Abstract
Multistage clonal expansion (MSCE) models of carcinogenesis are continuous-time Markov process models often used to relate cancer incidence to biological mechanism. Identifiability analysis determines what model parameter combinations can, theoretically, be estimated from given data. We use a systematic approach, based on differential algebra methods traditionally used for deterministic ODE models, to determine identifiable combinations for a generalized subclass of MSCE models with any number of pre-initation stages and one clonal expansion. Additionally, we determine the identifiable combinations of the generalized MSCE model with up to four clonal expansion stages, and conjecture the results for any number of clonal expansion stages. The results improve upon previous work in a number of ways and provide a framework to find the identifiable combinations for further variations on the MSCE models. Finally, our approach, which takes advantage of the Kolmogorov backward equations for the probability generating functions of the Markov process, demonstrates that identifiability methods used in engineering and mathematics for systems of ODES can be applied to continuous-time Markov processes.
Keywords: Multistage clonal expansion model, identifiability, continuous-time Markov process, differential algebra
1 Introduction
The two-stage clonal expansion (TSCE) model is a continuous-time Markov process proposed by Moolgavkar, Venzon, and Knudson [1, 2] to capture the initiation–promotion–progression hypothesis of carcinogenesis, wherein normal cells undergo a genetic transformation that causes clonal expansion, followed by progression to malignancy. The initiation–promotion–progression paradigm allows one to consider carcinogenic factors as initiators or promoters given their mechanism of action and their differential effects at different stages of life. The TSCE model formulation may be extended to three or more stages or other more complex variations, which are collectively called multistage clonal expansion (MSCE) models. Parameter estimation with multistage clonal expansion models has proven a valuable approach, and MSCE models have been successfully used to analyze and fit data from pancreatic, colorectal, esophageal, and oral cancer, among others [3–16].
Consideration of identifiability is the first step in estimation of model parameters from data. A model is said to be identifiable if all model parameters may be uniquely determined from given observed data [17-19]. Identifiability is a key step in ensuring successful parameter estimation and is often considered in two forms: structural identifiability, which considers the best-case scenario of noise-free, continuously measured data in order to uncover identifiability issues inherent in the model structure, and practical identifiability, which addresses issues such as noise, bias, and frequency of sampling [20]. While the best-case scenario is unrealistic, structural identifiability is necessary for practical identifiability and can often lead to useful insights for model reparameterization and data collection strategies.
For deterministic models, one often frames the identifiability problem as testing the injectivity of the map from the parameters to the output trajectories (implicitly defined by the corresponding ordinary differential equations (ODE) system) [21]. There are a wide range of approaches to answering questions of identifiability for such systems, including Laplace transformation, Taylor series, similarity transformation, and differential algebra [19, 21-28].
The identifiability of certain individual clonal expansion models, which are stochastic rather than deterministic, has been addressed primarily on a case-by-case basis and in no systematic way. Heidenreich et al. [29] determined the identifiability of the TSCE model with constant and piecewise-constant parameters when fitted to incidence data through derivation of closed form solutions of the corresponding hazard function. Luebeck and Moolgavkar [5] similarly analyzed the identifiability of MSCE models with multiple pre-initiation stages and constant parameters. Little et al. [30] developed bounds for the number of identifiable combinations for a class of stochastic cancer models with genomic instability—which includes MSCE models—through observing parameter combinations in the form of the cancer hazard in the model and numerical evaluations of the Fisher information matrix.
Here, we present a derivation of the identifiability of a generalized subclass of MSCE models with multiple pre-initiation steps when fitting to age-specific cancer incidence data, as is typical. We use a differential algebra approach that was developed for deterministic ODE models and which has not previously been brought to bear on this class of models [21, 26, 27, 31, 32]. We do this by leveraging the Kolmogorov backward equations for continuous-time Markov processes, which can be reduced to a system of differential equations. This approach has many advantages: it is analytical and systematic, returns explicit identifiable combinations rather than bounds, and is a global result over the parameter space. We additionally demonstrate the identifiability of the fully general case with multiple clonal expansions for models with up to four clonal expansion stages and conjecture that our framework could be extended to any number of stages. Our work demonstrates that approaches for identifiability in deterministic dynamical systems can be used in Markov branching processes and, more generally, continuous-time Markov processes.
2 Methods
2.1 Derivation of the MSCE model
Although the mathematics of multistage clonal expansion models has been detailed elsewhere [1-3, 11, 29, 33-39], we provide a sketch of the derivation in order to provide a basis for using the differential algebra method of identifiability with other continuous-time Markov processes. The n-stage clonal expansion model (Figure 1a) is characterized by a set of conditional probability generating functions, where Yk(t), 1 ≤ k ≤ n − 2, and Z(t) are as in Table I, and τ is a fixed time such that 0 ≤ τ ≤ t. If we define
| (1) |
for some dummy variables y1, ⋯ , yn−1, and z, then the conditional probability generation functions are as follows:
| (2) |
Figure 1.
