Summary
Adhesin-mediated bacterial interspecies interactions are important elements in oral biofilm formation. They often occur on a species-specific level, which could determine health- or disease association of a biofilm community. Among the key players involved in these processes are the ubiquitous fusobacteria that have been recognized for their ability to interact with numerous different binding partners. Fusobacterial interactions with Streptococcus mutans, an important oral cariogenic pathogen, have previously been described but most studies focused on binding to non-mutans streptococci and specific cognate adhesin pairs remain to be identified. Here, we demonstrated differential binding of oral fusobacteria to S. mutans. Screening of existing mutant derivatives indicated SpaP as the major S. mutans adhesin specific for binding to Fusobacterium nucleatum ssp. polymorphum but none of the other oral fusobacteria tested. We inactivated RadD, a known adhesin of F. nucleatum ssp. nucleatum for interaction with a number of gram-positive species, in F. nucleatum ssp. polymorphum and used a Lactococcus lactis heterologous SpaP expression system to demonstrate SpaP interaction with RadD of F. nucleatum ssp. polymorphum. This is a novel function for SpaP, which has mainly been characterized as adhesin for binding to host proteins including salivary glycoproteins. In conclusion, we describe an additional role for SpaP as adhesin in interspecies adherence with RadD-SpaP as the interacting adhesin pair for binding between S. mutans and F. nucleatum ssp. polymorphum. Furthermore, S. mutans attachment to oral fusobacteria appears to involve species- and subspecies-dependent adhesin interactions.
Keywords: Streptococcus mutans, Fusobacterium, adhesin, SpaP, RadD
INTRODUCTION
The bacterial species of the human oral cavity depend on their ability to attach to surfaces or each other for colonization and persistence in this nutritious ecological niche. Consequently, the proteins involved in adherence are important components enabling microorganisms to form and reside in complex oral biofilms, in which distinct groups of bacteria perform specific functions (Kolenbrander et al. 2010; Rickard et al. 2003). While microbial interactions within these biofilms trigger important physiological changes in partner species that influence many properties including virulence features, physical attachment via specific adhesins is key for successful initiation of surface colonization and biofilm integration (Guo et al. 2014; Wright et al. 2013).
Oral representatives of the ubiquitous fusobacteria have been noted for their binding to a diverse array of microbial species and are considered to be important for biofilm formation and architecture (Kolenbrander et al. 1993; Kolenbrander et al. 2010; Rickard et al. 2003). Fusobacteria enable their own integration into biofilms by adhering to surface-attached early colonizers such as streptococci and actinomyces. Moreover, fusobacteria recruit other bacterial species including important periodontal pathogens that cannot directly attach to surfaces or early colonizers. This feature can promote microbial community shifts and impacts polymicrobial pathogenesis.
Cultivable oral fusobacteria are predominantly comprised of the species F. periodonticum and F. nucleatum (Potts et al. 1983). While F. periodonticum contains a single species, F. nucleatum includes five subspecies nucleatum, polymorphum, fusiforme, animalis, and vincentii (Bolstad et al. 1996). Consolidation of the subspecies fusiforme and vincentii into one group was recently proposed based on their phylogenetic similarities (Kook et al. 2013). Fusobacteria not only thrive in subgingival biofilms (Aruni et al. 2015), they are present in supragingival plaque (Haffajee et al. 2008), predominant in early childhood caries (Corby et al. 2005), dentinal and root caries lesions (Lima et al. 2011), ecological niches that are also dominated by oral streptococci including the cariogenic species Streptococcus mutans. Streptococci are the most prevalent early colonizers and comprise the primary binding partner for recruitment of fusobacteria into oral biofilms (Kolenbrander et al. 2010; Rickard et al. 2003). While the streptococcal adhesin for interaction with fusobacteria remains to be identified, we previously characterized RadD as a major fusobacterial adhesin in F. nucleatum ssp nucleatum for the well-established physical attachment to S. sanguinis and S. gordonii (Kaplan et al. 2009). The binding of F. nucleatum to S. mutans, however, has been demonstrated (Bradshaw et al. 1998; Falkler et al. 1981) but largely remains to be investigated.
Surface fibrils have been implicated in the interaction of streptococcal species with host proteins, eukaryotic cells and other species including with F. nucleatum (Handley et al. 1985). Corncob formation of S. cristatus with Corynebacterium matruchotii as well as F. nucleatum was impaired in a mutant strain lacking long fibrils that were suggested to be encoded by the Sortase A (SrtA) recognition consensus containing SrpA protein. Other (SrtA) dependent cell wall-anchored proteins such as SspA/SspB of S. gordonii have been implicated in the interactions between oral streptococci and other oral species including Porphyromonas gingivalis, actinomyces and Candida (Back et al. 2015; Brooks et al. 1997; Daep et al. 2006; Demuth et al. 1996; Egland et al. 2001; Jakubovics et al. 2005). The SspA/SspB homolog of S. mutans, SpaP, has been demonstrated to bind to host salivary proteins (Demuth et al. 1990; Lee et al. 1989), host matrix proteins including type I collagen, fibronectin, laminin, or keratin as well as serum components such as fibrinogen (Beg et al. 2002; Busscher et al. 2008; Kelemen et al. 2004; Kishimoto et al. 1989; Petersen et al. 2002; Sciotti et al. 1997; Soell et al. 2010). Recognition of certain actinomyces strains was reported when SpaP is expressed in S. gordonii in conjunction with CshA, a protein typically not present in S. mutans (Jakubovics et al. 2005).
In this study, we characterized the adhesion of oral fusobacterial species to S. mutans and identified SpaP as the major adhesin of S. mutans that directly interacts with the adhesin RadD of the F. nucleatum subspecies polymorphum but none of the other oral fusobacteria tested.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions
All strains used in this study are listed in Table 1 and were grown as previously described (Jakubovics et al. 2005; Kaplan et al. 2009; Levesque et al. 2005; Zhu et al. 2009). In brief, fusobacteria were grown at 37°C on Columbia agar plates supplemented with 5 % sheep blood or in Columbia broth (CB) (Difco, Detroit, MI) under anaerobic conditions (5 % CO2, 5 % H2, 90 % N2). S. mutans wildtype and mutant strains were grown in Todd Hewitt (TH) broth (BD Difco, Detroit, MI) at 37°C in the presence of 5% CO2. The medium was supplemented with 15 μg/mL of erythromycin for growth of the respective srtA-, fruA-, wapA-, and wapE-deficient UA140 and UA159 derivatives. The gbpC- and spaP-deficient S. mutans strains were cultured in TH medium supplemented with spectinomycin (500 μg/mL). Lactococcus lactis MG1363 containing vector pUB1000 and the corresponding plasmids expressing spaP genes from different strains of S. mutans (Jakubovics et al. 2005) were grown statically in M17 medium supplemented with 0.5% (w/v) glucose and 5 μg/mL of erythromycin at 30°C.
Table 1.
