Abstract
Fezf1 and Fezf2 are highly conserved transcription factors that were first identified by their specific expression in the anterior neuroepithelium of Xenopus and zebrafish embryos. These proteins share an N-terminal domain with homology to the canonical engrailed repressor motif and a C-terminal DNA binding domain containing six C2H2 zinc-finger repeats. Over a decade of study indicates that the Fez proteins play critical roles during nervous system development in species as diverse as fruit flies and mice. Herein we discuss recent progress in understanding the functions of Fezf1 and Fezf2 in neurogenesis and cell fate specification during mammalian nervous system development. Going forward we believe that efforts should focus on understanding how expression of these factors is precisely regulated, and on identifying target DNA sequences and interacting partners. Such knowledge may reveal the mechanisms by which Fezf1 and Fezf2 accomplish both independent and redundant functions across diverse tissue and cell types.
Introduction
Development of the mammalian nervous system encompasses multiple steps including differentiation of immature neurons and glia from stem cell precursors, migration of these immature cells to their final locations, extension of axons and dendrites, and establishment of mature neural circuits. This process is highly dependent upon transcriptional regulators that function to determine a cell’s identity, position, and connections within the nervous system. In this review we discuss recent progresses in understanding the roles of the zinc-finger transcription factors Fezf1 and Fezf2 during neurogenesis and the specification of distinct neuronal fates. We provide an overview of their distinct and overlapping functions during development of the olfactory system and forebrain, and highlight some unresolved questions. Determining the functions of these transcription factors during neural development has important implications for understanding brain patterning, neurogenesis, and cell fate specification.
The Fez family of zinc-finger transcription factors is evolutionarily conserved
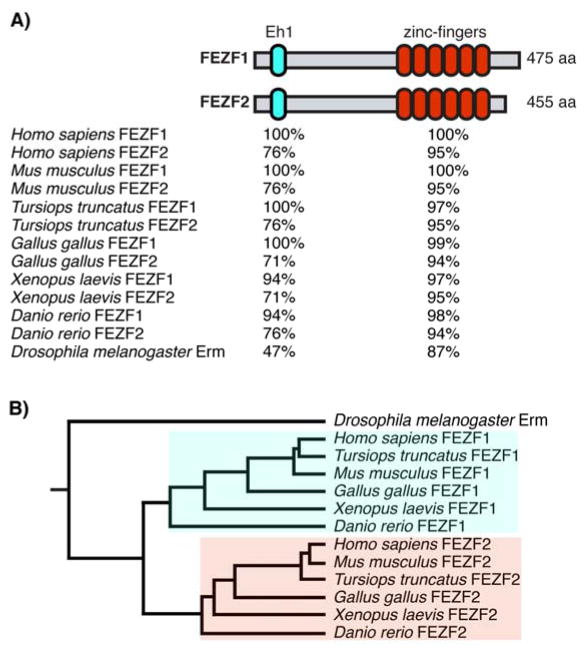
The Forebrain Embryonic Zinc Finger (Fez) family is a highly conserved family of transcription factors (Figure 1) that play roles in neurogenesis, developmental patterning, cell-fate specification and axon guidance [1]. The Drosophila genome contains a single Fez family homologue, Earmuff (erm), while in vertebrates there are two family members, Fezf1 (also known as Fez) and Fezf2 (also known as Fezl, and Zfp312) [2,3] (Figure 1B). These genes were first identified on the basis of their specific expression in the anterior neuroepitheulim of Xenopus and zebrafish embryos [4–6]. Both evolutionarily and within the same species, the Fez proteins exhibit high levels of homology, particularly within their DNA binding and protein-protein interaction domains (Figure 1A).
Figure 1.
The Fez family of transcription factors. A: Schematic of FEZF1 and FEZF2 proteins highlighting the strong evolutionary conservation of the Engrailed homology 1 (Eh1) and zinc-finger DNA binding domains. Conservation is reported as percent homology to Homo sapiens FEZF1. B: Phylogenetic tree of FEZ family transcription factors. A single duplication event appears to have created the two FEZ proteins from a common ancestor.
Interestingly, within these lower organisms the Fez family appears to play conserved roles in the development and fate specification of multiple neuronal structures (Table 1). In the developing Drosophila larval brain, erm is expressed in intermediate neural progenitor cells and is required to prevent their de-differentiation back into type II neuroblasts [2,7]. This function of erm is likely dependent on physical association with the Drosophila homologue of the SWI/SNF chromatin-remodeling complex [8]. The genomes of both Xenopus laevis and zebrafish contain homologues of both Fezf1 and Fezf2 (Figure 1A–B). In Xenopus these genes are expressed in the forebrain during early development [4,9]. However, aside from a conserved role in diencephalon patterning [9], little has been reported about the functions of these genes during development of the frog nervous system. In contrast, more extensive work in zebrafish has demonstrated that these factors are expressed in the forebrain and function in diencephalon patterning and neuronal differentiation [5,10–12]. Additionally, Fezf2 has been shown to play critical roles in the proper development of forebrain dopaminergic neurons [13–16].
Table 1.
Expression and functions of the Fez family in non-mammalian species.
| Organism | Gene Name(s) | Expression | Function(s) | References |
|---|---|---|---|---|
| D. melanogaster | erm (dfezf) | intermediate neural progenitors (INPs) | maintenance of INP differentiation | 2, 7–8 |
| X. laevis |
fezf1 (fez) fezf2 (fez-like) |
forebrain | diencephalon patterning | 4,9 |
| D. rerio |
fezf1 (fez) fezf2 (fez-like, too-few) |
forebrain | diencephalon patterning, neuronal differentiation, dopaminergic neuron specification | 5, 10–16 |
Fez Family members all share an N terminal Engrailed homology 1 (Eh1) repressor domain that is known to interact with the Groucho/TLE family of transcriptional co-repressors [5]. In addition, they contain six C2H2 zinc-finger DNA binding motifs at the C terminal. Previous work indicates that FEZF1 and FEZF2 can function redundantly to repress the transcription of a common set of target genes [3,17]. This observation should become increasingly important as new knowledge is gained about the genetic and molecular pathways in which these factors function.
