Abstract
The localization of prenylated Ras at the plasma membrane promotes activation of Ras by receptor tyrosine kinases and stimulates oncogenic signaling by mutant Ras. The Nogo-B receptor (NgBR) is a transmembrane receptor that contains a conserved hydrophobic pocket. Here, we demonstrate that the NgBR promotes the membrane accumulation of Ras by directly binding prenylated Ras at the plasma membrane. We show that NgBR knockdown diminishes the membrane localization of Ras in multiple cell types. NgBR overexpression in NIH-3T3 fibroblasts increases membrane-associated Ras, induces the transformed phenotype in vitro, and promotes the formation of fibrosarcoma in nude mice. NgBR knockdown in human breast cancer cells reduces Ras membrane localization, inhibits EGF-stimulated Ras signaling, and diminishes tumorigenesis of xenografts in nude mice. Our data demonstrate that NgBR is a unique receptor that promotes accumulation of prenylated Ras at the plasma membrane and promotes EGF pathways.
Keywords: Nogo-B receptor, Ras, tumorigenesis, receptor tyrosine kinase, Prenylation
Introduction
Ras GTPases and their mutants are well-characterized oncogenes causing cell transformation and tumorigenesis2, 23, 25. The events that promote membrane localization of these GTPases have translational importance, because oncogenic signaling by these small GTPases depends on their interactions with regulators and effectors at the plasma membrane11, 56. Galectin-1 and galectin-3 are cytosolic proteins with hydrophobic farnesyl-binding pockets that strengthen the association of GTP-bound H-Ras and K-Ras at the plasma membrane in association with non-lipid raft subdomains3, 52, 57. Bindings of PDEδ is essential to maintain the subcellular localization of several farnesylated Ras variants7, 18, 35. Caveolin-1 acts as a membrane docking site in lipid raft domains for binding GDP-loaded H-Ras38, 44, 50, 51, 53. However, transmembrane proteins that act as docking sites in non-lipid raft domains for GDP/GTP-loaded H-Ras and K-Ras have not been identified. Here, we demonstrate that NgBR, a transmembrane receptor outside of caveolae-rich lipid rafts, recruits both GDP- and GTP-bound H-Ras and K-Ras to the plasma membrane and is a major promoter of oncogenic signaling by these GTPases.
NgBR was identified as a receptor specific for Nogo-B33, and is involved in blood vessel development42, 47, 69. Genetic depletion of NgBR results in early stage embryonic lethality42, 47. NgBR also is highly expressed in late stages of invasive ductal carcinoma63 and promotes the epithelial-mesenchymal transition of breast tumor cells70 as well as the chemo-resistance of human hepatocellular carcinoma14. However, the underlying mechanism by which NgBR promotes endothelial cell migration and tumor cell growth is unknown. As discussed in our previous report33, the cytoplasmic domain of NgBR has a high degree of homology to the Cis-Isoprenyl Diphosphate Synthases (Cis-IPPS) family of lipid modifying enzymes, which have conserved hydrophobic pockets for binding isoprenyl lipids and/or prenylated proteins29. Similar hydrophobic pockets occur in other proteins that bind prenylated GTPases, such as PEDδ7, 18 and RhoGDI1, 20. Our previous results showed that NgBR has negligible lipid transferase activity33. Here, we demonstrate that the hydrophobic cytoplasmic domain of NgBR acts as a docking site for binding prenylated Ras and promotes the plasma membrane accumulation and activation of both H-Ras and K-Ras. Our findings will facilitate further understanding of the mechanisms that localize and activate Ras GTPases at the plasma membrane.
Results
NgBR binds farnesylated Ras and enhances Ras plasma membrane localization
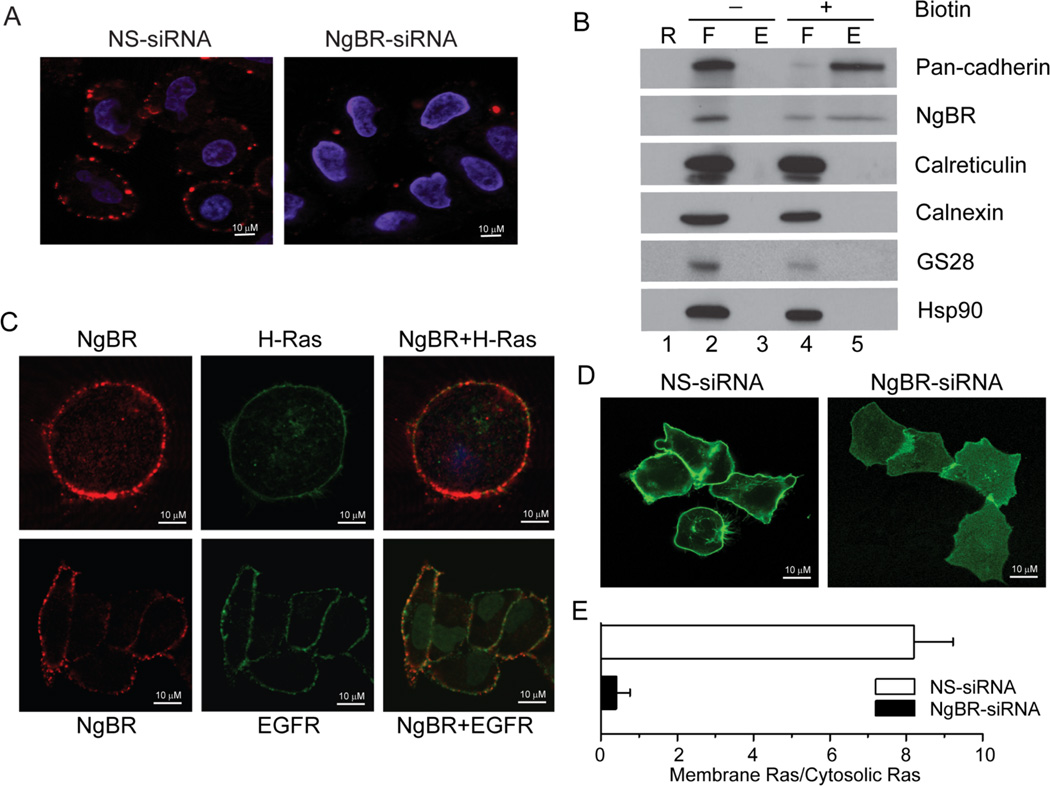
As shown in Figure 1A, NgBR is sporadically distributed at the plasma membrane of non-permeabilized HeLa cells. Specific immunofluorescence (IF) staining was diminished at the plasma membrane when the HeLa cells were transfected with validated NgBR siRNA (Fig.1A) or when the primary antibody was pre-incubated with blocking peptide (Figure S1A, upper panel). NgBR were also detected in the cell surface protein fraction (Figure 1B, lane 5) isolated by the cell surface biotinylation approach. Interestingly, IF staining indicated that NgBR can be partially co-localized with H-Ras and EGF receptors (EGFR) at the plasma membrane (Fig. 1C), but NgBR does not co-localize with caveolin-1 (Fig. S1B). These results indicate that the NgBR is excluded from the caveolae-rich lipid raft domain of the plasma membrane. The localization of EGFP-H-Ras at the plasma membrane (Fig 1D left panel) was diminished by depletion of NgBR with siRNA, causing most of the EGFP-H-Ras to accumulate in the cytosolic compartment (Fig. 1D, right panel). The distribution of EGFP fluorescence signal intensity across the three-dimensional reconstruction of the z-stack image is shown in Figure S2. The results clearly demonstrated that depletion of NgBR reduced the intensity of EGFP-H-Ras at the plasma membrane (Fig. S2). Quantitative imaging analysis indicated that EGFP-H-Ras was localized at the plasma membrane in 5% of the HeLa cells transfected with NgBR siRNA, compared to more than 80% of the HeLa cells transfected with non-targeting siRNA (Fig. 1E). Similarly, NgBR is also required for K-Ras translocation to the plasma membrane (Fig. S3A). The RNAi-mediated depletion of NgBR in HeLa cells not only reduces plasma membrane localization of wild-type H-Ras, but also diminishes the membrane localization of the constitutively activated H-Ras mutant (H-Ras G12V) and dominant negative H-Ras mutant (H-Ras S17N) (Fig. S3B). We found that depletion of NgBR does not diminish H-Ras farnesylation (Fig. S4A, lane 3), which is diminished by treatment with a farnesylation inhibitor (FTI277) (Fig. S4A, lane 5). As shown in Figure S4B, NgBR knockdown in HeLa cells does not change the expression levels of either Caveolin-1 or Galectin-1 and -3. These results suggest that NgBR regulates Ras plasma membrane accumulation via a different mechanism independent of caveolin-1 and galectin-1/-3.
Figure 1. NgBR is essential for H-Ras plasma membrane localization.
