 24-hour period. Since lysis of the tumor cells embedded in the 3D collagen microenvironment present in our microdevice is a net effect of both the chemotactic characteristics and intrinsic killing of the engineered TCR–T cells, this assay mimics more closely what is encountered physiologically during adoptive T cell therapy of solid tumors, in which these parameters can be a critical determinant of antitumor efficiency.
Using our microfluidic device, we were able to more accurately detect the different antitumor activity exerted by mRNA-TCR– and retroV-TCR–T cells, previously demonstrated in vivo in a xenograft animal model (19). We initially thought that the reduced antitumor activity observed in vivo was exclusively caused by the transient nature of TCR expression in mRNA-TCR–T cells. In reality, within the time limit of efficient TCR expression (24–48 hours after electroporation), mRNA-TCR–T cells possess a lower cytotoxic ability than the retroV-TCR–T cells (Figure 3A). Interestingly, when these engineered T cells were assessed in a 2D killing assay in which target cell killing was largely independent of T cell motility (Figure 6B), mRNA TCR T cells were more cytotoxic against tumor cells. A plausible explanation that reconciles these different observations is that, compared with retroV-TCR–T cells, mRNA-TCR–T cells are less motile, making them less efficient in situations in which cytotoxicity is influenced by cell motility.
However, we also observed that this reduced antitumor activity of mRNA-TCR–T cells was compensated for by a reduced dependency on IL-2, the presence of which is required for optimal function of retroviral TCR–T cells. Such differences are likely related to the differential in vitro manipulation and expansion time required to produce these two TCR–T cell preparations and the precise mechanisms responsible for such results are under investigation. Here, we further stress that such detailed information about the antitumor efficacy and differential IL-2 dependency of different TCR–T cell preparations has important clinical value, especially since IL-2 is often used as an adjuvant in immunotherapy clinical trials.
We are aware that our 3D microdevice-based system does not fully recapitulate the complexity present in natural tumors, e.g., it lacks the local milieu of other stromal and immune cells as well as a functional vasculature. Nevertheless, it can more closely reproduce the features of solid tumors than traditional 2D assays by allowing preformed cancer cell aggregates to be embedded in a 3D matrix, within which they interact with the invading T cells. Using such a system, we were not only able to demonstrate the destruction of tumor aggregates by TCR-engineered T cells, but we also acquired specific evidence of the importance of inflammatory cytokines and O2 levels for the optimum function of TCR-engineered T cells. This directly effects the antitumor efficiency of TCR-engineered T cells against primary tumors located in different anatomical compartments and also applies to circulating tumor cells and metastatic tumor clusters that have just begun the seeding process at secondary sites, all of which have varying physical and biochemical conditions.
One clear advantage of our 3D system over animal models is the fact that it can be easily customized to recreate crucial aspects of the tissue microenvironment present in solid tumors (e.g., inflammation, oxygen levels), allowing the dissection of complex phenomena into simpler distinct units. We can speculate that, for example, by modifying genes involved in the metabolic requirements of T cell function, we can design TCR–T cells that are better equipped to function in a hypoxic environment (25). Our 3D device can be used to precisely screen whether such modifications might lead to a gain of function in a low O2 environment. Here, we clearly show that the traditional 2D well plate assay neglects the migratory component of an in vivo setting and may result in the incorrect assessment of the antitumor efficacy of adoptively transferred engineered T cells, while the 3D microdevice can detect the reduced migratory capability of the engineered T cells in a low O2 environment. Though we have focused on the effects of hypoxia and inflammatory cytokines (Figure 6), comparative analysis of culture supernatants (Supplemental Figure 3) and HLA-I molecule expression on target cells (Supplemental Figure 4) also revealed differences between the 2D and 3D assay systems, illustrating the extensive functional changes associated with a 3D environment. For example, CXCL10, a chemokine produced by the target cells in the presence of activated T cells (26), was detected in dominant quantities only in the 2D assay. This could possibly result as a consequence of the gravity-mediated extended interaction between T cells and target cells in 2D, while, in 3D, the migratory component of the assay precludes such a scenario, leading to a different supernatant profile.