Fezf1 and Fezf2 are expressed in unique and overlapping domains during development of the olfactory system and forebrain
In mice, expression of Fezf1 is first detectable in the head fold on embryonic day 7.5 (E7.5) [18]. At E8.5 Fezf1 and Fezf2 exhibit similar and overlapping expression patterns in the developing forebrain, where Fezf1 is expressed across a slightly larger domain than Fezf2 [19]. Two days later at E10.5, Fezf1 and Fezf2 are expressed in similar and overlapping patterns in the forebrain and olfactory system (Figure 2A). Fezf1 is expressed in neural progenitors throughout the forebrain and olfactory pit [17,19,20]. Fezf2 is also broadly expressed in progenitor cells of the forebrain, but in contrast to Fezf1, its expression within the olfactory pit is restricted to the progenitor cells that will give rise to the vomeronasal organ (VNO) [17,19,20]. This early restriction of Fezf2 expression to the future VNO positions it as the earliest marker that distinguishes progenitor cells that will give rise to the VNO from those that will generate the main olfactory epithelium (MOE).
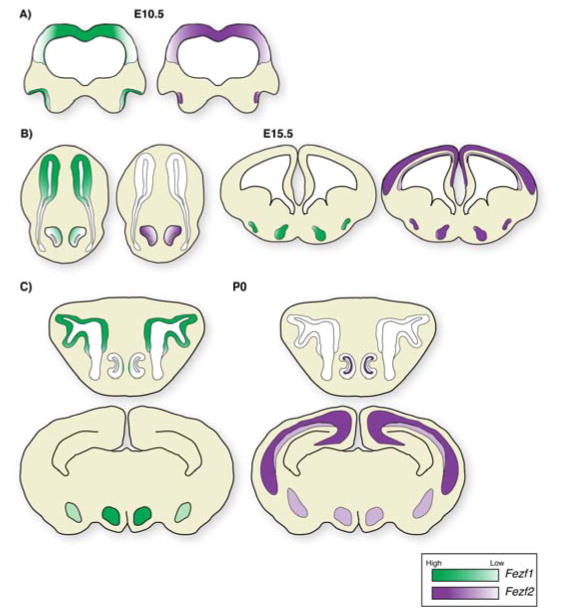
Figure 2.
Fezf1 and Fezf2 expression during embryogenesis. A: At E10.5 Fezf1 is expressed in progenitor cells in the forebrain and olfactory pit. Fezf2 is also expressed in forebrain progenitor cells; however within the olfactory pit its expression is restricted to the future VNO. B: By late gestation (E15.5), Fezf1 expression remains within the olfactory epithelium, but is absent from the cerebral cortex, and is decreasing within the VNO. In addition, it is expressed within the developing amygdala and hypothalamus. In contrast, Fezf2 expression remains high within the developing VNO and cerebral cortex. Within the cerebral cortex, Fezf2 is expressed at high levels in deep-layer postmitotic neurons and at a lower level in progenitor cells. Fezf2 is also expressed in the hypothalamus. C: At birth, expression of Fezf1 and Fezf2 largely mimics that at E15.5. Within the VNO, expression of Fezf2 has become restricted to the sustantacular cell layer. Within the cerebral cortex it maintains a high expression level in L5 SCPNs and lower levels in L6 CThPNs. Green represents Fezf1 expression, while purple represents Fezf2 expression.
As development proceeds, expression of these factors begins to demarcate distinct neuronal structures. At E15.5 Fezf1 is broadly expressed in progenitor cells and neurons of the developing MOE, but the expression declines within the developing VNO once it segregates away from the MOE [17,20,21] (Figure 2B). Within the brain, Fezf1 expression has subsided within the cerebral cortex, but it remains in the developing amygdala, ventral thalamus and hypothalamus [17–22] (Figure 2B). In contrast, Fezf2 is expressed within the VNO, neocortical progenitor cells, and newly born deep-layer projection neurons [17,20,21,23]. By this stage, Fezf2 expression begins to show a characteristic high expression level in layer five (L5) neurons, and lower levels of expression in L6 neurons and progenitors [23]. In addition, Fezf2 is expressed within the thalamic eminence, prethalamus, developing amygdala and hypothalamus [18,19,22] (Figure 2B).
By birth, expression of these factors becomes restricted to domains that largely mimic their adult expression patterns (Figure 2C). Expression of Fezf1 is confined to progenitor cells and sensory neurons of the MOE as well as neurons of the hypothalamus and amygdala [17,21,22]. In contrast, Fezf2 is expressed at high levels within L5 neurons of the cerebral cortex and at lower levels within L6 neurons and progenitors [23–26]. Additionally, it is expressed within the supporting cells of the vomeronasal organ, and neurons within the hypothalamus and amygdala [17,22]. The dynamic and overlapping expression patterns of these genes underscore their essential functions during development of multiple structures within the nervous system. However, the cellular and molecular mechanisms that regulate the expression of these genes remain largely unknown.
The Fez family is required for development of the main and accessory olfactory systems
During development of the murine olfactory system, Fezf1 is expressed in olfactory sensory neurons (OSNs) and their precursors. Genetic loss-of-function studies indicate that it is required for the maturation of OSNs and their innervation of the olfactory bulb [17,20,21]. OSNs that lack Fezf1 expressed decreased levels of mature OSN markers including olfactory receptors (ORs), G-protein subunit alpha, and olfactory marker protein (OMP) [17,20,21] (Figure 3A). Although the proneural gene Mash1 exhibited slightly lower levels of expression in the Fezf1 mutant MOE, neurogenesis appeared to be largely unaffected, as assayed by expression of NeuroD1, NeuroD6 and Ki67 [21]. In agreement with the failed maturation of Fezf1−/− OSNs, axons from these neurons failed to cross the cribriform plate and innervate the olfactory bulb [17,20,21] (Figure 3A). This defect was attributed to the inability of these axons to penetrate the meninges of the developing brain. Although the exact mechanisms remain to be explored, it seems likely that impaired protease function may at least in part underlie this defect [21].