(A) NgBR is localized in the plasma membrane of non-permeabilized HeLa cells. Immunofluorescence staining was performed using NgBR rabbit polyclonal antibody specially recognizing the ecto-domain of NgBR. (B) NgBR is detected in the fraction of biotinylated cell surface proteins. HeLa cell surface proteins were biotinylated under non-permeabilized conditions and isolated using streptavidin agarose resin from the Pierce Cell Surface Protein Isolation Kit. Proteins were detected by Western blot analysis. Pan-Cadherin, Calreticulin and GS28 are plasma membrane, ER membrane and Golgi membrane markers, respectively. The plus symbol (+) denotes results for cells treated with the Sulfo-NHS-SS-Biotin reagent; the minus symbol (−) denotes results for cells that were not treated with the biotin reagent but were otherwise carried through the kit procedure. The lane designated “R” shows control samples that were isolated in the absence of avidin-agarose resin, the lanes designated “F” show proteins that flowed through the columns because they did not bind the avidin-agarose resin, and the lanes designated “E” show proteins that were eluted from the columns after binding to the avidin-agarose resin. As expected, surface proteins bound to the avidin-agarose resin if the cells were labeled with biotin (“E”, lane 5), but surface proteins flowed through the columns if the cells were not labeled with biotin (“F”, lane 2). (C) NgBR is co-localized with EGFP-H-Ras in EGFP-H-Ras transfected HeLa cell (upper panel) and NgBR is partially co-localized with EGFR in HeLa cell plasma membrane (bottom panel). (D) NgBR knockdown impaired the plasma membrane localization of EGFP-H-Ras in HeLa cells. (E) Quantification results were presented as the ratio of cells showing EGFP-H-Ras plasma membrane location to cells showing EGFP-H-Ras cytoplasma location. Data are represented as mean ± SEM (* P< 0.05, n=3).
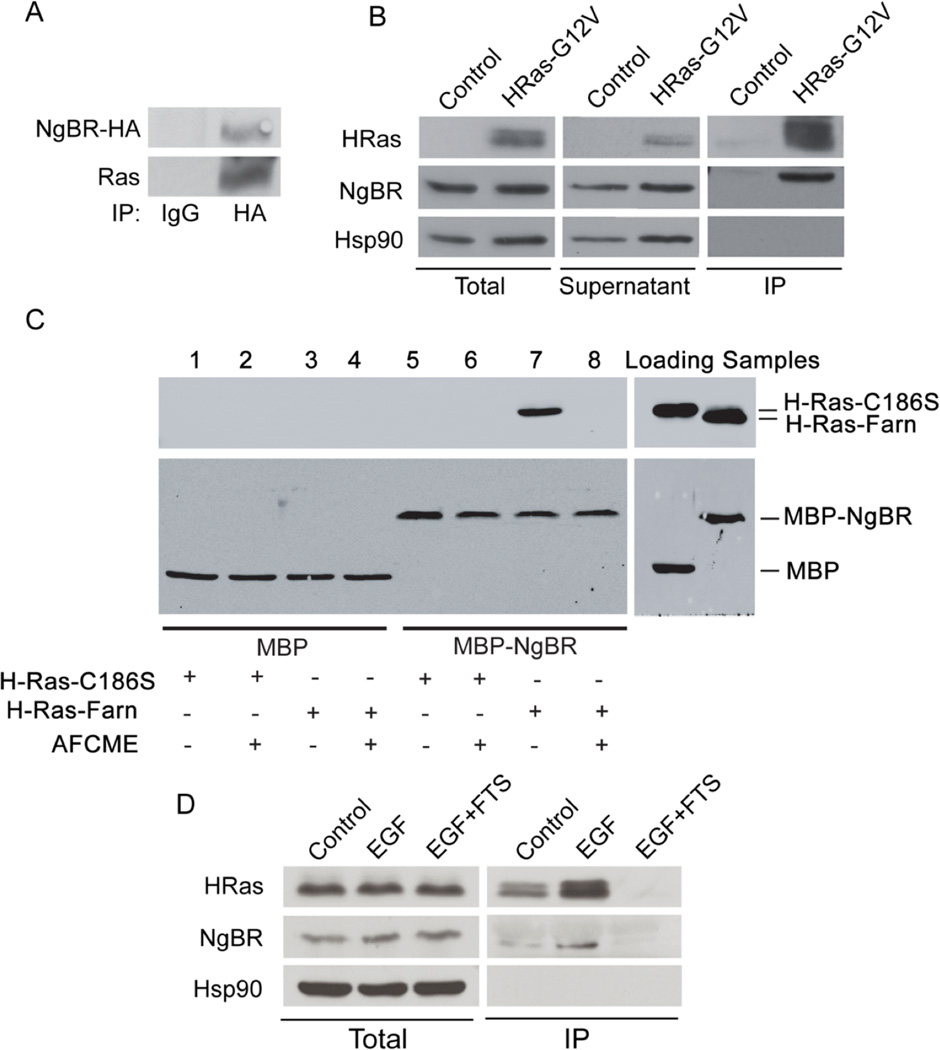
To assess the ability of NgBR to form a stable complex with H-Ras, we transfected either NgBR-HA plasmid DNA to HEK293T cells (Fig. 2A) or constitutively activated H-Ras-G12V mutant plasmid DNA to HeLa cells (Fig. 2B), and detected both NgBR and Ras in the complex of NgBR-HA immunoprecipitation (Fig. 2A) or in the activated Ras complex (Fig. 2B, right panel) pulled down by using beads conjugated to the GST-tagged Ras-binding domain (RBD) of Raf, which can specifically pull-down GTP-loaded Ras60. These results suggest that NgBR regulates H-Ras by forming a stable complex with H-Ras.
Figure 2. Prenylation of H-Ras is essential for NgBR-Ras interaction.
(A) Endogenous Ras was detected in the complex of NgBR-HA immunoprecipitated from 293T cells transfected with NgBR-HA plasmid DNA. (B) Endogenous NgBR interacts with the constitutively activated H-Ras. Plasmid DNA of H-Ras-G12V mutant was transfected to HeLa cells. The complex of activated Ras (GTP-loaded Ras) was precipitated from the quiescent cells using GST-RBD beads. Protein levels were detected by Western blotting. Both Ras and endogenous NgBR were detected in the complex precipitated by the Raf-pull-down method. (C) NgBR interacts with Ras directly in a cell-free system. AFCME: a farnesyl analog, N-Acetyl-S-farnesyl-L-cysteine-methyl ester. Protein levels were detected by Western blotting. Purified MBP (Maltose binding protein)-tagged NgBR (MBP-NgBR) was incubated with either unfarnesylated Ras (H-Ras-C186S, containing a mutation at the cysteine of CAAX box) or farnesylated Ras (H-Ras-Farn). The complex was immunoprecipitated using anti-MBP magnetic beads (New England BioLabs). (D) Blocking prenyl-dependent interactions of H-Ras diminishes its interactions with NgBR. HeLa cells were stimulated with 100 ng/ml EGF for 5 minutes. The complex of the GTP-loaded H-Ras was pulled down using GST-RBD beads. FTS pretreatment (20µM for overnight) abolished the interaction of GTP-loaded H-Ras and NgBR. FTS: S-trans, trans-farnesylthiosalicylic acid. Data are validated in 3 independent experiments.
Farnesylation of the CAAX box of H-Ras is generally required for H-Ras to localize at the plasma membrane38, 39, 45. To elucidate whether NgBR directly binds farnesylated H-Ras, purified NgBR tagged with Maltose binding protein (MBP-NgBR) was incubated with non-prenylated H-Ras (H-Ras-C186S, a mutation at the cysteine of CAAX box) or with farnesylated H-Ras (H-Ras-Farn). IP using anti-MBP magnetic beads indicated that MBP-NgBR forms strong complex with farnesylated H-Ras (Fig. 2C, lane 7), and the complex formation can be blocked using the farnesyl analogs N-Acetyl-S-farnesyl-L-cysteine-methyl-ester (AFCME) (Fig. 2C, lane 8). There was no complex formation between MBP-NgBR and unfarnesylated H-Ras-C186S (Fig. 2C, lane 5). Sequence alignment of the cytoplasmic domain of NgBR with human and bacterial Cis-Isoprenyl Diphosphate Synthases (hCIT and UPPS, respectively) (Fig. S5A) showed that several conserved hydrophobic residues, such as I117, L120, V122, are essential for binding farnesyl diphosphate (FPP), and that E262 are critical for maintaining the structure of the farnesyl group-binding pocket8, 9, 16. Site-directed mutagenesis of the three residues I117, L120 and V122 to alanine (designated the TM mutant) or E262 to alanine (E262A), but not L186 to alanine (L186A), diminished the ability of MBP-NgBR to form a stable complex with H-Ras-farn (Fig. S5B). These results suggest that NgBR preferentially binds farnesylated H-Ras, and the hydrophobic residues at the carboxyl terminus of NgBR are critical for binding farnesylated Ras. The ability of endogenous NgBR to form a complex with the activated H-Ras is further indicated by pull-down assays with GST-RBD beads (Fig. 2D). EGF stimulation in HeLa cells not only increases the activation of endogenous H-Ras, but also increases the amount of NgBR associated with the activated H-Ras (Fig. 2D). The specificity of NgBR for binding the activated H-Ras is validated by our finding that treatment with S-trans, trans-farnesylthiosalicylic acid (FTS), an inhibitor blocking translocation of farnesylated Ras to the plasma membrane40, diminishes the ability of endogenous NgBR to form a complex with endogenous H-Ras (Fig. 2D). Taken together, these findings demonstrate a strong interaction between NgBR and Ras that is dependent on Ras farnesylation.