Bacterial strains and plasmids used in this study
| Strain | Description | Antibiotic resistancea | Source |
|---|---|---|---|
| Fusobacteria | |||
| ATCC33693 | Wildtype F. periodonticum | Laboratory collection | |
| ATCC23726 | Wildtype F. nucleatum ssp nucleatum | Laboratory collection | |
| ΔFn1526 | ATCC23726 ΔradD mutant | Tap | Kaplan et al. 2009 |
| ATCC51191 | Wildtype F. nucleatum, ssp animalis | Laboratory collection | |
| ATCC49256 | Wildtype F. nucleatum ssp. vincentii | Laboratory collection | |
| ATCC10953 | Wildtype F. nucleatum ssp polymorphum | Laboratory collection | |
| ATCC10953 ΔradD | ATCC10953 ΔradD mutant | Tap | This study |
| Streptococcus mutans | |||
| UA159 | Wildtype S. mutans | Laboratory collection | |
| UA159 srtA | UA159 srtA mutant | Erm | Levesque et al. 2005 |
| UA159 fruA | UA159 fruA mutant | Erm | Levesque et al. 2005 |
| UA159 wapA | UA159 wapA mutant | Erm | Levesque et al. 2005 |
| UA159 wapE | UA159 wapE mutant | Erm | Levesque et al. 2005 |
| MZ159c | UA159 gbpC mutant | Spc | Zhu et al. 2009 |
| UA140 | Wildtype S. mutans | Laboratory collection | |
| UA140 srtA | UA140 srtA mutant | Erm | This study |
| UA140 fruA | UA140 fruA mutant | Erm | This study |
| UA140 wapA | UA140 wapA mutant | Erm | This study |
| UA140 wapE | UA140 wapE mutant | Erm | This study |
| UA140 gbpC | UA140 gbpC mutant | Spc | This study |
| UA140 spaP | UA140 spaP mutant | Spc | This study |
| Lactococcus lactis | |||
| MG1363 | Wildtype L. lactis | Jakubovics et al. 2005 | |
| Plasmids | |||
| pFW5 | S. mutans gene inactivation vector | Spc | Podbielski et al. 1996 |
| pFW5-spaP | pFW5 carrying spaP66–1100 | Spc | This study |
| pJET1.2/blunt | PCR fragment cloning vector | Amp | |
| pHS31 | F. nucleatum gene inactivation vector | Tap | Kaplan et al. 2009 |
| pBS24 | pHS31 carrying radD4931–5962 | Tap | This study |
| pUB1000 | L. lactis expression vector | Erm | Jakubovics et al. 2005 |
| pUB1559 | pUB1000 pac-S. mutans NG8 | Erm | Jakubovics et al. 2005 |
| pUB1660 | pUB1000 spaP-S. mutans Guy’s | Erm | Jakubovics et al. 2005 |
| pUB1661 | pUB1000 spaP-S. mutans Ingbritt 162 | Erm | Jakubovics et al. 2005 |
Amp – Ampicillin (50μg/ml); Erm – erythromycin (15μg/ml for S. mutans, 5μg/ml for L. lactis); Spc –spectinomycin (500μg/ml); Tap – Thiamphenicol (5μg/ml)
Strain Construction
S. mutans
Genomic DNA was prepared from S. mutans UA159 srtA-, fruA-, wapA-, and wapE-defective strains (Levesque et al. 2005) and S. mutans UA159 strain lacking gbpC (Zhu et al. 2009) using the MasterPure™ DNA purification kit (Epicentre, Madison, WI, USA). Between 1 and 5 μg of this genomic DNA was directly transformed into S. mutans strain UA140 via competence-stimulating peptide (CSP)-induced natural transformation to generate the corresponding mutant derivatives in this background. Transformants were selected on TH agar containing 15 μg/mL erythromycin (srtA-, fruA-, wapA-, and wapE) or 500 μg/mL spectinomycin (gbpC) and confirmed via PCR and DNA sequencing. To generate an insertional gene inactivation mutant derivative of S. mutans UA140 in spaP, an internal 1035-bp fragment (nucleotide 66 to nucleotide 1100) was amplified with primer pair SPAP-F (CGCGGATCCTCTAGGAACAGTAGCAGCAGTCT) containing a BamHI site (underlined) and SPAP-R (CTAGTCTAGATAAGTCGCCTTAGCATTCTCATT) containing a XbaI site (underlined) using Pfu polymerase (Stratagene) with standard amplification protocols and inserted into suicide vector pFW5 (Podbielski et al. 1996). The resulting plasmid pFW5-spaP was confirmed by restriction analysis, PCR amplification and DNA sequencing prior to transformation into S. mutans UA140 as described above. Transformants were selected on TH agar containing 500 μg/mL spectinomycin and confirmed by PCR, sequencing and Western Blot (Supplemental Fig. 1).
F. nucleatum ssp. polymorphum
A ΔradD mutant derivative of F. nucleatum ssp. polymorphum ATCC10953 was constructed by inactivating the radD encoding gene (FNP_1046) via single homologous recombination as described earlier (Kaplan et al. 2009). In brief, a 1032 bp internal gene fragment (nucleotide 4931 to nucleotide 5962) was amplified using the primer pair RADD-F (GCGGCTGAATTCCTGGAACAGGAATGTATTTAACAGGTAACAGC) and RADD-R (GCGGAGGGATCCCATTAGCTGCTTTATTATATCCAGATTTTGTATAAATACC) appended with EcoRI and BamHI, respectively, from genomic DNA of F. nucleatum ssp. polymorphum ATCC10953 and sub-cloned into the pJET1.2/blunt vector. The resulting plasmid pBS24 was digested with EcoRI/BamHI, ligated into EcoRI/BamHI digested pHS31 vector (Kaplan et al. 2009) and transformed into Escherichia coli. After confirmation of the integration plasmid by restriction analysis and sequencing, the plasmid DNA was electroporated into F. nucleatum ssp. polymorphum ATCC10953 and plated on selective medium containing 5μg/ml thiamphenicol. The insertional mutant was confirmed via PCR, sequencing.
Coaggregation Assay
Interspecies coaggregation was performed in modified coaggregation buffer (CAB) and quantified as described (Kaplan et al. 2009). For measuring coaggregation in the presence of saliva, saliva was collected from several volunteers, pooled and treated according to published procedures (Kitada et al. 2012). Briefly, the pooled saliva was clarified by centrifugation at 2100xg for 10 min. The supernatant was then removed and filtered through 0.22um (Millipore) filters. The filter-sterilized saliva was added to a final concentration of 50% to the individual coaggregation partners prior to combining the strains. The coaggregation index (C.I.), representing the coaggregation efficiency between two species, was calculated as follows: ((OD600(A)+OD600(B))/2-OD600(A+B))/(OD600(A)+OD600(B)/2). OD600(A) and OD600(B) represent the optical density of each individual species at 600nm, and OD600(A+B) represents the optical density of the mixture supernatant after 10 min incubation or as indicated if different from the standard procedure.
Biofilm integration assay
Biofilm Growth
Overnight cultures of S. mutans were diluted into TH medium containing 25% saliva, 0.5% mannose and 0.5% sucrose to a concentration of 2×105 cells/ml and 500 ul each of this suspension was inoculated into wells of a 48 well culture plate prior to incubation under anaerobic conditions (10% H2, 10% CO2, 80% N2) at 37°C for 18 hrs to allow for biofilm formation. The medium was removed and the wells were washed for three times with 1X PBS. Overnight cultures of F. nucleatum ssp polymorphum were adjusted to 5×108 cells in fresh CB containing 50% saliva and 50 ul were added into each well and incubated under anaerobic conditions for 4 hrs prior to harvesting and DNA extraction. Triplicate wells were inoculated for each experiment, which were combined for DNA extraction. At least three biological replicates were performed per condition.