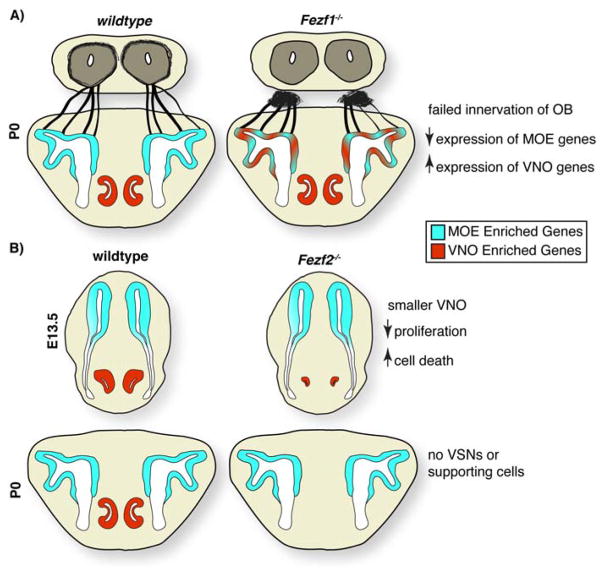
Figure 3.
Functions of Fezf1 and Fezf2 during olfactory system development. A: In Fezf1−/− mice, olfactory sensory neurons express decreased levels of MOE enriched genes and instead up-regulate VNO enriched genes. This is accompanied by the failure of OSN axons to cross the cribriform plate and innervate the olfactory bulb. B: The VNO initially segregates away from the developing olfactory pit in Fezf2−/− mice; however, it is substantially smaller. Because of decreased proliferation and increased apoptosis, VNO sensory neurons and sustantacular cells are completely absent at birth.
Fezf1 also plays a crucial role in the proper specification of OSN versus vomeronasal sensory neuron (VSN) identity [17]. Expression profiling of Fezf1−/− OSNs demonstrated that these neurons expressed genes normally enriched within VSNs. These included vomeronasal receptors class 1 and 2 (V1Rs, V2Rs), trace amine associated receptors (TAARS), and the VNO-specific ion channel Trpc2 [17] (Figure 3A). The ectopic expression of genes normally transcribed at high levels within the VNOs suggests that Fezf1 is a major cell fate determinant for specifying OSN identity. In addition, this raises the possibility that defects in OSN maturation and innervation of the OB may in part be due to the mixed cellular identity of the Fezf1−/− OSNs.
Fezf2 is expressed at high levels specifically within the developing VNO and is excluded from OSNs and progenitor cells of the MOE [17]. Within the VNO, Fezf2 is initially expressed in progenitor cells. However, by E16.5 its expression becomes restricted to sustantacular cells. In contrast to OSNs lacking Fezf1, Fezf2−/− VSNs exhibited decreased proliferation and underwent apoptotic cell death beginning around E12.5 (Figure 3B). These defects in cell proliferation, combined with increased cell death, ultimately resulted in the complete loss of VSNs and supporting cells by birth [17]. The early degeneration of the VNO in Fezf2 mutant animals precluded the analysis of cellular identity. However it seems probable that - similarly to up-regulation of VSN genes in the Fezf1−/− OSNs - the sustantacular cells in the VNO of Fezf2−/− mice may exhibit a mixed cellular identity. Supporting this, in utero electroporation of Fezf1 cDNA into upper-layer cortical neurons had the same effect on cortical neuron development as mis-impression of Fezf2 [17], hence indicating that these transcription factors can regulate the expression of a common set of genes. This observation raises the question of whether FEZF1 and FEZF2 are functionally interchangeable during the development of the mammalian nervous system. Additionally, it is unclear why some cell types express both Fezf1 and Fezf2 while others express only one of these factors.
Fezf1 and Fezf2 function redundantly during patterning of the diencephalon
The developing diencephalon can be subdivided into prethalaus, thalamus, hypothalamus, and pretectum. No defect in diencephalic patterning was observed in either Fezf1−/− or Fezf2−/− mice. However, examination of Fezf1−/−; Fezf2−/− double mutant mice revealed that these factors play redundant roles in patterning of these structures [19]. In double mutant animals, the prethalamus was completely lost and this was accompanied by a rostral expansion of the pretectum and a marked reduction in thalamic size [19]. Analysis of regional markers such as Gbx2 and Lhx1 during early development demonstrated that the rostral diencephalon failed to be specified, thus allowing a rostral expansion of the caudal diencephalon. These defects were accompanied by a loss of the zona limitans intrathalamica (ZLI) [19], an important patterning center located between the boundry of the prethalamus and thalamus [27]. Both Fezf1 and Fezf2 are expressed rostral to the ZLI boundry, which is demarcated by the expression of the transcription factor Irx1. Indeed, misexpression of either Fezf1 or Fezf2 caudual to the ZLI was sufficicent to repress formation of caudual diencephalon structures [19]. These results suggest that Fezf1 and Fezf2 function redundantly during early diencephalon patterning to repress a caudual diencephalon fate within the rostral diencephalon; furthermore, they raise the possibility that these factors may play hitherto unrecognized roles in the patterning of other neural strucutres.
Fezf1 and Fezf2 play unique and redundant roles in forebrain neurogenesis
Fezf1 and Fezf2 have both been implicated in controlling the neurogenic programs of neocortical progenitors during development of the cerebral cortex [3,28]. These factors were shown to bind the promoter of the basic helix-loop-helix transcription factor Hes5 and repress its transcription during early cortical neurogenesis [3]. In Fezf1−/− Fezf2−/− double mutant mice, expression of Hes5 was increased throughout the forebrain [3]. This led to decreased neurogenesis as assayed by expression of the proneural gene Neurogenin2 and the postmitotic neuron marker Tuj1 [3] (Figure 4A). Additionally, the TBR2-expressing basal progenitor cell population was reduced (Figure 4A). These neurogenic defects ultimately resulted in decreased numbers of projection neurons born during early corticogenesis (Figure 4A). Notably, defects in cortical neurogenesis were not observed in Fezf1 or Fezf2 single mutants, suggesting that these factors function redundantly in the regulation of Hes5 transcription and that loss of a single Fez family member is not sufficient to impair early neurogenesis [3]. Recent work has extended these observations, and suggests that the histone H2B ubiquitylation factor Bre1a functions upstream of Fezf1 and Fezf2 in the control of Hes5 expression [28]. Accordingly, perturbation of Bre1a expression in neural stem cells resulted in altered Hes5 levels and defects in neurogenesis [28].
