Abstract
Enterococcus faecalis is an important opportunistic pathogen which is frequently detected in mineral water and spring water for human consumption and causes human urinary tract infections, endocarditis and neonatal sepsis. The aim of this study was to determine the prevalence, virulence genes, antimicrobial resistance and genetic diversity of E. faecalis from mineral water and spring water in China. Of 314 water samples collected from January 2013 to January 2014, 48 samples (15.3%) were contaminated E. faecalis. The highest contamination rate occurred in activated carbon filtered water of spring water (34.5%), followed by source water of spring water (32.3%) and source water of mineral water (6.4%). The virulence gene test of 58 E. faecalis isolates showed that the detection rates of asa1, ace, cylA, gelE and hyl were 79.3, 39.7, 0, 100, 0%, respectively. All 58 E. faecalis isolates were not resistant to 12 kinds of antibiotics (penicillin, ampicillin, linezolid, quinupristin/dalfopristin, vancomycin, gentamicin, streptomycin, ciprofloxacin, levofloxacin, norfloxacin, nitrofurantoin, and tetracycline). Enterobacterial repetitive intergenic consensus-PCR classified 58 isolates and three reference strains into nine clusters with a similarity of 75%. This study is the first to investigate the prevalence of E. faecalis in mineral water and spring water in China. The results of this study suggested that spring water could be potential vehicles for transmission of E. faecalis.
Keywords: Enterococcus faecalis, mineral water, spring water, ERIC-PCR, virulence genes
Introduction
Enterococci mainly inhabits in human and animal faces, ham sausage, pasteurized milk and drinking water (du Toit et al., 2000; Franz et al., 2003; Zou et al., 2011). Although some Enterococcal species are considered relevant for their technological properties (such as ripening, aroma development and inhibition of pathogens), they are not, unlike other lactic acid bacteria, recognized as probiotics (Kuriyama et al., 2003; Emaneini et al., 2008; Jamet et al., 2012). Indeed, enterococci are a major cause of nosocomial infections, such as urinary tract infections, endocarditis and neonatal sepsis (Franz et al., 2011; Sanchez Valenzuela et al., 2012; Werner et al., 2013). The main enterococcal isolates involved in nosocomial infections is Enterococcus faecalis (Giraffa, 2002; Kayser, 2003). E. faecalis is an important opportunistic pathogen, which is frequently detected in mineral water and spring water for human consumption (Nicas et al., 1989; Martin-Platero et al., 2009; Buhnik-Rosenblau et al., 2013). Several studies have indicated that E. faecalis is a suitable indicator of the presence of pathogens in mineral water and spring water (Švec and Sedláček, 1999; Davis et al., 2005; Meng, 2007). The Natural Mineral Water National Standard GB 8537 (the National Food safety Standards of China) has short-listed E. faecalis as a microorganism indicator in mineral water and spring water factory in China.
For surveillance or tracing sources of E. faecalis, molecular typing methods such as pulsed field gel electrophoresis (PFGE) (Weng et al., 2013), amplified fragment length polymorphism (AFLP) (Bruinsma et al., 2002), random amplified polymorphic DNA (RAPD) (Martin-Platero et al., 2009), multi-locus sequence typing (MLST) (Homan et al., 2002; Werner et al., 2012) and enterobacterial repetitive intergenic consensus (ERIC-PCR) (Martin-Platero et al., 2009), can be used in E. faecalis isolates. Among these molecular typing approaches, PFGE is the most effective technology or typing of E. faecalis isolates due to its high reproducibility and discriminatory ability. However, PFGE is labor intensive and time-consuming (Weng et al., 2013). In contrast, ERIC-PCR is a relatively simple and cost-effective method, which has been successfully used for genotyping of different bacterial pathogens and for tracking the bacterial source of contaminated water products (Martin-Platero et al., 2009).
Enterococcus faecalis can produce dozens of virulence substances including hemolysin and surface adhesion substances (Aslam et al., 2012; Choi and Woo, 2013). The pathogenesis of five virulence genes including asa1, ace, cylA, gelE and hyl has been systematically studied. The asa1gene encodes surface adhesion substances by which E. faecalis can be fixed on a eukaryotic cell; the ace gene encodes surface adhesion proteins by which E. faecalis are resistant to immune function of the host cells; the cylA gene encodes hemolysin which can leads to the death of host cells; the gelE gene encodes gelatinase, which can hydrolyze gelatin leading to the proliferation of bacteria in the host cell; the hyl gene encodes hyaluronidase which can hydrolyze the tissue of host cell (Coque et al., 1995; Eaton and Gasson, 2001; Cosentino et al., 2010; Yang et al., 2015).
