Abstract
The whitefly Bemisia tabaci is a phloem-feeding pest that lives predominantly on herbaceous species and causes serious damage to hosts. Whitefly saliva is thought to contain proteins that modulate plant defences and facilitate feeding. A predicted secreted protein, laccase 1 (LAC1), was found in the salivary gland transcriptome of B. tabaci and might be existed in the watery saliva of B. tabaci. As LAC1 has a potential role in detoxification of secondary plant compounds in insects, we speculated that it may participate in the insect’s response to plant defences. Here, we cloned the complete cDNA of LAC1 and found that (1) LAC1 was highly expressed in the salivary gland (SG) and midgut; (2) LAC1 transcript level in head (containing SG) was 2.1 times higher in plant-fed than in diet-fed whiteflies and 1.6 times higher in the head and 23.8 times higher in the midgut of whiteflies that fed on jasmonic acid (JA)-sprayed plants than on control plants; and (3) silencing LAC1 decreased the survival rate of plant-fed whiteflies but had a marginal effect on whiteflies raised on an artificial diet. These results indicate that LAC1 enables whiteflies to overcome the chemical defences of host plants and might act as an effector in saliva.
Introduction
Throughout the history of co-evolution, both host plants and herbivorous insects have developed a wide variety of complex defence and anti-defence strategies1, 2. Plants have evolved strategies to defend themselves against insect herbivory that are based on physical barriers, constitutive chemical defences, and directly and indirectly inducible defences3. Conversely, insects have developed sophisticated mechanisms to overcome these plant defences. Their response takes the form of changes in gene expression and the protein repertoire in cells lining the alimentary tract, the first line of defence. To survive, insect herbivores adjust their feeding habits, digestive physiology, and gene expression to optimally adapt to their hosts4.
The whitefly Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae) is an efficient plant-feeding insect with a global distribution that causes serious damage to crops by its direct feeding or by the transmission of plant viruses5–7. B. tabaci is a complex species with at least 36 morphologically indistinguishable cryptic species5, 8. Among them, B. tabaci MED (commonly known as biotype Q) has now spread to most parts of China and has caused significant damage to host plants8–11.
Importantly, the whitefly can successfully and persistently feed on phloem sap from sieve elements12, and its saliva is thought to contain effector proteins that overcome plant defences13–15. Saliva injected into plant tissues has long been considered to play crucial roles in aiding penetration and nutrient ingestion and modulating plant responses, and some secreted proteins have been identified as effectors that inhibit plant defences, prevent phloem sieve-element occlusion, and otherwise promote the unique phloem feeding style16. Scientists have found several effectors in aphids, such as C002, Mp10, Mp55, Me10, Me23, and Armet, which play an important role in overcoming plant defences and helping the herbivore establish a population on host plants3, 16–20. However, there are few reports on effectors in whiteflies.
We found a predicted secreted protein, laccase 1 (benzenediol:oxygen oxidoreductase; EC 1.10.3.2, LAC1), in the salivary gland transcriptome of B. tabaci (data unpublished). Though several laccase isoforms have been identified in insects21–27, less is known about insect LAC1. LAC1 belongs to the multicopper (MCO) family28, it has been proposed that insect LAC1 may play an important role in metal ion metabolism, lignocellulose digestion and detoxification of secondary plant compounds23, 26. As LAC1 has many potential catalytic and oxidative functions, we speculated that it may participate in the insect’s response to plant defences.
The goal of this study was to elucidate the LAC1 function in B. tabaci and confirm its putative functions as an effector protein that modulates whitefly–plant interactions. In this study, we first cloned the complete cDNA sequence and determined the temporal and spatial expression pattern of LAC1. Next, to clarify whether LAC1 is related to the insect’s response to plant defences, we compared the expression differences between plant-fed and diet-fed B. tabaci and between jasmonic acid (JA)-sprayed plant-fed and control plant-fed B. tabaci. Finally, using RNAi, we examined whether the presence of LAC1 influences B. tabaci survival rates on tomato plants. This study helps us to understand the mechanisms by which whiteflies overcome plant defences.
Results
Characteristics of the LAC1 gene and protein
The complete cDNA of LAC1 contained an open reading frame (ORF) of 2733 bases encoding a protein of 911 amino acid residues with a predicted molecular weight of 103.03 kDa (Figure S1), the GenBank accession number of LAC1 is KY643659. SignalP predicted an N-terminal signal secretion peptide with a cleavage site between Pro19 and Thr20. No transmembrane domain was found, suggesting that LAC1 is a secreted protein. SOSUI predicted the average hydrophobicity to be −0.483388, which suggests that LAC1 is a soluble protein. The encoded protein has three typical Cu-oxidase domains in the polyphenol oxidase family, indicating that LAC1 belongs to the blue copper-containing polyphenol oxidase family.
The full-length LAC1 also showed a high level of amino acid sequence identity with LAC sequences from other insects found in GenBank. An alignment of the protein sequences of 18 laccases is shown in Figure S2. The origin of the LAC genes and their GenBank accession numbers are listed in Table S2. Using BLAST analysis, transcripts of Cu-oxidase domains homologous to B. tabaci LAC1 were identified in many insects. The Cu-oxidase domains seemed to be relatively conserved among all the studied insect laccases.
Sequence retrieval and phylogenetic analysis
Phylogenetic analyses were conducted to examine the relationships among B. tabaci MED LAC1 and the LAC genes of other insects, bacteria, fungus and plants (Fig. 1). The LAC genes of B. tabaci, Diaphorina citri, Acyrthosiphon pisum, Nilaparvata lugens and Nephotettix cincticeps, which are all Homoptera insects, are closely related. However, BT MEAM1 LAC2 and BT MEAM1 LAC4 are quite genetically distant from LAC1, indicating that LAC1 is functionally distinct from other LACs. The phylogenetic analysis also revealed that laccases from these groups (viz., insects, bacteria, fungi and plants) form independent clades consistent with their taxonomical classification.
