Abstract
Salmonella serovars are important reservoirs of antimicrobial resistance. Recently, we reported on multidrug-resistant (MDR) Salmonella enterica serovar Typhimurium strains among pigs with resistance to ampicillin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline (resistance [R] type AKSSuT) and resistance to amoxicillin-clavulanic acid, ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (R type AxACSSuT). In the present study, 67 isolates (39 from humans and 28 from pigs) of clinically important Salmonella serovar Muenchen were characterized. Among the porcine isolates, 75% showed resistance to seven antimicrobials: ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, amoxicillin-clavulanic acid, and kanamycin (R type ACSSuTAxK). One isolate from humans showed resistance to 10 of the 12 antimicrobials: ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, tetracycline, amoxicillin-clavulanic acid, kanamycin, gentamicin, cephalothin, and ceftriaxone (R type ACSSuTAxKGCfCro). Pulsed-field gel electrophoresis revealed no clonality between the porcine and the human strains. The porcine and the human MDR strains carried class 1 integrons of 2.0 and 1.0 kb, respectively. Genes specific to the porcine strain included aadA2, aphA1-Iab, and tetA(B). DNA sequencing revealed that the porcine isolates carried blaOXA-30 on a class 1 integron. Genes specific to the human strain included blaTEM, strA, strB, cmlA, tetA(A), and aadA2. No blaCMY-2 gene was detected. Serovar Muenchen strains of porcine and human origin were able to transfer resistance genes to laboratory strain Escherichia coli MG1655 by conjugation. Plasmid restriction with four restriction enzymes, EcoRI, BamHI, HindIII, and PstI, showed that the conjugative plasmids from porcine Salmonella serovar Muenchen and Typhimurium R-type MDR strains isolated from the same farms at the same time were similar on the basis of the sizes and the numbers of bands and Southern hybridization. The plasmid profiles among the Salmonella serovar Muenchen isolates from the two host species were different. This is the first report to show a high frequency of MDR Salmonella serovar Muenchen strains from pigs and a human strain that is similar to the MDR isolates with the AmpC enzyme previously reported among Salmonella serovars Newport and Typhimurium strains. The MDR strains from the two host species independently represent public health concerns, as Salmonella serovar Muenchen is among the top 10 causes of salmonellosis in humans.
More than 2,500 serovars of nontyphoidal Salmonella are known to exist and are considered potential food-borne pathogens. However, most human infections are caused by a few serovars. Salmonella enterica serovar Muenchen ranks among the top 10 serovars that cause clinical salmonellosis in humans in the United States. According to recent reports from the Centers for Disease Control and Prevention (CDC) (10), this serovar accounts for 2% of salmonellosis cases (ranked eighth among the serovars that cause clinical salmonellosis) in the United States. Historically, outbreaks due to this serovar in humans were reported in as early as 1961 (32). Three multistate outbreaks caused by this serovar have been reported in the past decade. These were associated with the consumption of food products, including alfalfa sprouts and unpasteurized orange juice, and the use of marijuana (8, 28, 33). Point sources of each of these products were implicated in the spread of the outbreaks to more than four states. Information on the potential role of commercial swine production in the dissemination of multidrug resistant (MDR) salmonellae in the United States is very limited. In addition, the role that food-producing animals play as a primary source of MDR Salmonella has often been questionable (36).
The occurrence of MDR Salmonella strains has been best documented in Salmonella serovar Typhimurium, but MDR is extremely rare in many other relatively common serovars, including Salmonella serovar Muenchen. One major concern to public health has been the emergence of MDR strains such as definitive type 104 (DT104), which was first recognized in the United Kingdom in 1984 (35) and which was later identified in other parts of the world (4, 5, 21, 22, 29). This phage type commonly exhibits resistance to five antimicrobial agents: ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (resistance [R] type ACSSuT). An MDR serovar Newport strain that carries AmpC that has recently emerged and that shows resistance to multiple classes of antimicrobials, including expanded-spectrum cephalosporins, has also been increasing in prevalence and is often associated with dairy products (16). Antimicrobial resistance in various other serovars, including Salmonella serovar Muenchen, may play an important role in the ecology of antimicrobial resistance and may have significance in the development of clinical disease. In the few reported outbreaks attributed to Salmonella serovar Muenchen, either antimicrobial susceptibility testing was not done or the strains responsible for the outbreak were rarely resistant (20). According to a recent report of the human arm of the National Antimicrobial Resistance Monitoring System (23), 86% of Salmonella serovar Muenchen isolates tested in the United States in 2001 were pansusceptible, and none of the isolates showed pentaresistance.