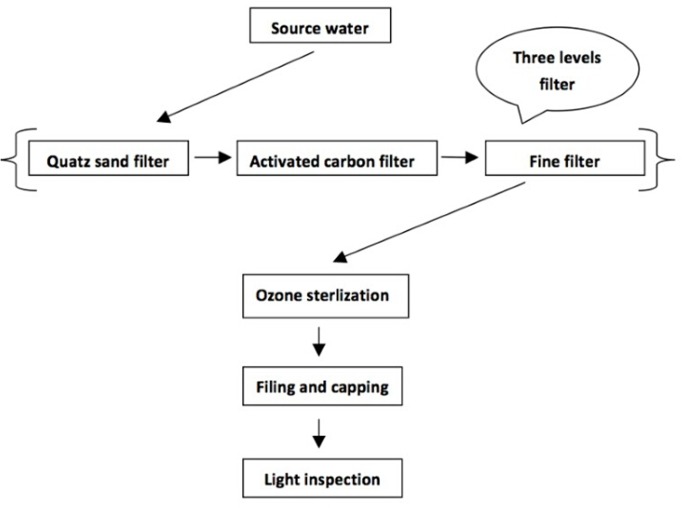
As shown in Figure 1, mineral water and spring water are generally produced using the same process, including three level filter (quartz sand filter, activated carbon filter, and fine filter), ozone sterilization, filling and capping, and light inspection of finished product. For manufacturers, the quality of source water, activated carbon filtered water and finished product are important. Source water reflects raw quality of mineral and spring water, and finished product is for human consumption. Activated carbon filter with a lot of pores and large surface area is a kind of common water treatment equipment, which processes a strong physical adsorption capacity to absorb organic pollutants and microbes. Recent studies have shown that activated carbon filter has become a gathering place for microbes and is the most serious in microbial contamination in whole production process of mineral water and spring water.
FIGURE 1.
Production flow chart of mineral water and spring water in China.
Systematic contamination survey of E. faecalis in mineral water and spring water has not yet been conducted. The aim of this study was for the first time to determine the prevalence, virulence genes, antimicrobial resistance and genetic diversity of E. faecalis from mineral water and spring water in China. The information generated in this study will provide insights into the prevalence and differentiation of E. faecalis isolates in mineral water and spring water.
Materials and Methods
Sample Collection
From January 2013 to January 2014, a total of 314 water Samples were collected from 101 mineral water and spring water factories in 10 provinces of China (Guangdong, Guangxi, Fujian, Hainan, Hubei, Shanghai, Beijing, Yunnan, Guizhou and Sichuan). Samples of source water (112), activated carbon filtered water (101) and finished product (101) of spring water and mineral water were collected from each water factories. Samples of source water (112) include Surface water (14) and Groundwater (98). All water samples were maintained at 4°C during transportation and testing was conducted within 1 h after receiving the samples.
Isolation and Enumeration of E. faecalis
Briefly, 250 ml of water sample was filtered through a 0.45 μm membrane (Millipore Co., Billerica, MA, United States) in a stainless steel multi-line filter system (Huankai Co., Guangzhou, China). The membrane was placed on KF agar medium (Huankai Co., Guangzhou, China), a selective medium for E. faecalis and then cultured at 36°C for 2 days. Presumptive colonies with red color were selected for catalase test and cultured in brain heart infusion broth at 45°C for 2 days and Bile broth at 36°C for 3 days, respectively. Colonies positive for the three confirmation tests were considered presumed E. faecalis.
Enterococcus faecalis Identification
All the presumed E. faecalis were identified by the API 20 STREP biochemical identification system (Biomerieux Co., Lyon, France) and specific PCR for the E. faecalis species. According to seven code of API 20 Strep biochemical identification system, coincidence rate of E. faecalis isolates can be tested. Further identification of E. faecalis was determined by PCR using sodA species-specific primers. Genomic DNA was extracted from collected E. faecalis isolates by using a Bacterial Genomic DNA Purification kit (Dongsheng Biotech, Guangzhou, China) according to the manufacturer’s instruction. E. faecalis were identified by amplification of 210 bp fragments with primer pairs EFS1 (5′ CTGTAGAAGACCTAATTTCA)/EFS2 (5′ CAGCTGTTTTGA AAGCAG) (Jamet et al., 2012).
ERIC-PCR Analysis
Total DNA from E. faecalis isolates was extracted as previously described. Genomic DNA concentration was determined at 260 nm by using a Nano Drop ND 1000 UVe-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). The ERIC primers used were referred from the study of Versalovic et al. (1991). ERIC-PCR typing was performed for the collected E. faecalis isolates by using the protocol described by Li (2013) with some modifications. The PCR mixture (25 μl) contained 1.5 unit Hot start polymerase (Promega, Madison, WI, United States), 0.5 μmol/l each primer, 2.5 μmol/l MgCl2, 200 μmol/l each dNTP, and 1 μl of the template genomic DNA (50 ng). Amplifications were performed with a DNA thermocycler (Applied Biosystems, Foster City, CA, United States) under the following temperature profiles: an initial denaturation at 95°C for 5 min; 35 cycles of 1 min at 94°C, 1 min at 36°C and 2 min at 72°C; and a final extension at 72°C for 8 min (Martin-Platero et al., 2009; Li, 2013). The ERIC-PCR products were separated by electrophoresis in a 1.5% agarose gel with Goldview staining (0.005%, v/v) and then photographed using a UV Imaging System (GE Healthcare, Milwaukee, WI, United States). The images were captured in TIFF file format for further analysis.