Figure 1.
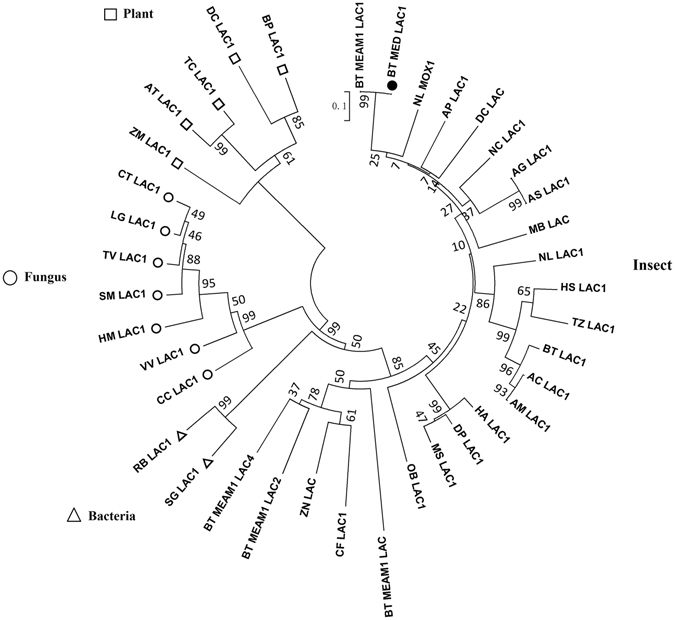
Phylogenetic trees comparing B. tabaci MED LAC1 and the known insect LAC genes. The phylogenetic tree was generated by MEGA 5.05 based on the neighbour-joining method according to amino acid sequences. Bootstrap support values calculated on 1000 replications are shown on the branches. The laccase sequences used to generate the tree are listed in Table S3.
Temporal and spatial expression of LAC1
RNA from the salivary gland, midgut and ovary was analysed using real-time quantitative PCR (qPCR) to determine the transcript levels of LAC1. LAC1 is most highly expressed in the salivary gland and midgut (Fig. 2A) and is expressed at a higher level in female adults than male adults (Fig. 2B). LAC1 mRNA is expressed consistently in all stages (Fig. 2C).
Figure 2.
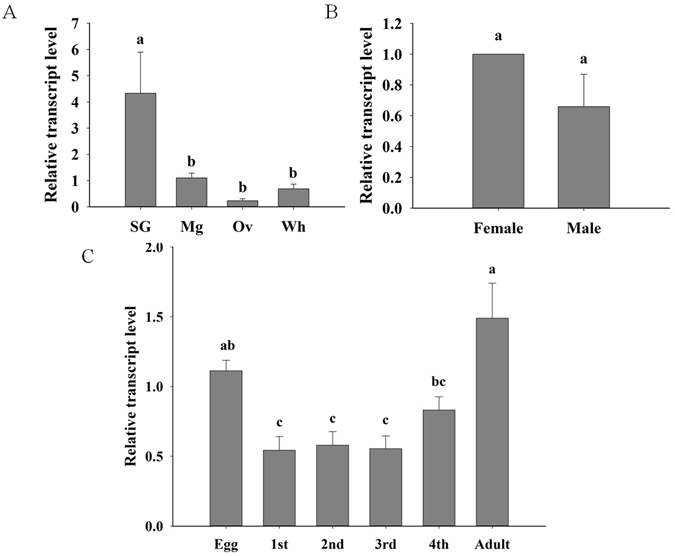
Temporal and spatial expression of B. tabaci LAC1 measured by real-time qPCR. (A) Expression in salivary gland (SG), midgut(Mg), ovary(Ov) and whole body (Wh). (B) Expression in female and male. (C) Expression in egg, larval and adult stages. Values are represented as the mean ± SEM. The letters above the columns indicate the comparison among groups evaluated, p < 0.05.
LAC was found in the watery saliva of B. tabaci
LAC activity was examined in B. tabaci fed a sucrose diet through a membrane (Table 1). ABTS is thought to be the best substrate to detect laccase21. ABTS only reacts with LAC at pH 5; therefore, the results measured in whitefly saliva mixed with ABTS represented the enzyme activity of LAC. This experiment demonstrated that LAC exists in B. tabaci watery saliva.
Table 1.
Laccase activities in B. tabaci flies fed a sucrose diet.
| Substrate (2 mM) | Absorbance at 420 nm | ||
|---|---|---|---|
| After feeding | Unfed control | ||
| ABTS, pH 5 | 0.0447 ± 0.0004 | 0.0427 ± 0.0002 | p = 0.0012 |
Two hundred B. tabaci adults were exposed to 200 μL of a 10% sucrose solution for 24 h, and subsequently, 50 μL of the recycled feeding liquid was analysed in a microplate reader at 28 °C for 5 min. Mean ± SEM, n = 3.
LAC1 expression was higher in plant-fed than diet-fed whiteflies and higher in JA-sprayed plant-fed than control plant-fed whiteflies
To explore whether LAC1 is necessary in plant feeding, the transcript levels of LAC1 in B. tabaci heads, which contain the salivary glands, were compared between plant-fed and artificial diet-fed B. tabaci using qPCR. The normalized level of the LAC1 transcript was approximately 2.2-fold higher in plant-fed B. tabaci than in artificial diet-fed B. tabaci (Fig. 3). This result suggests that more LAC1 was required by B. tabaci during interactions with host plants.
Figure 3.