Our laboratory recently reported on common MDR strains in various Salmonella serovars from pigs, particularly Salmonella serovar Typhimurium (including variety Copenhagen). The two most common MDR R types identified were ampicillin, kanamycin, streptomycin, sulfamethoxazole, and tetracycline (R type AKSSuT) and amoxicillin-clavulanic acid, ampicillin, chloramphenicol, streptomycin, sulfamethoxazole, and tetracycline (R type AxACSSuT) (14, 15). In the present study, we investigated MDR strains of Salmonella serovar Muenchen from pigs and humans, identified and compared the sequences of their resistance genes and class 1 integrons, and compared the clonalities of isolates from humans with those of porcine origin. We also performed a conjugation assay and plasmid restriction pattern analysis to identify and compare the sizes of the conjugative plasmids in Salmonella serovar Muenchen. We then compared the genotypic characteristics with those detected in Salmonella serovar Typhimurium from the same groups of pigs to determine the likelihood of horizontal dissemination of resistance factors between Salmonella serovars Muenchen and Typhimurium.
MATERIALS AND METHODS
Origin and isolation of Salmonella serovar Muenchen.
The swine isolates evaluated in the present study were collected from 24 pig farms as part of a longitudinal study conducted between 1998 and 2001 (13). The farms belonged to two predominant production systems in North Carolina, and convenience sampling was done. Both systems use a form of pig production commonly known as all in, all out, in which groups of pigs of similar ages are moved into a clean and disinfected barn, reared without mixing with new groups of pigs, and marketed together. The antimicrobial use patterns among the farms were recorded, and a report on these patterns was published previously (15). The pigs in different finishing farms in both production systems originated from the same nursery sources. Within a production system, farm workers commonly serve more than one farm. In addition, a total of 39 Salmonella serovar Muenchen isolates collected from humans from various regions of North Carolina during the same time period that the porcine isolates were collected were received from the North Carolina State Laboratory of Public Health (NCSLPH) and were included in the present study.
Antimicrobial susceptibility testing.
The isolates were tested for their susceptibilities to 12 antimicrobial agents by the Kirby-Bauer disk diffusion method. The antimicrobials tested (as well as the abbreviations used for the R types and the disk potencies) were as follows: ampicillin (A; 10 μg), amoxicillin-clavulanic acid (Ax; 30 μg), amikacin (An; 30 μg), ceftriaxone (Cro; 30 μg), cephalothin (Cf; 30 μg), chloramphenicol (C; 30 μg), ciprofloxacin (Cip; 5 μg), gentamicin (G; 10 μg), kanamycin (K; 30 μg), streptomycin (S; 10 μg), sulfamethoxazole (Su; 250 μg), and tetracycline (T; 30 μg). In order to confirm the phenotypes of resistance to some antimicrobials, such as ceftriaxone, the epsilometric test (Etest) was used according to the guidelines of the manufacturer (AB Biodisk, Piscataway, N.J.). The results were interpreted according to the criteria of the NCCLS. Escherichia coli ATCC 25922 and ATCC 35218, Enterococcus faecalis ATCC 29212, Staphylococcus aureus ATCC 29213, and Pseudomonas aeruginosa ATCC 27853 were routinely used as quality control organisms, according to the recommendations of the NCCLS (24, 25).
PFGE.
Pulsed-field gel electrophoresis (PFGE) macrorestriction analysis was done as recommended by CDC (9). Briefly, cells from 200 μl of an overnight culture were lysed, and the intact genomic DNA was embedded in agarose plugs and digested with the XbaI restriction enzyme. The digested DNA was then separated with a contour-clamped homogeneous electric field (CHEF)-DRIII apparatus (Bio-Rad Laboratories, Hercules, Calif.) with the following reagents and conditions: 0.5× TBE (Tris-borate-EDTA), 1% Seakem Gold agarose (FMC BioProducts, Rockland, Maine), 14°C, and 6 V/cm for 17 h, with switch times ranging from 2.2 to 63 s. The S. enterica serovar Braenderup universal marker (kindly provided by Leslie Wolf, NCSLPH) was used as the reference marker. The gels were stained with ethidium bromide for 30 min, destained three times for 20 min each with distilled water, and photographed with an Alpha imager (Alpha Innotech Corporation, San Leandro, Calif.). Analysis of the PFGE data was performed with Bionumerics software (Applied Maths, Kortrijik, Belgium) by using different band clusterings and the ward tree-building approach with an optimization of 1% and a position tolerance of 0.75%. Visual inspection of the patterns was done as a final step in the analysis.