Detection of Virulence Genes
Five virulence genes, asa1, ace, cylA, gelE and hyl were individually detected in all the collected E. faecalis isolates with the PCR technique (Eaton and Gasson, 2001; Zou et al., 2011). All primers were synthesized by Sangon Biotech company (Shanghai, China). The primers used to identify virulence genes are shown in Table 1. E. faecalis isolates CMCC 32219 (Guangdong culture collection centre) was used as positive control and distilled water was used as the negative control.
Table 1.
PCR primers for virulence gene detection.
| No. | Primers | Sequence (5’→3’) | bp | Function |
|---|---|---|---|---|
| 1 | asa1-F | CACGCTATTACGAACTATGA | 375 | Surface adhesion |
| asa1-R | TAAGAAAGAACATCACCACGA | substances | ||
| 2 | ace-F | GGAATGACCGAGAACGATGGC | 616 | Surface adhesion |
| ace-R | GCTTGATGTTGGCCTGCTTCCG | proteins | ||
| 3 | cylA-F | ACTCGGGGATTGATAGGC | 688 | Hemolysin |
| cylA-R | GCTGCTAAAGCTGCGCTT | |||
| 4 | gelE-F | TATGACAATGCTTTTTGGGAT | 213 | Gelatinase |
| gelE-R | AGATGCACCCGAAATAATATA | |||
| 5 | hyl-F | ACAGAAGAGCTGCAGGAAATG | 276 | Hyaluronidase |
| hyl-R | GACTGACGTCCAAGTTTCCAA |
Antibiotic Resistance
According to the Clinical and Laboratory Standards Institute (CLSI) standards, all the collected E. faecalis isolates were tested by the disk diffusion method (Clinical and Laboratory Standards Institute [CLSI] (2006)). E. faecalis isolates CMCC 32219 (Guangdong culture collection centre) was used as positive control. A panel of antibiotics at the specific concentration per disk were tested: penicillin G (10 U), ampicillin (10 μg), linezolid (30 μg), quinupristin/dalfopristin (15 μg), vancomycin (30 μg), gentamicin (120 μg), streptomycin (10 μg), ciprofloxacin (5 μg) levofloxacin (5 μg), norfloxacin (10 μg), nitrofurantoin (300 μg), tetracycline (30 μg) (Oxoid Co., Hampshire, United Kingdom) (Jamet et al., 2012; Sanchez Valenzuela et al., 2013). The isolates were classified as sensitive, intermediate, and resistant using the breakpoints specified by the CLSI.
Fingerprint Data Analysis
ERIC-PCR fingerprint patterns were analyzed using a Gel-Pro analyser (version 6.0) according to the manufacturer’s instructions. The observed bands in the gels were evaluated based on the presence (code 1) or absence (code 0) of polymorphic fragments for the ERIC products. The cluster analysis was performed using NTSYSpc (version 2.10e), and similarities between ERIC-PCR profiles were calculated based on the simple-matching similarity matrix and unweighted pair group method with arithmetic.
Results
Contamination of E. faecalis in Mineral Water and Spring Water
Of the 314 water samples tested, 48 (15.3%) were positive for E. faecalis, including 24 (21.4%) of 112 source water, 22 (21.8%) of 101 activated carbon filtered water samples and 2 (1.9%) of 101 finished product samples. From the contaminated samples, 58 E. faecalis isolates were obtained (Supplementary Table S1). As shown in Table 2, the rate of E. faecalis contamination in all mineral water samples was 3.8%, and the contamination rates of source water, activated carbon filtered water and finished product were 6.4, 4.7, and 0%, respectively. The rate of E. faecalis contamination in all spring water samples was 23.8%, and the contamination rates of source water, activated carbon filtered water and finished product were 32.3, 34.5, and 7.5%, respectively. In all the 48 contaminated samples, the contamination levels in spring water samples are significantly higher than those in mineral water samples. The contamination level of source water and activated carbon filtered water were 48.0 and 26.4 CFU/250 ml in spring water samples.
Table 2.
Prevalence of E. faecalis from mineral water and spring water in China.
| Source water | Activated carbon filtered water | Finish product | Total | |||||
|---|---|---|---|---|---|---|---|---|
| Samples | CR (%) | CL (CFU/250 ml) | CR (%) | CL (CFU/250 ml) | CR (%) | CL (CFU/250 ml) | CR (%) | CL (CFU/250 ml) |
| M water | 6.4 | 2.3 | 4.7 | 3.5 | 0 | 0 | 3.8 | 2.8 |
| S water | 32.3 | 48.0 | 34.5 | 26.4 | 3.4 | 7.5 | 23.8 | 36.0 |
| Average | 21.4 | 42.2 | 21.8 | 24.3 | 1.9 | 7.5 | 15.3 | 32.5 |
M and S water represent mineral and spring water, respectively; CR and CL represent contamination rate and level in positive samples, respectively.
All the 112 source water Samples include 14 surface source water samples and 98 underground source water samples. As shown in Table 3, 16 (16.3%) samples of underground source water were positive in all 98 samples. Meanwhile, eight (57.1%) samples of surface source water were positive in all 14 samples.
Table 3.