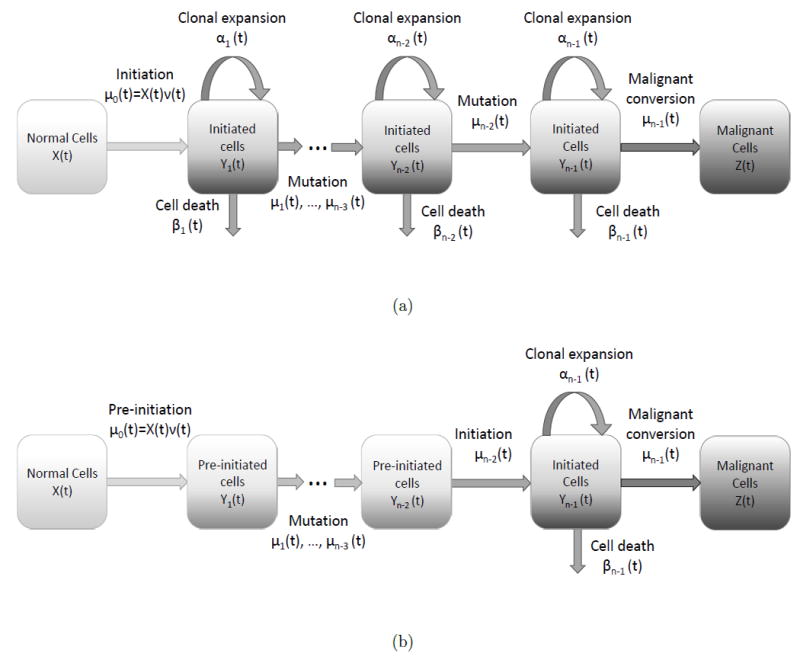
Generalized MSCE models. (a) The fully generalized model with clonal expansion at each pre-malignant step. (b) The standard model with several pre-initiation steps and one clonal expansion.
Table I.
Variables and parameters of the generalized multistage clonal expansion (MSCE) model
| Variables | |
|---|---|
| X(t) | Number of normal cells, treated deterministically or set to be constant X(t) = X |
| Yk(t) | Number of cells in initiated stage k |
| Z(t) | Number of malignant cells |
| Parameters | |
| ν(t) | Per cell mutation rate for normal cells (asymmetric division) |
| μ0(t) | := ν(t)X(t), a notational convenience |
| μk(t) | Mutation rate at the kth stage (asymmetric division) |
| αk(t) | Clonal expansion rate at the kth stage (symmetric division) |
| βk(t) | Cell death rate at the kth stage |
These probability functions satisfy the Kolmogorov backward equations. Here, we assume that the parameters, which are listed in Table I, are constant in time (age). These equations are
| (3) |
with initial conditions
| (4) |
The usual data in this context are age-specific incidence curves (e.g. as are available in the Surveillance, Epidemiology and End Results (SEER) cancer registries). The age-specific incidence curve corresponds to a model hazard. The hazard and survival contain equivalent information , so, for simplicity of analysis, we consider the survival to be known. For this model, the survival can be related to Ψ in the following way:
| (5) |
Let s = t − τ and define x(s) = Ψ (1, ⋯ , 1, 1, t − s, t), x1(s) = Φ1(1, ⋯ , 1, 1, t − s, t), ⋯ , xn−1(s) = Φn−1(1, ⋯ , 1, 1, t − s, t). Then x(t) = S(t). Let ẋk denote derivative of xk with respect to s. Then the following set of differential equations, 1 ≤ k ≤ n − 2, governs the survival:
| (6) |
with initial conditions x(0) = 1, xk(0) = 1, and xn−1(0) = 1.
2.2 Differential algebra approach to identifiability
As noted earlier, structural identifiability focuses on examining the inherent, structural estimation properties of a given model and data, assuming a best-case scenario in which the model output (i.e. the observed variable(s)) is perfectly observed and the model is correctly specified. While this is unrealistic for real data, structural identifiability is a necessary condition for practical estimation from real-world data that many times goes unchecked, and in fact many mathematical models used in practice turn out to be structurally unidentifiable. Structural identifiability allows us to resolve these issues and can help in designing data collection or estimation strategies.
Here we give an overview of structural identifiability definitions and the differential algebra approach for deterministic dynamical systems. For more details, the reader is referred to Saccomani et al. [21] and Audoly et al. [26]. For simplicity, here we consider the case where we have only one measured variable υ and one input function u, although the same definitions and approach can be used for multiple inputs and outputs as well. Consider a vector of states x(t) (unobserved), vector of parameters to be estimated ρ, and observed (known) input u(t) and output υ(t) in the ODE model
| (7) |
Structural identifiability analysis addresses the following question: given the model, states x, known input u, and known output υ, is it possible to uniquely identify the model parameters ρ? This can be framed as an injectivity question: is the map (implicitly defined by f and g) from parameter values (ρ) to output trajectories (υ) injective? [21]. Structural identifiability is a global property, but, because there may be some degenerate parameters or initial conditions for which an otherwise identifiable model may be unidentifiable (e.g. if all initial conditions or parameters are zero), it is typically defined almost everywhere over parameter and initial-condition space.
Definition 1
Parameter ρi in the model given in Eq. (7) is uniquely structurally identifiable if, for almost all values and initial conditions, the observation of an output trajectory (υ(t) = υ* (t)) uniquely determines the parameter value , i.e. if only one value of ρi could have resulted in the observed output.
Definition 2
The model given in Eq. (7) is structurally identifiable if each ρi is structurally identifiable.
If a model is not structurally identifiable, it is said to be unidentifiable, and there exists a set of identifiable combinations of parameters that represents the parametric information available in the data (except in degenerate cases where the model is reducible or has insensitive parameters). Such a set is not unique; any set of combinations that generate the same field is an equivalent set of identifiable combinations, e.g. {ab, c/b} and {ab, ac} are equivalent sets of identifiable combinations.
We must emphasize that identifiability is an assessment that is dependent on both what quantities are observed (i.e. the data u(t) and υ(t)) and on the parameterization of the model. A model is unidentifiable if even one parameter cannot be uniquely determined from the available data. An unidentifiable model can sometimes be rendered identifiable by reparameterization (i.e. in terms of identifiable combinations) or by changing what data are measured.