The CAAX motif of H-Ras is critical for binding NgBR
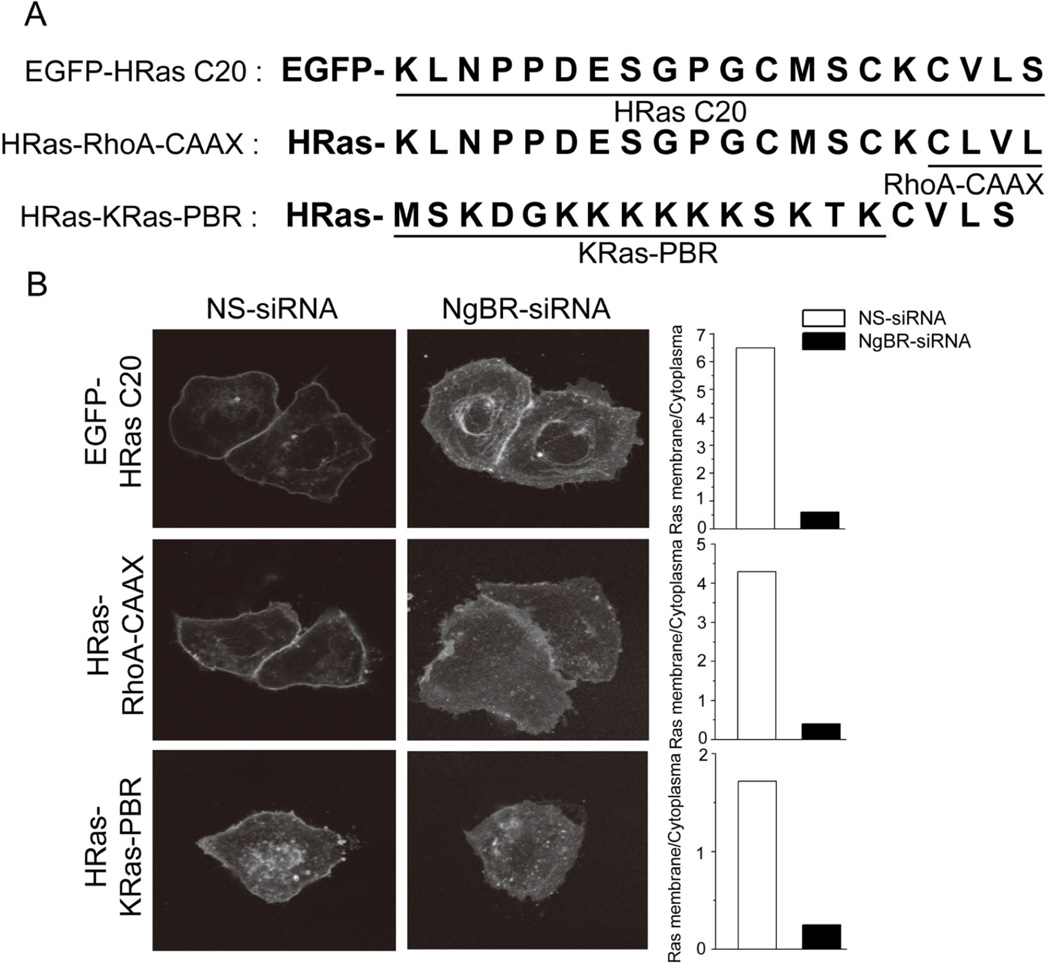
To elucidate which domain of H-Ras is important for binding with NgBR, we examined the intracellular localization of EGFP fused to 1) the C-terminal 20 residues of H-Ras (H-RasC20), 2) full-length H-Ras that has its CAAX motif (CVLS) replaced with the RhoA CAAX motif (CLVL) (H-Ras-RhoA-CAAX), and 3) full-length H-Ras that has its aa 170 – 185 (klnppdesgpgcmsck) replaced with aa 170–185 of K-Ras constituting the polybasic region (mskdgkkkkkksktkc) (H-Ras-K-Ras-PBR)25, 43, 66, 71. The diagram of these mutants is presented in Figure 3A. As shown in Figure 3B, EGFP-H-RasC20 localizes at the plasma membrane just as EGFP-H-Ras does (Fig. 1D). NgBR knockdown prevents the accumulation of EGFP-H-RasC20 at the plasma membrane, indicating that the interaction between NgBR and H-Ras is dependent on the C20 domain of H-Ras and independent of the N-terminal catalytic domain of H-Ras. We found that the H-Ras-RhoA-CAAX mutant that is geranylgeranylated also localizes at the plasma membrane, and this localization is diminished by NgBR knockdown, indicating that the NgBR can recruit geranylgeranylated GTPases just as it recruits farnesylated GTPases. Previous reports49, 54 demonstrated that palmitoylation of C181 and C184 residues in H-Ras enhances H-Ras binding affinity to lipid raft microdomains of the plasma membrane, and loss of palmitoylation results in the localization of H-Ras at intracellular compartments such as the Golgi. As shown in Figure 3B, the H-Ras-K-Ras-PBR mutant possessing the K-Ras PBR localizes at both the plasma membrane and Golgi/ER regions. NgBR knockdown significantly abolished the plasma membrane localization of H-Ras-K-Ras-PBR, but not the Golgi/ER location of H-Ras-K-Ras-PBR. This finding suggests that NgBR-regulated H-Ras plasma membrane translocation is dependent on prenylation of the CAAX motif but is independent of the palmitoylation of H-Ras. Collectively, these results indicate that NgBR binds prenylated Ras, which is critical for Ras plasma membrane accumulation.
Figure 3. The CAAX motif of H-Ras is critical for binding NgBR.
(A) The diagram shows the H-Ras domain mutants. EGFP-HRas C20: EGFP fused to the C-terminal 20 residues of H-Ras; HRas-RhoA-CAAX: EGFP fused to full-length H-Ras that has its CAAX motif (CVLS) replaced with the RhoA CAAX motif (CLVL); HRas-KRas-PBR: EGFP fused to full-length H-Ras that has its aa 170 – 185 (klnppdesgpgcmsck) replaced with aa 170–185 of K-Ras constituting the polybasic region (mskdgkkkkkksktkc). (B) NgBR knockdown inhibited membrane localization of EGFP-HRasC20 and HRas-KRas-PBR. Images of EGFP-HRas membrane localization (left panel), and quantification of the results (right panel) are shown.
NgBR overexpression promotes Ras membrane localization and recapitulates the tumorigenic functions of Ras GTPases
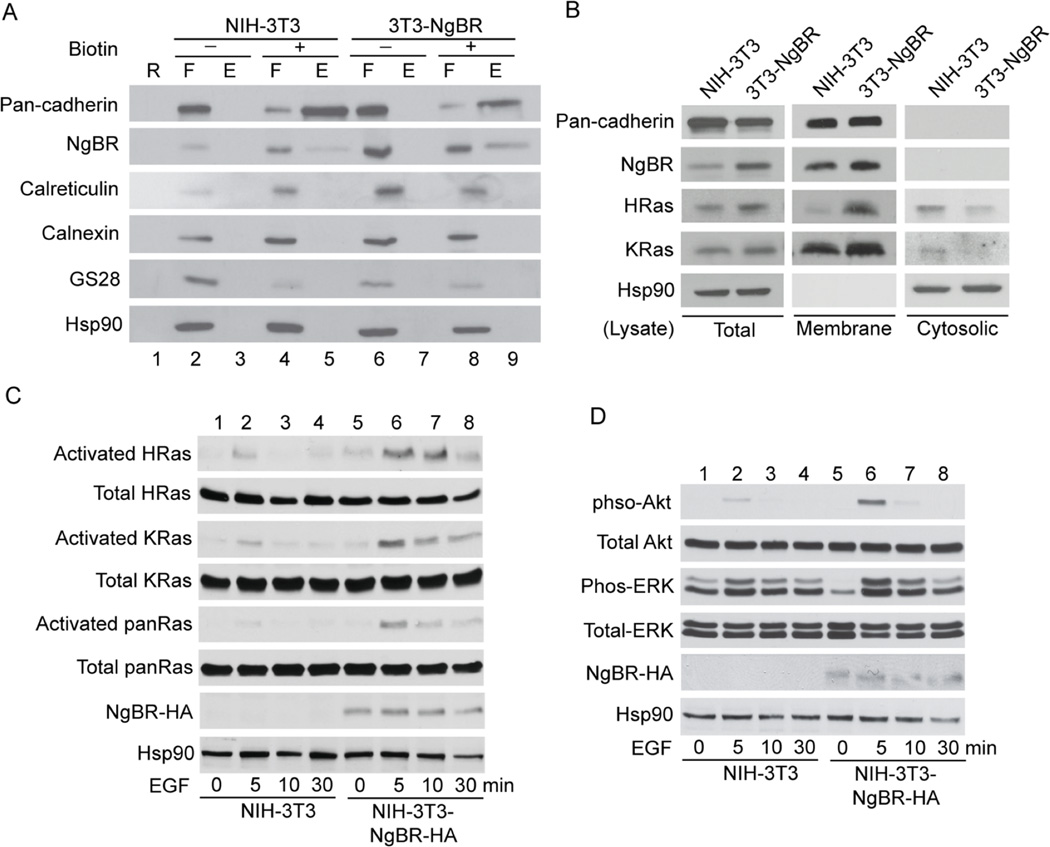
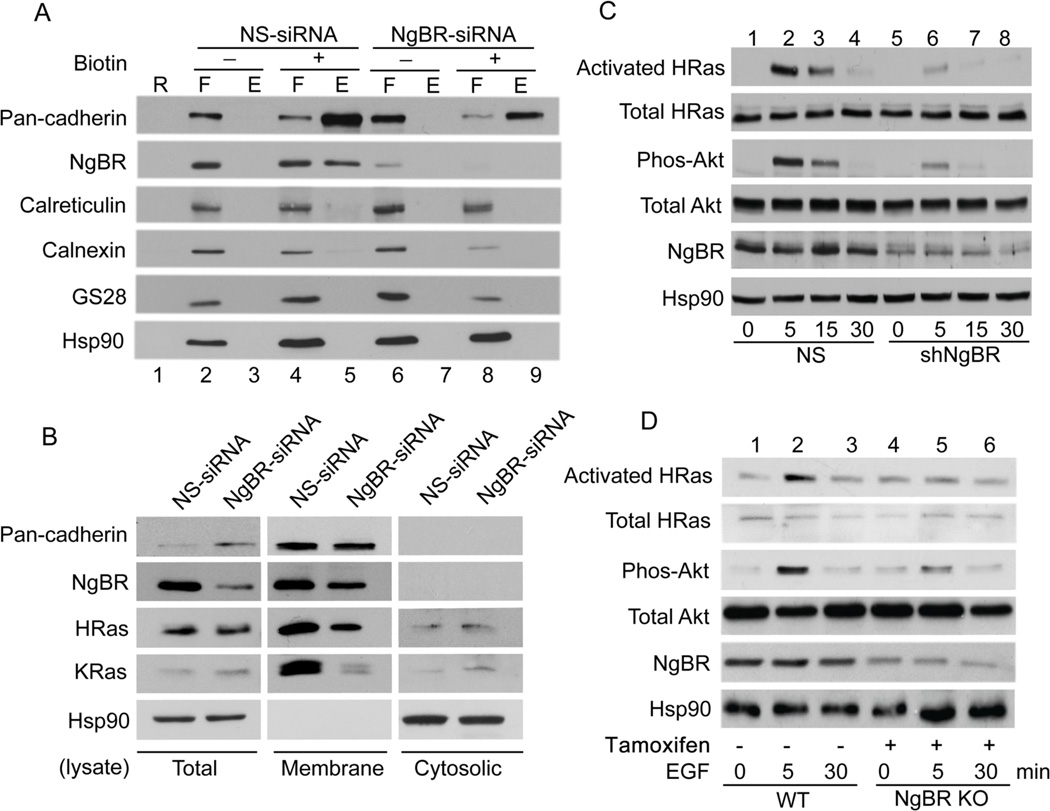
To further evaluate the plasma membrane localization of Ras by NgBR, we established an NIH-3T3 stable cell line overexpressing NgBR-HA. The localization of NgBR-HA in the plasma membrane of NIH-3T3-NgBR-HA cells was confirmed using a cell surface biotinylation assay. The levels of biotinylated NgBR in the cell surface fraction of NIH-3T3-NgBR-HA cells (Fig. 4A, lane 9) are much higher than that in NIH-3T3 cells (Fig. 4A, lane 5). The purity of the isolated cell surface membrane fraction was confirmed by no contamination of endoplasmic reticulum (Calreticulin) and Golgi (GS28) membrane proteins, respectively. As shown in Fig. 4B, NgBR overexpression increases the levels of membrane-associated H-Ras and K-Ras without significantly increasing the total protein levels of H-Ras and K-Ras. NgBR overexpression does not increase the levels of membrane-associated pan-cadherin, indicating that the increased membrane association of Ras in NIH-3T3-NgBR-HA cells