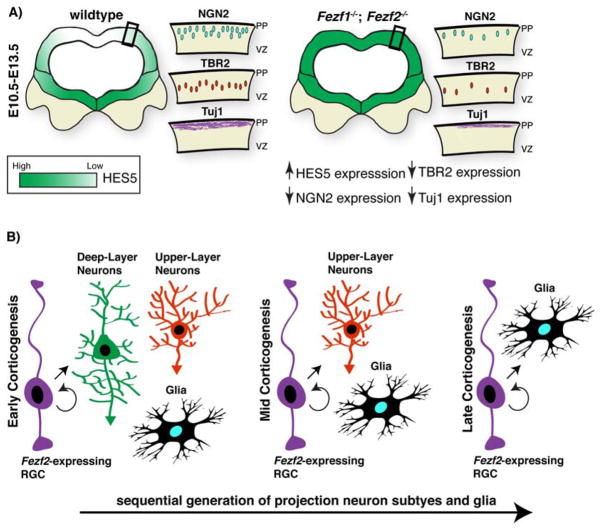
Figure 4.
Fezf1 and Fezf2 functions during forebrain neurogenesis. A: Loss of both Fezf1 and Fezf2 during early forebrain neurogenesis results in an increase in HES5 expression. This leads to decreased neurogenesis and intermediate progenitor generation and decreases in the number of deep-layer projection neurons at birth. B: Fezf2 is expressed in multipotent RGCs that sequentially generate all major cortical projection neuron subtypes and glia.
In contrast to Fezf1, which is only expressed in the cortex during its early stages of development, Fezf2 is expressed in neocortical progenitors throughout cortical neurogenesis [23]. Analyses of Fezf2−/− mice indicate that it is required for the generation and fate specification of subcerebral projection neurons in layer 5 (L5) [24–26,29,30] (Figure 5A). However, since expression of Fezf2 is also enriched in these post-mitotic neurons of L5, whether Fezf2 is required in L5 neurons or in neocortical progenitors has remained unclear. Indeed, it was speculated that expression of Fezf2 in early neocortical progenitors may direct these cells to generate deep-layer neurons, and thus Fezf2-expressing progenitors were predicted to be lineage-restricted to generate early-born, deep-layer neurons [31,32]. Recent in vivo lineage tracing experiments using the Fezf2 locus indicate that this is not that case, and that -- at both the population and clonal level -- Fezf2-expressing progenitor cells sequentially generate deep- and upper-layer projection neurons and glia in accordance with their birthdates [33] (Figure 4B). This observation suggests that Fezf2 likely has divergent functions within cortical progenitor cells and postmitotic neurons. Given that mis-expression of Fezf2 in late cortical progenitor cells (see below) is sufficient to generate ectopic subcerebrally-projecting neurons [24,26,29], the above finding indicates that simple expression of Fezf2 alone is not sufficient to promote a deep-layer neuron identity. To further our understanding of the mechanisms by which Fezf2 controls projection neuron fates, it will be important to dissect its precise functions in progenitor cells versus postmitotic neurons.
Figure 5.
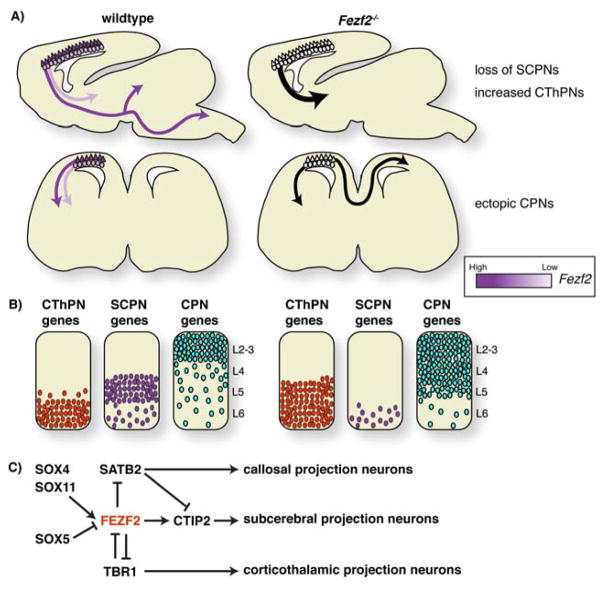
Control of neocortical projection neuron fate by Fezf2. A: In wildtype mice Fezf2 is expressed at high levels in L5 SCPNs that send axons to the midbrain, hindbrain and spinal cord and at lower levels in CThPNs that project to the thalamus. Fezf2−/− mice fail to generate subcerebral projections, and instead increased projections to the thalamus are observed. In the absence of Fezf2, L5 neurons extend axons across the corpus callosum, similar to CPNs. B: Gene expression changes after loss of Fezf2 suggest that it represses alternate CPN and CThPN fates. C: A model of the genetic interactions involving Fezf2 during generation of distinct cortical projection neuron subtypes.
Fezf2 is required for the proper fate specification of multiple classes of cortical projection neurons
The mammalian neocortex contains 6 layers of projection neurons. Neurons within the same layer tend to share similar morphology, axonal projection patterns, and gene expression profiles [34]. Based upon their axonal projection patterns, cortical projection neurons can be broadly divided into two classes: corticofugal projection neurons (CFuPNs) and cortico-cortical projection neurons. CFuPNs can be further divided into corticothalamic projection neurons (CThPNs) and subcerebral projection neurons (SCPNs). CThPNs send axons into the thalamus and their cell bodies predominantly reside within L6. In contrast, SCPN cell bodies are highly enriched in L5 and these neurons extend axons into subcerebral targets including the spinal cord, superior colliculus and brainstem. Cortico-cortical projection neurons send axons to other cortical areas, either within the ipsi- or contralateral cortical hemisphere (callosal projection neurons, CPNs). These neurons are present throughout layers 2–6, but are most concentrated within upper cortical layers (L2–3).