The customizability of the 3D system further allows us to mimic clinical scenarios by introducing pharmacological agents. We have chosen to illustrate this with a clinically adopted mTOR inhibitor used at doses observed in the sera of patients with organ transplantation (24) and by pretreating HepG2-Env cancer cells, recreating the stable immunosuppression achieved after transplantation. Such a scenario is often encountered, since allogeneic organ transplantation remains an important therapeutic option, especially for advanced stages of various cancers. Using our 3D system, we showed that retroV-TCR T cells maintained their antitumor function, despite exposure to immunosuppressive rapamycin. The exact mechanism remains to be elucidated, but previous reports showing increased HBsAg expression in hepatoma cells treated with mTOR inhibitors (27) and the improved survival in low-risk transplanted patients immunosuppressed with rapamycin (28) provided an immunological rationale for our observation.
Currently, our experiments have been limited to only two different cell types, cancer hepatocytes and engineered TCR–T cells. However, it will be possible to increase the complexity of the system by incorporating other immune cells, such as NK cells and macrophages, to observe their roles in eradicating the tumor. The possibility of measuring cytokine/chemokines released in the supernatants from the microdevice provides a further opportunity to evaluate the conditions induced in the system by the addition of TCR–T cells. Moreover, it will be possible to combine patient tissue specimens and autologous engineered T cells to build a totally patient-specific preclinical screening tool. By incorporating this test into routine clinical practice, we believe it will be possible to reduce the negative effect of cell inefficiency or incompatibility.
In conclusion, we demonstrate that a 3D microfluidic model can be used to decipher the factors that alter T cell function in a solid tumor assay that can be customized to replicate certain features found clinically or conditions present in the tumor microenvironment. We propose that the use of this model, with proper validation from in vivo experiments, can boost the development of TCR–T cell preparations to identify the most efficient immunotherapy protocol for a specific therapeutic goal.
Methods
Chip design and fabrication
The detailed design and fabrication procedure of the poly(dimethylsiloxane) (PDMS) device (Figure 1), comprising a gel region with adjoining media channels separated from the gel channel by trapezoidal posts (100 μm spacing) was tested and validated in our previous publications (16, 29, 30). The height of the microfluidic channels is 120 μm, the culture media channel width is 500 μm, and the gel region width is 1 mm. Briefly, the device was fabricated by PDMS soft lithography on a photoresist (SU8-50, MicroChem) mould obtained by stereo lithography (31). PDMS was prepared at a 10:1 ratio of base/curing agent (Sylgard 184, Dow Chemical) and cured at 80°C for 2 hours. Access holes in the PDMS devices for gel (1-mm diameter) and fluid (4-mm diameter) injection were created with biopsy punches (Ted pella). The devices were autoclaved, dried overnight at 80°C, and subsequently bonded after plasma surface activation for 2.5 minutes to a glass coverslip according to a previously described protocol (32). To facilitate the collagen gel adhesion to the surrounding material (glass and PDMS), devices were coated with poly-D-lysine (1 mg/ml in distilled H2O) for 1 hour, washed 3 times with water, and dried overnight at 80°C in order to restore the original hydrophobic characteristic of the PDMS and glass surfaces.
Cell engineering process
Retroviral transduction and expansion of TCR–transduced T cells.
The HBV Env183-91–specific TCR (Env183-TCR) Vα34 and Vβ3 chains were cloned and inserted into vector MP-71 (a gift from Hans Stauss, University College London, London, United Kingdom). Retrovirus was produced and bulk T cells were transduced as previously described (18); 72 hours after transduction, expression of TCR transgenes was determined using HLA-A201-Env183-91 pentamer and analyzed by flow cytometry. Bulk transduced T cells were expanded and restimulated using 5 × 105 TCR-transduced cells, 2 × 105 irradiated (40 Gy) T2 cells pulsed with 1 μg/ml Env183-91 peptide (FLLTRILTI), and 1.8 × 106 irradiated (25 Gy) peripheral blood mononuclear cells (PBMCs) as feeders and grown in AIM-V (Thermo Fisher Scientific) supplemented with 2% human AB serum (Thermo Fisher Scientific) and 100 IU/ml rIL-2 (R&D Systems) for 10 days. Mock-transduced T cells were expanded with 1 μg/ml phytohemagglutinin (Sigma-Aldrich) and irradiated feeders. Expanded TCR-transduced T cells were analyzed for TCR expression using specific HLA-0201 Env183-91 (FLLTRILTI) peptide pentameric complexes (Proimmune) before freezing in aliquots for experiments.