Differential algebra offers one approach for evaluating the structural identifiability of rational-function differential-equation models. Technical details of the differential algebra approach to identifiability may be found elsewhere [21, 32], but this method is built on the idea of treating the differential equations as elements of a differential polynomial ring, that is, a polynomial ring in the variables and their derivatives, with an additional derivative operation. Once framed in this algebraic perspective, reduction techniques such as characteristic sets or Gröbner bases can be used to reduce the model to a form in which the identifiability properties can be determined, called the input–output equation [26, 40].
The input–output equation is central to the differential algebra technique [41]. It is a monic differential polynomial only in terms of u and υ, their derivatives, and the parameters ρ. In the case of multiple outputs, there will be as many of these monic differential polynomials—input–output equations—as there are observed output variables. The solutions of the input–output equation are precisely the possible input-output pairs for the system; in other words, the input–output equation is an equivalent differential equation where the unobserved variables have been eliminated, so that every solution trajectory for the model (in terms of x, u, υ) corresponds to a solution for the input–output equation (in terms of only u and υ), though we note that multiple model trajectories may correspond to the same input–output solution. The coefficients of the input–output equation are a complete, though typically not minimal (redundancies are usual), set of identifiable combinations, and testing for structural identifiability can thus be reduced to testing the injectivity of the map from the parameters to the identifiable combinations. We illustrate the differential algebra technique and the input–utput equation for a simple example in Appendix A.
The input–output equation must be monic—the choice of variable ranking is arbitrary, though u < u̇ < ü < ⋯ < υ < υ̇ < ϋ < ⋯ is traditional [26]—or the set of identifiable combinations may not be uniquely determined. For example, the following are equivalent differential polynomials,
but the map from {a, b, c} to is injective while that to {1, ab, ac} is not. The input–output equation is required to be monic to identify the correct set of identifiable combinations.
Finally, we note that, in the notation of this section, the MSCE model (Eq. 6) has states x = (x(t), x1(t), … , xn−1(t)), output (data) υ(t) = x(t), and has no input u(t).
3 Results
3.1 Two-stage clonal expansion (TSCE) model
Although the identifiability of the TSCE model is well-known [29], this model provides a tractable test-case for the differential algebra approach to identifiability in this context.
Theorem 1
If cancer survival (or, equivalently, age-specific incidence) is perfectly measured, the two-stage clonal expansion model with constant parameters (ν, X, α, β, μ1) is unidentifiable but has three identifiable parameter combinations, which may be represented as μ0μ1, α1μ1, and α1 − β1 − μ1, where μ0 = ν X.
Proof
From Eqs. (6), the following equations contain all information of the the two-stage clonal expansion model:
| (8) |
We assume that the survival function x is perfectly measured. The goal here is to determine the identifiable parameter combinations from the input–output equation for the system, which will be a monic polynomial of the observed output x and its derivatives.
We solve for x1 in terms of x and its derivatives,
| (9) |
Plug this in to the ẋ1 equation,
| (10) |
simplifying to
| (11) |
This last equation is a monic polynomial of x and its derivatives, is equivalent to the original differential equations, and is thus an input–output equation. We can read a set of identifiable parameter combinations from the equation coefficients: μ0μ1,α1 − β1 − μ1, and α1μ1.
Remark
The two-stage clonal expansion is often parameterized [5] as
| (12) |
It is easy to see that {r, p, q} is an equivalent set of identifiable parameter combinations.
Remark
Although the initial conditions can, generally, provide additional identifiable combinations, they do not in this case. At the initial conditions, x(0) = 1 and x1(0) = 1,
| (13) |
As the data is x, we can identify ẋ(0), which, in this case, is identically equal to 0 and thus does not provide any additional parametric information. We do not observe x1, so ẋ1(0) = −μ1 is not observed.
3.2 Generalized MSCE model with multiple pre-initiation steps
We extend the result and method for the two-stage model to an n-stage model in which only the final non-malignant compartment has clonal expansion (Figure 1b). This model, unlike the fully generalized MSCE model, is often used in the literature to model cancer progression (e.g. [5, 9, 11]). The differential equations defining the survival x—and implicitly the hazard—of this model may be found by setting each of α1, … , αn−2, β1, … , βn−2 to zero in Eqs. (6):
| (14) |
for 1 ≤ k ≤ n − 2 and with initial conditions x(0) = 1, xk(0) = 1, and xn−1(0) = 1.
Theorem 2
If cancer survival (or, equivalently, age-specific incidence) is perfectly measured, the n-stage (n ≥ 3) multistage clonal expansion (MSCE) model with only one, final clonal expansion and n + 3 constant parameters (ν, X, α, β, μ1, ⋯ , μn−1) is unidentifiable but has n identifiable parameter combinations, which may be represented by μ0, … , μn−3, μn−1μn−2, αn−1μn−1, αn−1 − βn−1 − μn−1, where μ0 = νX.
In order to highlight the result and its implications without the distraction of technical details, we leave the proof to Appendix B. This is a global result over parameter space, and there are no degenerate parameter values of interest: when μk = 0, the problem is no longer of biological interest, and, when excluding those cases, αk = 0 and βk = 0 are not degenerate values for the theorem.
3.3 Generalized MSCE model with multiple clonal expansions
Here, we consider the full model (Eqs. (6), Figure 1a), allowing clonal expansion to occur at each pre-malignant stage.
Proposition 1
If cancer survival (or, equivalently, age-specific incidence) is perfectly measured, the n-stage (n ≥ 3) multistage clonal expansion (MSCE) model with 3n − 1 constant parameters (ν, X, α1, … , αn−1, β1, … , βn−1, μ1, … , μn−1) is unidentifiable.
As above, we leave the proof to Appendix B.