Specification of subcerebral projection neurons
Recent studies indicate that Fezf2 is an essential cell-fate determining gene for SCPNs. Fezf2 is expressed in radial glia cells (RGCs) in the ventricular zone and deep-layer neurons of the neocortex, exhibiting a high expression level in L5 and lower expression in L6 [23,35,36] (Figure 5A). In Fezf2 mutant mice, deep-layer projection neurons were not properly specified; SCPNs in L5 failed to send axons to subcerebral targets including the pons and spinal cord, and instead switched their fate to become CThPNs or CPNs [24–26,29,30,37]. Electrophysiological recording from individual L5 neurons in Fezf2 mutant mice demonstrated that mutant neurons changed their electrophysiology properties from that of a non-adapting cell type (subcerebral neuron identity) to an adapting cell type (callosal neuron identity) [29]. Further, expression of the transcription factor SATB2, which marks callosal projection neurons, was significantly increased in deep cortical layers [29] (Figure 5B).
In addition to the subcerebral-to-callosal identity switch, some L5 neurons in Fezf2 mutant mice changed their identity to become CThPNs. Expression of TBR1, a transcription factor that is specifically expressed in CThPNs, and is essential for their identity, was expanded into L5 [24,30]. A combination of retrograde tracing and birthdating experiments demonstrated that neurons born during the peak-time of L5 neurogenesis projected axons into the thalamus instead of their normal subcerebral targets. Collectively, these results indicate that Fezf2 is a crucial cell-fate determinant for subcerebral projection neurons, and suggest that subcerebral neuron fate is achieved through the repression of alternate callosal and corticothalamic fates (Figure 5C).
Genetic cross-repression specifies distinct neuronal fates in the developing cortex
Additional studies aimed at identifying cell-fate determining genes for other cortical projection neuron subtypes provide further support for the model that cortical projection neuron identity is achieved through the repression of alternate neuronal fates. SATB2, a DNA binding protein involved in chromatin remodeling, is expressed at high levels in callosal projection neurons, and was reported to be essential for the specification and execution of a callosal neuron fate [38,39]. In Satb2−/− mice, the mutant neurons did not send axons across the corpus callosum, and instead projected axons to subcortical targets [38,39]. Moreover, expression of the subcerebral neuron marker CTIP2 was significantly increased in upper cortical layers, suggesting a partial callosal to subcerebral neuron identity switch [38,39].
Similarly, analysis of Tbr1−/− mice demonstrated that it is required for the specification of corticothalamic neuron identity [30,40–42]. In Tbr1−/− mice, corticothalamic projections were absent and L6 neurons switched their identity to become SCPNs; they expressed SCPN markers and projected axons into subcerebral targets [30,40,41]. Among the observed gene expression changes was an increase in Fezf2 expression, and chromatin-immunoprecipitation experiments demonstrated that TBR1 binds directly to a conserved region in the 3′ end of the Fezf2 gene [30,40]. This interaction may function to repress high-levels of Fezf2 expression and a subcerebral neuron identity within L6 neurons. In support of this, misexpression of TBR1 in L5 neurons prevented these neurons from sending axons into the brainstem [30,40]. These results demonstrate that TBR1 is a major cell-fate determinant for CThPNs, and that it achieves this, at least in part, through repression of subcerebral neuron identity.
A common theme emerging from studies of Fezf2−/−, Satb2−/− and Tbr1−/− mice is that cortical projection neuron fate is achieved through the inhibition of alternate fates [43]. Indeed, analysis of Fezf2−/−; Tbr1−/− mice demonstrated that some of the defects observed in Fezf2−/− animals -- including development of corticospinal projections -- was partly restored [30]. In the future, it will be important to identify the downstream pathways of FEZF2, TBR1 and SATB2, and to determine whether, in addition to repressing alternate neuronal identities, these genes also activate the pathways required for subcerebral, corticothalamic and callosal neuron differentiation, respectively.
Fezf2 is sufficient to reprogram cortical projection neurons
Recent studies suggest that mis-expression of Fezf2 is sufficient to generate ectopic SCPNs. Initial experiments utilized in utero electroporation to introduce high-levels of Fezf2, driven from an extrachromosomal plasmid, into late-stage neocortical RGCs [24,26,29]. At this developmental time point, these RGCs normally generate upper-layer CPNs. However, projection neurons derived from electroporated RGCs extended axons to subcortical targets including the thalamus, pons and spinal cord [24,26,29]. In some cases, the induction of genes normally associated with deep-layer SCPNs was also reported, including TBR1 and CTIP2 [24,26]. Interestingly, electroporated neurons migrated to their normal positions in upper cortical layers, indicating that laminar position can be functionally dissociated from axonal and gene expression patterns [26,29].
Reprograming of ventral forebrain progenitors
Building upon these observations, more recent work focused on the ability of Fezf2 to reprogram ventral forebrain progenitors into CFuPNs [44]. Rouaux and Arlotta (2010) reported that mis-expression of Fezf2 in striatal progenitors by in utero electroporation was sufficient to induce the expression of genes enriched in deep-layer cortical projection neurons. These included SOX5, TBR1, BHLHB5 and FOG2 (also known as ZFPM2). The induction of CFuPN markers correlated with changes in cellular morphology from a stellate-like morphology (medium-spiny neuron) to a pyramidal-like morphology (cortical projection neuron). In addition, electroporated cells were reported to extend axons to subcerebral targets including the thalamus, cerebral peduncle and substantia nigra. The authors also reported that progenitors that had been electroporated with a Fezf2-expressing plasmid maintained expression of the ventral progenitor cell markers Mash1 and GSH2, and did not turn on expression of dorsal progenitor markers such as PAX6 or TBR2. This suggests that the reprograming does not transition through a dorsal-forebrain like progenitor cell stage. However, whether the reported reprograming was initiated at the progenitor cell or postmitotic stage remains unclear.