is not due to a general increase in membrane-associated proteins. The interaction between activated Ras and NgBR in NIH-3T3-NgBR-HA cells is shown in Figure S6. Activation of EGF receptors in NIH-3T3-NgBR-HA cells increases the amount of NgBR being pulled down with H-Ras and K-Ras using GST-RBD beads, most likely because EGF stimulates Ras activation at the plasma membrane as described in previous reports6, 23, 31, 32. As shown in Fig. 4C, EGF-stimulated activation of both H-Ras and K-Ras significantly increases in NIH-3T3-NgBR-HA cells as compared to control NIH-3T3 cells. Consequently, overexpression of NgBR-HA enhances EGF-stimulated phosphorylation of Akt and ERK in NIH-3T3 cells (Fig. 4D). However, overexpression of NgBR-HA does not increase the phosphorylation of EGF receptor (EGFR) (Fig. S7, lane 4).
Figure 4. NgBR over-expression promotes Ras membrane accumulation and EGF-stimulated Ras activation.
(A) Over-expression of NgBR increases the NgBR protein levels in the fraction of biotinylated cell surface proteins. NIH-3T3 cell surface proteins were biotinylated under non-permeabilized conditions and isolated using streptavidin agarose resin from the Pierce Cell Surface Protein Isolation Kit as described in Figure 1B. Proteins were determined by Western blot analysis using antibodies that detect endogenous proteins. (B) Over-expression of NgBR in NIH-3T3 cells promotes H-Ras and K-Ras membrane localization. The plasma membrane proteins were isolated by the ultracentrifugation method. The protein levels were determined by Western blotting using antibodies that detect endogenous proteins. (C) Over-expression of NgBR in NIH-3T3 cells increases the EGF-induced H-Ras, K-Ras and pan-Ras activation. The activated Ras proteins were isolated using GST-RBD beads and protein levels were determined by Western blotting. (D) Over-expression of NgBR in NIH-3T3 cells increases the EGF induced phosphorylation of Akt and ERK. Phosphorylation of Akt and ERK was determined by Western blotting using phosphorylation specific antibodies. Data are validated in 3 independent experiments.
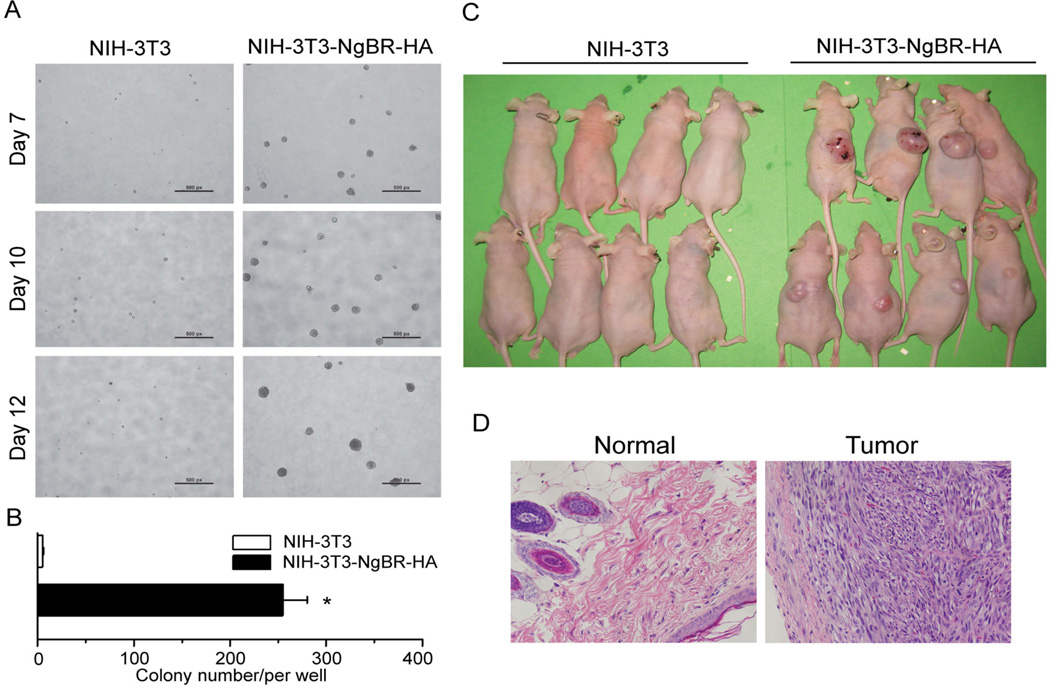
Like the typical functions of Ras overexpression such as transformation and tumorigenesis of NIH-3T3 cells as reported in previous publications13, 27, 36, NgBR overexpression transforms NIH-3T3 cells and significantly increases colony formation in soft agar, indicating anchorage independent growth (Fig. 5A and 5B). To determine the contribution of NgBR-mediated Ras recruitment to tumorigenesis, we determined the effects of NgBR mutants on the EGF-stimulated Ras activation and signaling. As shown in Figure S7, unlike wild-type NgBR-HA (Fig. S7, lane 4) or NgBR(L186)-HA mutant (Fig. S7, lane 6), which does not lose the binding to Ras (Fig. S5B, lane 2 and 3), NgBR(E262)-HA and NgBR(TM)-HA mutants (Fig. S7, lane 8 and 10), which lose the binding to Ras (Fig. S5B, lane 4 and 5), cannot increase EGF-stimulated Ras activation and phosphorylation of Akt and ERK in NIH-3T3 cells (Fig. S7, lane 8 and 10). Consequently, unlike wild-type NgBR-HA or NgBR (L186)-HA mutant, NgBR(E262)-HA and NgBR(TM)-HA mutants, cannot increase colony formation in soft agar (Fig. S8). To examine tumorigenesis in vivo, NIH-3T3 stable cells overexpressing either NgBR-HA or empty vector were injected subcutaneously into nude mice. After 8 weeks, tumors of various sizes appeared in 8 mice receiving NIH-3T3-NgBR-HA cells but not in mice injected with control NIH-3T3 cells (Fig. 5C). Histological analysis showed that NgBR overexpression results in fibrosarcoma (Fig. 5D). These results demonstrate that NgBR overexpression increases membrane-associated Ras and promotes EGF-stimulated activation of Ras and its downstream kinases such as Akt and ERK. Importantly, NgBR overexpression in NIH-3T3 cells recapitulates the similar cellular functions of cell transformation and tumorigenesis that activated Ras induces.