Conjecture 1
If cancer survival (or, equivalently, age-specific incidence) is perfectly measured, the n-stage (n ≥ 3) multistage clonal expansion (MSCE) model with 3n − 1 constant parameters has 3n − 3 identifiable parameter combinations, which may be represented as α1, … , αn−2, β1, … , β n−2, μ0, … , μn−3, μn−1μn−2, αn−1μn−1, αn−1 − βn−1 − μn−1, where μ0 = νX.
The conjecture is true for n ≤ 5; the proof, left to Appendix B, is an extension of that of Proposition 1. We believe that the method developed in the proof of Theorem 1 could be used to prove this conjecture in general, though additional combinatorial results will likely be needed to deal with the added complexity.
In Figure 2, we plot the hazards for the full model with four to eight stages using two different sets of parameters. For each model with n stages, the plotted points are generated using parameter values μk−1 = 10−2, αk = 3, βk = 2.8 for k = 1, … , n− 2 and μn−2 = 10−3, αn−1 = 3, βn−1 = 2.5 + 10−6, and μn−1 = 10−6. The corresponding lines use the parameters μk−1 = 10−2, αk = 3, βk= 2.8 for k = 1, … , n − 2 and μn−2 = 10−2, αn−1 = 30, βn−1 = 29.5 + 10−7, and μn−1 = 10−7. The indistinguishability of the hazards generated with each of the two parameters sets is consistent with the conjecture.
Figure 2.
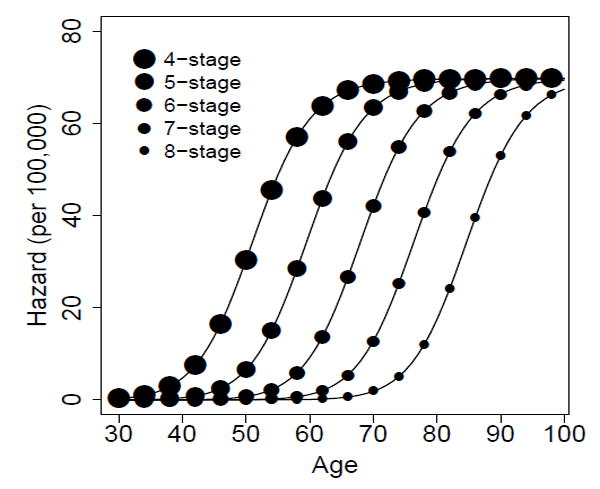
Hazards of multistage clonal expansion models with four to eight stages under two different parameter sets each (points vs. lines). See text for parameter details.
4 Discussion
Structural identifiability analysis is necessary for accurate estimation of model parameters from data, a fact that merits wider appreciation. Failure to verify the identifiable combinations in one’s model given one’s data may result in specious parameter estimates. Conversely, knowing the identifiable combinations can lead to insight and helpful model reparameterizations (e.g. [42]). This is true for the two-stage clonal expansion model. Using the r, p, q parameterization (Eqs. (12)), the survival and hazard can be expressed succinctly, and, observing that r = μ0/α, p ≈ −(α − β) and [43], one can identify multiplicative effects (e.g. temporal effects) on initiation, promotion (net cell proliferation), and malignant conversion respectively, as in Brouwer et al. [16].
The identifiability of MSCE models has been previously considered by Heidenreich et al. [29] (two stage model), Luebeck and Moolgavkar [5] (MSCE models with up to three pre-initiation steps), and Little et al. [30] (bounds on the maximum number of identifiable combinations in a generalized class of models that includes the MSCE model with any number of clonal expansion steps). Some of these previous results have relied on the form of the hazard function, which can only bound the identifiable combinations, or numerical evaluations of the rank of the Fisher information matrix, which, although strong evidence of local identifiability, is not formal proof. We offer an analytical proof of the exact identifiable combinations for MSCE models with any number of pre-initiation steps and one clonal expansion. This is a global result over the parameter space. Additionally, we provide a framework and conjecture for considering the exact identifiable combinations for the model with any number of clonal expansion stages, which we prove for n ≤ 5. For practical purposes, parsimonious carcinogenesis models are unlikely to need this many clonal expansion stages, let alone more. Moreover, this framework extends easily to variations of MSCE models that future work may consider, such as those incorporating disease precursors, e.g. gastroesophageal reflux disease (GERD) for esophageal cancer [15] or human papillomavirus (HPV) infection for anogenital or oral cancer [39].
Our methods and results are important in a larger context as well. We expand the differential algebra approach for structural identifiability, which has been primarily been used in the field of biological, deterministic ODE models (though is of course applicable to models in other fields), into the realm of stochastic branching processes and, more generally, continuous-time Markov processes. Once one is able to write a continuous-time Markov process as a system of differential equations of probability generating functions, a variety of identifiability techniques become available (e.g. Taylor series expansion [24] or similarity transform [23]). Of course, use of these techniques requires that one’s data relate to the probability generating functions in some way, so it is as of yet unclear exactly how widely applicable this framework will be. However, our approach to identifiability is applicable to at least one broad class of continuous-time Markov chain models, those that relate data to survival methods (i.e. time-to-event processes), which is true of many carcinogenesis and other health-outcome models.
This work sets the stage for several important problems. We have considered constant parameters, but time varying and piecewise-constant parameters are of great interest in the context of time-varying exposures [44-47]. The results given here address the piecewise constant case in part, since the problem can be expressed as multiple instances of the case with constant parameters, although additional analysis of initial conditions will be needed. Further, as data for each constant-parameter model will be limited (a full trajectory for each constant-parameter model is not observed), practical identifiability considerations arise. For more general time-varying parameters additional analysis is needed, though if the functional forms of the time varying parameters are known and if they are rational functions or approximable as such, then a similar approach as used here could be taken. Future work may also be able to see the conjecture given in this work proved beyond n = 5 using the differential algebra framework, but strong combinatorial tools may be necessary to disentangle the complexity of the coefficients of the input–output equation of the full model. Additionally, as mentioned above, future work that considers variations of the MSCE model will greatly benefit from this adaptable framework.