Preparation of activated and resting T cells for mRNA electroporation.
For T cell activation, PBMCs were stimulated with 600 IU/ml IL-2 and 50 ng/ml anti-CD3 (OKT-3; eBioscience) in AIM-V 2% human AB serum for 8 days, and rIL-2 was increased to 1,000 IU/ml 1 day before electroporation. For resting T cells, primary human T cells were enriched from PBMCs using the Pan T Cell Isolation Kit (Miltenyi Biotec GmbH) and cultured overnight in AIM-V 2% human AB serum plus 100 IU/ml rIL-2 before electroporation.
Production of HBV Env183-191 TCR mRNA and electroporation procedures.
Purified DNA of the HBV Env183-91–specific TCR subcloned into the pVAX1 vector, as previously described (19), was linearized using the FastDigest XbaI restriction enzyme (Thermo Fisher Scientific) and used to produce the TCR mRNA using the Ambion mMESSAGE mMACHINE T7 Ultra kit (Thermo Fisher Scientific). Briefly, T7 RNA polymerase was added to start transcription, and RNA was capped with Anti-Reverse Cap Analog (Thermo Fisher Scientific). Then, poly(A)-tail was added by E. coli poly(A) polymerase and ATP. The resulting product was concentrated by lithium chloride precipitation and redissolved in buffer. For electroporation using the 4D-Nucleofector device (Lonza), 10 × 106 activated or resting T cells, as described above, were suspended in 100 μl Nucleofector Solution and supplement (Lonza) and TCR mRNA was added at 200 μg/ml. The mixture was placed in a certified cuvette (Lonza) and electroporated. After electroporation, cells were resuspended in AIM-V 10% human AB serum plus 100 IU/ml rIL-2 and cultured at 37°C and 5% CO2 for 24 hours prior to analysis of TCR expression.
HBV antigen–expressing HepG2 target cells
The human liver carcinoma cell line, HepG2 (ATCC), was transduced with a construct containing either the full genotype D HBV core (HepG2-Core) or envelope (HepG2-Env) gene covalently linked to GFP using the Lenti-X HTX packaging system (Clontech) according to the manufacturer’s instructions. These transduced target cells were maintained in R10 culture media (RPMI 1640 supplemented with 10% heat-inactivated FBS, 20 mM HEPES, 0.5 mM sodium pyruvate, 100 IU/ml penicillin, 100 μg/ml streptomycin, MeM amino acids with L-glutamine, MeM nonessential amino acids [Thermo Fisher Scientific], 5 μg/ml Plasmocin [InvivoGen] with 5 μg/ml of puromycin [Clontech]) to select for transgene-expressing target cells. To pretreat HepG2-Env cells with rapamycin (InvivoGen), the cells were cultured as above with the addition of 5 nM rapamycin for 2 weeks. The concentration of rapamycin was maintained with every media change.
3D microdevice-based assay
Dissociated liver carcinoma target cells.