Figure 5. NgBR over-expression recapitulates the tumorigenesis function of Ras GTPases.
Over-expression of NgBR in NIH-3T3 cells causes transformation of NIH-3T3 cells. Images of colony formation of NIH-3T3-NgBR-HA cells (A) and quantification of the results (B) are shown. Data are represented as mean ± SEM (* P< 0.05, n=3). (C) High expression of NgBR in NIH-3T3 cells promotes tumorigenesis of NIH-3T3 cells. NIH-3T3 stable cell lines were subcutaneously implanted into the flank region of nude mice. Images of the nude mice were taken 5 weeks after cell implantation (n=8). (D) HE staining of normal skin tissues and NIH-3T3-NgBR-HA tumor tissues. NIH-3T3: NIH-3T3 cells containing the control empty vector. NIH-3T3-NgBR-HA: NIH-3T3 cells highly expressing NgBR-HA.
NgBR is essential for EGF signaling in breast cancer cells and contributes to the growth of breast tumor xenografts
To elucidate the role of NgBR-mediated Ras recruitment in breast cancer cells, we examined the effects of NgBR knockdown on EGF signaling in MDA-MB-231 cells, an invasive breast carcinoma cell line26. As shown in Figure 6A, the levels of NgBR at the plasma membrane of MDA-MB-231 cells treated with NgBR siRNA was significantly reduced, which was determined using a cell surface biotinylation assay. NgBR knockdown in these MDA-MB-231 cells significantly reduced membrane-associated H-Ras and K-Ras, but did not change the levels of membrane protein pan-cadherin (Fig. 6B). In addition, we established a subline of MDA-MB-231 cells with knockdown of NgBR by infecting the cells with lentivirus-mediated small hairpin interfering RNA (shRNA) targeting the 3’UTR region of NgBR (shNgBR). Lentivirus carrying non-silencing (NS) shRNA was used as a negative control. As shown in Fig. 6C, EGF stimulation robustly activated H-Ras and its preferred downstream kinase Akt, and NgBR knockdown significantly abolished the EGF-stimulated H-Ras activation and Akt phosphorylation. In contrast, EGF stimulation only moderately increased the ERK phosphorylation and K-Ras activation, and this reponse was not altered by NgBR knockdown (Fig. S9). This results might occur because MDA-MB-231 cells have constitutively activated K-Ras and Raf-1 mutants21, 22. We also examined the effects of NgBR knockdown on EGF-mediated signaling pathways in T24 bladder carcinoma cells, which have wild-type K-Ras and constitutively activated H-Ras4. As shown in Figure S10, the regulatory effects of NgBR in T24 cells are opposite to what happened in MDA-MB-231 cells. NgBR knockdown significantly abolished EGF-stimulated K-Ras activation and ERK phosphorylation, but had no inhibitory effects on H-Ras activation. In addition, we isolated primary cultured murine embryonic fibroblast (MEF) cells from NgBR inducible knockout mice. NgBR knockout was induced by adding 4-hydroxy tamoxifen in the culture medium. As shown in Figure 6D, NgBR genetic knockout also inhibits EGF-stimulated H-Ras activation and Akt phosphorylation (Fig. 6D, lane 6). These findings strongly suggest that NgBR have diverse regulatory roles that are dependent on the status of H-Ras and K-Ras mutations in different types of carcinoma cells.
Figure 6. NgBR expression in breast cancer cells is essential for EGF signaling.
(A) Knockdown of NgBR decreases the NgBR protein levels in the fraction of biotinylated cell surface proteins. MDA-MB-231 cell surface proteins were biotinylated under non-permeabilized conditions and isolated using streptavidin agarose resin from Pierce Cell Surface Protein Isolation Kit as described in Figure 1B. Proteins were detected by Western blot analysis. (B) NgBR knockdown diminishes H-Ras and K-Ras membrane localization in MDA-MB-231 cells. NgBR was knocked down by using siRNA. The plasma membrane proteins were isolated by the ultracentrifugation method. The protein levels were determined by Western blotting. (C) NgBR knockdown reduces the EGF-induced activation of H-Ras and phosphorylation of Akt in MDA-MB-231 cells. NgBR was knocked down by using shRNA. The activated Ras proteins were isolated using GST-RBD beads and protein levels were determined by Western blotting. Phosphorylation of Akt and ERK was determined by Western blotting using phosphorylation specific antibodies. (D) NgBR genetic knockout reduces the EGF-induced activation of H-Ras and phosphorylation of Akt in MEF cells. Mouse embryonic fibroblast (MEF) cells were isolated from NgBR inducible knockout mice. NgBR was genetically knocked out by culture with tamoxifen. After stimulation with 10 ng/ml EGF for 5 minutes in the quiescent cells, the activated Ras proteins were isolated using GST-RBD beads. Protein levels were detected by Western blotting. Data are validated in 3 independent experiments.
As shown in Figures S11A and S11B, NgBR knockdown decreases colony formation of MDA-MB-231 cells. NgBR knockdown also moderately reduces the growth of MDA-MB-231 cells, as determined by the WST-1 assay59(Fig. S11C), but does not cause apoptosis (Fig. S11D), as determined by TUNEL staining24. As a positive control, we detected TUNEL staining in MDA-MB-231 cells treated with cisplatin (Fig. S11D). Wound healing assays (Fig. S11E) indicated that NgBR knockdown does not significantly alter cell motility28. In summary, NgBR knockdown decreases the colony formation and cell growth of MDA-MB-231 cells.
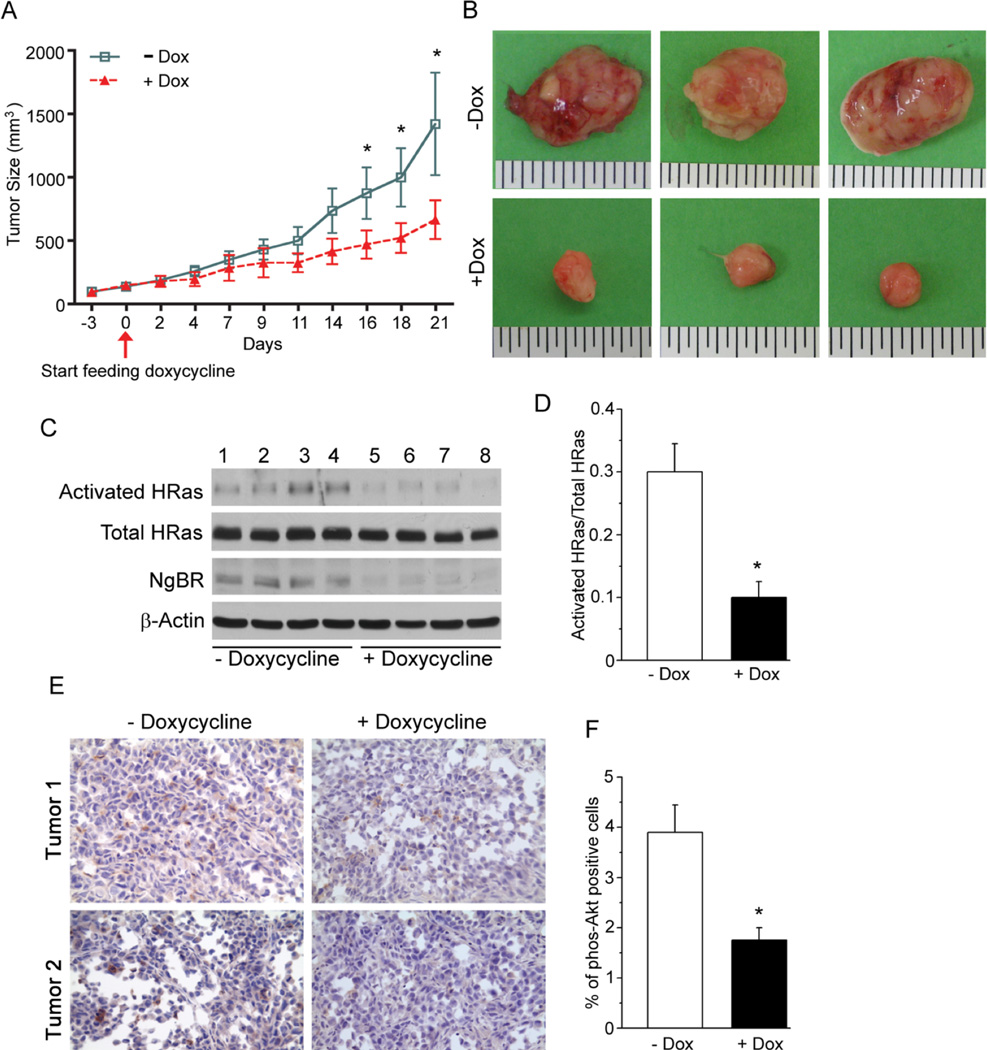
To further evaluate the in vivo function of NgBR knockdown on breast tumor growth, we established NgBR inducible knockdown MDA-MB-231 stable cell lines using TRIPZ inducible lentivrial shRNA58 (Thermo Scientific). As shown in Figure S12, doxycycline diminished the expression of NgBR in the shNgBR group (lanes 4 and 6) but not in the NS control group (lanes 3 and 5). Inducible knockdown of NgBR abolished EGF-stimulated phosphorylation of Akt (lane 6). After removing doxycycline from the culture medium for 48 hours, the NgBR expression in the shNgBR group recovered to the same endogenous levels as that in the NS group, and consequently restored the phosphorylation of Akt in response to EGF-stimulation (lane 8). These results confirmed that the inducible knockdown system works well in response to doxycycline, and indicated that the NgBR is essential for EGF-stimulated phosphorylation of Akt in MDA-MB-231 cells. The results of the colony formation assay (Fig. S12B/C) showed that doxycycline administration (Fig. S12B, right bottom panel) reduces colony formation of MDA-MB-231 cells carrying shRNA-NgBR (shNgBR) as compared to the same cells in the absence of doxycycline (Fig. S12B, left bottom panel), but does not affect the colony formation of MDA-MB-231 cells having non-silencing shRNA (NS) (Fig. S12B, upper panels). This finding demonstrates that doxycycline-induced specific knockdown of NgBR abolishes the in vitro malignant phenotype of MDA-MB-231 cells. We implanted the MDA-MB-231 cells carrying shNgBR in the flank region of nude mice. When tumors grew to the size of about 200 mm3, the nude mice bearing the tumors were divided into 2 groups; one group was supplied with 1% sucrose drinking water and the other group was supplied with 1% sucrose drinking water plus 5% doxycycline for 3 weeks. Analysis of tumor size over three weeks (Fig. 7A and 7B) indicated that the growth of the xenografts was slower in the mice given doxycycline. Real-time PCR analysis confirmed that NgBR transcript levels (Fig. S13A) and protein levels (Fig. 7C) were decreased in the mice given doxycycline. Importantly, doxycycline administration diminished H-Ras activity in the tumor xenografts (Fig. 7C and 7D). Immunohistological analysis of the tumor xenografts demonstrate that doxycycline-induced NgBR reduction (Fig. 7E, right panel; Fig. 7F) reduced the number of cells staining positive for phosphorylated Akt, but did not have any significant effects on the phosphorylation of ERK in the tumor xenografts (Fig. S13B). These results demonstrated that NgBR promotes Ras plasma membrane accumulation in tumor cells and NgBR recapitulates the oncogenic function of Ras in cell transformation and tumor growth.