Crystal violet Assay
Biofilm formation of S. mutans was evaluated via Crystal Violet (CV) staining according to published procedures (Zmantar et al. 2010). In brief, supernatants were removed from each well and rinsed once with 250 μl of sterile phosphate-buffered saline (PBS). Plates were inverted and dried. Next, attached bacteria were fixed at room temperature for 15 min by adding 200 μl of methanol into each well. The plates were stained with a 100 μl aqueous solution of 0.5% crystal violet (Thermo FisherScientific, Waltham, MA) for 15 min at room temperature. The plates were then carefully rinsed with Millipore water until there was no visible trace of the stain. Bound stain was dissolved by adding 160 μl of 95% ethanol. The optical density (OD) of each well was measured at 570 nm and was represented as relative to negative control wells that only contained TH.
Extraction of DNA from Biofilms
Prior to DNA isolation, the supernatant was carefully removed from each well and the wells were rinsed twice with 1XPBS. Genomic DNA was isolated directly from biofilm cells attached to the wells using QIAamp DNA Micro kit (Qiagen) according to manufacturer’s instructions with modification of the final elution to 30 μl. Lysis buffer was directly added to the wells, biofilms were scraped off and added directly into a 0.5 ml screw cap microtube containing 0.1mm silica beads. The samples were treated with bead beating for 30 sec three times at 1 minute intervals. After centrifugation of the samples at 13000xg for 5 min, the supernatant was transferred to a fresh tube and incubated with Proteinase K for 1 hr at 56°C. The samples were treated according to manufacturer’s instructions.
Quantitative (Real-Time) Polymerase Chain Reaction (qPCR)
Relative proportions of F. nucleatum ssp polymorphum integrated into S. mutans biofilms were determined by quantitative assessment of DNA with species-specific primer pairs similar to our published procedures (Park et al. 2016). Gene specific primer pairs gtfBF 5′ GCCTACAGCTCAGAGATGCTATTC and gtfB R 5′ GCCATACACCACTCATGAATTGA 3′ were used to amplify the S. mutans-specific gtfB gene, while fomA of F. nucleatum was amplified with primer pair fomA-F 5′ GTTGCTCCAGCTTGGAGACCAAAT and fomA-R 5′AAGTTTACTTTTGTTAAAGTTTGTAATCTTCC (Park et al. 2016) for specific detection of Fusobacterium nucleatum. Amplification and signal detection by qPCR on iCycler Thermal Cycler (Bio-Rad, Hercules, CA) was performed in a total volume of 20 μl containing 2 μl of 10× iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), 1ul of 0.5 μM each of forward and reverse primers, 7 μl of Millipore water and 1 μl (10ng) of template in 96-well optical plates (Thermo Fisher Scientific, Waltham, MA). Each PCR run was carried out with an initial incubation of 10 min at 95°C followed by 40 cycles of denaturing at 95°C for 15 sec; annealing and elongation at 60°C for 1 min. After the 40 cycles of amplification, an additional denaturing step was performed at 95°C for 1 min followed by annealing and elongation at 60°C for 1 min. A melting curve analysis was completed after each run. The DNA concentrations (ng ml −1) were calculated with standard curves obtained by tenfold serial dilutions of bacterial genomic DNA. All standards were run in duplicate to generate a standard curve to determine the efficiency of each primer set. Three independent qPCR runs were performed with three technical replicates for each sample to assess reproducibility and inter-run variability. Relative ratios of the tested species to each other were calculated as previously described (Park et al. 2016).
Western Blotting
Whole cell lysates of S. mutans UA140 and ΔspaP UA140 (1×107 cells) midlog phase cultures were obtained by boiling the samples in 1× non-reducing Laemmli buffer (without BME), which were resolved on 4–12% precast gradient gels (NuPAGE™ Novex™). The proteins on the gel were transferred to a nitrocellulose membrane (BIO-RAD) using the Trans-Blot Semi-Dry Electrophoretic Transfer cell (BIO-RAD) at 15V for 1 hr. The blot was then processed according to standard protocols. Monoclonal antibody 4–10A8C was used at a 1:100 dilution (Brady et al. 1992). Anti-mouse HRP conjugated secondary antibody was detected by using a SuperSignal West Dura Extended Duration Substrate kit (Thermo Fisher Scientific, Waltham, MA) according to manufacturer’s instructions.
Statistical Analysis
Statistical significance (p<0.05) of differences was evaluated via one-way analysis of variance (ANOVA) with post hoc Tukey’s test.
RESULTS
S. mutans UA140 Adheres Differentially to Fusobacteria
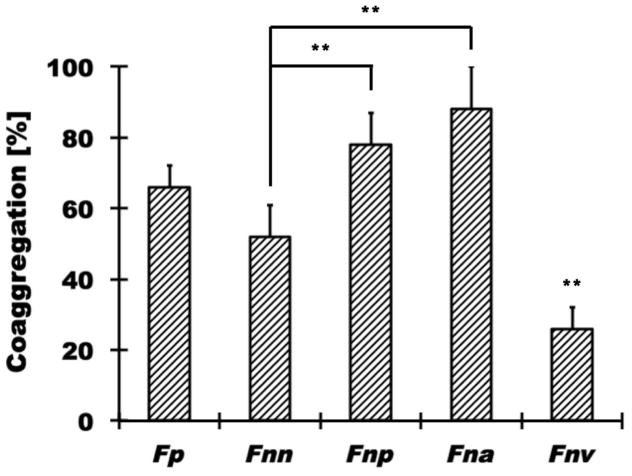
All experiments in this study were performed with S. mutans UA140, since this strain exhibited noticeably better binding to fusobacteria than the S. mutans reference strain UA159 (Supplemental Fig. 2). S. mutans UA140 adhered to F. periodonticum and three of the four F. nucleatum subspecies tested (Fig. 1). F. nucleatum ssp. polymorphum (C.I. = 78±9) and F. nucleatum ssp. animalis (C.I. = 88±12) exhibited the highest extent of co-aggregation with S. mutans, followed by F. periodonticum (C.I. = 66±6) and F. nucleatum ssp. nucleatum (C.I. = 52±9). The F. nucleatum subspecies vincentii did not display substantial coaggregation with S. mutans (C.I. = 26±6) and was therefore not further included in our analysis.
Fig. 1. Quantitative coaggregation assays between S. mutans and different oral fusobacteria.
The coaggregation of F. periodonticum [Fp] and several F. nucleatum subspecies (ssp. nucleatum [Fnn], ssp. polymorphum [Fnp], ssp. animalis [Fna] and ssp. vincentii [Fnv]) with S. mutans was evaluated in a quantitative coaggregation assay as described in Materials and Methods. Fnv coaggregation with S. mutans was significantly different (** p<0.01) from all other fusobacteria tested, while Fnn was significantly different from Fnp, Fna and Fnv but not Fp. Values represent the average of at least three independent experiments, and the error bars correspond to the standard deviations.