To prepare 200 μl of a 2.5 mg/ml type-I collagen gel solution containing homogenously dissociated HepG2 targets, 20 μl 10× PBS was mixed with 4 μl NaOH (0.5 N), 129.2 μl collagen type I (Corning), 20 μl freshly trypsinized and dissociated HepG2 targets at 50 × 106 cells/ml, and 22.9 μl cell culture water (31). The final pH of the gel solution was approximately 7, as determined using a pH indicator strip. The collagen gel solution containing the HepG2 target cells was then injected into the dedicated gel region of the device and polymerized for 40 minutes in the cell culture incubator at 37°C and 5% CO2. Immediately after gel polymerization, the media channels were filled with R10 media in order to hydrate the gel and keep the HepG2 target cells vital. The cell-impermeable nuclear dye DRAQ7 (Biolegend) was also added in the R10 media at a concentration of 3 μM to discriminate between live and dead cells. The devices were then incubated for 24 hours to permit the interaction of the HepG2 target cells with the collagen matrix. Devices with gel only (no embedded cells, control) were prepared similarly by adding collagen gel solution containing 20 μl 10× PBS, 4 μl NaOH (0.5 N), 129.2 μl collagen I, and 42.8 μl of cell culture water. Prior to the injection of the T cells in the device, R10 medium in the device was supplemented with DRAQ7 containing AIM-V 2% human AB serum plus 100 IU/ml rIL-2 with or without the addition of 1,000 IU/ml of IFN-γ (R&D Systems) and 100 ng/ml of TNF-α (R&D Systems) (to mimic the inflammation status) where applicable. In experiments involving rapamycin, the drug was added to the microdevice at the stated concentrations together with the introduction of the engineered T cells.
In order to visualize the spatial position of the engineered T cells, the cells were stained with 3 μM CellTracker Violet BMQC (Thermo Fisher Scientific) in RPMI 1640 for 30 minutes at 37°C. T cell suspensions were then washed with AIM-V 2% human AB serum, followed by another 30-minute incubation at 37°C. The stained T cells were then washed and resuspended in the corresponding media at 3 × 106 cells/ml. 30 μl of the T cell suspension was added into one of the two culture media channels flanking the central gel region of each device. Finally, the devices were incubated overnight in the stated conditions.
Live-imaging (time-lapse) experiments were performed using either a LSM7800 confocal microscope (Zeiss) or a FV1200 confocal microscope (Olympus) equipped with an environmental chamber set at 37°C and 5% CO2. The microscope was programmed in order to acquire Z-stacks of the selected regions at the stated time intervals. For static imaging experiments, confocal images of the same region of interest were acquired before T cell addition and after 15-hour incubation.
Liver carcinoma aggregate target cells.
Cell aggregates were prepared by using a customized Petri dish as described previously (21). Briefly, a laser cutter (Versa Laser, Universal Laser Systems) was used to control the melting of the Petri dish bottom surface, creating an array of microwells in which cells were allowed to settle and grow as aggregates. Pluronic solution (2 ml at 0.2% in PBS solution) was used to treat the surface of the modified Petri dish for 1 hour at room temperature in order to prevent cell adhesion and facilitate the formation of the cell aggregates. In order to obtain cell aggregates with the appropriate size (40- to 100-μm diameter) for use within the microdevice, HepG2 target cells were seeded at a concentration of 120,000 cells/ml and cultured for 3 consecutive days. Aggregates were collected and filtered in two consecutive filtration steps: (a) 40-μm filtration, in order to exclude all the cell aggregates smaller than 40 μm, and (b) 100-μm filtration, in order to exclude all the elements bigger than 100 μm. The cell aggregates were then mixed with the collagen solution in order to inject with the 3D matrix as described above.
2D well-based cytotoxicity assay
HepG2-Env target cells expressing luciferase were plated overnight in a 96-well flat bottom plate to permit adherence. Cells were washed with HBSS and cocultured with retroV-TCRe– or mRNA-TCRe–T cells at various E/T ratios in duplicates in AIM-V 2% human AB serum plus 100 IU/ml rIL-2 for 15 hours. Cytotoxicity was measured by quantifying luciferase expression in remaining target cells after coculture. Briefly, culture medium was discarded and 100 μl of Steady-Glo reagent (Promega) was added to each well and incubated for 5 minutes to allow cell lysis. Luminescence was measured with a microplate reader (Tecan). Target cells without effectors were used as a reference for maximum luminescence.