Figure 7. NgBR expression in breast cancer cells contributes to the growth of breast tumor xenografts.
(A) NgBR knockdown reduces the growth rate and size of MDA-MB-231 tumor xenografts. MDA-MB-231 stable cells expressing doxycycline-inducible NgBR shRNA were subcutaneously implanted into the flank region of the nude mice. NgBR was knocked down by feeding the mice with doxycycline in drinking water when the size of the tumor xenografts reached 200 mm3. Data are represented as mean ± SEM (* P< 0.05, n=6). (B) Representative images of tumor xenografts isolated from the nude mice are shown. NgBR knockdown reduces the size of MDA-MB-231 tumor xenografts in the nude mice. (C, D) NgBR knockdown reduces the activation of H-Ras in MDA-MB-231 tumor xenografts, as indicated by isolating the activated H-Ras proteins using GST-RBD beads and detecting the proteins by Western blotting (C), and quantifying the O.D. of the proteins in the Western blots (D). Data are represented as mean ± SEM (* P< 0.05, n=4). (E, F) NgBR knockdown reduces the phosphorylation of Akt in MDA-MB-231 tumor xenografts. Immunostaining of phosphorylated Akt in MDA-MB-231 tumor xenograft (E) and quantification results (F) are shown. Data are represented as mean ± SEM (* P< 0.05, n=4).
Discussion
In this study, we demonstrate that the Nogo-B receptor (NgBR) plays a vital role in oncogenic signaling and malignant transformation induced by Ras GTPases by recruiting these GTPases to the plasma membrane. The three Ras isoforms (H-Ras, N-Ras and K-Ras) interact with receptor tyrosine kinases at the plasma membrane, but exhibit different abilities to activate downstream kinases such as phosphatidylinositol-3-OH kinase (PI-3K) and Raf-1 kinase6, 17, 38. H-Ras preferentially activates PI-3K kinase and is a weak activator for Raf-1, but K-Ras has the converse activation profile38, 62, 68. These functional differences have been attributed to the different locations of the Ras isoforms in distinct subdomains of the plasma membrane17, 38. Farnesylation and palmitoylation of H-Ras in the ER and Golgi, respectively67, target H-Ras to caveolae-rich lipid rafts17, 38, 50, 53, 54. Activation of H-Ras redistributes it from lipid rafts into non-lipid raft regions, where H-Ras activates downstream kinases17, 38, 44, 50. In contrast, K-Ras is farnesylated but not palmitoylated, and is located outside lipid rafts irrespective of its activation status17, 38, 50. However, based on current models45, 50, 51, H-Ras traffics to lipid raft subdomains first and then re-distributes to non-lipid raft subdomains after H-Ras is activated and in the GTP-bound state. According to this model, only H-Ras that is in the GTP-bound state will be located in non-lipid raft subdomains. Somewhat surprisingly, our results indicate that the depletion of NgBR in the non-caveolae-rich lipid raft subdomain abolishes the majority of H-Ras plasma membrane localization. Based on findings5, 65 from the model membrane system that does not include NgBR, it indicates that NgBR is not required for the trafficking and nanoclusters of H-Ras in the lipid-enriched plasma membrane. Thus, our findings suggest that NgBR is a novel binding partner that provides a docking site for GDP- or GTP-bound H-Ras outside of lipid rafts, contrary to current models proposing that only GTP-bound H-Ras localizes outside of lipid rafts. Further studies are needed to reconcile these findings with current models.
NgBR was initially identified as the receptor for Nogo-B33. High affinity of Nogo-B binding to NgBR is sufficient for Nogo-B-mediated chemotaxis33 and is essential for developmental angiogenesis42, 47, 69. Recent reports also demonstrated that NgBR is involved in the epithelial-mesenchymal transition of breast tumor cells70 and the resistance of human hepatocellular carcinoma to chemotherapy14. However, the underlying mechanism of NgBR signaling in cancer is still unclear. Analysis of the NgBR sequence reveals its C-terminal domain has high homology with cis-isoprenyltransferase (cis-IPTase)33. Further studies revealed that NgBR is a critical subunit of cis-IPTase, which is involved in the synthesis of dolichol that is an intermediate product required for protein glycosylation19, 41. Our results demonstrate that the hydrophobic domain of NgBR binds farnesylated Ras independent of cis-IPTase because purified MBP-NgBR alone binds farnesylated H-Ras (Fig. 2C). Findings from our studies demonstrate that NgBR-mediated Ras membrane accumulation may contribute to these identified roles of NgBR in tumor cells14, 70.
Although Ras mutation rarely occurs in breast cancer (less than 10%)23, 46, oncogenic signaling by Ras contributes to the tumorigenic and invasive potential of breast epithelial cells23, 30. Therefore, upregulation of normal Ras activity by receptor tyrosine kinases (RTK), such as the EGF receptor family, is a major focus for breast cancer research23, 30. Introduction of activated H-Ras into certain mouse or human breast epithelial cells results in acquisition of a transformed phenotype in vitro (such as increased anchorage independent growth) and in vivo, as shown by the ability to form tumors in nude mice10, 15, 48. In addition, a large number of clinical studies have demonstrated overexpression of Ras in breast cancers12, 34, 64. There is a significant trend for tumors to be lymph node positive with increasing expression of Ras proteins, and Ras overexpression is related to early disease recurrence and death in lymph node negative patients64. Therefore, understanding how the Ras activation pathway is regulated in breast cancer cells will be critical for developing new therapeutic approaches. The EGF signaling pathway is one of the major therapeutic targets for cancer therapy37, 61. As shown in Figure S14, EGF binds the EGF receptor (EGFR) to recruit RasGEFs such as SOS via its SHC domain and Grb2 adapter protein6, 23, 31, 32. Membrane associated Ras is one of the critical substrates for activated EGFR and is required for further activation of both the PI3K-Akt and Raf1-ERK pathways, which contribute to tumorigenesis and tumor resistance31, 32. Our results showed NgBR overexpression in NIH-3T3 cells recapitulates the similar cellular functions of cell transformation and tumorigenesis that activated Ras induces. NgBR is essential for Ras plasma membrane accumulation in tumor cells and NgBR recapitulates the oncogenic function of Ras in cell transformation and tumor growth. Furthermore, we found that NgBR mainly regulates the EGF-stimulated activation of wild type H-Ras and K-Ras because they must translocate to the plasma membrane to be activated by GEFs recruited by the EGFR6, 23, 55. It is possible that constitutively activated H-Ras and K-Ras can activate their downstream kinases not only at the plasma membrane but also at other cellular compartments such as the ER and Golgi17, 45. These results also suggest that NgBR is a novel therapeutic target for controlling Ras signaling mediated by many receptor tyrosine kinases such as EGF, FGF and VEGF receptors.
Materials and methods
Tumor implantation
106 MDA-MB-231 shNgBR stable cells were subcutaneously implanted to the flank region of 6–8 weeks male nude mice (Charles River Laboratories). When the average tumor volume reached about 200 mm3, mice were separated to two groups randomly and then fed with or without 2mg/ml doxycycline in 1% sucrose drinking water. The diameters of the tumors were measured every other day with a caliper. The tumor volume was calculated as Tumor volume=0.5 × length × width2. After three weeks of doxycycline administration, mice were sacrificed and tumors were isolated. Half of the tumor tissues were fixed in 4%PFA/PBS for histology analysis and another half were frozen in liquid nitrogen for gene expression analysis. For NIH-3T3 cell implantation, 106 NIH-3T3 control cells or highly expressing NgBR-HA cells (NIH-3T3-NgBR-HA) were subcutaneously implanted to the flank region of 6–8 weeks male nude mice. After 6–8 weeks, mice were sacrificed and tumor tissues were isolated as above described. Tumors from animals that died before the endpoint were excluded from the analysis. All of the animal experiments were approved by the Institutional Animal Care Use Committee of the Medical College of Wisconsin.