SpaP Acts as S. mutans Adhesin for Binding to F. nucleatum ssp. polymorphum
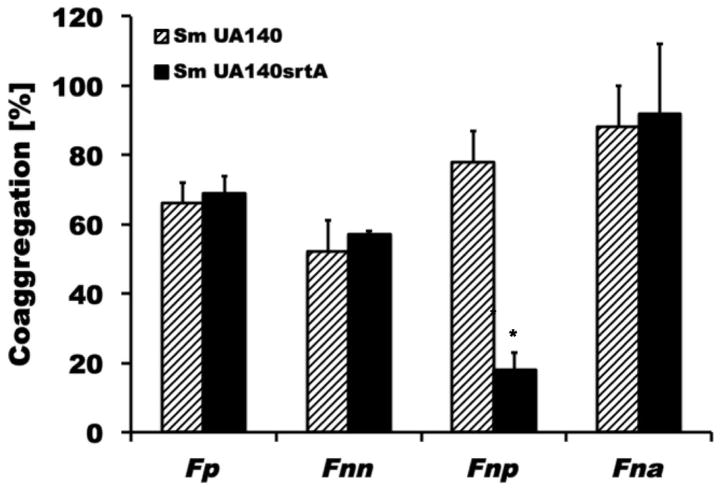
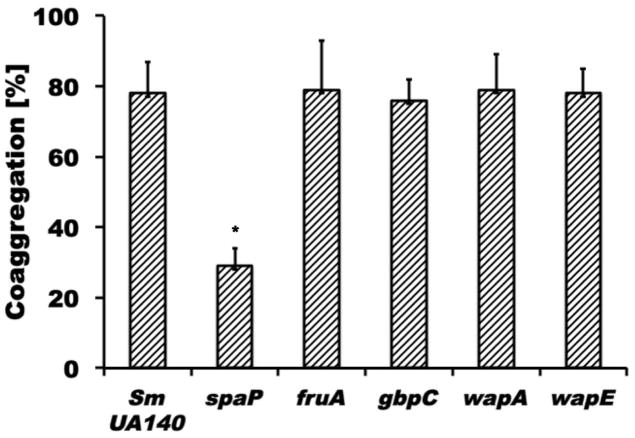
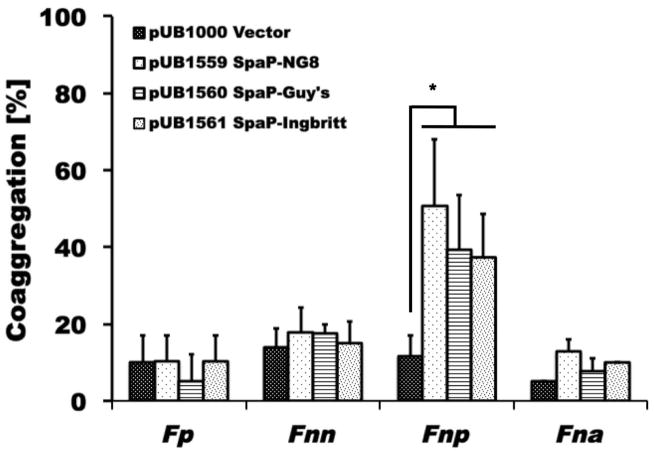
Many surface proteins in gram-positive microorganism including S. mutans depend on sortase A (SrtA) for proper cell wall anchoring and surface display (Marraffini et al. 2006; Paterson et al. 2004). To investigate if SrtA-dependent proteins play a role in adherence between S. mutans and fusobacteria, a srtA mutant of S. mutans was generated (see Material and Methods for details) and examined for binding to the fusobacterial species and subspecies tested in this study (Fig. 2). F. nucleatum ssp. polymorphum demonstrated a significant reduction in coaggregation with the srtA derivative of S. mutans UA140 (C.I. = 18±5; p<0.01), while none of the other fusobacteria exhibited any notable difference in adherence. To further investigate which one of the known SrtA-dependent proteins played a role in this process, we generated spaP-, fruA-, wapA-, wapE-, and gbpC-deficient S. mutans derivatives of UA140 for testing of coaggregation with F. nucleatum ssp. polymorphum. Among these mutant strains, only the spaP-deficient strain displayed significantly reduced coaggregation compared to wildtype (Fig. 3). To further confirm SpaP as the S. mutans adhesin mediating binding to F. nucleatum ssp. polymorphum, we employed a previously established system in which streptococcal membrane proteins including SpaP derived from three different strains of S. mutans (S. mutans NG8, Ingbritt 162 and Guy’s) are functionally displayed on the surface of L. lactis (Jakubovics et al. 2005). While F. nucleatum ssp. polymorphum recognized all three SpaP proteins from the different S. mutans strains, none of the other oral fusobacterial species did (Fig. 4). Comparison of the SpaP sequences expressed in the L. lactis system with those of S. mutans UA140 and UA159 revealed that SpaP of UA140 is most similar to SpaP of NG8, while SpaP of UA159 is more closely related to those expressed by S. mutans Ingbritt 162 and Guy’s (Supplemental Fig. 3). Specifically, SpaP of NG8 and SpaP of UA140 are both lacking alanine-753, which is present in the other SpaP sequences included in the comparison, and contain a five amino acid long insertion after position 797 that is absent in the SpaP proteins encoded by strains UA 159, Ingbritt 162 and Guy’s. Furthermore, the spaP gene of Ingbritt 162 contains a frameshift mutation that results in a truncation of the protein after position 1214 (Jakubovics et al. 2005). This SpaP derivative is still predominantly associated with the membrane despite missing the cell wall anchorage region and part of the C-region.
Fig. 2. Quantitative coaggregation assays between S. mutans derivatives defective in srtA with oral fusobacteria.
Coaggregation of different oral fusobacteria with wildtype S. mutans UA140 (diagonally striped bars) and the corresponding srtA mutant derivative (solid black bars). Fnp coaggregation with the srtA derivative of S. mutans was significantly different (* p<0.05) from the interaction with the wildtype parent. Values represent the average of at least three independent experiments, and the error bars correspond to the standard deviations.
Fig. 3. Quantitative coaggregation assays between S. mutans derivatives defective in srtA dependent surface proteins with F. nucleatum ssp. polymorphum.
Coaggregation of F. nucleatum ssp. polymorphum and mutant derivatives of S. mutans UA140 defective in spaP, gbpC, wapA, wapE or fruA. Fnp coaggregation with the spaP derivative of S. mutans was significantly different (* p<0.05) from the interaction with the wildtype parent and the other mutant derivatives tested. Values represent the average of at least three independent experiments, and the error bars correspond to the standard deviations.
Fig. 4. Quantitative coaggregation of SpaP-producing L. lactis strains with different fusobacteria.
Quantitative coaggregation of Fp, Fnn, Fnp, and Fna with L. lactis/pUB1000 (vector – black dotted bars), L. lactis/pUB1559 (producing SpaP of S. mutans NG8 – white dotted bars), L. lactis/pUB1660 (producing SpaP of S. mutans Guy’s – striped bars), and L. lactis/pUB1660 (producing SpaP of S. mutans Ingbritt 162 – irregularly dotted bars). Only binding of Fnp to the SpaP derivatives expressing L. lactis strains was significantly higher (* p<0.05) than all other interactions tested. Values represent the average of at least three independent experiments, and the error bars correspond to the standard deviations.