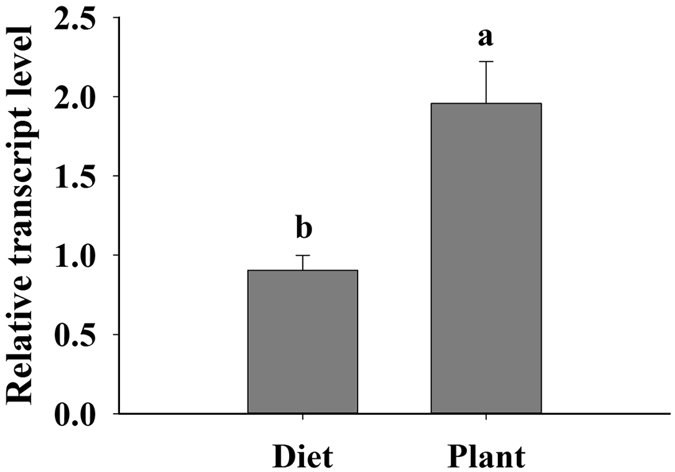
Transcript levels of LAC1 in the heads (containing the salivary glands) of diet- and plant-fed B. tabaci. Values are represented as the mean ± SEM. The letters above the columns indicate the comparison among groups, p < 0.05.
To explore whether LAC1 is related to the insect’s response to plant defences, we compared the difference in LAC1 expression between JA-sprayed plant-fed and control plant-fed whiteflies. Tomato plant defences can be induced by spraying them with exogenous JA29, 30. When B. tabaci flies fed on tomato plants that were sprayed with JA for 2 days, the LAC1 expression in the head increased approximately 1.64-fold (Fig. 4A), and the LAC1 expression in the midgut increased approximately 23.81-fold (Fig. 4B). These results show that LAC1 is required to counteract the defence reaction of plants.
Figure 4.
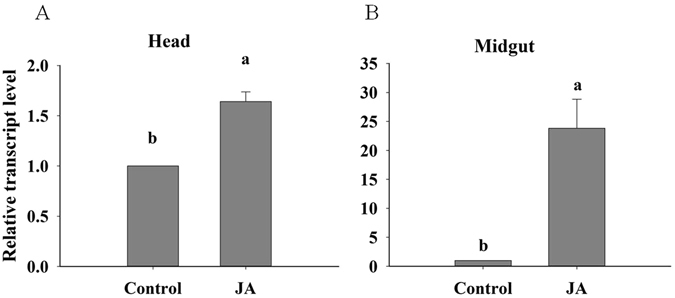
Transcript levels of LAC1 in JA-sprayed plant-fed or control plant-fed B. tabaci. (A) Expression in heads (containing the salivary glands). (B) Expression in the midgut. Values are represented as the mean ± SEM. The letters above the columns indicate the comparison among groups, p < 0.05.
LAC1 is indispensable for B. tabaci survival on tomato plants
To test whether the presence of LAC1 influences B. tabaci survival rates on tomato plants and whether this influence is related to the effect of LAC1 on the ability of B. tabaci to reach the phloem, we compared the performance of B. tabaci adults on different food matrices. LAC1 silencing generally reduced the survival rate (Fig. 5), and the effect was most pronounced on tomato plants. When B. tabaci was fed LAC1 dsRNA, the LAC1 transcript level in the B. tabaci whole bodies was reduced by approximately 55% and 74% at 2 and 4 days after feeding, respectively (Fig. 6). B. tabaci flies were fed LAC1 dsRNA for 2 days and then moved onto Lycopersicon esculentum or fed an artificial diet. Compared with a control GFP dsRNA, the LAC1 dsRNA significantly reduced the survival rates of B. tabaci that fed on host plants (P < 0.001) (Fig. 5A) but had a smaller influence on B. tabaci given an artificial diet (P = 0.1179) (Fig. 5B). The survival rate of B. tabaci adults with silenced LAC1 was much higher in insects raised on an artificial diet than in those raised on plants. These results demonstrate that LAC1 contributes to the survival rate of B. tabaci adults raised on tomato plants.
Figure 5.
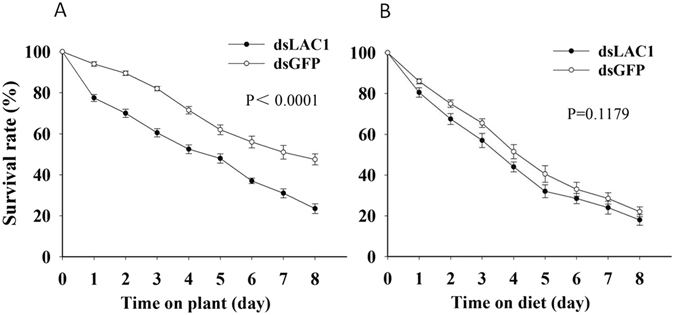
Knocking down LAC1 decreases the survival rates of B. tabaci adults. (A and B) Mean survival rates of B. tabaci adults that were fed dsRNA against LAC1 (dsLAC1) or GFP (dsGFP) and then fed on tomato plants (A) or an artificial diet (B). P values of the difference between the curves of the interference and control groups are indicated, p < 0.05.
Figure 6.

Transcript levels of B. tabaci LAC1 after dsRNA feeding. Values are represented as the mean ± SEM. The letters above the columns indicate the comparison among groups evaluated by an ANOVA using SAS 8.1, p < 0.05.
Discussion
Though there have been some reports on insect laccases, the physiological functions of insect LAC1 have not been established. Our experiments demonstrate that LAC1 is required for B. tabaci survival on tomato plants and is important in enabling whiteflies to overcome the chemical defences of host plants.