Prevalence of E. faecalis in surface water and groundwater.
| Samples | Positive amounts | Total amounts | Contamination rates% |
|---|---|---|---|
| Surface water | 8 | 14 | 57.1 |
| Groundwater | 16 | 98 | 16.3 |
Detection of Virulence Genes in E. faecalis Isolates
In this study, the presence of asa1, ace, cylA, gelE and hyl genes was detected in 58 E. faecalis isolates. As shown in Table 4, all the 58 E. faecalis isolates (100%) harbored the gelE gene, among which 46 (79.3%) and 23 (39.7%) also had the asa1 and ace genes, respectively. However, no cylA or hyl gene was detected from all the isolates.
Table 4.
Virulence genes of 58 E. faecalis isolates.
| Virulence gene | No. of positive sample (%) |
|---|---|
| asa1 | 46 (79.3) |
| ace | 23 (39.7) |
| cylA | 0 (0) |
| gelE | 58 (58) |
| hyl | 0 (0) |
Antibiotic Resistance
According the diameter of zone of inhibition, all the 58 E. faecalis isolates were classified as sensitive, intermediate, and resistant using the breakpoints specified by the CLSI. The result of antibiotic resistance showed that all 58 isolates were sensitive to 12 kinds of antibiotic (penicillin, ampicillin, linezolid, quinupristin/dalfopristin, vancomycin, gentamicin, streptomycin, ciprofloxacin levofloxacin, norfloxacin, nitrofurantoin, and tetracycline) which were selected according to the standard of CLSI. No resistant E. faecalis isolate was found.
Biochemical Identification
With E. faecalis ATCC 29212 (a), CMCC 32219 (b) and CMCC 32223 (c) (Guangdong culture collection centre) as positive controls, according to seven code of API 20 Strep biochemical identification system, biotypes of E. faecalis isolates can be tested. As shown in Table 5, biotypes of 44 E. faecalis isolates and two control isolates are 5143711. Biotypes of four E. faecalis isolates and one control isolate are 5153711, and biotypes of ten E. faecalis isolates are 7143711.
Table 5.
Biochemical profiles of 58 E. faecalis isolates.
| Biotypes | No. of isolates | 7 code |
|---|---|---|
| A | a, b, 2∼5, 8∼11, 14∼36, 38∼41, 44, 46, 49∼52, 54, 56, 58 | 5143711 |
| B | 1, 6∼7, 37, 45, 47∼48, 53, 55, 57 | 7143711 |
| C | c, 12∼13, 42∼43 | 5153711 |
a, b and c represent E. faecalis ATCC 29212, CMCC 32219 and CMCC 3222, respectively.
ERIC-PCR
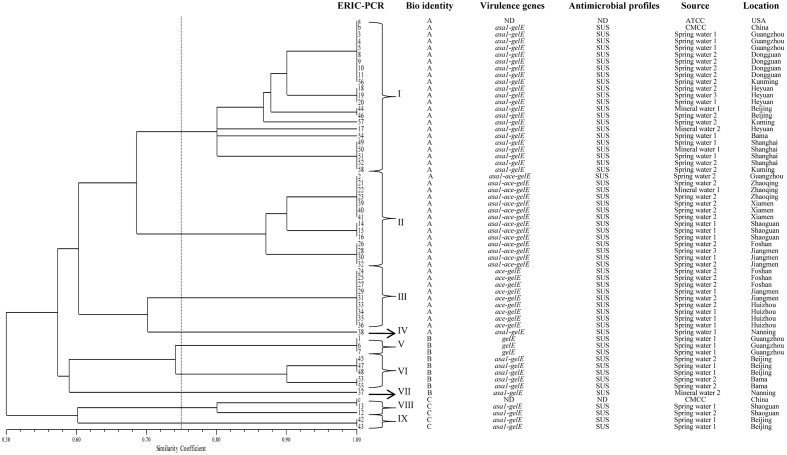
The ERIC-PCR patterns of three control isolates (a, b, c) and 58 collected E. faecalis isolates are shown in Supplementary Figure S1. The ERIC of DNA profiles consisting of 5 to 15 bands ranged from 230 bp to approximately 4,000 bp. As shown in Figure 2, three control isolates (a, b, c) and 58 collected E. faecalis isolates were grouped into nine clusters at similarity coefficient of 75%. Control isolates ATCC 29212 (a), CMCC 32219 (b) and 21 collected E. faecalis were distributed among the cluster I, a prevalent cluster. Control isolate CMCC 32223 (c) and two collected E. faecalis isolates were distributed among cluster VIII. Cluster IV has only one isolate (NO.38) and Cluster VII has only one isolate (NO. 37).
FIGURE 2.
ERIC-PCR DNA fingerprint analysis of E. faecalis isolates from mineral water and spring water in China. a, b and c represent E. faecalis ATCC 29212, CMCC 32219 and CMCC 32223, respectively; ND, not detect; SUS, susceptibility; 1, 2 and 3 represent source water, activated carbon filtered water and finish product, respectively.