F. nucleatum ssp. polymorphum RadD Recognizes S. mutans SpaP
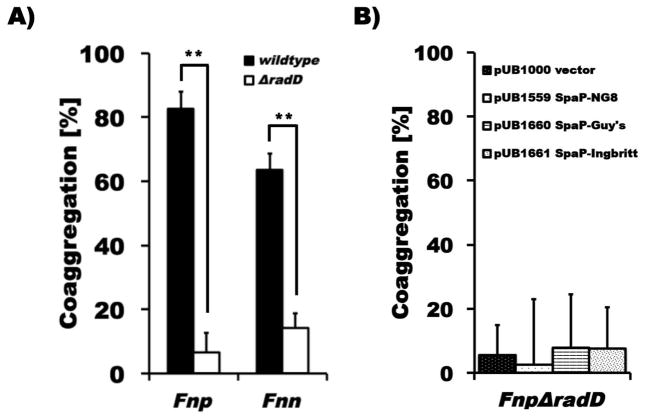
Since we previously found that RadD is a major adhesin for binding of F. nucleatum ssp. nucleatum to a variety of Gram-positive oral species (Kaplan et al. 2009), we generated a ΔradD derivative of F. nucleatum ssp. polymorphum (See Materials and Methods for details) to investigate if this large outer membrane protein is involved in mediating the interaction of this fusobacterial subspecies with S. mutans. We found that binding of this ΔradD mutant strain to S. mutans was completely abolished compared to the interaction displayed by the parent strain (Fig. 5a). The ΔradD derivative of F. nucleatum ssp. nucleatum was also defective in binding to S. mutans (Fig. 5a), even though this subspecies of F. nucleatum does not recognize SpaP as cognate adhesin (Fig. 4). We then tested attachment of the different SpaP variants expressing L. lactis strains used above (Fig. 4) with the ΔradD derivative of F. nucleatum ssp. polymorphum and confirmed that RadD was essential for effective binding to SpaP (Fig. 5b).
Fig. 5. Quantitative coaggregation F. nucleatum ssp. polymorphum and F. nucleatum ssp. nucleatumΔradD derivative with S. mutans and SpaP-producing L. lactis strains.
A) Quantitative coaggregation of Fnp (black bars) and Fnn (white bars) wildtype and the respective ΔradD mutant derivatives, which bind significantly less (** p<0.01) to wildtype S. mutans strain UA140, and B) FnpΔradD with the same SpaP-producing L. lactis strains tested in Fig. 4. Values represent the average of at least three independent experiments, and the error bars correspond to the standard deviations.
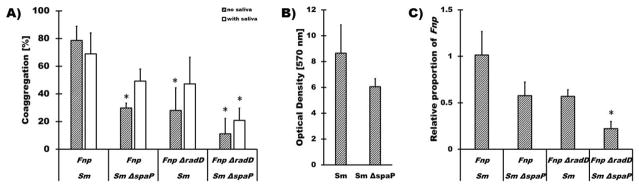
Since SpaP has been well established to bind to the salivary agglutinin complex consisting primarily of the high molecular weight scavenger protein gp340 (Ericson et al. 1983; Kishimoto et al. 1989; Oho et al. 1998), we tested if the presence of saliva altered the co-aggregation properties of S. mutans and F. nucleatum ssp polymorphum wildtype and mutant strains (Fig. 6A). Confirming the specificity of the interaction identified here, when SpaP as well as RadD were eliminated co-aggregation of S. mutans and F. nucleatum ssp polymorphum was significantly decreased in both the presence and absence of saliva. The impact of eliminating only a single binding partner was not as apparent in the presence of saliva and may reflect additional molecular interactions involving both salivary and/or bacterial constituents. Although deletion of spaP did not significantly decrease the ability of S. mutans alone to produce biofilms in the presence of sucrose and saliva (Fig. 6B), incorporation of F. nucleatum ssp polymorphum into similarly grown S. mutans biofilms was significantly decreased when both SpaP and RadD were eliminated (Fig. 6C). Consistent with the co-aggregation results in the presence of saliva, this effect was less apparent when only one of the two binding partners was missing.
Fig. 6. Quantitative coaggregation and biofilm integration of F. nucleatum ssp. polymorphum and itsΔradD derivative with S. mutans and its ΔspaP derivative.
A) Quantitative coaggregation of Fnp wildtype and the respective ΔradD mutant derivative with S. mutans strain UA140 (Sm) and its ΔspaP derivative (SmΔspaP) in the absence (striped bars) and presence (white bars) of 50% saliva; B) Quantitative crystal violet staining of biofilms formed by S. mutans strain UA140 (Sm) and its ΔspaP derivative (SmΔspaP) in the presence of 0.5% sucrose and 25% saliva; C) binding of Fnp wildtype and the respective ΔradD mutant derivative to S. mutans strain UA140 (Sm) and the ΔspaP derivative (SmΔspaP) biofilms in the presence (white bars) of 25% saliva. Significant differences to the respective combination of wildtype strains for each condition are indicated (* p<0.05).
DISCUSSION
Fusobacteria establish themselves as prevalent members of oral biofilms via their ability to bind to a large variety of early-colonizing species that attach directly to oral surfaces (Kolenbrander et al. 1993; Kolenbrander et al. 2010). While research has highlighted the interaction between F. nucleatum and non-mutans streptococci (Kaplan et al. 2009; Kolenbrander et al. 1993), co-adherence with mutans streptococci such as the cariogenic S. mutans, has been reported (Bradshaw et al. 1998; Falkler et al. 1981) but largely remains to be investigated. Here, we evaluated the interaction of F. periodonticum and different subspecies of F. nucleatum with S. mutans and identified RadD and SpaP as the cognate adhesin pair for the specific interaction between F. nucleatum ssp. polymorphum and S. mutans.
Interspecies adherence between oral bacteria is often species- or even strain-dependent (Kolenbrander et al. 1993) and the differential binding of the fusobacteria tested in this study to S. mutans (Fig. 1) is consistent with previous observations of variability in the attachment of F. nucleatum strains to different isolates of S. mutans (Falkler et al. 1981). Species-specificity is not limited to interbacterial interactions but was also observed for the attachment of F. nucleatum subspecies to eukaryotic cells (Xie et al. 1991). Furthermore, certain subspecies of F. nucleatum colonize distinct ecological niches of the oral cavity (Bolstad et al. 1996; Eren et al. 2014) and their relative abundance is correlated with health and disease (Uzel et al. 2011). Specific interspecies recognition plays an important role in the sophisticated oral biofilm structure, architecture and communication (Guo et al. 2014; Kolenbrander et al. 2010; Rickard et al. 2003). Fusobacteria significantly contribute to this important aspect of oral microbial community formation by connecting early colonizing species with the many pathogenic members containing late colonizers.
Our results suggest that the large outer membrane protein RadD, which we previously identified as the major adhesin for binding of F. nucleatum ssp. nucleatum to a number of early-colonizing Gram-positive species (Kaplan et al. 2009), also functions as adhesin for the interaction with S. mutans for at least two subspecies of F. nucleatum (Fig. 5A). RadD of F. periodonticum and F. nucleatum ssp. animalis were not tested, because only F. nucleatum ssp. nucleatum and F. nucleatum ssp. polymorphum can currently be genetically manipulated. Interestingly, however, only RadD of F. nucleatum ssp. polymorphum recognizes S. mutans SpaP as its cognate counterpart. Despite containing RadD homologues (Supplemental Fig. 4AC) and our finding that the ΔradD derivative of F. nucleatum ssp. nucleatum is deficient in interaction with S. mutans (Fig. 5A), none of the other fusobacterial species and subspecies tested bind to S. mutans via SpaP (Figs. 3 and 4) or any other SrtA-dependent surface protein (Fig. 2). Thus, in contrast to F. nucleatum ssp. polymorphum, the RadD-mediated interaction between S. mutans and F. nucleatum ssp. nucleatum appears to involve a binding partner that is not linked to the cell surface by SrtA. This disparity in RadD-specificity could be due to the extensive variability in the N-terminal part of the RadD protein between the F. nucleatum subspecies (Supplemental Fig. 4A). Further detailed RadD sequence comparison indicates that a SpaP-binding motif might be located in the highly variable regions between amino acids 285 to 625 or 1716 and 1811, which contain a number of residues that are uniquely present in F. nucleatum ssp. polymorphum (Supplemental Fig. 4B). Alternatively, observed sequence variations might affect the formation or accessibility of a conformational-depending binding site.