Data analysis
All images acquired from the confocal microscope systems (live and static imaging) were visualized and analyzed using Imaris (Bitplane). The quantity of the dissociated HepG2 target cells was detected based on their expression of GFP using the spots function within the software. To perform the analysis, the mean diameter of the dissociated HepG2 target cells was first determined by analyzing the maximum intensity projection images of GFP in Cell Profiler (33). The calculated mean diameter was then used as the estimated diameter for spots analysis in Imaris. By plotting the mean fluorescence intensity of DRAQ7 and GFP of the detected HepG2 target cells, one could clearly differentiate between live and dead cells (Figure 2B). The percentage of killed target cells was calculated by comparing the number of live HepG2 target cells at 0 hours and after incubation under various conditions.
For the experiments involving liver carcinoma cell aggregates, killing was quantified based on the volume of GFP and DRAQ7 expression. The data were determined using the surface function in Imaris. Surface reconstruction was first performed using GFP expression, followed by the surface reconstruction of DRAQ7 expression within the reconstructed GFP volume. Percentage cell death was calculated as the percentage of DRAQ7 volume of total GFP volume, while the percentage of killing was obtained by comparing the percentage of cell death before and after incubation with engineered T cells under the various conditions.
Supernatant analysis
Culture media from each individual device was collected directly from the microfluidic channels flanking the central collagen gel region (~160 μl total volume). Residual T cells in the supernatant were removed by centrifugation and the supernatants were stored at –80°C prior to analysis. Concentrations of secreted factors in the supernatants were assessed via cytokine multiplex bead-based array according to the manufacturer’s instructions (Luminex). Only secreted factors with at least 1 sample having a concentration above 10 pg/ml were analyzed.
HLA-I expression
Primary monoclonal antibody EMR8-5 (Abcam) against HLA class I molecule was used in combination with immunofluorescence to analyze the HLA expression on the cell surface. Cells were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 15 minutes at room temperature, permeabilized with 0.1% Triton X-100 in PBS for 10 minutes, and blocked with 3% BSA/5% goat serum in PBS for 2 hours. Cells were then incubated with primary antibody (1: 100) in PBS overnight at 4°C. The devices were rinsed 5 times with PBS and incubated in secondary antibodies (goat anti-mouse, Alexa Fluor 594 conjugate, 1:200) and NucBlue (Thermo Fisher Scientific) in 0.5% BSA/0.5% goat serum and PBS for 3 hours at room temperature. After incubation, the devices were washed with PBS and imaged using a Zeiss LSM780 confocal microscope (Carl Zeiss). The signal intensity and the cell number were analyzed with IMARIS (Bitplane). The plotted normalized value represents the total signal intensity divided by the number of cells present in the region of interest.
Statistics
Statistical significance was evaluated with 2-tailed t test and, where appropriate, 1-way ANOVA with Dunnett’s multiple comparisons test or 2-way ANOVA with Tukey’s multiple comparisons test using the data analysis software Prism 6. Only P values and adjusted P values (ANOVA) of less than 0.05 were considered significant and displayed in the figures.
Study approval
The present study using human PBMCs was reviewed and approved by the Institutional Review Board of the National University of Singapore. PBMCs were collected following informed consent from healthy donors.
Author contributions
Experiments were designed by AP, ATT, RDK, and AB and performed by AP, AT, SK, AC, MC, EA, CM, EC, GA, and MTR. The manuscript was written by AP and ATT and edited by RDK and AB. All authors discussed and interpreted the results.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation, Prime Minister’s Office, Singapore, under its CREATE programme; Singapore-MIT Alliance for Research and Technology BioSystems and Micromechanics IRG; and a Singapore Translational Research (STaR) Investigator Award (National Medical Research Council/STaR/013/2012) to AB.