Finally, another important consideration is that of practical identifiability. In the context of real data, this structural identifiability analysis provides upper bounds on the number of identifiable parameter combinations, but there may be less parametric information available in real data. Such problems have been identified for MSCE models [11], but further analysis will be needed to address these issues more broadly.
Acknowledgments
This work was supported by NIH grant U01CA182915.
Appendix A
To illustrate these differential algebra approach to identifiability, we consider the classic example of a linear two-compartment model, commonly used in pharmacokinetics; the unidentifiability of this model is well-established through a range of methods [17, 26]. The model equations are given by
| (15) |
where x1(t) and x2(t) are the masses of a drug/substance in the plasma and tissue respectively, u(t) is a known input function (e.g. an intravenous injection or constant infusion at a known dose), the κij are unknown parameters to be estimated, and the output equation υ(t) is the plasma concentration, where ψ is the plasma volume (another unknown parameter to be estimated). Then our input–output equation should be a differential equation in terms of the parameters, input u, output υ, and their derivatives. This can be generated as follows—we substitute x1 = ψυ into the ẋ1 equation above, and solve for x2 to give
| (16) |
Plugging this in to the ẋ2 equation yields the following (taking a derivative of Eq. (16) to substitute for ẋ2),
| (17) |
Clearing denominators and combining terms yields
| (18) |
This differential polynomial is monic and thus an input-output equation for the system under a ranking of the variables that places u as higher ranked than υ. However, the ranking u < u̇ < ü < ⋯ < υ < υ̇ < ϋ < ⋯ is traditional [26], so we take
| (19) |
as our input-output equation. The coefficients of Eq. (19) are the set of identifiable combinations for the model. The importance of making the input-output equation monic (or otherwise clearing the coefficient of one of the terms) can be seen here—if we did not include such a restriction, we could multiply Eq. (19) by an arbitrary parameter combination, which would then be the coefficient of the ϋ term and appear to be identifiable. From these coefficients, we can see immediately that the model is unidentifiable—there are only four identifiable combinations, but there are five parameters. Moreover, we can see from the coefficient of u̇ that the parameter ψ is identifiable (since if 1/ψ is known, then ψ is known).
More broadly, testing for identifiability is usually accomplished by testing injectivity of the map from the parameters to the coefficients, i.e. evaluating each coefficient at two (symbolic) points, setting the two equal (e.g. ), and then testing whether it is possible to solve the resulting equations for each parameter in the form . In this case, it is apparent that the parameters are not identifiable. However we can find simpler representations of the identifiable combinations than the coefficients of Eq. (19): by noting that ψ is identifiable, we see that the coefficient for u shows that (κ12+κ02) is also identifiable (since both ψ and (κ12+κ02)/ψ are). Continuing in this fashion yields a simplified set of identifiable combinations: ψ, (κ12 + κ02), κ21 + κ01, and κ12κ21. Further examination shows that we can reparameterize the model in terms of the identifiable combinations by rescaling x̃2 = κ12x2, resolving the identifiability problem for the model (discussed further in [26]).
This example is simple enough to permit by-hand computation of the input–output equations and identifiable combinations. However, many models (even relatively simple nonlinear models) can result in extremely lengthy input output equations (e.g. terms numbering in the hundreds) or complicated combination structures which are not feasible to calculate by hand [27, 31]. Thus, it is common to use computational algebra techniques such as characteristic sets or Gröbner bases for many of the above steps [26, 27, 48], such as elimination of the unobserved variables x to generate an input–output equation or calculation of the identifiability results from the coefficients of the input–output equation. These approaches typically reduce a given set of polynomials/differential polynomials using some sort of ranking of the variables, typically ranking u < υ < x typically [26].
Appendix B
To prove Theorem 2, we begin with a series of lemmas.
Lemma 1
For 1 ≤ k < n − 1, xk is a rational function of x and its derivatives and may be written in the form , where qk and uk are polynomials of x and its derivatives and qk is monic.
Proof
We proceed by induction. Observe that
| (20) |
Next, assume that xk, for some 1 ≤ k < n − 2, may be written in the form , where qk and uk are polynomials of x and its derivatives and qk is monic. Then, from the ẋk equation, we find
| (21) |
where
| (22) |
| (23) |
Because qk is monic, is also monic. Further, qk and uk are clearly polynomials in x and its derivatives. Hence the result.
Lemma 2
The highest power of x in the polynomial qk is 2k−1, and the highest order derivative of x is k − 1. In particular, qk contains the term x2k−1, which is the only term with this power of x. The only terms in qk of with the power 2k−1 − 1 of x are, for 0 ≤ m ≤ k − 1,
The highest power of x in the polynomial uk is 2k−1 −1 and the highest order derivative is k. In particular, uk contains the term
which is the only term in uk with this power of x.
Proof
The relevant terms in qk and uk for the first few k are written out in Table II for convenience. We have q1 = x and , so the base case is—partly vacuously—true. Now, suppose that the hypotheses are true. Let qk+1 = (qk + uk)qk. Then, its term with the highest power of x is . Since qk contains the terms , 1 ≤ m ≤ k − 1, and x2k−1 , contains the terms, for 1 ≤ m ≤ k − 1,
Table II.