Postmitotic reprograming of cortical projection neurons
The reprograming ability of Fezf2 was further explored in two studies published at the beginning of 2013, which investigated the effects of mis-expressing Fezf2 in newly born postmitotic neurons [45,46]. De la Rosa et al. (2013) used a variant of in utero electroporation (iontoporation) to deliver Fezf2-expressing plasmids into postmitotic neurons of the L4 barrel cortex. These cells are the principal recipients of projections from the thalamus; they exhibit a characteristic spiny morphology and extend axonal projections within the cortex. When Fezf2 was mis-expressed postmitotically in L4 spiny neurons, the authors reported a change in cellular morphology to a pyramidal-like morphology, a redirection of axons to subcortical targets and a change in electrophysiological properties that were reminiscent of L5 neurons. In addition, the electroporated neurons were reported to shut off the expression of L4 markers including Rorβ, SATB2 and CUX1, and turn on the expression of the L5 marker ER81. Notably, the induction of the more broadly expressed deep-layer markers CTIP2 and SOX5 was not reported.
Using a similar in utero electroporation based strategy, Rouaux and Arlotta (2013) demonstrated that during a short temporal window following exit from cell cycle, newly-born cortical neurons destined for layers 2–4 could be coaxed into extending axons subcortically to the thalamus, cerebral peduncle and spinal cord. Additionally, the authors reported the induction of genes normally enriched in deep layers including ER81, SOX5, CRYM, CRIM1, TBR1 and FEZF2 itself. The molecular changes associated with the reprogramming lasted through at least the first postnatal month. Both of these studies reported that reprogramming of postmitotic neurons is still possible several weeks after birth; however, the efficiency dropped dramatically within the first 3 postnatal days. This suggests that the ability of newly-born neurons to respond to ectopic Fezf2 decreases rapidly following their exit from the cell cycle.
Although the ability to redirect axons to subcortical regions remains consistent across all of the studies described above, the molecular changes associated with the reported fate-switch are variable. In one study only changes in axonal targeting were reported [29]. However, additional reports indicate that the expression of genes normally associated with CSMNs may be induced -- though the extent of this induction remains unclear [24,44–46]. Notably, the induction of broadly expressed deep-layer markers such as CTIP2 is variable between different studies [45,46]. In future, it will be crucial to repeat these experiments using less invasive methods for mis-expression of Fezf2 such as inducible transgenic strategies. Combining stably expressed transgenes with the tamoxifen inducible Cre-Lox system would allow precise spatial and temporal control of Fezf2 expression within defined cell types. Finally, identifying additional genes that function with Fezf2 in controlling projection neuron fate specification should prove beneficial in understanding the full potential of Fezf2 to reprogram the fate of projection neurons within the developing forebrain.
Developmental regulation
Fezf1 was first identified through a screen in Xenopus embryos for genes that were induced in response to overexpression of Noggin [4]. Similarly, Fezf2 was isolated by screening for genes that were up-regulated in zebrafish in response to overexpression of the Wnt inhibitor Dkk1 [5,6]. Less is known about the control of these genes by secreted factors in mammals. However, recent work using differentiated human embryonic stem cells demonstrated that application of exogenous Wnt3a to the culture medium resulted in decreased Fezf2 expression while application of DKK1 increased Fezf2 expression [47]. This indicates that at least some of the same regulatory mechanisms that control transcription of these factors are conserved throughout evolution.
What are the regulatory mechanisms that spatially and temporally control Fezf2 transcription during development of the neocortex? Previous work indicates that multiple transcription factors directly bind to the Fezf2 locus to regulate its expression. SOX5, an Sry box-containing transcription factor enriched in L6 CThPNs, was shown to bind a highly-conserved noncoding element downstream of Fezf2, enhancer 434 (also known as E4) [36,48]. Binding of SOX5 to this enhancer was sufficient to repress transcription of a luciferase reporter, and Sox5−/− mice exhibit increased levels of Fezf2 expression within L6 of the cortex [48]. More recent work has identified two additional SOX family members, SOX4 and SOX11 that bind to enhancer 434 and, in contrast to SOX5, promote Fezf2 transcription [36]. Deletion of enhancer 434 resulted in a dramatic reduction of Fezf2 expression in the cortex and a loss of the corticospinal tract, phenocopying Fezf2−/− mice. Similarly, Sox4−/−; Sox11−/− double mutant mice displayed a loss of the corticospinal tract and reduced expression of deep-layer markers [36]. The temporal and molecular mechanisms by which these SOX family members function at enhancer 434 remain unknown; however a competition based model in which SOX5 and SOX4/SOX11 vie for occupancy of enhancer 434 to regulate Fezf2 transcription seems probable.
More recent work using transgenic reporter mice has demonstrated that, when isolated from its endogenous locus, enhancer 434 drove strong reporter activity in cortical progenitor cells [35]. Additionally, this study uncovered, a minimal promoter element consisting of 2.7 kilobases (kb) upstream of the Fezf2 start codon that was sufficient to drive reporter gene expression within both progenitor cells and postmitotic neurons of the cerebral cortex, throughout development. The authors also investigated the activity of a third non-coding element upstream of Fezf2, enhancer 1316. In contrast to enhancer 434, when assayed in isolation, enhancer 1316 was strictly active in postmitotic neurons. Collectively, this study concluded that the combined actions of enhancer 434, enhancer 1316, and the 2.7 kb promoter may function cooperatively to coordinate Fezf2 expressions across multiple spatial and temporal domains [35]. However, none of the isolated promoter and enhancer elements drove reporter gene expression in a pattern identical to that of endogenous Fezf2. This suggests that these elements may represent only a fraction of the overall cis-regulatory program controlling Fezf2 expression, similar to observations made in Drosophila through the study of pair-rule gene expression [49,50].
Additional work is needed to fully dissect the mechanisms that control Fezf2 expression. For instance, how the activity of enhancers 434 and 1316 are integrated within additional regulatory programs during cortical development, such as binding of TBR1 to the 3′ UTR of Fezf2 is unclear. Multiple transcription factors that play important roles in cortical development have been shown to bind around the Fezf2 locus [30,35,40,48]. However, the temporal order in which these factors occupy their binding sites as well as how they interact with one another is unknown. The recent identification of Fezf2-expressing radial glial cells as the progenitors for both deep- and upper-layer neurons and glia indicates that Fezf2 expression levels in progenitor cells and different types of cortical projection neurons are precisely controlled and thus highlights the need to finely dissect the regulatory mechanisms that control its expression. As additional cis-regulatory elements and trans-acting factors are uncovered it will become increasingly important to understand the extent to which they independently, and in corporation, regulate Fezf2 transcription.