SpaP, the cognate S. mutans adhesin for F. nucleatum ssp. polymorphum (Fig. 3) is a SrtA-dependent surface protein of the Antigen I/II family, whose members are highly conserved among most oral streptococci and function as multiligand-binding proteins in eukaryotic cell and protein recognition, surface binding and attachment to other microorganisms (Nobbs et al. 2009). Previous studies suggest an important role for SpaP in disease development: Lack of SpaP decreased S. mutans cariogenicity in a gnotobiotic rat model (Crowley et al. 1999) and a close association between childhood caries prevalence and a high proportion of SpaP in S. mutans was reported (Duran-Contreras et al. 2011). SpaP has been shown to recognize a number of host ligand proteins, which include salivary proteins, extracellular matrix (ECM) proteins, and certain serum components (Beg et al. 2002; Brady et al. 2010; Busscher et al. 2008; Kelemen et al. 2004; Love et al. 1997; Nakai et al. 1993; Petersen et al. 2002; Sciotti et al. 1997; Soell et al. 2010; Switalski et al. 1993). Specifically, the N-terminal alanine-rich region (A-region) as well as parts of the variable region (V-region) mediate binding to the ECM proteins type I collagen, fibronectin, keratin and laminin, while distinct parts of the A-region and the C-terminal region are involved in binding of salivary glycoproteins such as the salivary agglutinin among others.
RadD of F. nucleatum ssp polymorphum appears to be recognized by parts of SpaP that are not involved in interaction with salivary glycoproteins, since addition of saliva did not interfere with the ability of S. mutans to bind to this fusobacterial subspecies between planktonic as well as biofilm cells (Fig. 6). Furthermore, the C-terminally truncated SpaP of S. mutans Ingbritt 162 that is lacking the binding site for salivary agglutinin, still binds to F. nucleatum ssp polymorphum (Figs. 4 and 5). Similarly, lack of the alanine-753 and a five amino acid insertion after position 797 in the SpaP proteins encoded by strains NG8 and UA140, do not seem to be important for RadD recognition, since SpaP of NG8 displays binding comparable to SpaP of Ingritt 162 and Guy’s in the L. lactis display system (Fig. 4 and Supplemental Fig. 3).
While SpaP was also found to participate in binding to certain actinomyces when expressed in S. gordonii, this interaction requires the presence of CshA, a protein typically not present in S. mutans (Jakubovics et al. 2005). In contrast, SspA and SspB, the SpaP homologues in S. gordonii, have been identified as adhesins for a variety of interspecies interactions including Actinomyces naeslundii (Egland et al. 2001; Jakubovics et al. 2005), Porphyromonas gingivalis (Brooks et al. 1997; Daep et al. 2006) and Candida albicans (Demuth et al. 1996). Distinct binding motifs that are localized in different parts of the molecule mediate adherence to the various species: Adhesion to A. naeslundii involves the A-region, which participates in formation of the fibrillar stalk, and binding to P. gingivalis has been attributed to the globular domain forming the C-terminal region. Demuth and coworkers (Demuth et al. 2001) demonstrated that the corresponding region in SpaP is unable to mediate binding to P. gingivalis, suggesting the importance of species-specific sequences for the recognition of interacting partner species. While our comparison of available RadD sequences suggest a potential role of certain highly variable N-terminal regions in SpaP recognition, further studies are needed to confirm this possibility and to explore if a linear motif of SpaP mediates its selective recognition of F. nucleatum ssp. polymorphum, or whether the interaction is dependent on a conformational determinant
Our finding, that effective binding of F. nucleatum ssp. polymorphum to S. mutans is mediated by the interaction of the adhesin pair RadD-SpaP adds further functionality and potential virulence attributes to these two multifactorial proteins. Interaction between these two species could broaden their options for integration into the supragingival microbial communities. Since few studies examine oral fusobacterial distribution on a subspecies level (Eren et al. 2014; Uzel et al. 2011), we can only speculate about the possible biological relevance of the apparent interaction between fusobacteria and S. mutans. Stable colonization of the oral cavity by fusobacteria has been demonstrated prior to the eruption of teeth (Kononen 1999), while S. mutans is generally considered to require tooth surfaces for efficient establishment, even though it has been detected in edentulous infants prior to tooth eruption (Berkowitz 2006). The ability of S. mutans to attach to fusobacteria including F. nucleatum ssp. polymorphum could thus constitute an additional opportunity for oral biofilm integration of cariogenic S. mutans. In contrast, attachment of F. nucleatum ssp. polymorphum to S. mutans in the supragingival plaque could be beneficial, since this fusobacterial subspecies was found to have acid-neutralizing abilities (Takahashi et al. 1997).
In summary, most of the predominant oral fusobacterial species and subspecies are able to adhere to the early colonizer S. mutans albeit at varying degrees. Furthermore, we identified SpaP of S. mutans as a specific adhesin for recognition of the F. nucleatum ssp. polymorphum adhesin RadD. This is the first time a cognate adhesin pair was identified for attachment of a fusobacterial species to another organism. While S. mutans does not interact with a large number of other bacterial species (Wang et al. 2011), it is an interesting phenomenon that the ubiquitous fusobacteria, which are known to bind to numerous other microorganisms recognize at least two different surface structures on S. mutans for attachment. This could possibly expand their own ability as well as that of S. mutans to effectively colonize available oral surfaces.
Supplementary Material
Whole cell extracts of S. mutans UA140 (lane 1) and its mutant derivative lacking spaP (lane 2) were separated on a SDS gel and probed with SpaP specific monoclonal antibody 4–10A as described (Brady et al. 1992).
Binding of with S. mutans strains UA159 and UA140 with F. nucleatum ssp. nucleatum (striped bars) and F. nucleatum ssp. polymorphum (dotted bars) was evaluated in a quantitative coaggregation assay as described in Materials and Methods. The level of attachment of both F. nucleatum subspecies to UA140 was significantly (** p<0.01) higher than to UA140.
(A) Phylogenetic relationship and (B) protein sequence comparison of SpaP from S. mutans strains UA140, UA159, NG8, Ingbritt 162 and Guy’s (Jakubovics et al. 2005). The phylogenetic tree was constructed using Phylogeny.fr (Dereeper et al. 2008). # indicates the respective truncated versions of the proteins expressed by the corresponding plasmids of the L. lactis system used in the study (Jakubovics et al. 2005).
(A) Schematic of RadD indicating homologies between subspecies: high – 90–100% (dark green), medium – 60–90% (yellow), and low – <60% (light green); (B) homology plot for RadD of all oral F. nucleatum subspecies using averages of 100 amino acid long windows; dots symbolize unique amino acids for Fnp; box 1 (aa 285 to 650) and box 2 (aa 1716 to 1811) highlight the two regions in which most of these unique amino acids are localized; (C) sequence alignment of RadD of Fna, Fnn, Fnp and Fnv; identical (*) and conserved (: and .) amino acids are indicated.
Acknowledgments
We thank Dr. Dennis G. Cvitkovitch (Toronto, Ontario, Canada) and Dr. Justin Merrit (Portland, Oregon, USA) for kindly providing the different S. mutans UA159 mutant derivatives, and Dr. Howard Jenkinson (Bristol, UK) for the gracious gift of SpaP-expressing L. lactis strains. We also would like to thank Dr. Jeannine Brady (University of Florida, Florida, USA) for the kind gift of monoclonal SpaP-specific antibodies. Purified CSP was a gift from C3 Jian, Inc. This work was supported in part by NIH/NIDCR grants R01-DE020102, R01-DE021108 and R01-DE018276.