Data analysis
All in vitro experiments presented are representative and were replicated at least three times. Sample sizes for relevant experiments were determined by power analysis. The results of quantitative assays were analyzed using statistical software SPSS 16.0 for Windows. Data are presented as mean ± the standard error of the mean (SEM). Barlett’s test was performed prior to ANOVA. The statistical significance of differences was evaluated with the ANOA analysis followed by Dunnett’s post-hoc analysis. Significance was defined as P < 0.05.
Supplementary Material
Highlights.
A conceptual advance of the mechanisms controlling membrane localization of Ras.
Nogo-B receptor is a cell surface protein with hydrophobic domains.
Nogo-B recptor binds farnesylated Ras and increases Ras membrane accumulation.
Nogo-B receptor promotes Ras activation and Ras oncogenic signaling.
Acknowledgments
Dr. John Hancock at University of Texas Health Science Center at Houston generously provided pEGFP-C1 vector. This work is supported in part by start-up funds from Division of Pediatric Surgery and Division of Pediatric Pathology, Medical College of Wisconsin (MCW) and Advancing a Healthier Wisconsin endowment to MCW, AHA SDG 0730079N, NIH R01HL108938, Wisconsin Breast Cancer Showhouse (WBCS), Institutional Research Grant # 86-004-26 from the American Cancer Society, We Care Fund, Kathy Duffey Fogarty Award for breast cancer research, State of Wisconsin Tax Check-off program for breast & prostate cancer research and Children’s Hospital of Wisconsin Research Institute Pilot Innovative Research Grant to Q.R.M., AHA postdoctoral fellowship 13POST13940002 and 11POST5690035, China State Key Basic Research Program Grant 2016YFA0501401 and the support from the Hundred Talents Program of CAS to B.Z. Additional support was provided by NIH R01 CA188871 (CLW), the Rock River Cancer Research Foundation (CLW), and the Nancy Laning Sobczak, Ph.D., Breast Cancer Research Award (CLW), NIH R01HL128647 (ZY).
Footnotes
Author Contributions:
B.Z., W.H., S.K., P.G., U.R., Z.L., B.W., W.Q.D, Q.R.M. conducted the experiments; Q.R.M., B.Z., W.H. designed the experiments and wrote the paper; Z.Y. provided computer modeling; C.W. provided reagents and edited the paper; Q.R.M. was responsible for overall integration and execution of the scientific approaches.
References
- 1.Abramovitz A, Gutman M, Nachliel E. Structural coupling between the Rho-insert domain of Cdc42 and the geranylgeranyl binding site of RhoGDI. Biochemistry. 2012;51:715–723. doi: 10.1021/bi201211v. [DOI] [PubMed] [Google Scholar]
- 2.Ahearn IM, Haigis K, Bar-Sagi D, Philips MR. Regulating the regulator: post-translational modification of RAS. Nature reviews Molecular cell biology. 2012;13:39–51. doi: 10.1038/nrm3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belanis L, Plowman SJ, Rotblat B, Hancock JF, Kloog Y. Galectin-1 is a novel structural component and a major regulator of h-ras nanoclusters. Molecular biology of the cell. 2008;19:1404–1414. doi: 10.1091/mbc.E07-10-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard EJ, McKenna WG, Hamilton AD, Sebti SM, Qian Y, Wu JM, et al. Inhibiting Ras prenylation increases the radiosensitivity of human tumor cell lines with activating mutations of ras oncogenes. Cancer research. 1998;58:1754–1761. [PubMed] [Google Scholar]
- 5.Brunsveld L, Waldmann H, Huster D. Membrane binding of lipidated Ras peptides and proteins - The structural point of view. Bba-Biomembranes. 2009;1788:273–288. doi: 10.1016/j.bbamem.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 6.Buday L, Downward J. Many faces of Ras activation. Biochimica et biophysica acta. 2008;1786:178–187. doi: 10.1016/j.bbcan.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, et al. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nature cell biology. 2012;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- 8.Chang SY, Ko TP, Chen AP, Wang AH, Liang PH. Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies. Protein Sci. 2004;13:971–978. doi: 10.1110/ps.03519904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen AP, Chang SY, Lin YC, Sun YS, Chen CT, Wang AH, et al. Substrate and product specificities of cis-type undecaprenyl pyrophosphate synthase. The Biochemical journal. 2005;386:169–176. doi: 10.1042/BJ20040785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark R, Stampfer MR, Milley R, O'Rourke E, Walen KH, Kriegler M, et al. Transformation of human mammary epithelial cells by oncogenic retroviruses. Cancer research. 1988;48:4689–4694. [PubMed] [Google Scholar]
- 11.Cox AD, Der CJ, Philips MR. Targeting RAS Membrane Association: Back to the Future for Anti-RAS Drug Discovery? Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:1819–1827. doi: 10.1158/1078-0432.CCR-14-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dati C, Muraca R, Tazartes O, Antoniotti S, Perroteau I, Giai M, et al. c-erbB-2 and ras expression levels in breast cancer are correlated and show a co-operative association with unfavorable clinical outcome. International journal of cancer Journal international du cancer. 1991;47:833–838. doi: 10.1002/ijc.2910470607. [DOI] [PubMed] [Google Scholar]
- 13.DeFeo D, Gonda MA, Young HA, Chang EH, Lowy DR, Scolnick EM, et al. Analysis of two divergent rat genomic clones homologous to the transforming gene of Harvey murine sarcoma virus. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:3328–3332. doi: 10.1073/pnas.78.6.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong C, Zhao B, Long F, Liu Y, Liu Z, Li S, et al. Nogo-B receptor promotes the chemoresistance of human hepatocellular carcinoma via the ubiquitination of p53 protein. Oncotarget. 2016;7:8850–8865. doi: 10.18632/oncotarget.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunzburg WH, Salmons B, Schlaeffli A, Moritz-Legrand S, Jones W, Sarkar NH, et al. Expression of the oncogenes mil and ras abolishes the in vivo differentiation of mammary epithelial cells. Carcinogenesis. 1988;9:1849–1856. doi: 10.1093/carcin/9.10.1849. [DOI] [PubMed] [Google Scholar]
- 16.Guo RT, Ko TP, Chen AP, Kuo CJ, Wang AH, Liang PH. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. The Journal of biological chemistry. 2005;280:20762–20774. doi: 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- 17.Hancock JF. Ras proteins: different signals from different locations. Nature reviews Molecular cell biology. 2003;4:373–384. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 18.Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The complex of Arl2-GTP and PDE delta: from structure to function. The EMBO journal. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrison KD, Park EJ, Gao N, Kuo A, Rush JS, Waechter CJ, et al. Nogo-B receptor is necessary for cellular dolichol biosynthesis and protein N-glycosylation. The EMBO journal. 2011;30:2490–2500. doi: 10.1038/emboj.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman GR, Cerione RA. Flipping the switch: the structural basis for signaling through the CRIB motif. Cell. 2000;102:403–406. doi: 10.1016/s0092-8674(00)00045-3. [DOI] [PubMed] [Google Scholar]
- 21.Hollestelle A, Elstrodt F, Nagel JH, Kallemeijn WW, Schutte M. Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol Cancer Res. 2007;5:195–201. doi: 10.1158/1541-7786.MCR-06-0263. [DOI] [PubMed] [Google Scholar]
- 22.Ikediobi ON, Davies H, Bignell G, Edkins S, Stevens C, O'Meara S, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5:2606–2612. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nature reviews Molecular cell biology. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly KJ, Sandoval RM, Dunn KW, Molitoris BA, Dagher PC. A novel method to determine specificity and sensitivity of the TUNEL reaction in the quantitation of apoptosis. American journal of physiology Cell physiology. 2003;284:C1309–C1318. doi: 10.1152/ajpcell.00353.2002. [DOI] [PubMed] [Google Scholar]
- 25.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nature reviews Drug discovery. 2007;6:541–555. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 26.Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 27.Land H, Parada LF, Weinberg RA. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature. 1983;304:596–602. doi: 10.1038/304596a0. [DOI] [PubMed] [Google Scholar]
- 28.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nature protocols. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 29.Liang PH, Ko TP, Wang AH. Structure, mechanism and function of prenyltransferases. European journal of biochemistry / FEBS. 2002;269:3339–3354. doi: 10.1046/j.1432-1033.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 30.Malaney S, Daly RJ. The ras signaling pathway in mammary tumorigenesis and metastasis. Journal of mammary gland biology and neoplasia. 2001;6:101–113. doi: 10.1023/a:1009572700317. [DOI] [PubMed] [Google Scholar]