Relevant terms in qk and uk for k ≤ 4
| k | Relevant terms in qk | Relevant term in uk | ||
|---|---|---|---|---|
| 1 | x |
|
||
| 2 |
|
|
||
| 3 |
|
|
||
| 4 |
|
|
Since we have identified all of the terms with a power on x of 2k−1 and 2k−1 − 1 in qk, we have identified all of terms of power 2k−1 − 1 in . Additionally, there can be only one such term from qkuk: since qk contains x2k−1 and uk contains , qkuk contains . Hence qk+1 contains the terms, for 1 ≤ m ≤ k,
Further, since the highest order derivative of x in uk is x(k), the term in uk+1 of order k + 1 must come from . In particular, u̇k contains . Then, , uk+1 contains the term
Hence the result.
Now, we prove Theorem 2.
Proof
For ease of notation, let q :=qn−1 and u :=un−1. Now, we replace xn−1 with in the ẋn−1 equation to find an input–output equation.
| (24) |
| (25) |
| (26) |
| (27) |
| (28) |
Viewed as a function of x, this last equation is an input–output equation. Under an appropriate ranking, it is monic because of the x2n−1 term in q2. As in the proof of the previous lemma, q2 also contains the terms, for 1 ≤ m ≤ n − 2,
From the qu term, we get
Next, from , as in the proof of the lemma, we get
From , we get
A term of the same kind arrives from . Noting that the derivative of contains ,
We have identified n + 1 coefficients in the input–output equation. They are, for 1 ≤ m ≤ n − 2,
and
Thus, we can identify μ0, μ1, … , μn−3 (n > 3), μn−2μn−1, αn−1μn−1, αn−1 − βn−1 − μn−1.
However, there may be additional terms in the input–output equations. Thus, a priori, it is possible that smaller combinations making up these terms could be identifiable (or even that the model itself might be). So, we must show that the overall model is unidentifiable, and, moreover, that none of these combinations can be broken down into smaller identifiable pieces. To this end, we find a model equivalent to the original model (Eq. (14)) that can be parameterized using only the above identifiable combinations. To do so, solve ẋn−2 for , and plug this into the ẋn−1 equation to arrive at the following set of equations:
| (29) |
for 1 ≤ k ≤ n − 3 and with initial conditions x(0) = 1, xk(0) = 1, xn−2(0) = 1, and ẋn−2(0) = 0. Because the parameters μn−2, μn−1, αn−1, and βn−1 appear only in the combinations μn−2μn−1, αn−1μn−1, and αn−1 − βn−1 − μn−1 in the model equations, specifying values for these parameter combinations fully describes the model. Because a product is the smallest unit in a combination, it is clear that μn−2, μn−1, and αn−1 are not individually identifiable. Because βn−1 appears only in a sum with αn−1 and μn−1, it too is unidentifiable.
Hence, the result.
Next, we prove Proposition 1.
Proof
That the full model is unidentifiable, generally, can be seen as follows. The model below is equivalent to that in described by Eqs. (6).
| (30) |
for 1 ≤ k ≤ n − 3 with initial conditions x(0) = 1, xk(0) = 1 , and xn−3(0) = 1, xn−2(0) = 1, and ẋn−2(0) = 0. As in the previous proof, parameters μn−2, μn−1, αn−1, and βn−1 appear only in the combinations μn−2μn−1, αn–1 μn−1, and αn−1 − βn−1 − μn−1 in Eqs. (30). So, the full model is indeed unidentifiable.
Finally, we sketch the proof of Conjecture 1 for n ≤ 5. Calculations were carried out in Mathematica 10.2.
Proof
Solve the ẋ equation for x1. Take a derivative to find ẋ1. We now have x1 and ẋ1 as a function of x and its derivatives. Plug these into the ẋ1 equation so that it becomes an equation of x3, x, and derivatives of x. Solve for x2 as a function of x and its derivatives, and compute ẋ2. Continue in this manner until we have xn as a function of x and its derivatives. Substitute xn and ẋn into the final equation. We now have a single equation of x and its derivatives that contains all of the information of the system. Divide the equation by , which makes the equation monic under the appropriate ranking. This is an input–output equation. The equation has the following number of coefficients: 11 for n = 3, 48 for n = 4, 365 for n = 5. Determine the identifiable combinations from the list of coefficients by setting the coefficients equal to copies of themselves with placeholder parameter values and finding a Gröbner basis.