Identification of LAC1
Currently, information on the possible LAC1 substrates in the whitefly is lacking, and LAC1 in B. tabaci may perform more than one function. The physiological substrates of LACs that are hydrolysed in vivo have not been fully elucidated in insects. To perform its catalytic function, laccase depends on Cu atoms that are distributed at three different copper centres. These copper centres in laccases are categorized into three groups: Type-1 (T1), Type-2 (T2) and Type-3 (T3)31. The laccase genes typically have three to four of these copper domains32, the structure of which can vary greatly between different organisms. This extensive structural variability results in a corresponding variation in catalytic properties and oxidative function33–36, such as their involvement in lignin biosynthesis (in plants) as well as lignin degradation (in fungi and bacteria)31. Laccases catalyse the oxidation of a wide array of substrates during four-electron transfer processes while reducing O2 to water37. We found that LAC1 also has three different copper domains: T1, T2 and T3. This structural characteristic demonstrates that LAC1 belongs to the laccase family and has catalytic properties as well as oxidative function. In contrast to most enzymes, which catalyse substrate-specific reactions, laccases oxidize a wide range of compounds that vary from di-, poly-, and substituted phenols to diamines, aromatic amines, and benzenethiols31, 38. As laccases have so many kinds of substrates, LAC1 in B. tabaci may perform more than one function in vivo.
Phylogenetic analyses comparing the B. tabaci MED LAC1 protein sequence to 37 other LACs from insects, fungi, bacteria, and plants showed that the B. tabaci laccases are evolutionarily unique and distinct from other insect LAC2 proteins with known or suspected roles in integument formation23, 26, 39. According to the phylogenetic analysis, B. tabaci LAC1 is most similar to N. lugens MOX1 (AKN21378.1), D. citri LAC1 (XP_008487811.1), N. cincticeps LAC1 (BAJ06132.1) and A. pisum LAC1 (XP_001948070.1) (Fig. 1). This result illustrated that laccases of plant-sucking insects may have similar physiological functions and be evolutionarily related.
ABTS is specifically oxidized by laccase40. We observed a significant difference in laccase activity between saliva and the control solution, indicating that the ABTS-oxidizing enzyme is laccase. So far, LAC1 is the only laccase that has been identified in insect saliva; for example, LAC1 has been shown to be expressed in the salivary glands and secreted into the saliva of green rice leafhoppers and termites21–23. Therefore, we concluded that the laccase we found in B. tabaci saliva might be LAC1.
Functional analysis
The temporal and spatial expression of LAC1 was determined and indicated that LAC1 mRNA is expressed consistently in all stages and is most highly expressed in the salivary gland and midgut. This result suggested that as a secretory protein, LAC1 may be produced by the salivary gland and midgut epithelial cells and secreted into the salivary channel and lumen, respectively. Our results indicate that LAC1 is expressed throughout development and growth and plays a crucial role in biological processes. Interestingly, LAC1 was also found to be expressed in the ovaries of B. tabaci, and a higher expression level was observed in female adults than in male adults. This suggests that LAC1 may have other biological functions. For instance, LAC1 produced in the ovaries may be secreted into the plants by the ovipositor of the female adult to counteract plant responses and assist in egg deposition. Further research is necessary to elucidate this role of LAC1.
Hattori et al. (2005)21 detected LAC1 in the watery saliva and salivary glands of N. cincticeps and proposed that one of the functions of LAC1 is the promotion of rapid oxidative gelling of the stylet sheath by the quinine tanning reaction, as laccase is hypothesized to be involved in insect cuticle sclerotization24, 25, 41. As a phloem-feeding insect like N. cincticeps, B. tabaci may also use LAC1 to promote the oxidative gelling of the stylet sheath. Huang et al. (2016)42 found the survival rate of dssalivap-3 treated insect on plant was decreased, and salivap-3 is involved in the formation of the salivary sheath. Moreover, the different survival rate of dsNlShp treated insect on plant and artificial diet was also observed in salivary sheath-deficient brown planthoppers, similar with the result of dsLAC1 treatment. The NlShp acts as the main component of both salivary sheath and salivary flange43. So we speculate that LAC1 is synthesized in primary salivary glands and transported out through the salivary ducts, promoting rapid oxidative gelling of the stylet sheath and salivary flange to harden.
LAC1, as a secretory protein, is released into the salivary channel and then secreted via the mouth parts into plant tissues. Laccase catalyses the oxidation of phenolic lignin units as well as a wide variety of phenolic compounds and aromatic amines38. Penetrating plant tissues intracellularly, the stylets of whiteflies rupture the walls of the epidermal and mesophyll cells to access the phloem. The LAC1 ejected along with the watery saliva may play a role in the oxidation of phenolic compounds in plant tissues. This function helps B. tabaci successfully feed by inhibiting the defence reaction of plants.
When comparing the LAC1 expression level in whiteflies fed either plants or an artificial diet, we found that whiteflies required additional LAC1 when they fed on plants than on an artificial diet. Furthermore, the LAC1 transcript level was several times higher in the salivary gland and midgut when the whiteflies fed on L. esculentum sprayed with JA than when they fed on control plants. Exogenous JA can activate plant defence genes either via the octadecanoid pathway or by acting directly on the genes29, 30, 44. Activation of defence genes leads to metabolic reconfiguration. Secondary metabolites such as phenols, terpenoids and alkaloids involved in plant defence response are released into phloem sap and transported throughout the plant45, 46, and the phloem sap is then consumed by B. tabaci. Coy et al. (2010)23 found that laccases are secreted from the salivary gland into the Reticulitermes flavipes termite midgut and play a role in lignocellulose-related phenol oxidation in the termite gut. We found that LAC1 was expressed both in the salivary gland and midgut, so we hypothesize that, similar to R. flavipes, B. tabaci LAC1 is secreted from the salivary gland into the midgut and plays a role in detoxifying phenolic compounds and other toxic compounds. Furthermore, Liu et al. (2016)47 found that engineered pseudomonas putida cells expressing surface-immobilized laccases could completely eliminate chlorpyrifos via direct biodegradation. LAC1 may also help B. tabaci detoxify pesticides consumed with the plant phloem sap.