Footnotes
Conflict of interest: A. Bertoletti collaborates and receives research support from Gilead Sciences to test the effect of HBV antigens on immune cell function. He acted as a consultant and served on the advisory boards of Gilead Sciences, Janssen-Cilag, Novartis, Ionis, MedImmune, and Abivax. A. Bertoletti is also a cofounder of Lion TCR Pte Ltd, a biotech company developing T cell receptors for treatment of virus-related cancers and chronic viral diseases. R.D. Kamm is a cofounder and board member of AIM Biotech Pte Ltd, which markets 3D microfluidic systems. A. Pavesi is a consultant for AIM Biotech Pte Ltd. A.T. Tan is a consultant for Lion TCR Pte Ltd.
Reference information:JCI Insight. 2017;2(12):e89762. https://doi.org/10.1172/jci.insight.89762.
Contributor Information
Andrea Pavesi, Email: andreap@imcb.a-star.edu.sg.
Anthony T. Tan, Email: anthony.tan@duke-nus.edu.sg.
Sarene Koh, Email: sarene_koh@sics.a-star.edu.sg.
Adeline Chia, Email: adeline.chia@duke-nus.edu.sg.
Marta Colombo, Email: marta3.colombo@mail.polimi.it.
Emanuele Antonecchia, Email: emanuele.antonecchia@mail.polimi.it.
Carlo Miccolis, Email: carlo.miccolis@mail.polimi.it.
Erica Ceccarello, Email: ceccarelloe@student.imcb.a-star.edu.sg.
Giulia Adriani, Email: giulia.adriani@smart.mit.edu.
Antonio Bertoletti, Email: antonio@sics.a-star.edu.sg.
References
- 1.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348(6230):62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischbach MA, Bluestone JA, Lim WA. Cell-based therapeutics: the next pillar of medicine. Sci Transl Med. 2013;5(179):179ps7. doi: 10.1126/scitranslmed.3005568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kershaw MH, Westwood JA, Darcy PK. Gene-engineered T cells for cancer therapy. Nat Rev Cancer. 2013;13(8):525–541. doi: 10.1038/nrc3565. [DOI] [PubMed] [Google Scholar]
- 4.Brentjens RJ, et al. CD19-targeted T cells rapidly induce molecular remissions in adults with chemotherapy-refractory acute lymphoblastic leukemia. Sci Transl Med. 2013;5(177):177ra38. doi: 10.1126/scitranslmed.3005930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kochenderfer JN, et al. Chemotherapy-refractory diffuse large B-cell lymphoma and indolent B-cell malignancies can be effectively treated with autologous T cells expressing an anti-CD19 chimeric antigen receptor. J Clin Oncol. 2015;33(6):540–549. doi: 10.1200/JCO.2014.56.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan RA, et al. Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother. 2013;36(2):133–151. doi: 10.1097/CJI.0b013e3182829903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkhurst MR, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol Ther. 2011;19(3):620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blank CU, Haanen JB, Ribas A, Schumacher TN. CANCER IMMUNOLOGY. The “cancer immunogram”. Science. 2016;352(6286):658–660. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 9.Caruana I, et al. Heparanase promotes tumor infiltration and antitumor activity of CAR-redirected T lymphocytes. Nat Med. 2015;21(5):524–529. doi: 10.1038/nm.3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang W, et al. Potentiating the antitumour response of CD8(+) T cells by modulating cholesterol metabolism. Nature. 2016;531(7596):651–655. doi: 10.1038/nature17412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menger L, et al. TALEN-mediated inactivation of PD-1 in tumor-reactive lymphocytes promotes intratumoral T-cell persistence and rejection of established tumors. Cancer Res. 2016;76(8):2087–2093. doi: 10.1158/0008-5472.CAN-15-3352. [DOI] [PubMed] [Google Scholar]
- 12.Adriani G, Pavesi A, Tan AT, Bertoletti A, Thiery JP, Kamm RD. Microfluidic models for adoptive cell-mediated cancer immunotherapies. Drug Discov Today. 2016;21(9):1472–1478. doi: 10.1016/j.drudis.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung KJ, Ewald AJ. A collective route to metastasis: Seeding by tumor cell clusters. Science. 2016;352(6282):167–169. doi: 10.1126/science.aaf6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joyce JA, Fearon DT. T cell exclusion, immune privilege, and the tumor microenvironment. Science. 2015;348(6230):74–80. doi: 10.1126/science.aaa6204. [DOI] [PubMed] [Google Scholar]