References
- 1.Moolgavkar SH, Venzon DJ. Two-event models for carcinogenesis: incidence curves for childhood and adult tumors. Mathematical Biosciences. 1979;47(1-2):55–77. [Google Scholar]
- 2.Moolgavkar SH, Knudson AG. Mutation and cancer: a model for human carcinogenesis. Journal of the National Cancer Institute. 1981;66(6):1037–52. doi: 10.1093/jnci/66.6.1037. [DOI] [PubMed] [Google Scholar]
- 3.Little MP. Are two mutations sufficient to cause cancer? Some generalizations of the two-mutation model of carcinogenesis of Moolgavkar, Venzon, and Knudson,and of the Multistage Model of Armitage and Doll. Biometrics. 1995;4:1278–1291. [PubMed] [Google Scholar]
- 4.Little MP, Haylock RGE, Muirhead CR. Modelling lung tumour risk in radon-exposed uranium miners using generalizations of the two-mutation model of Moolgavkar, Venzon and Knudson. International journal of radiation biology. 2002;78(1):49–68. doi: 10.1080/09553000110085797. [DOI] [PubMed] [Google Scholar]
- 5.Luebeck EG, Moolgavkar SH. Multistage carcinogenesis and the incidence of colorectal cancer. Proceedings of the National Academy of Sciences. 2002;99(23):15095–15100. doi: 10.1073/pnas.222118199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meza R, Luebeck EG, Moolgavkar SH. Gestational mutations and carcinogenesis. Mathematical biosciences. 2005;197(2):188–210. doi: 10.1016/j.mbs.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Hazelton WD, Moolgavkar SH, Curtis SB, Zielinski JM, Ashmore JP, Krewski D. Biologically based analysis of lung cancer incidence in a large Canadian occupational cohort with low-dose ionizing radiation exposure, and comparison with Japanese atomic bomb survivors. Journal of Toxicology and Environmental Health Part A. 2006;69(11):1013–38. doi: 10.1080/00397910500360202. [DOI] [PubMed] [Google Scholar]
- 8.Jeon J, Luebeck EG, Moolgavkar SH. Age effects and temporal trends in adenocarcinoma of the esophagus and gastric cardia (United States) Cancer Causes & Control. 2006;17(7):971–81. doi: 10.1007/s10552-006-0037-3. [DOI] [PubMed] [Google Scholar]
- 9.Jeon J, Meza R, Moolgavkar SH, Luebeck EG. Evaluation of screening strategies for pre-malignant lesions using a biomathematical approach. Mathematical Biosciences. 2008;213(1):56–70. doi: 10.1016/j.mbs.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luebeck EG, Moolgavkar SH, Liu AY, Boynton A, Ulrich CM. Does folic acid supplementation prevent or promote colorectal cancer? Results from model-based predictions. Cancer Epidemiology, Biomarkers & Prevention. 2008;17(6):1360–7. doi: 10.1158/1055-9965.EPI-07-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meza R, Jeon J, Moolgavkar SH, Luebeck EG. Age-specific incidence of cancer: Phases, transitions, and biological implications. Proceedings of the National Academy of Sciences. 2008;105(42):16284–9. doi: 10.1073/pnas.0801151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meza R, Jeon J, Moolgavkar S. Quantitative Cancer Risk Assessment of Nongenotoxic Carcinogens. In: Hsu CH, Stedeford T, editors. Cancer Risk Assessment. John Wiley & Sons, Inc.; 2010. pp. 636–658. [Google Scholar]
- 13.Meza R, Jeon J, Renehan AG, Luebeck EG. Colorectal cancer incidence trends in the United States and United kingdom: evidence of right- to left-sided biological gradients with implications for screening. Cancer research. 2010;70(13):5419–29. doi: 10.1158/0008-5472.CAN-09-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dewanji A, Jeon J, Meza R, Luebeck EG. Number and size distribution of colorectal adenomas under the multistage clonal expansion model of cancer. PLOS Computational Biology. 2011;7(10):e1002213. doi: 10.1371/journal.pcbi.1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazelton WD, Curtius K, Inadomi JM, Vaughan TL, Meza R, Rubenstein JH, et al. The role of gastroesophageal reflux and other factors during progression to esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev. 2015;24(7):1–6. doi: 10.1158/1055-9965.EPI-15-0323-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brouwer AF, Eisenberg MC, Meza R. Age Effects and Temporal Trends in HPV-Related and HPV-Unrelated Oral Cancer in the United States: A Multistage Carcinogenesis Modeling Analysis. PLOS One. 2016;11(3):e0151098. doi: 10.1371/journal.pone.0151098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bellman R, Åström KJ. On structural identifiability. Mathematical Biosciences. 1970;7:329–339. [Google Scholar]
- 18.Rothenberg TJ. Identification in Parametric Models. Econometrica. 1971;39(3):577–591. [Google Scholar]
- 19.Cobelli C, DiStefano JJ. Parameter and structural identifiability concepts and ambiguities: a critical review and analysis. American Journal of Physiology. 1980;239:R7–R24. doi: 10.1152/ajpregu.1980.239.1.R7. [DOI] [PubMed] [Google Scholar]
- 20.Raue A, Kreutz C, Maiwald T, Bachmann J, Schilling M, Klingmüller U, et al. Structural and practical identifiability analysis of partially observed dynamical models by exploiting the profile likelihood. Bioinformatics. 2009;25(15):1923–1929. doi: 10.1093/bioinformatics/btp358. [DOI] [PubMed] [Google Scholar]
- 21.Saccomani MP, Audoly S, Bellu G, D’Angio L. A new differential algebra algorithm to test identifiability of nonlinear systems with given initial conditions. Proceedings of the 40th IEEE Conference on Decision and Control; 2001. pp. 3108–3113. [Google Scholar]
- 22.Pohjanpalo H. System identifiability based on the power series expansion of the solution. Mathematical Biosciences. 1978;41(1-2):21–33. [Google Scholar]
- 23.Vajda S, Godfrey KR, Rabitz H. Similarity transformation approach to identifiability analysis of nonlinear compartmental models. Mathematical Biosciences. 1989;93:217–248. doi: 10.1016/0025-5564(89)90024-2. [DOI] [PubMed] [Google Scholar]