- 31.McCubrey JA, Steelman LS, Abrams SL, Lee JT, Chang F, Bertrand FE, et al. Roles of the RAF/MEK/ERK and PI3K/PTEN/AKT pathways in malignant transformation and drug resistance. Advances in Enzyme Regulation. 2006;46:249–279. doi: 10.1016/j.advenzreg.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 32.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochimica et biophysica acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miao RQ, Gao Y, Harrison KD, Prendergast J, Acevedo LM, Yu J, et al. Identification of a receptor necessary for Nogo-B stimulated chemotaxis and morphogenesis of endothelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10997–11002. doi: 10.1073/pnas.0602427103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyakis S, Sourvinos G, Spandidos DA. Differential expression and mutation of the ras family genes in human breast cancer. Biochemical and biophysical research communications. 1998;251:609–612. doi: 10.1006/bbrc.1998.9527. [DOI] [PubMed] [Google Scholar]
- 35.Nancy V, Callebaut I, El Marjou A, de Gunzburg J. The delta subunit of retinal rod cGMP phosphodiesterase regulates the membrane association of Ras and Rap GTPases. The Journal of biological chemistry. 2002;277:15076–15084. doi: 10.1074/jbc.M109983200. [DOI] [PubMed] [Google Scholar]
- 36.Newbold RF, Overell RW. Fibroblast immortality is a prerequisite for transformation by EJ c-Ha-ras oncogene. Nature. 1983;304:648–651. doi: 10.1038/304648a0. [DOI] [PubMed] [Google Scholar]
- 37.Nyati MK, Morgan MA, Feng FY, Lawrence TS. Integration of EGFR inhibitors with radiochemotherapy. Nature reviews. 2006;6:876–885. doi: 10.1038/nrc1953. [DOI] [PubMed] [Google Scholar]
- 38.Omerovic J, Laude AJ, Prior IA. Ras proteins: paradigms for compartmentalised and isoform-specific signalling. Cell Mol Life Sci. 2007;64:2575–2589. doi: 10.1007/s00018-007-7133-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Omerovic J, Prior IA. Compartmentalized signalling: Ras proteins and signalling nanoclusters. The FEBS journal. 2009;276:1817–1825. doi: 10.1111/j.1742-4658.2009.06928.x. [DOI] [PubMed] [Google Scholar]
- 40.Parish CA, Brazil DP, Rando RR. On the mechanism of the inhibition of transducin function by farnesylcysteine analogs. Biochemistry. 1997;36:2686–2693. doi: 10.1021/bi961844r. [DOI] [PubMed] [Google Scholar]
- 41.Park EJ, Grabinska KA, Guan Z, Stranecky V, Hartmannova H, Hodanova K, et al. Mutation of Nogo-B Receptor, a Subunit of cis-Prenyltransferase, Causes a Congenital Disorder of Glycosylation. Cell metabolism. 2014;20:448–457. doi: 10.1016/j.cmet.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park EJ, Grabinska KA, Guan Z, Sessa WC. NgBR is essential for endothelial cell glycosylation and vascular development. EMBO reports. 2016;17:167–177. doi: 10.15252/embr.201540789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pechlivanis M, Kuhlmann J. Hydrophobic modifications of Ras proteins by isoprenoid groups and fatty acids--More than just membrane anchoring. Biochimica et biophysica acta. 2006;1764:1914–1931. doi: 10.1016/j.bbapap.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 44.Prior IA, Harding A, Yan J, Sluimer J, Parton RG, Hancock JF. GTP-dependent segregation of H-ras from lipid rafts is required for biological activity. Nature cell biology. 2001;3:368–375. doi: 10.1038/35070050. [DOI] [PubMed] [Google Scholar]
- 45.Prior IA, Hancock JF. Ras trafficking, localization and compartmentalized signalling. Seminars in cell & developmental biology. 2012;23:145–153. doi: 10.1016/j.semcdb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Cancer research. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rana U, Liu Z, Kumar SN, Zhao B, Hu W, Bordas M, et al. Nogo-B receptor deficiency causes cerebral vasculature defects during embryonic development in mice. Dev Biol. 2016;410:190–201. doi: 10.1016/j.ydbio.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Redmond SM, Reichmann E, Muller RG, Friis RR, Groner B, Hynes NE. The transformation of primary and established mouse mammary epithelial cells by p21-ras is concentration dependent. Oncogene. 1988;2:259–265. [PubMed] [Google Scholar]
- 49.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 50.Rocks O, Peyker A, Bastiaens PI. Spatio-temporal segregation of Ras signals: one ship, three anchors, many harbors. Current opinion in cell biology. 2006;18:351–357. doi: 10.1016/j.ceb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 51.Rotblat B, Prior IA, Muncke C, Parton RG, Kloog Y, Henis YI, et al. Three separable domains regulate GTP-dependent association of H-ras with the plasma membrane. Molecular and cellular biology. 2004;24:6799–6810. doi: 10.1128/MCB.24.15.6799-6810.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rotblat B, Belanis L, Liang H, Haklai R, Elad-Zefadia G, Hancock JF, et al. H-Ras nanocluster stability regulates the magnitude of MAPK signal output. PloS one. 2010;5:e11991. doi: 10.1371/journal.pone.0011991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roy S, Luetterforst R, Harding A, Apolloni A, Etheridge M, Stang E, et al. Dominant-negative caveolin inhibits H-Ras function by disrupting cholesterol-rich plasma membrane domains. Nature cell biology. 1999;1:98–105. doi: 10.1038/10067. [DOI] [PubMed] [Google Scholar]
- 54.Roy S, Plowman S, Rotblat B, Prior IA, Muncke C, Grainger S, et al. Individual palmitoyl residues serve distinct roles in H-ras trafficking, microlocalization, and signaling. Molecular and cellular biology. 2005;25:6722–6733. doi: 10.1128/MCB.25.15.6722-6733.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scaltriti M, Baselga J. The epidermal growth factor receptor pathway: a model for targeted therapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:5268–5272. doi: 10.1158/1078-0432.CCR-05-1554. [DOI] [PubMed] [Google Scholar]
- 56.Schmick M, Kraemer A, Bastiaens PI. Ras moves to stay in place. Trends Cell Biol. 2015;25:190–197. doi: 10.1016/j.tcb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 57.Shalom-Feuerstein R, Plowman SJ, Rotblat B, Ariotti N, Tian T, Hancock JF, et al. K-ras nanoclustering is subverted by overexpression of the scaffold protein galectin-3. Cancer research. 2008;68:6608–6616. doi: 10.1158/0008-5472.CAN-08-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature genetics. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 59.Sylvester PW. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol Biol. 2011;716:157–168. doi: 10.1007/978-1-61779-012-6_9. [DOI] [PubMed] [Google Scholar]
- 60.Taylor SJ, Resnick RJ, Shalloway D. Nonradioactive determination of Ras-GTP levels using activated ras interaction assay. Methods Enzymol. 2001;333:333–342. doi: 10.1016/s0076-6879(01)33067-7. [DOI] [PubMed] [Google Scholar]
- 61.Toulany M, Kasten-Pisula U, Brammer I, Wang S, Chen J, Dittmann K, et al. Blockage of epidermal growth factor receptor-phosphatidylinositol 3-kinase-AKT signaling increases radiosensitivity of K-RAS mutated human tumor cells in vitro by affecting DNA repair. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:4119–4126. doi: 10.1158/1078-0432.CCR-05-2454. [DOI] [PubMed] [Google Scholar]
- 62.Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. The Journal of biological chemistry. 2001;276:15609–15615. doi: 10.1074/jbc.M010573200. [DOI] [PubMed] [Google Scholar]
- 63.Wang B, Zhao B, North P, Kong A, Huang J, Miao QR. Expression of NgBR is highly associated with estrogen receptor alpha and survivin in breast cancer. PloS one. 2013;8:e78083. doi: 10.1371/journal.pone.0078083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watson DM, Elton RA, Jack WJ, Dixon JM, Chetty U, Miller WR. The H-ras oncogene product p21 and prognosis in human breast cancer. Breast Cancer Res Treat. 1991;17:161–169. doi: 10.1007/BF01806365. [DOI] [PubMed] [Google Scholar]
- 65.Weise K, Huster D, Kapoor S, Triola G, Waldmann H, Winter R. Gibbs energy determinants of lipoprotein insertion into lipid membranes: the case study of Ras proteins. Faraday Discuss. 2013;161:549–561. doi: 10.1039/c2fd20100c. [DOI] [PubMed] [Google Scholar]
- 66.Williams CL. The polybasic region of Ras and Rho family small GTPases: a regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003;15:1071–1080. doi: 10.1016/s0898-6568(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 67.Wright LP, Philips MR. Thematic review series: lipid posttranslational modifications. CAAX modification and membrane targeting of Ras. Journal of lipid research. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 68.Yan J, Roy S, Apolloni A, Lane A, Hancock JF. Ras isoforms vary in their ability to activate Raf-1 and phosphoinositide 3-kinase. The Journal of biological chemistry. 1998;273:24052–24056. doi: 10.1074/jbc.273.37.24052. [DOI] [PubMed] [Google Scholar]
- 69.Zhao B, Chun C, Liu Z, Horswill MA, Pramanik K, Wilkinson GA, et al. Nogo-B receptor is essential for angiogenesis in zebrafish via Akt pathway. Blood. doi: 10.1182/blood-2010-02-271577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao B, Xu B, Hu W, Song C, Wang F, Liu Z, et al. Comprehensive proteome quantification reveals NgBR as a new regulator for epithelial-mesenchymal transition of breast tumor cells. Journal of proteomics. 2014;112C:38–52. doi: 10.1016/j.jprot.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zohn IM, Campbell SL, Khosravi-Far R, Rossman KL, Der CJ. Rho family proteins and Ras transformation: the RHOad less traveled gets congested. Oncogene. 1998;17:1415–1438. doi: 10.1038/sj.onc.1202181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.