References
- Aruni AW, Dou Y, Mishra A, Fletcher HM. The Biofilm Community-Rebels with a Cause. Curr Oral Health Rep. 2015;2:48–56. doi: 10.1007/s40496-014-0044-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back CR, Douglas SK, Emerson JE, et al. Streptococcus gordonii DL1 adhesin SspB V-region mediates coaggregation via receptor polysaccharide of Actinomyces oris T14V. Mol Oral Microbiol. 2015;30:411–24. doi: 10.1111/omi.12106. [DOI] [PubMed] [Google Scholar]
- Beg AM, Jones MN, Miller-Torbert T, Holt RG. Binding of Streptococcus mutans to extracellular matrix molecules and fibrinogen. Biochem Biophys Res Commun. 2002;298:75–9. doi: 10.1016/s0006-291x(02)02390-2. [DOI] [PubMed] [Google Scholar]
- Berkowitz RJ. Mutans streptococci: acquisition and transmission. Pediatr Dent. 2006;28:106–9. discussion 192–8. [PubMed] [Google Scholar]
- Bolstad AI, Jensen HB, Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55–71. doi: 10.1128/cmr.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw DJ, Marsh PD, Watson GK, Allison C. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect Immun. 1998;66:4729–32. doi: 10.1128/iai.66.10.4729-4732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Maddocks SE, Larson MR, et al. The changing faces of Streptococcus antigen I/II polypeptide family adhesins. Mol Microbiol. 2010;77:276–86. doi: 10.1111/j.1365-2958.2010.07212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady LJ, Piacentini DA, Crowley PJ, et al. Differentiation of salivary agglutinin-mediated adherence and aggregation of mutans streptococci by use of monoclonal antibodies against the major surface adhesin P1. Infect Immun. 1992;60:1008–17. doi: 10.1128/iai.60.3.1008-1017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks W, Demuth DR, Gil S, Lamont RJ. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect Immun. 1997;65:3753–8. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busscher HJ, van de Belt-Gritter B, Dijkstra RJ, et al. Streptococcus mutans and Streptococcus intermedius adhesion to fibronectin films are oppositely influenced by ionic strength. Langmuir. 2008;24:10968–73. doi: 10.1021/la8016968. [DOI] [PubMed] [Google Scholar]
- Corby PM, Lyons-Weiler J, Bretz WA, et al. Microbial risk indicators of early childhood caries. J Clin Microbiol. 2005;43:5753–9. doi: 10.1128/JCM.43.11.5753-5759.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley PJ, Brady LJ, Michalek SM, Bleiweis AS. Virulence of a spaP mutant of Streptococcus mutans in a gnotobiotic rat model. Infect Immun. 1999;67:1201–6. doi: 10.1128/iai.67.3.1201-1206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daep CA, James DM, Lamont RJ, Demuth DR. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect Immun. 2006;74:5756–62. doi: 10.1128/IAI.00813-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth DR, Duan Y, Brooks W, et al. Tandem genes encode cell-surface polypeptides SspA and SspB which mediate adhesion of the oral bacterium Streptococcus gordonii to human and bacterial receptors. Mol Microbiol. 1996;20:403–13. doi: 10.1111/j.1365-2958.1996.tb02627.x. [DOI] [PubMed] [Google Scholar]
- Demuth DR, Irvine DC, Costerton JW, et al. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect Immun. 2001;69:5736–41. doi: 10.1128/IAI.69.9.5736-5741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth DR, Lammey MS, Huck M, et al. Comparison of Streptococcus mutans and Streptococcus sanguis receptors for human salivary agglutinin. Microb Pathog. 1990;9:199–211. doi: 10.1016/0882-4010(90)90022-i. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran-Contreras GL, Torre-Martinez HH, de la Rosa EI, et al. spaP gene of Streptococcus mutans in dental plaque and its relationship with early childhood caries. Eur J Paediatr Dent. 2011;12:220–4. [PubMed] [Google Scholar]
- Egland PG, Du LD, Kolenbrander PE. Identification of independent Streptococcus gordonii SspA and SspB functions in coaggregation with Actinomyces naeslundii. Infect Immun. 2001;69:7512–6. doi: 10.1128/IAI.69.12.7512-7516.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA. 2014;111:E2875–84. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson T, Rundegren J. Characterization of a salivary agglutinin reacting with a serotype c strain of Streptococcus mutans. Eur J Biochem. 1983;133:255–61. doi: 10.1111/j.1432-1033.1983.tb07456.x. [DOI] [PubMed] [Google Scholar]
- Falkler WA, Jr, Burger BW. Microbial surface interactions: reduction of the haemagglutination activity of the oral bacterium Fusobacterium nucleatum by absorption with Streptococcus and Bacteroides. Arch Oral Biol. 1981;26:1015–25. doi: 10.1016/0003-9969(81)90112-6. [DOI] [PubMed] [Google Scholar]
- Guo L, He X, Shi W. Intercellular communications in multispecies oral microbial communities. Front Microbiol. 2014;5:328. doi: 10.3389/fmicb.2014.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffajee AD, Socransky SS, Patel MR, Song X. Microbial complexes in supragingival plaque. Oral Microbiol Immunol. 2008;23:196–205. doi: 10.1111/j.1399-302X.2007.00411.x. [DOI] [PubMed] [Google Scholar]
- Handley PS, Carter PL, Wyatt JE, Hesketh LM. Surface structures (peritrichous fibrils and tufts of fibrils) found on Streptococcus sanguis strains may be related to their ability to coaggregate with other oral genera. Infect Immun. 1985;47:217–27. doi: 10.1128/iai.47.1.217-227.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics NS, Stromberg N, van Dolleweerd CJ, et al. Differential binding specificities of oral streptococcal antigen I/II family adhesins for human or bacterial ligands. Mol Microbiol. 2005;55:1591–605. doi: 10.1111/j.1365-2958.2005.04495.x. [DOI] [PubMed] [Google Scholar]
- Kaplan CW, Lux R, Haake SK, Shi W. The Fusobacterium nucleatum outer membrane protein RadD is an arginine-inhibitable adhesin required for inter-species adherence and the structured architecture of multispecies biofilm. Mol Microbiol. 2009;71:35–47. doi: 10.1111/j.1365-2958.2008.06503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen L, Rizk S, Debreczeny M, et al. Streptococcal antigen I/II binds to extracellular proteins through intermolecular beta-sheets. FEBS Lett. 2004;566:190–4. doi: 10.1016/j.febslet.2004.04.029. [DOI] [PubMed] [Google Scholar]
- Kishimoto E, Hay DI, Gibbons RJ. A human salivary protein which promotes adhesion of Streptococcus mutans serotype c strains to hydroxyapatite. Infect Immun. 1989;57:3702–7. doi: 10.1128/iai.57.12.3702-3707.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitada K, Oho T. Effect of saliva viscosity on the co-aggregation between oral streptococci and Actinomyces naeslundii. Gerodontology. 2012;29:e981–7. doi: 10.1111/j.1741-2358.2011.00595.x. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Ganeshkumar N, Cassels FJ, Hughes CV. Coaggregation: specific adherence among human oral plaque bacteria. FASEB J. 1993;7:406–13. doi: 10.1096/fasebj.7.5.8462782. [DOI] [PubMed] [Google Scholar]