- 24.Chappell MJ, Godfrey KR, Vajda S. Global identifiability of the parameters of nonlinear systems with specified inputs: A comparison of methods. Mathematical Biosciences. 1990;102:41–73. doi: 10.1016/0025-5564(90)90055-4. [DOI] [PubMed] [Google Scholar]
- 25.Evans ND, Chappell MJ. Extensions to a procedure for generating locally identifiable reparameterisations of unidentifiable systems. Mathematical Biosciences. 2000;168:137–159. doi: 10.1016/s0025-5564(00)00047-x. [DOI] [PubMed] [Google Scholar]
- 26.Audoly S, Bellu G, D’Angiò L, Saccomani MP, Cobelli C. Global identifiability of nonlinear models of biological systems. IEEE Transactions on Biomedical Engineering. 2001;48(1):55–65. doi: 10.1109/10.900248. [DOI] [PubMed] [Google Scholar]
- 27.Meshkat N, Eisenberg M, Distefano JJ. An algorithm for finding globally identifiable parameter combinations of nonlinear ODE models using Gröbner Bases. Mathematical biosciences. 2009;222(2):61–72. doi: 10.1016/j.mbs.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Raue A, Karlsson J, Saccomani MP, Jirstrand M, Timmer J. Comparison of approaches for parameter identifiability analysis of biological systems. Bioinformatics. 2014;30:1440–1448. doi: 10.1093/bioinformatics/btu006. [DOI] [PubMed] [Google Scholar]
- 29.Heidenreich WF, Luebeck EG, Moolgavkar SH. Some properties of the hazard function of the two-mutation clonal expansion model. Risk Analysis. 1997;17(3):391–9. doi: 10.1111/j.1539-6924.1997.tb00878.x. [DOI] [PubMed] [Google Scholar]
- 30.Little MP, Heidenreich WF, Li G. Parameter identifiability and redundancy in a general class of stochastic carcinogenesis models. PLOS One. 2009;4(12):1–6. doi: 10.1371/journal.pone.0008520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisenberg MC, Robertson SL, Tien JH. Identifiability and estimation of multiple transmission pathways in cholera and waterborne disease. Journal of theoretical biology. 2013;324:84–102. doi: 10.1016/j.jtbi.2012.12.021. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg M. Generalizing the differential algebra approach to input–output equations in structural identifiability. arXiv. 2013:1–11. arXiv:1302.5484v1. [Google Scholar]
- 33.Dewanji A, Venzon DJ, Moolgavkar SH. A stochastic two-stage model for cancer risk assessment. II. The number and size of premalignant clones. Risk analysis : an official publication of the Society for Risk Analysis. 1989;9(2):179–187. doi: 10.1111/j.1539-6924.1989.tb01238.x. [DOI] [PubMed] [Google Scholar]
- 34.Moolgavkar S, Luebeck G. Two-event model for carcinogenesis: Biological, mathematical, and statistical considerations. Risk Analysis. 1990;10(2):323–341. doi: 10.1111/j.1539-6924.1990.tb01053.x. [DOI] [PubMed] [Google Scholar]
- 35.Tan WY. Stochastic Models of Carcinogenesis. New York: Marcel Dekker; 1991. [Google Scholar]
- 36.Heidenreich WF. On the parameters of the clonal expansion model. Radiation and Environmental Biophysics. 1996;35(2):127–129. doi: 10.1007/BF02434036. [DOI] [PubMed] [Google Scholar]
- 37.Crump KS, Subramaniam RP, Van Landingham CB. A numerical solution to the nonhomogeneous two-stage MVK model of cancer. Risk Analysis. 2005;25(4):921–6. doi: 10.1111/j.1539-6924.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 38.Meza R. Some Extensions and Applications of Multistage Carcinogenesis Models. University of Washington. 2006 [Google Scholar]
- 39.Brouwer AF. Models of HPV as an Infectious Disease and as an Etiological Agent of Cancer. University of Michigan. 2015 [Google Scholar]
- 40.Meshkat N, Anderson C, DiStefano JJ. Alternative to Ritt’s pseudodivision for finding the input-output equations of multi-output models. Mathematical Biosciences. 2012;239(1):117–123. doi: 10.1016/j.mbs.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Ljung L, Glad T. On global identifiability for arbitrary model parametrizations. Automatica. 1994;30(2):265–276. [Google Scholar]
- 42.Luebeck E, Curtius K, Jeon J, Hazelton W. Impact of tumor progression on cancer incidence curves. Cancer research. 2013;73(3):1086–1096. doi: 10.1158/0008-5472.CAN-12-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moolgavkar SH, Meza R, Turim J. Pleural and peritoneal mesotheliomas in SEER: age effects and temporal trends, 1973-2005. Cancer Causes & Control. 2009;20(6):935–44. doi: 10.1007/s10552-009-9328-9. [DOI] [PubMed] [Google Scholar]
- 44.Luebeck EG, Heidenreich WF, Hazelton WD, Paretzke HG, Moolgavkar SH. Biologically based analysis of the data for the Colorado uranium miners cohort: age, dose and dose-rate effects. Radiation research. 1999;152(4):339–51. [PubMed] [Google Scholar]
- 45.Hazelton WD, Luebeck EG, Heidenreich WF, Moolgavkar SH. Analysis of a historical cohort of Chinese tin miners with arsenic, radon, cigarette smoke, and pipe smoke exposures using the biologically based two-stage clonal expansion model. Radiation research. 2001;156(1):78–94. doi: 10.1667/0033-7587(2001)156[0078:aoahco]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 46.Meza R, Hazelton WD, Colditz GA, Moolgavkar SH. Analysis of lung cancer incidence in the Nurses’ Health and the Health Professionals’ Follow-Up Studies using a multistage carcinogenesis model. Cancer causes & control : CCC. 2008;19(3):317–28. doi: 10.1007/s10552-007-9094-5. [DOI] [PubMed] [Google Scholar]
- 47.Richardson DB. Multistage modeling of leukemia in benzene workers: a simple approach to fitting the 2-stage clonal expansion model. American Journal of Epidemiology. 2009 Jan;169(1):78–85. doi: 10.1093/aje/kwn284. [DOI] [PubMed] [Google Scholar]
- 48.Bellu G, Saccomani MP, Audoly S, D’Angiò L. DAISY: A new software tool to test global identifiability of biological and physiological systems. Computer Methods and Programs in Biomedicine. 2007;88(1):52–61. doi: 10.1016/j.cmpb.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]