Conclusions and Outlook
Fezf1 and Fezf2 are conserved zinc-finger transcription factors that play important roles during development of the forebrain and olfactory system. In the generation of both tissues, they appear to have distinct as well as redundant functions. How, and why, they function redundantly in some cases but not others remains unclear. However, it is possible that these factors may cooperate on essential tasks such as early patterning and neurogenesis in the forebrain, and as development proceeds they increasingly perform unique functions.
Going forward, we believe that pressing issues are to identify the cofactors that function with Fezf1 and Fezf2, to uncover their direct binding targets and to understand how transcription of these factors is spatially and temporally controlled. Given the high level of homology between Fezf1 and Fezf2 it seems likely that distinct protein-protein interactions may help to govern their activities in a tissue specific manner. Combining this knowledge with the identification of the regulatory elements and transcriptional regulators that control Fezf1 and Fezf2 transcription should clarify the mechanisms by which these factors control neurogenesis and neuronal fate specification across multiple cell and tissue types.
Acknowledgments
Research in the Chen lab was supported by grants from the California Institute of Regenerative Medicine (RN1-00530-01), the NIMH (R01-MH094589), and a Basil O’Connor Starter Scholar Research Award from the March of Dimes Foundation.
References
- 1.Shimizu T, Hibi M. Formation and patterning of the forebrain and olfactory system by zinc-finger genes Fezf1 and Fezf2. Development, growth & differentiation. 2009;51:221–31. doi: 10.1111/j.1440-169X.2009.01088.x. [DOI] [PubMed] [Google Scholar]
- 2.Weng M, Golden KL, Lee CY. dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Developmental cell. 2010;18:126–35. doi: 10.1016/j.devcel.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu T, Nakazawa M, Kani S, Bae YK, et al. Zinc finger genes Fezf1 and Fezf2 control neuronal differentiation by repressing Hes5 expression in the forebrain. Development. 2010;137:1875–85. doi: 10.1242/dev.047167. [DOI] [PubMed] [Google Scholar]
- 4.Matsuo-Takasaki M, Lim JH, Beanan MJ, Sato SM, et al. Cloning and expression of a novel zinc finger gene, Fez, transcribed in the forebrain of Xenopus and mouse embryos. Mechanisms of development. 2000;93:201–4. doi: 10.1016/s0925-4773(00)00264-1. [DOI] [PubMed] [Google Scholar]
- 5.Hashimoto H, Yabe T, Hirata T, Shimizu T, et al. Expression of the zinc finger gene fez-like in zebrafish forebrain. Mechanisms of development. 2000;97:191–5. doi: 10.1016/s0925-4773(00)00418-4. [DOI] [PubMed] [Google Scholar]
- 6.Hashimoto H, Itoh M, Yamanaka Y, Yamashita S, et al. Zebrafish Dkk1 functions in forebrain specification and axial mesendoderm formation. Developmental biology. 2000;217:138–52. doi: 10.1006/dbio.1999.9537. [DOI] [PubMed] [Google Scholar]
- 7.Irimia M, Pineiro C, Maeso I, Gomez-Skarmeta JL, et al. Conserved developmental expression of Fezf in chordates and Drosophila and the origin of the Zona Limitans Intrathalamica (ZLI) brain organizer. EvoDevo. 2010;1:7. doi: 10.1186/2041-9139-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koe CT, Li S, Rossi F, Wong JJ, et al. The Brm-HDAC3-Erm repressor complex suppresses dedifferentiation in Drosophila type II neuroblast lineages. eLife. 2014;3:e01906. doi: 10.7554/eLife.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Seguel E, Alarcon P, Gomez-Skarmeta JL. The Xenopus Irx genes are essential for neural patterning and define the border between prethalamus and thalamus through mutual antagonism with the anterior repressors Fezf and Arx. Developmental biology. 2009;329:258–68. doi: 10.1016/j.ydbio.2009.02.028. [DOI] [PubMed] [Google Scholar]
- 10.Yang ZG, Liu NG, Lin S. A zebrafish forebrain-specific zinc finger gene can induce ectopic dlx2 and dlx6 expression. Developmental biology. 2001;231:138–48. doi: 10.1006/dbio.2000.0139. [DOI] [PubMed] [Google Scholar]
- 11.Jeong JY, Einhorn Z, Mathur P, Chen L, et al. Patterning the zebrafish diencephalon by the conserved zinc-finger protein Fezl. Development. 2007;134:127–36. doi: 10.1242/dev.02705. [DOI] [PubMed] [Google Scholar]
- 12.Yang N, Dong Z, Guo S. Fezf2 regulates multilineage neuronal differentiation through activating basic helix-loop-helix and homeodomain genes in the zebrafish ventral forebrain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2012;32:10940–8. doi: 10.1523/JNEUROSCI.2216-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong JY, Einhorn Z, Mercurio S, Lee S, et al. Neurogenin1 is a determinant of zebrafish basal forebrain dopaminergic neurons and is regulated by the conserved zinc finger protein Tof/Fezl. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:5143–8. doi: 10.1073/pnas.0600337103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Levkowitz G, Zeller J, Sirotkin HI, French D, et al. Zinc finger protein too few controls the development of monoaminergic neurons. Nature neuroscience. 2003;6:28–33. doi: 10.1038/nn979. [DOI] [PubMed] [Google Scholar]
- 15.Russek-Blum N, Gutnick A, Nabel-Rosen H, Blechman J, et al. Dopaminergic neuronal cluster size is determined during early forebrain patterning. Development. 2008;135:3401–13. doi: 10.1242/dev.024232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blechman J, Borodovsky N, Eisenberg M, Nabel-Rosen H, et al. Specification of hypothalamic neurons by dual regulation of the homeodomain protein Orthopedia. Development. 2007;134:4417–26. doi: 10.1242/dev.011262. [DOI] [PubMed] [Google Scholar]
- 17.Eckler MJ, McKenna WL, Taghvaei S, McConnell SK, et al. Fezf1 and Fezf2 are required for olfactory development and sensory neuron identity. The Journal of comparative neurology. 2011;519:1829–46. doi: 10.1002/cne.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirata T, Suda Y, Nakao K, Narimatsu M, et al. Zinc finger gene fez-like functions in the formation of subplate neurons and thalamocortical axons. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;230:546–56. doi: 10.1002/dvdy.20068. [DOI] [PubMed] [Google Scholar]
- 19.Hirata T, Nakazawa M, Muraoka O, Nakayama R, et al. Zinc-finger genes Fez and Fez-like function in the establishment of diencephalon subdivisions. Development. 2006;133:3993–4004. doi: 10.1242/dev.02585. [DOI] [PubMed] [Google Scholar]