Feeding whiteflies with either LAC1 dsRNA or a GFP control generally resulted in a reduced survival rate on plants or an artificial diet; this phenomenon may have been caused by man-made mechanical damage when transferring the insects. However, because the treated and control insects were handled in the same manner, this damage did not affect the results of the experiments. LAC1 silencing generally reduced the survival rate, and the effect was most pronounced on tomato plants. This result demonstrates that LAC1 contributes to the survival of B. tabaci adults raised on tomato plants.
In conclusion, we speculate that LAC1 in the whitefly saliva, salivary gland and midgut may enable the insect to overcome the chemical defences of host plants. LAC1 might be an effector involved in modulating B. tabaci feeding and survival on plants. This study suggests a potentially important role for LAC1 in determining the patterns of whitefly–plant relationships through detoxifying defensive phytochemicals.
Materials and Methods
Insects and host plants
B. tabaci MED whiteflies were maintained on tomato plants (L. esculentum, cv. Zhongza 9) in separate greenhouses at 25 ± 1 °C with a photoperiod of 14:10 h light:dark and 60–70% relative humidity. Tomato plants were individually grown in plastic pots 9 cm in diameter in greenhouses under the same conditions as previously mentioned. Tomato plants at the five-to-six true leaf stage were used in these experiments.
Detection of LAC 1 activity in B. tabaci watery saliva
Active adult B. tabaci raised on the tomato plants were chosen for the experiment. The method of Peng et al. (2013)48 was used with some small modifications. Briefly, the whiteflies were collected and starved for 4 h to eliminate the possible effect of the ingested plant laccase. Then, we randomly placed 200 B. tabaci adults into a glass tube with two transparent ends (3 cm in diameter and 8 cm in height), and the top of each glass tube was covered with two pieces of parafilm for artificial feeding. After that, we injected 200 μL of artificial diet solution into the space between the two pieces of parafilm. The solution was a dissolved mixture of 10% sucrose plus 10 mM MES (2-morpholinoethanesulfonic acid) in distilled water. A whitefly-free tube was used as the control.
After 24 h, a disposable syringe was used to recover the diet solution, which now contained the saliva of the whiteflies (hereafter referred to as recycled liquid). Care was taken to avoid breaking the basement membrane in the aspiration process. The control treatment was also recovered in a similar manner. We dispensed 50 μL of the recycled liquid into a 96-well microplate (Eppendorf, China) and then added 150 μL of ABTS (3-benzothiazole ethyl benzene sulfonic acid) substrate solution (see details for preparation below) and mixed it with the recycled liquid by pipetting up and down. The kinetics of the 200 μL ABTS substrate-enzyme was determined by measuring the absorbance of light at 240 nm, and the measurements were taken 0 min and 5 min after the solutions were mixed together. Each treatment had three analytical replicates on the microplate. The reaction was carried out at 28 °C, and the enzyme activity was calculated as the change in light absorbance per litre of solution per unit of time (OD240nm·min−1·L−1).
The ABTS substrate solution was prepared as follows: (1) A 100 mM NaAc (sodium acetate) solution was made by dissolving 0.8203 g of NaAc in 100 mL of distilled water. (2) A 2.0 mM ABTS solution (pH 5.0) was made by dissolving 0.1097 g of ABTS in 100 mL of the 100 mM NaAc solution. The pH was adjusted to 5, and the solution was stored in a brown bottle49.
Cloning of the B. tabaci LAC1 transcript
Total RNA was extracted from B. tabaci using Trizol (Invitrogen, Carlsbad, CA, USA) and then reverse transcribed into cDNA with a PrimeScript® 1st Strand cDNA Synthesis Kit (TaKaRa, Japan). The template for rapid amplification of cDNA ends (RACE) was synthesized using the Clontech SMARTerTM RACE cDNA Amplification Kit (TaKaRa, Japan).
Degenerate PCR and nested PCR were performed in a TP600 Thermal Cycler (TaKaRa, Japan). The reactions were catalysed using Ex TaqTM and Tks GflexTM (TaKaRa, Japan). The PCR program consisted of an initial denaturation for 2 min at 98 °C, followed by 35 cycles of 94 °C for 30 s, 55–60 °C (based on the primer annealing temperatures) for 30 s, 72 °C for 1–2 min and a final extension at 72 °C for 10 min. The sequences of the primers designed to obtain the full-length LAC1 are shown in Table 1.
The amplified PCR fragments were gel purified with the QIAquick Gel Extraction Kit (Qiagen, Germany) and then ligated into the pClone007 vector (Beijing Tsingke Biotech Co., Ltd., China) and transferred into DH5α cells for sequencing.
Sequence retrieval and phylogenetic analysis
Protein sequences were deduced from the sequenced ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) and analysed using the SignalP (www.cbs.dtu.dk/services/SignalP) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui) servers, which identify the signal peptide and predict membrane proteins, respectively. The molecular weight and isoelectric points of the deduced protein sequences were calculated by the ExPASy Proteomics Server (http://cn.expasy.org/tools/pi_tool.html). Transmembrane helices were analysed using TMHMM v.2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0).
LAC sequences of B. tabaci MEAM1 were found in the whitefly genome database (http://www.whiteflygenomics.org/cgi-bin/bta/index.cgi). Homologous proteins from other species were identified using the NCBI BlastP software NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and were aligned using DNAMAN6.0. (see Table S2 for accession numbers and related information).