Discussion
In accordance with the Natural Mineral Water National Standard GB 8537, E. faecalis must be absent from 250 ml of water samples. In this study, 48 (15.3%) E. faecalis-positive samples from a total of 314 water samples were found. This result was consistent with previous investigations conducted in China (17.0%) (Li, 2013) and Greece (17.4%) (Grammenou et al., 2006). E. faecalis has not been systematically studied from mineral water and spring water, and our results were obtained from a large number of samples in China. Therefore, these nationwide data are more beneficial for risk assessment. The E. faecalis contamination rates in spring water samples were significantly higher than those in mineral water samples. The high prevalence of E. faecalis in the spring water indicated poor hygiene practices during manufacture process. The finished product of spring water presented a contamination rate of 3.4%, which can adversely affect the health of costumers. Therefore, the Chinese food safety management should implement further supervision for spring water products as well as implementation of Good Hygiene Practices (GHP). In addition, contamination rate of source water in surface was significantly higher than those in groundwater. Based on the contamination rate of E. faecalis, groundwater is better than surface water as source water.
In this study, E. faecalis contamination rate of activated carbon filtered in spring water reached 26.4%, which was the highest among all water samples tested. Activated carbon filter system is commonly used for mineral water treatment to ensure equipment life, improve water quality and prevent pollution. Activated carbon filter which has a lot of pores and large surface area processes a strong physical adsorption capacity to absorb organic pollutants and microbes (Feng et al., 2013). Recent studies have shown that activated carbon filter has become a gathering place for microbes and is the most serious in microbial contamination in whole production process of mineral water and spring water (Camper et al., 1986). Hence, manufacturers of mineral water and spring water must establish measures for monitoring the activated carbon filtration system, as well as ensure timely cleaning and regular replacement of activated carbon (da Silva Fernandes et al., 2015; Fernandes et al., 2015).
Highly virulent E. faecalis isolates may cause diseases even at relatively low concentrations. In this study, up to 100% of E. faecalis isolates were gelE-positive and 79.3% were asa1-positive, which is consistent with previous studies and provides further evidence that these virulence genes are widely distributed among E. faecalis (Moraes et al., 2012; Anderson et al., 2015). The presence of these genes indicates pathogenic potential and probability to cause diseases. However, the expression of virulence genes is mainly related to quorum sensing. Further studies must be performed to examine whether the E. faecalis isolates in this study are pathogenic (Huebner et al., 1999; Delpech et al., 2012). All the 58 collected E. faecalis isolates were cylA-negative and hyl-negative, which is not consistent with previous studies (Creti et al., 2004; Medeiros et al., 2014). All the 58 collected E. faecalis isolates were sensitive to 12 kinds of antibiotic, which is different from previous investigation about drink water (Macedo et al., 2011), food (Gaglio et al., 2016) and clinical samples (Dahlén et al., 2012; Gilmore et al., 2013). The difference of the results may be due to different sources of the samples. The antibiotic resistance of E. faecalis come from different sources have huge difference (Abriouel et al., 2008). In this study, most of collected E. faecalis isolates derived from groundwater. So far, there is almost no research on antibiotic resistance of E. faecalis isolated from the groundwater.
ERIC-PCR is one of the most widely adopted PCR typing methods and is chosen for analyses of genetic diversity (Chen et al., 2014; Xie et al., 2015; Zhang et al., 2015). This method provides discriminatory value and is a rapid method for E. faecalis typing. In this study, the ERIC-PCR results provided a better overview of E. faecalis diversity. Most of collected E. faecalis isolates in the same area belong to the same cluster, which agrees with the results of previous studies (Martin-Platero et al., 2009). Three isolates (18, 19 and 20) obtained from spring water in Heyuan city showed 100% similarity. Two isolates (44 and 46) from spring water in Beijing city also showed identical ERIC patterns. Additionally, 10 isolates (a, b, 3, 4, 5, 8, 9, 10, 11 and 58) from different sources yielded an identical pattern, suggesting that they were highly homogenous and had a close genetic relationship. A correlation between the genomic profiles and the virulence genes was observed in these strains. Most of the isolates that carried asa1, ace and gelE were grouped in cluster B. In this cluster, a good correlation among ERIC patterns, virulence profiles, and the sample source was found in some isolates. In this study, no antibiotic resistance isolate was found, so no correlation was observed between the ERIC-PCR profiles and the antibiotic resistance profiles of the isolates.
In summary, our study for the first time revealed the high prevalence of E. faecalis from mineral water and spring water in China, which should have a potentially pathogenic effect on the health of consumers. The results of this study suggested that spring water product could be potential vehicles for transmission of E. faecalis. Meanwhile mineral water and spring water manufacturing factories must pay high attention to the contamination of activate carbon filters. These data may provide useful information for the development of public health policies and effective strategies to ensure the safety of our drinking water products.
Ethics Statement
All procedures performed in studies involving human participants were in accordance with the ethical standards.
Author Contributions
Conceived and designed the experiments: QW, JZ, and LW. Performed the experiments: LW and WG. Analyzed the data: LW, MC, and LX. Contributed reagents/materials/analysis tools: JW and LM. Contributed to the writing of the manuscript: LWand QW.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to acknowledge the financial support of the Science and Technology Planning Project of Guangdong Province (2012A032300018).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01109/full#supplementary-material
Dendrogram of ERIC-PCR patterns of collected E. faecalis isolates. M and CK represents marker and control check, respectively; a, b and c represent E. faecalis ATCC 29212, CMCC 32219 and CMCC 3222, respectively.