- 20.Hirata T, Nakazawa M, Yoshihara S, Miyachi H, et al. Zinc-finger gene Fez in the olfactory sensory neurons regulates development of the olfactory bulb non-cell-autonomously. Development. 2006;133:1433–43. doi: 10.1242/dev.02329. [DOI] [PubMed] [Google Scholar]
- 21.Watanabe Y, Inoue K, Okuyama-Yamamoto A, Nakai N, et al. Fezf1 is required for penetration of the basal lamina by olfactory axons to promote olfactory development. The Journal of comparative neurology. 2009;515:565–84. doi: 10.1002/cne.22074. [DOI] [PubMed] [Google Scholar]
- 22.Kurrasch DM, Cheung CC, Lee FY, Tran PV, et al. The neonatal ventromedial hypothalamus transcriptome reveals novel markers with spatially distinct patterning. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2007;27:13624–34. doi: 10.1523/JNEUROSCI.2858-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inoue K, Terashima T, Nishikawa T, Takumi T. Fez1 is layer-specifically expressed in the adult mouse neocortex. The European journal of neuroscience. 2004;20:2909–16. doi: 10.1111/j.1460-9568.2004.03763.x. [DOI] [PubMed] [Google Scholar]
- 24.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, et al. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–31. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 25.Chen B, Schaevitz LR, McConnell SK. Fezl regulates the differentiation and axon targeting of layer 5 subcortical projection neurons in cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17184–9. doi: 10.1073/pnas.0508732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JG, Rasin MR, Kwan KY, Sestan N. Zfp312 is required for subcortical axonal projections and dendritic morphology of deep-layer pyramidal neurons of the cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17792–7. doi: 10.1073/pnas.0509032102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends in neurosciences. 2010;33:373–80. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishino Y, Hayashi Y, Naruse M, Tomita K, et al. Bre1a, a Histone H2B Ubiquitin Ligase, Regulates the Cell Cycle and Differentiation of Neural Precursor Cells. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2014;34:3067–78. doi: 10.1523/JNEUROSCI.3832-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen B, Wang SS, Hattox AM, Rayburn H, et al. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:11382–7. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKenna WL, Betancourt J, Larkin KA, Abrams B, et al. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:549–64. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, et al. Molecular logic of neocortical projection neuron specification, development and diversity. Nature reviews Neuroscience. 2013;14:755–69. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franco SJ, Muller U. Shaping our minds: stem and progenitor cell diversity in the mammalian neocortex. Neuron. 2013;77:19–34. doi: 10.1016/j.neuron.2012.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo C, Eckler MJ, McKenna WL, McKinsey GL, et al. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80:1167–74. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kwan KY, Sestan N, Anton ES. Transcriptional co-regulation of neuronal migration and laminar identity in the neocortex. Development. 2012;139:1535–46. doi: 10.1242/dev.069963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eckler MJ, Larkin KA, McKenna WL, Katzman S, et al. Multiple conserved regulatory domains promote Fezf2 expression in the developing cerebral cortex. Neural Development. 2014 doi: 10.1186/1749-8104-9-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shim S, Kwan KY, Li M, Lefebvre V, et al. Cis-regulatory control of corticospinal system development and evolution. Nature. 2012;486:74–9. doi: 10.1038/nature11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komuta Y, Hibi M, Arai T, Nakamura S, et al. Defects in reciprocal projections between the thalamus and cerebral cortex in the early development of Fezl-deficient mice. The Journal of comparative neurology. 2007;503:454–65. doi: 10.1002/cne.21401. [DOI] [PubMed] [Google Scholar]
- 38.Britanova O, de Juan Romero C, Cheung A, Kwan KY, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–92. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 39.Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, et al. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–77. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Han W, Kwan KY, Shim S, Lam MM, et al. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3041–6. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bedogni F, Hodge RD, Elsen GE, Nelson BR, et al. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13129–34. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hevner RF, Shi L, Justice N, Hsueh Y, et al. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–66. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasan K, Leone DP, Bateson RK, Dobreva G, et al. A network of genetic repression and derepression specifies projection fates in the developing neocortex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:19071–8. doi: 10.1073/pnas.1216793109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rouaux C, Arlotta P. Fezf2 directs the differentiation of corticofugal neurons from striatal progenitors in vivo. Nature neuroscience. 2010;13:1345–7. doi: 10.1038/nn.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De la Rossa A, Bellone C, Golding B, Vitali I, et al. In vivo reprogramming of circuit connectivity in postmitotic neocortical neurons. Nature neuroscience. 2013;16:193–200. doi: 10.1038/nn.3299. [DOI] [PubMed] [Google Scholar]
- 46.Rouaux C, Arlotta P. Direct lineage reprogramming of post-mitotic callosal neurons into corticofugal neurons in vivo. Nature cell biology. 2012;15:214–21. doi: 10.1038/ncb2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kmet M, Guo C, Edmondson C, Chen B. Directed differentiation of human embryonic stem cells into corticofugal neurons uncovers heterogeneous Fezf2-expressing subpopulations. PloS one. 2013;8:e67292. doi: 10.1371/journal.pone.0067292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwan KY, Lam MM, Krsnik Z, Kawasawa YI, et al. SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16021–6. doi: 10.1073/pnas.0806791105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:13570–5. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrioli LP, Vasisht V, Theodosopoulou E, Oberstein A, et al. Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development. 2002;129:4931–40. doi: 10.1242/dev.129.21.4931. [DOI] [PubMed] [Google Scholar]