- Kolenbrander PE, Palmer RJ, Jr, Periasamy S, Jakubovics NS. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8:471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- Kononen E. Oral colonization by anaerobic bacteria during childhood: role in health and disease. Oral Dis. 1999;5:278–85. doi: 10.1111/j.1601-0825.1999.tb00090.x. [DOI] [PubMed] [Google Scholar]
- Kook JK, Park SN, Lim YK, et al. Fusobacterium nucleatum subsp. fusiforme Gharbia and Shah 1992 is a later synonym of Fusobacterium nucleatum subsp. vincentii Dzink et al. 1990. Curr Microbiol. 2013;66:414–7. doi: 10.1007/s00284-012-0289-y. [DOI] [PubMed] [Google Scholar]
- Lee SF, Progulske-Fox A, Erdos GW, et al. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II) Infect Immun. 1989;57:3306–13. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque CM, Voronejskaia E, Huang YC, et al. Involvement of sortase anchoring of cell wall proteins in biofilm formation by Streptococcus mutans. Infect Immun. 2005;73:3773–7. doi: 10.1128/IAI.73.6.3773-3777.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima KC, Coelho LT, Pinheiro IV, et al. Microbiota of dentinal caries as assessed by reverse-capture checkerboard analysis. Caries Res. 2011;45:21–30. doi: 10.1159/000322299. [DOI] [PubMed] [Google Scholar]
- Love RM, McMillan MD, Jenkinson HF. Invasion of dentinal tubules by oral streptococci is associated with collagen recognition mediated by the antigen I/II family of polypeptides. Infect Immun. 1997;65:5157–64. doi: 10.1128/iai.65.12.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai M, Okahashi N, Ohta H, Koga T. Saliva-binding region of Streptococcus mutans surface protein antigen. Infect Immun. 1993;61:4344–9. doi: 10.1128/iai.61.10.4344-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–50. doi: 10.1128/MMBR.00014-09. Table of Contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oho T, Yu H, Yamashita Y, Koga T. Binding of salivary glycoprotein-secretory immunoglobulin A complex to the surface protein antigen of Streptococcus mutans. Infect Immun. 1998;66:115–21. doi: 10.1128/iai.66.1.115-121.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Shokeen B, Haake SK, Lux R. Characterization of Fusobacterium nucleatum ATCC 23726 adhesins involved in strain-specific attachment to Porphyromonas gingivalis. Int J Oral Sci. 2016 in press. [Google Scholar]
- Paterson GK, Mitchell TJ. The biology of Gram-positive sortase enzymes. Trends Microbiol. 2004;12:89–95. doi: 10.1016/j.tim.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Petersen FC, Assev S, van der Mei HC, et al. Functional variation of the antigen I/II surface protein in Streptococcus mutans and Streptococcus intermedius. Infect Immun. 2002;70:249–56. doi: 10.1128/IAI.70.1.249-256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podbielski A, Spellerberg B, Woischnik M, et al. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS) Gene. 1996;177:137–47. doi: 10.1016/0378-1119(96)84178-3. [DOI] [PubMed] [Google Scholar]
- Potts TV, Holdeman LV, Slots J. Relationships among the oral fusobacteria assessed by DNA-DNA hybridization. J Dent Res. 1983;62:702–5. doi: 10.1177/00220345830620060101. [DOI] [PubMed] [Google Scholar]
- Rickard AH, Gilbert P, High NJ, et al. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- Sciotti MA, Yamodo I, Klein JP, Ogier JA. The N-terminal half part of the oral streptococcal antigen I/IIf contains two distinct binding domains. FEMS Microbiol Lett. 1997;153:439–45. doi: 10.1111/j.1574-6968.1997.tb12608.x. [DOI] [PubMed] [Google Scholar]
- Soell M, Hemmerle J, Hannig M, et al. Molecular force probe measurement of antigen I/II-matrix protein interactions. Eur J Oral Sci. 2010;118:590–5. doi: 10.1111/j.1600-0722.2010.00785.x. [DOI] [PubMed] [Google Scholar]
- Switalski LM, Butcher WG, Caufield PC, Lantz MS. Collagen mediates adhesion of Streptococcus mutans to human dentin. Infect Immun. 1993;61:4119–25. doi: 10.1128/iai.61.10.4119-4125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Saito K, Schachtele CF, Yamada T. Acid tolerance and acid-neutralizing activity of Porphyromonas gingivalis, Prevotella intermedia and Fusobacterium nucleatum. Oral Microbiol Immunol. 1997;12:323–8. doi: 10.1111/j.1399-302x.1997.tb00733.x. [DOI] [PubMed] [Google Scholar]
- Uzel NG, Teles FR, Teles RP, et al. Microbial shifts during dental biofilm re-development in the absence of oral hygiene in periodontal health and disease. J Clin Periodontol. 2011;38:612–20. doi: 10.1111/j.1600-051X.2011.01730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RK, He XS, Hu W, et al. Analysis of interspecies adherence of oral bacteria using a membrane binding assay coupled with polymerase chain reaction-denaturing gradient gel electrophoresis profiling. Int J Oral Sci. 2011;3:90–7. doi: 10.4248/IJOS11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright CJ, Burns LH, Jack AA, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28:83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Gibbons RJ, Hay DI. Adhesive properties of strains of Fusobacterium nucleatum of the subspecies nucleatum, vincentii and polymorphum. Oral Microbiol Immunol. 1991;6:257–63. doi: 10.1111/j.1399-302x.1991.tb00488.x. [DOI] [PubMed] [Google Scholar]
- Zhu M, Ajdic D, Liu Y, et al. Role of the Streptococcus mutans irvA gene in GbpC-independent, dextran-dependent aggregation and biofilm formation. Appl Environ Microbiol. 2009;75:7037–43. doi: 10.1128/AEM.01015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zmantar T, Kouidhi B, Miladi H, et al. A microtiter plate assay for Staphylococcus aureus biofilm quantification at various pH levels and hydrogen peroxide supplementation. New Microbiol. 2010;33:137–45. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Whole cell extracts of S. mutans UA140 (lane 1) and its mutant derivative lacking spaP (lane 2) were separated on a SDS gel and probed with SpaP specific monoclonal antibody 4–10A as described (Brady et al. 1992).
Binding of with S. mutans strains UA159 and UA140 with F. nucleatum ssp. nucleatum (striped bars) and F. nucleatum ssp. polymorphum (dotted bars) was evaluated in a quantitative coaggregation assay as described in Materials and Methods. The level of attachment of both F. nucleatum subspecies to UA140 was significantly (** p<0.01) higher than to UA140.
(A) Phylogenetic relationship and (B) protein sequence comparison of SpaP from S. mutans strains UA140, UA159, NG8, Ingbritt 162 and Guy’s (Jakubovics et al. 2005). The phylogenetic tree was constructed using Phylogeny.fr (Dereeper et al. 2008). # indicates the respective truncated versions of the proteins expressed by the corresponding plasmids of the L. lactis system used in the study (Jakubovics et al. 2005).
(A) Schematic of RadD indicating homologies between subspecies: high – 90–100% (dark green), medium – 60–90% (yellow), and low – <60% (light green); (B) homology plot for RadD of all oral F. nucleatum subspecies using averages of 100 amino acid long windows; dots symbolize unique amino acids for Fnp; box 1 (aa 285 to 650) and box 2 (aa 1716 to 1811) highlight the two regions in which most of these unique amino acids are localized; (C) sequence alignment of RadD of Fna, Fnn, Fnp and Fnv; identical (*) and conserved (: and .) amino acids are indicated.