- 15.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zervantonakis IK, Hughes-Alford SK, Charest JL, Condeelis JS, Gertler FB, Kamm RD. Three-dimensional microfluidic model for tumor cell intravasation and endothelial barrier function. Proc Natl Acad Sci U S A. 2012;109(34):13515–13520. doi: 10.1073/pnas.1210182109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qasim W, et al. Immunotherapy of HCC metastases with autologous T cell receptor redirected T cells, targeting HBsAg in a liver transplant patient. J Hepatol. 2015;62(2):486–491. doi: 10.1016/j.jhep.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Gehring AJ, et al. Engineering virus-specific T cells that target HBV infected hepatocytes and hepatocellular carcinoma cell lines. J Hepatol. 2011;55(1):103–110. doi: 10.1016/j.jhep.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 19.Koh S, et al. A practical approach to immunotherapy of hepatocellular carcinoma using T cells redirected against hepatitis B virus. Mol Ther Nucleic Acids. 2013;2:e114. doi: 10.1038/mtna.2013.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.June CH. Principles of adoptive T cell cancer therapy. J Clin Invest. 2007;117(5):1204–1212. doi: 10.1172/JCI31446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tu TY, et al. Rapid prototyping of concave microwells for the formation of 3D multicellular cancer aggregates for drug screening. Adv Healthc Mater. 2014;3(4):609–616. doi: 10.1002/adhm.201300151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, et al. Engineering human tumor-specific cytotoxic T cells to function in a hypoxic environment. Mol Ther. 2008;16(3):599–606. doi: 10.1038/sj.mt.6300391. [DOI] [PubMed] [Google Scholar]
- 23.Savoldo B, et al. Treatment of solid organ transplant recipients with autologous Epstein Barr virus-specific cytotoxic T lymphocytes (CTLs) Blood. 2006;108(9):2942–2949. doi: 10.1182/blood-2006-05-021782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stenton SB, Partovi N, Ensom MH. Sirolimus: the evidence for clinical pharmacokinetic monitoring. Clin Pharmacokinet. 2005;44(8):769–786. doi: 10.2165/00003088-200544080-00001. [DOI] [PubMed] [Google Scholar]
- 25.Chang CH, Pearce EL. Emerging concepts of T cell metabolism as a target of immunotherapy. Nat Immunol. 2016;17(4):364–368. doi: 10.1038/ni.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan AT, et al. A longitudinal analysis of innate and adaptive immune profile during hepatic flares in chronic hepatitis B. J Hepatol. 2010;52(3):330–339. doi: 10.1016/j.jhep.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 27.Teng CF, Wu HC, Tsai HW, Shiah HS, Huang W, Su IJ. Novel feedback inhibition of surface antigen synthesis by mammalian target of rapamycin (mTOR) signal and its implication for hepatitis B virus tumorigenesis and therapy. Hepatology. 2011;54(4):1199–1207. doi: 10.1002/hep.24529. [DOI] [PubMed] [Google Scholar]
- 28.Geissler EK, et al. Sirolimus Use in Liver Transplant Recipients With Hepatocellular Carcinoma: A Randomized, Multicenter, Open-Label Phase 3 Trial. Transplantation. 2016;100(1):116–125. doi: 10.1097/TP.0000000000000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pavesi A, et al. Engineering a 3D microfluidic culture platform for tumor-treating field application. Sci Rep. 2016;6:26584. doi: 10.1038/srep26584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai J, et al. Contact-dependent carcinoma aggregate dispersion by M2a macrophages via ICAM-1 and β2 integrin interactions. Oncotarget. 2015;6(28):25295–25307. doi: 10.18632/oncotarget.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald JC, et al. Fabrication of microfluidic systems in poly(dimethylsiloxane) Electrophoresis. 2000;21(1):27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 32.Shin Y, et al. Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels. Nat Protoc. 2012;7(7):1247–1259. doi: 10.1038/nprot.2012.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.