References
- Abriouel H., Omar N. B., Molinos A. C., López R. L., Grande M. J., Martínez-Viedma P., et al. (2008). Comparative analysis of genetic diversity and incidence of virulence factors and antibiotic resistance among enterococcal populations from raw fruit and vegetable foods, water and soil, and clinical samples. Int. J. Food Microbiol. 123 38–49. 10.1016/j.ijfoodmicro.2007.11.067 [DOI] [PubMed] [Google Scholar]
- Anderson A. C., Jonas D., Huber I., Karygianni L., Wölber J., Hellwig E., et al. (2015). Enterococcus faecalis from food, clinical specimens, and oral sites: prevalence of virulence factors in association with biofilm formation. Front. Microbiol. 6:1534 10.3389/fmicb.2015.01534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M., Diarra M. S., Masson L. (2012). Characterization of antimicrobial resistance and virulence genotypes of Enterococcus faecalis recovered from a pork processing plant. J. Food Prot. 75 1486–1491. 10.4315/0362-028X.JFP-11-524 [DOI] [PubMed] [Google Scholar]
- Bruinsma N., Willems R. J., Van Den Bogaard A. E., Van Santen-Verheuvel M., London N., Driessen C., et al. (2002). Different levels of genetic homogeneity in vancomycin-resistant and-susceptible Enterococcus faecium isolates from different human and animal sources analyzed by amplified-fragment length polymorphism. Antimicrob. Agents Chemother. 46 2779–2783. 10.1128/AAC.46.9.2779-2783.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhnik-Rosenblau K., Matsko-Efimov V., Danin-Poleg Y., Franz C. M., Klein G., Kashi Y. (2013). Biodiversity of Enterococcus faecalis based on genomic typing. Int. J. Food Microbiol. 165 27–34. 10.1016/j.ijfoodmicro.2013.04.009 [DOI] [PubMed] [Google Scholar]
- Camper A. K., Lechevallier M. W., Broadaway S. C., Mcfeters G. A. (1986). Bacteria associated with granular activated carbon particles in drinking water. Appl. Environ. Microbiol. 52 434–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wu Q., Zhang J., Yan Z. A., Wang J. (2014). Prevalence and characterization of Listeria monocytogenes isolated from retail-level ready-to-eat foods in South China. Food Control 38 1–7. 10.1016/j.foodcont.2013.09.061 [DOI] [Google Scholar]
- Choi J. M., Woo G. J. (2013). Molecular characterization of high-level gentamicin-resistant Enterococcus faecalis from chicken meat in Korea. Int. J. Food Microbiol. 165 1–6. 10.1016/j.ijfoodmicro.2013.02.016 [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute [CLSI] (2006). Performance Standards for Antimicrobial disk Susceptibility Tests. Wayne, PA: CLSI. [Google Scholar]
- Coque T. M., Patterson J. E., Steckelberg J. M., Murray B. E. (1995). Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171 1223–1229. 10.1093/infdis/171.5.1223 [DOI] [PubMed] [Google Scholar]
- Cosentino S., Podda G. S., Corda A., Fadda M. E., Deplano M., Pisano M. B. (2010). Molecular detection of virulence factors and antibiotic resistance pattern in clinical Enterococcus faecalis strains in Sardinia. J. Prev. Med. Hyg. 51 31–36. [PubMed] [Google Scholar]
- Creti R., Imperi M., Bertuccini L., Fabretti F., Orefici G., Di Rosa R., et al. (2004). Survey for virulence determinants among Enterococcus faecalis isolated from different sources. J. Med. Microbiol. 53 13–20. 10.1099/jmm.0.05353-0 [DOI] [PubMed] [Google Scholar]
- da Silva Fernandes M., Kabuki D. Y., Kuaye A. Y. (2015). Biofilms of Enterococcus faecalis and Enterococcus faecium isolated from the processing of ricotta and the control of these pathogens through cleaning and sanitization procedures. Int. J. Food Microbiol. 200 97–103. 10.1016/j.ijfoodmicro.2015.02.004 [DOI] [PubMed] [Google Scholar]
- Dahlén G., Blomqvist S., Almståhl A., Carlén A. (2012). Virulence factors and antibiotic susceptibility in enterococci isolated from oral mucosal and deep infections. J. Oral Microbiol. 4:10855 10.3402/jom.v3404i3400.10855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis K., Anderson M. A., Yates M. V. (2005). Distribution of indicator bacteria in Canyon Lake, California. Water Res. 39 1277–1288. 10.1016/j.watres.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Delpech G., Pourcel G., Schell C., De Luca M., Basualdo J., Bernstein J., et al. (2012). Antimicrobial resistance profiles of Enterococcus faecalis and Enterococcus faecium isolated from artisanal food of animal origin in Argentina. Foodborne Pathog. Dis. 9 939–944. 10.1089/fpd.2012.1192 [DOI] [PubMed] [Google Scholar]
- du Toit M., Franz C. M., Dicks L. M., Holzapfel W. H. (2000). Preliminary characterization of bacteriocins produced by Enterococcus faecium and Enterococcus faecalis isolated from pig faeces. J. Appl. Microbiol. 88 482–494. 10.1046/j.1365-2672.2000.00986.x [DOI] [PubMed] [Google Scholar]