The B. tabaci LAC1 sequence was used as a query in BLASTp searches to identify similar laccases in the GenBank database. A phylogenetic tree was constructed with 37 laccase sequences (see Table S3 for accession numbers and related information) using the neighbour-joining method with a matrix of pair-wise distances estimated by a poisson model for amino acid sequences through the MEGA 5.05 software. Bootstrap values were calculated on 1000 replications.
qPCR analysis of LAC1 expression patterns in different tissues and developmental stages
Three tissues (salivary glands, midgut and ovaries) were collected from approximately 200 B. tabaci adults for RNA extraction. Three replicates for each tissue were prepared. RNA was also isolated from the first to fourth instar nymphs and from adult B. tabaci flies. Three replicates and 20 individuals per replicate were prepared for each developmental stage. RNA extracted from the whole insect body was used as a reference. qPCR was performed to quantify the transcript levels of LAC1 in the three tissues and in various developmental stages. The amplification conditions consisted of an initial denaturation at 95 °C for 30 s, followed by 40 cycles of denaturation at 95 °C for 5 s and 60 °C for 30 s. The reactions were performed on a qTOWER 2.2 Real-Time Thermal Cycler (Analytikjena, Germany). The Qlac-qF, Qlac-qR and β-actin qPCR primer sequences can be found in Table S1. Differences in transcript levels were analysed using a one-way ANOVA for multiple comparisons with SAS 8.1 software. The results were presented as the mean ± SEM. The relative expression levels were calculated by the 2−ΔΔCt method.
Comparison of LAC1 transcripts in diet- and plant-fed B. tabaci
Sixty 0-day-old (within 1 h after eclosion) B. tabaci adults were placed on L. lycopersicum Mill plants for 96 h of feeding. Another group was fed an artificial diet (20% sucrose in distilled water) between two layers of parafilm stretched over the top of a glass tube (3 cm in diameter and 8 cm in height). The transcript levels of LAC1 in the heads (containing the salivary glands) of diet- and plant-fed B. tabaci were compared using qPCR. Twenty B. tabaci heads were included in each replicate, and three replicates were prepared. The values were reported as the mean ± SEM, and the differences were evaluated with a t-test using SAS 8.1.
Comparison of LAC1 transcripts in exogenous JA-sprayed plant-fed and control plant-fed B. tabaci
The JA was first dissolved in 1 mL of ethanol, and then distilled water was added to obtain a JA concentration of 1 mM. The JA solution was sprayed on tomato leaves. Next, 0-day-old B. tabaci adults were collected and transferred onto the plants 48 h after the JA spraying. Tomato plants sprayed with distilled water with 1 mL of ethanol were used as the control. The head and midgut were collected from approximately 200 B. tabaci adults for RNA extraction. Three replicates for each tissue were prepared. Differences were evaluated by a t-test using SAS 8.1.
Gene interference by feeding double-stranded RNA (dsRNA) and survival curve analysis
PCR primers with T7 promoter sequences were used to prepare the dsRNA of the LAC1 gene. Two primers for the specific amplification of dsLAC1, LAC1-dsRNA-F and LAC1-dsRNA-R, were designed (Table S1). A dsRNA for green fluorescent protein (GFP) was amplified using the primers GFP-dsRNA-F and GFP-dsRNA-R and used as a negative control (Table S1). dsRNA was generated and purified using a TranscriptAid T7 High Yield Transcription Kit (Thermo Scientific, USA) following the manufacturer’s protocols. The dsRNA was resuspended in RNase-free water and analysed by spectrophotometric quantification and agarose gel electrophoresis. The dsRNA was stored at −80 °C until further use.
Zero-day-old B. tabaci adults were fed a diet containing dsRNA diluted to 100 ng·μL−1 in an RNase-free 20% sucrose solution for 48 h. Two hundred 0-day-old B. tabaci adult females were collected and placed in a glass tube (3 cm in diameter, 5 cm high); this was repeated with 200 males. The tube opening was covered with two layers of parafilm, and 500 μL of the dsRNA solution was injected into the gap between the two layers. The other end of the tube was covered with gauze for ventilation. Ten groups of B. tabaci flies, with 20 individuals (10 females and 10 males) in each group, were fed and subsequently reared on L. esculentum plants or on an artificial diet for 8 days.
Three groups of B. tabaci flies, with 20 individuals in each group, were collected on the second and fourth days after dsRNA feeding to test the inhibition efficiency of LAC1 transcription in B. tabaci whole bodies.
The survival rate of whiteflies that fed on plants or an artificial diet was recorded every 24 h after dsRNA feeding and were reported as the mean ± SEM. The survival curves of the RNAi and control groups on a plant or artificial diet were statistically compared using the log-rank (Mantel-Cox) test.
Electronic supplementary material
Acknowledgements
We thank all the sample providers. This research was supported by International Science and Technology Cooperation Program of China (2015DFG32300), The National Key Research and Development Project of China (2016YFC1201200), Common Wealth Special Fund for The Agricultural Industry (201303019) and the Taishan Mountain Scholar Constructive Engineering Foundation of Shandong, China.
Author Contributions
F.H.W., D.F.C. and J.Y.G. contributed to experiments design and management. C.H.Y. performed the experiments, carried out the data analysis and drafted the manuscript. T.B.D. and K.K.W. helped with the experiments. D.C. and J.Y.G. edited and revised the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-03765-y
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Deng-Fa Cheng, Email: dfcheng@ippcaas.cn.
Fang-Hao Wan, Email: wanfanghao@caas.cn.