- Eaton T. J., Gasson M. J. (2001). Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67 1628–1635. 10.1128/AEM.67.4.1628-1635.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emaneini M., Aligholi M., Aminshahi M. (2008). Characterization of glycopeptides, aminoglycosides and macrolide resistance among Enterococcus faecalis and Enterococcus faecium isolates from hospitals in Tehran. Pol. J. Microbiol. 57 173–178. [PubMed] [Google Scholar]
- Feng S., Chen C., Wang Q. F., Zhang X. J., Yang Z. Y., Xie S. G. (2013). Characterization of microbial communities in a granular activated carbon–sand dual media filter for drinking water treatment. Int. J. Environ. Sci. Technol. 10 917–922. 10.1007/s13762-013-0188-1 [DOI] [Google Scholar]
- Fernandes M. S., Fujimoto G., De Souza L. P., Kabuki D. Y., Da Silva M. J., Kuaye A. Y. (2015). Dissemination of Enterococcus faecalis and Enterococcus faecium in a ricotta processing plant and evaluation of pathogenic and antibiotic resistance profiles. J. Food. Sci. 80 M765–M775. 10.1111/1750-3841.12824 [DOI] [PubMed] [Google Scholar]
- Franz C. M., Huch M., Abriouel H., Holzapfel W., Gálvez A. (2011). Enterococci as probiotics and their implications in food safety. Int. J. Food Microbiol. 151 125–140. 10.1016/j.ijfoodmicro.2011.08.014 [DOI] [PubMed] [Google Scholar]
- Franz C. M., Stiles M. E., Schleifer K. H., Holzapfel W. H. (2003). Enterococci in foods–a conundrum for food safety. Int. J. Food Microbiol. 88 105–122. 10.1016/S0168-1605(03)00174-0 [DOI] [PubMed] [Google Scholar]
- Gaglio R., Couto N., Marques C., De Fatima Silva Lopes M., Moschetti G., Pomba C., et al. (2016). Evaluation of antimicrobial resistance and virulence of enterococci from equipment surfaces, raw materials, and traditional cheeses. Int. J. Food Microbiol. 236 107–114. 10.1016/j.ijfoodmicro.2016.07.020 [DOI] [PubMed] [Google Scholar]
- Gilmore M. S., Lebreton F., Van Schaik W. (2013). Genomic transition of enterococci from gut commensals to leading causes of multidrug-resistant hospital infection in the antibiotic era. Curr. Opin. Microbiol. 16 10–16. 10.1016/j.mib.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraffa G. (2002). Enterococci from foods. FEMS Microbiol. Rev. 26 163–171. 10.1111/j.1574-6976.2002.tb00608.x [DOI] [PubMed] [Google Scholar]
- Grammenou P., Spiliopoulou I., Sazakli E., Papapetropoulou M. (2006). PFGE analysis of enterococci isolates from recreational and drinking water in Greece. J. Water Health 4 263–269. [PubMed] [Google Scholar]
- Homan W. L., Tribe D., Poznanski S., Li M., Hogg G., Spalburg E., et al. (2002). Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40 1963–1971. 10.1128/JCM.40.6.1963-1971.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner J., Wang Y., Krueger W. A., Madoff L. C., Martirosian G., Boisot S., et al. (1999). Isolation and chemical characterization of a capsular polysaccharide antigen shared by clinical isolates of Enterococcus faecalis and vancomycin-resistant Enterococcus faecium. Infect. Immun. 67 1213–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamet E., Akary E., Poisson M. A., Chamba J. F., Bertrand X., Serror P. (2012). Prevalence and characterization of antibiotic resistant Enterococcus faecalis in French cheeses. Food Microbiol. 31 191–198. 10.1016/j.fm.2012.03.009 [DOI] [PubMed] [Google Scholar]
- Kayser F. H. (2003). Safety aspects of enterococci from the medical point of view. Int. J. Food Microbiol. 88 255–262. 10.1016/S0168-1605(03)00188-0 [DOI] [PubMed] [Google Scholar]
- Kuriyama T., Williams D. W., Patel M., Lewis M. A., Jenkins L. E., Hill D. W., et al. (2003). Molecular characterization of clinical and environmental isolates of vancomycin-resistant Enterococcus faecium and Enterococcus faecalis from a teaching hospital in Wales. J. Med. Microbiol. 52 821–827. 10.1099/jmm.0.05123-0 [DOI] [PubMed] [Google Scholar]
- Li F. (2013). The distribution Regularity and Genetic Diversity of Food-Borne Pathogens in Packaging Drinking Nature Spring Water. Guangzhou: South China University of Technology. [Google Scholar]
- Macedo A. S., Freitas A. R., Abreu C., Machado E., Peixe L., Sousa J. C., et al. (2011). Characterization of antibiotic resistant enterococci isolated from untreated waters for human consumption in Portugal. Int. J. Food Microbiol. 145 315–319. 10.1016/j.ijfoodmicro.2010.11.024 [DOI] [PubMed] [Google Scholar]