References
- 1.Bi JL, Felton GW. Foliar oxidative stress and insect herbivory: Primary compounds, secondary metabolites, and reactive oxygen species as components of induced resistance. J. Chem. Ecol. 1995;21:1511–1530. doi: 10.1007/BF02035149. [DOI] [PubMed] [Google Scholar]
- 2.Herde M, Koo AJ, Howe GA. Elicitation of jasmonate-mediated defense responses by mechanical wounding and insect herbivory. Methods Mol. Biol. 2013;1011:51–61. doi: 10.1007/978-1-62703-414-2_5. [DOI] [PubMed] [Google Scholar]
- 3.Hogenhout SA, Bos JI. Effector proteins that modulate plant-insect interactions. Curr. Opin. Plant Biol. 2011;14:422–428. doi: 10.1016/j.pbi.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Zhu-Salzman K, Zeng R. Insect response to plant defensive protease inhibitors. Annu. Rev.Entomol. 2015;60:233–252. doi: 10.1146/annurev-ento-010814-020816. [DOI] [PubMed] [Google Scholar]
- 5.De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. Bemisia tabaci: A statement of species status. Ann. Rev. Entomol. 2011;56:1–19. doi: 10.1146/annurev-ento-112408-085504. [DOI] [PubMed] [Google Scholar]
- 6.Wan FH, Yang NW. Invasion and management of agricultural alien insects in China. Ann. Rev. Entomol. 2016;61:77–98. doi: 10.1146/annurev-ento-010715-023916. [DOI] [PubMed] [Google Scholar]
- 7.Boykin LM, et al. Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using bayesian analysis of mitochondrial COI DNA sequences. Mol. phylogenet. Evol. 2007;44:1306–1319. doi: 10.1016/j.ympev.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 8.Chu D, Gao CS, De Barro P, Wan FH, Zhang YJ. Investigation of the genetic diversity of an invasive whitefly (Bemisia tabaci) in China using both mitochondrial and nuclear DNA markers. Bull. Entomol. Res. 2011;101:467–475. doi: 10.1017/S0007485311000022. [DOI] [PubMed] [Google Scholar]
- 9.Chu D, Wan FH, Zhang YJ, Brown JK. Change in the biotype composition of Bemisia tabaci in shandong province of China from 2005 to 2008. Environ. Entomol. 2010;39:1028–1036. doi: 10.1603/EN09161. [DOI] [PubMed] [Google Scholar]
- 10.Pan H, et al. Further spread of and domination by Bemisia tabaci (Hemiptera: Aleyrodidae) biotype Q on field crops in China. J. Econ. Entomol. 2011;104:978–985. doi: 10.1603/EC11009. [DOI] [PubMed] [Google Scholar]
- 11.Chu D, Tao YL, Chi H. Influence of plant combinations on population characteristics of Bemisia tabaci biotypes B and Q. J. Econ. Entomol. 2012;105:930–935. doi: 10.1603/EC10373. [DOI] [PubMed] [Google Scholar]
- 12.Tjallingii WF, Esch TH. Fine structure of aphid stylet routes in plant tissues in correlation with epg signals. Physiol. Entomol. 1993;18:317–328. doi: 10.1111/j.1365-3032.1993.tb00604.x. [DOI] [Google Scholar]
- 13.Miles PW. Aphid saliva. Biol. Rev. 1999;74:41–85. doi: 10.1017/S0006323198005271. [DOI] [Google Scholar]
- 14.Tjallingii WF. Salivary secretions by aphids interacting with proteins of phloem wound responses. J. Exp. Bot. 2006;57:739–745. doi: 10.1093/jxb/erj088. [DOI] [PubMed] [Google Scholar]
- 15.Carolan JC, et al. Predicted effector molecules in the salivary secretome of the pea aphid (Acyrthosiphon pisum): A dual transcriptomic/proteomic approach. J. Proteome Res. 2011;10:1505–1518. doi: 10.1021/pr100881q. [DOI] [PubMed] [Google Scholar]
- 16.Elzinga DA, Jander G. The role of protein effectors in plant-aphid interactions. Curr. Opin. Plant Biol. 2013;16:451–456. doi: 10.1016/j.pbi.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Hogenhout SA, V. der Hoorn RA, Terauchi R, Kamoun S. Emerging concepts in effector biology of plant-associated organisms. Mol. Plant Microbe Interact. 2009;22:115–122. doi: 10.1094/MPMI-22-2-0115. [DOI] [PubMed] [Google Scholar]
- 18.Bos JI, et al. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid) PLoS Genet. 2010;6:e1001216. doi: 10.1371/journal.pgen.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson AJ, et al. Differential expression of candidate salivary effector proteins in field collections of Hessian fly, Mayetiola destructor. Insect Mol. Biol. 2015;24:191–202. doi: 10.1111/imb.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, et al. Armet is an effector protein mediating aphid-plant interactions. Faseb J. 2015;29:2032–2045. doi: 10.1096/fj.14-266023. [DOI] [PubMed] [Google Scholar]
- 21.Hattori M, Konishi H, Tamura Y, Konno K, Sogawa K. Laccase-type phenoloxidase in salivary glands and watery saliva of the green rice leafhopper. Nephotettix cincticeps. J. Insect physiol. 2005;51:1359–1365. doi: 10.1016/j.jinsphys.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Hattori M, et al. Molecular characterization and expression of laccase genes in the salivary glands of the green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae) Insect Biochem. Mol. Biol. 2010;40:331–338. doi: 10.1016/j.ibmb.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Coy MR, et al. Phenol-oxidizing laccases from the termite gut. Insect Biochem. Mol. Biol. 2010;40:723–732. doi: 10.1016/j.ibmb.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Yatsu J, Asano T. Cuticle laccase of the silkworm, Bombyx mori: Purification, gene identification and presence of its inactive precursor in the cuticle. Insect Biochem. Mol. Biol. 2009;39:254–262. doi: 10.1016/j.ibmb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 25.Arakane Y, Muthukrishnan S, Beeman RW, Kanost MR, Kramer KJ. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc. Natl. Acad. Sci. USA. 2005;102:11337–11342. doi: 10.1073/pnas.0504982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dittmer NT, et al. Characterization of cDNAs encoding putative laccase-like multicopper oxidases and developmental expression in the tobacco hornworm, Manduca sexta, and the malaria mosquito, Anopheles gambiae. Insect Biochem. Mol. Biol. 2004;34:29–41. doi: 10.1016/j.ibmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Futahashi R, et al. Laccase2 is required for cuticular pigmentation in stinkbugs. Insect Biochem. Mol. Biol. 2011;41:191–196. doi: 10.1016/j.ibmb.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Ye YX, Pan PL, Kang D, Lu JB, Zhang CX. The multicopper oxidase gene family in the brown planthopper. Nilaparvata lugens. Insect Biochem. Mol. Biol. 2015;63:124–132. doi: 10.1016/j.ibmb.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 29.Alam MM, Nahar K, Hasanuzzaman M, Fujita M. Exogenous jasmonic acid modulates the physiology, antioxidant defense and glyoxalase systems in imparting drought stress tolerance in different brassica species. Plant Biotechnol. Rep. 2014;8:279–293. doi: 10.1007/s11816-014-0321-8. [DOI] [Google Scholar]
- 30.Kaya A, Doganlar ZB. Exogenous jasmonic acid induces stress tolerance in tobacco (nicotiana tabacum) exposed to imazapic. Ecotox. Environ. Safe. 2016;124:470–479. doi: 10.1016/j.ecoenv.2015.11.026. [DOI] [PubMed] [Google Scholar]
- 31.Dwivedi UN, Singh P, Pandey VP, Kumar A. Structure-function relationship among bacterial, fungal and plant laccases. J. Mol. Catalysis B: Enzymatic. 2011;68:11. doi: 10.1016/j.molcatb.2010.11.002. [DOI] [Google Scholar]
- 32.Nakamura K, Go N. Function and molecular evolution of multicopper blue proteins. Cell Mol. Life Sci. 2005;62:2050–2066. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Claus H. Laccases: Structure, reactions, distribution. Micron. 2004;35:93–96. doi: 10.1016/j.micron.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 34.Sharma KK, Kuhad RC. Laccase: Enzyme revisited and function redefined. Indian J. Microbiol. 2008;48:309–316. doi: 10.1007/s12088-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon JR, Baldrian P, Murugesan K, Chang YS. Laccase-catalysed oxidations of naturally occurring phenols: From in vivo biosynthetic pathways to green synthetic applications. Microb. Biotechnol. 2012;5:318–332. doi: 10.1111/j.1751-7915.2011.00273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moin SF, Omar MN. Laccase enzymes: Purification, structure to catalysis and tailoring. Protein Peptide Lett. 2014;21:707–713. doi: 10.2174/09298665113209990058. [DOI] [PubMed] [Google Scholar]
- 37.Pezzella C, Guarino L, Piscitelli A. How to enjoy laccases. Cell Mol. Life Sci. 2015;72:923–940. doi: 10.1007/s00018-014-1823-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giardina P, et al. Laccases: A never-ending story. Cell Mol. Life Sci. 2010;67:369–385. doi: 10.1007/s00018-009-0169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lang M, Braun CL, Kanost MR, Gorman MJ. Multicopper oxidase-1 is a ferroxidase essential for iron homeostasis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2012;109:13337–13342. doi: 10.1073/pnas.1208703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niku-Paavola ML, Raaska L, Itavaara M. Detection of white-rot fungi by a non-toxic stain. Mycol. Res. 1990;94:27–31. doi: 10.1016/S0953-7562(09)81260-4. [DOI] [Google Scholar]
- 41.Sugumaran M, Giglio L, Kundzicz H, Saul S, Semensi V. Studies on the enzymes involved in puparial cuticle sclerotization in Drosophila melanogaster. Arch. Insect Biochem. Physiol. 1992;19:271–283. doi: 10.1002/arch.940190406. [DOI] [PubMed] [Google Scholar]
- 42.Huang HJ, et al. Screening and functional analyses of Nilaparvata lugens salivary proteome. J. proteome res. 2016;15:1883–1896. doi: 10.1021/acs.jproteome.6b00086. [DOI] [PubMed] [Google Scholar]
- 43.Huang HJ, et al. A salivary sheath protein essential for the interaction of the brown planthopper with rice plants. Insect Biochem. Mol. Biol. 2015;66:77–87. doi: 10.1016/j.ibmb.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 44.Thaler JS, Stout MJ, Karban R, Duffey SS. Exogenous jasmonates simulate insect wounding in tomato plants (Lycopersicon esculentum) in the laboratory and field. J. Chem. Ecol. 1996;22:1767–1781. doi: 10.1007/BF02028503. [DOI] [PubMed] [Google Scholar]
- 45.Walling LL. Avoiding effective defenses: Strategies employed by phloem-feeding insects. Plant Physiol. 2008;146:859–866. doi: 10.1104/pp.107.113142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kehr J. Phloem sap proteins: Their identities and potential roles in the interaction between plants and phloem-feeding insects. J. Exp. Bot. 2006;57:767–774. doi: 10.1093/jxb/erj087. [DOI] [PubMed] [Google Scholar]
- 47.Liu J, et al. Complete biodegradation of chlorpyrifos by engineered pseudomonas putida cells expressing surface-immobilized laccases. Chemosphere. 2016;157:200–207. doi: 10.1016/j.chemosphere.2016.05.031. [DOI] [PubMed] [Google Scholar]
- 48.Peng L, et al. Identification, comparison, and functional analysis of salivary phenol-oxidizing enzymes in Bemisia tabaci B and Trialeurodes vaporariorum. Entomol. Exp. Appl. 2013;147:282–292. doi: 10.1111/eea.12068. [DOI] [Google Scholar]
- 49.Collins PJ, Dobson ADW, Field JA. Reduction of the 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) cation radical by physiological organic acids in the absence and presence of manganese. Appl. Environ. Microbiol. 1998;64:2026–2031. doi: 10.1128/aem.64.6.2026-2031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