- Martin-Platero A. M., Valdivia E., Maqueda M., Martinez-Bueno M. (2009). Characterization and safety evaluation of enterococci isolated from Spanish goats’ milk cheeses. Int. J. Food Microbiol. 132 24–32. 10.1016/j.ijfoodmicro.2009.03.010 [DOI] [PubMed] [Google Scholar]
- Medeiros A. W., Pereira R. I., Oliveira D. V., Martins P. D., D’azevedo P. A., Van Der Sand S., et al. (2014). Molecular detection of virulence factors among food and clinical Enterococcus faecalis strains in South Brazil. Braz. J. Microbiol. 45 327–332. 10.1590/S1517-83822014005000031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F. (2007). The Basic Research of National Standard for Natural Mineral Water and Quality of Cultural Media. Beijing: Chinese Academy of Sciences. [Google Scholar]
- Moraes P. M., Perin L. M., Todorov S. D., Silva A., Franco B. D. G. M., Nero L. A. (2012). Bacteriocinogenic and virulence potential of Enterococcus isolates obtained from raw milk and cheese. J. Appl. Microbiol. 113 318–328. 10.1111/j.1365-2672.2012.05341.x [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Wu C. Y., Hobbs J. N., Jr., Preston D. A., Allen N. E. (1989). Characterization of vancomycin resistance in Enterococcus faecium and Enterococcus faecalis. Antimicrob. Agents Chemother. 33 1121–1124. 10.1128/AAC.33.7.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Valenzuela A., Benomar N., Abriouel H., Perez Pulido R., Martinez Canamero M., Galvez A. (2012). Characterization of Enterococcus faecalis and Enterococcus faecium from wild flowers. Antonie Van Leeuwenhoek 101 701–711. 10.1007/s10482-011-9684-9 [DOI] [PubMed] [Google Scholar]
- Sanchez Valenzuela A., Lavilla Lerma L., Benomar N., Galvez A., Perez Pulido R., Abriouel H. (2013). Phenotypic and molecular antibiotic resistance profile of Enterococcus faecalis and Enterococcus faecium isolated from different traditional fermented foods. Foodborne Pathog. Dis. 10 143–149. 10.1089/fpd.2012.1279 [DOI] [PubMed] [Google Scholar]
- Švec P., Sedláček I. (1999). Occurrence of Enterococcus spp. in waters. Folia Microbiol. 44 3–10. 10.1007/BF02816212 [DOI] [PubMed] [Google Scholar]
- Versalovic J., Koeuth T., Lupski J. R. (1991). Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19 6823–6831. 10.1093/nar/19.24.6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng P. L., Ramli R., Shamsudin M. N., Cheah Y. K., Hamat R. A. (2013). High genetic diversity of Enterococcus faecium and Enterococcus faecalis clinical isolates by pulsed-field gel electrophoresis and multilocus sequence typing from a hospital in Malaysia. Biomed. Res. Int. 2013:938937 10.1155/2013/938937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner G., Coque T. M., Franz C. M. A. P., Grohmann E., Hegstad K., Jensen L., et al. (2013). Antibiotic resistant enterococci—Tales of a drug resistance gene trafficker. Int. J. Med. Microbiol. 303 360–379. 10.1016/j.ijmm.2013.03.001 [DOI] [PubMed] [Google Scholar]
- Werner G., Fleige C., Fessler A. T., Timke M., Kostrzewa M., Zischka M., et al. (2012). Improved identification including MALDI-TOF mass spectrometry analysis of group D streptococci from bovine mastitis and subsequent molecular characterization of corresponding Enterococcus faecalis and Enterococcus faecium isolates. Vet. Microbiol. 160 162–169. 10.1016/j.vetmic.2012.05.019 [DOI] [PubMed] [Google Scholar]
- Xie T., Wu Q., Xu X., Zhang J., Guo W. (2015). Prevalence and population analysis of Vibrio parahaemolyticus in aquatic products from South China markets. FEMS Microbiol. Lett. 362:fnv178 10.1093/femsle/fnv178 [DOI] [PubMed] [Google Scholar]
- Yang J. X., Li T., Ning Y. Z., Shao D. H., Liu J., Wang S. Q., et al. (2015). Molecular characterization of resistance, virulence and clonality in vancomycin-resistant Enterococcus faecium and Enterococcus faecalis: a hospital-based study in Beijing, China. Infect. Genet. Evol. 33 253–260. 10.1016/j.meegid.2015.05.012 [DOI] [PubMed] [Google Scholar]
- Zhang S., Zhu X., Wu Q., Zhang J., Xu X., Li H. (2015). Prevalence and characterization of Escherichia coli O157 and O157:H7 in retail fresh raw meat in South China. Ann. Microbiol. 65 1993–1999. 10.1016/j.meatsci.2009.10.011 [DOI] [Google Scholar]
- Zou L. K., Wang H. N., Zeng B., Li J. N., Li X. T., Zhang A. Y., et al. (2011). Erythromycin resistance and virulence genes in Enterococcus faecalis from swine in China. New Microbiol. 34 73–80. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dendrogram of ERIC-PCR patterns of collected E. faecalis isolates. M and CK represents marker and control check, respectively; a, b and c represent E. faecalis ATCC 29212, CMCC 32219 and CMCC 3222, respectively.