Detection of resistance genes and class 1 integrons.
PCR was used to amplify various antimicrobial resistance genes, genes for quaternary ammonium compound resistance, and class 1 integrons. The genes tested, the respective primers, the expected amplicon sizes, and the origins of the primers are shown in Table 1. Amplification reactions were carried out with 1 μl of purified DNA with a DNAeasy tissue kit (Qiagen, Valencia, Calif.), 300 μM deoxynucleoside triphosphates, 1.5 mM MgCl2, 50 pmol of primers, and 0.5 U of Taq polymerase (Perkin-Elmer, Foster City, Calif.). Distilled water was added to bring the final volume to 20 μl. The PCR cycle included initial denaturation at 95°C for 5 min and 30 cycles of denaturation for 1 min at 95°C, primer annealing for 1 min at 54°C, and extension for 1 min at 72°C, with a final extension of 7 min at 72°C. Known positive control strains, strains UW05 (phage type DT104) and UT30 (phage type DT193), were used throughout the PCR analysis. Strain 530, generously provided by P. Winokur (University of Iowa), was used as a positive control for blaCMY-2.
TABLE 1.
Oligonucleotides used as primers for PCR amplification
| Gene | Forward primer sequence (5′ to 3′) | Reverse primer sequence (5′ to 3′) | Expected product size (bp) | Reference or Genbank accession no. |
|---|---|---|---|---|
| blaPSE1 | TTTGGTTCCGCGCTATCTG | TACTCCGAGCACCAAATCCG | 150 | 7 |
| blaTEM | GCACGAGTGGGTTACATCGA | GGTCCTCCGATCGTTGTCAG | 310 | 7 |
| blaCMY-2 | ATGATGAAAAAATCGTTATGC | TTGCAGCTTTTCAAGAATGCGC | 1143 | 37 |
| aphA1-Iab | AAACGTCTTGCTCGAGGC | CAAACCGTTATTCATTCGTGA | 500 | 12 |
| aadA | GTGGATGGCGGCCTGAAGCC | AATGCCCAGTCGGCAGCG | 528 | 19 |
| aadA2 | CGGTGACCATCGAAATTTCG | CTATAGCGCGGAGCGTCTCGC | 250 | AF071555 |
| strA | CCTGGTGATAACGGCAATTC | CCAATCGCAGATAGAAGGC | 548 | 19 |
| tetA(A) | GCTACATCCTGCTTGCCTTC | CATAGATCGCCGTGAAGAGG | 210 | X61367 |
| tetA(B) | TTGGTTAGGGGCAAGTTTTG | GTAATGGGCCAATAACACCG | 659 | 18 |
| tetA(G) | GCTCGGTGGTATCTCTGC | AGCAACAGAATCGGGAAC | 464 | S52437 |
| sulI | CTTCGATGAGAGCCGGCGGC | GCAAGGCGGAAACCCGCGCC | 435 | 31 |
| Class 1 integron | GGCATCCAAGCACAAGC | AAGCAGACTTGACTGAT | Variablea | 18 |
| qacE | ATCGCAATAGTTGGCGAAGT | CAAGCTTTTGCCCATGAAGC | 210 | 31 |
The amplicon size of class 1 integron may be variable, depending on the sizes and the numbers of the gene inserts. However, if no resistance gene cassette is inserted, the expected size is 153 bp.
DNA sequence analysis.
The amplicons of the class 1 integrons of MDR Salmonella serovar Muenchen isolates from swine obtained by PCR were sequenced. DNA from the amplified products was run on an agarose gel with 1% agarose, and a QIAquick gel extraction kit (Qiagen) was used to purify the DNA. The purified DNA samples were submitted to the University of North Carolina Sequencing Facility for sequencing.
Conjugation assay and plasmid restriction fragment analysis.
Candidate MDR donor strains, Salmonella serovar Muenchen from pigs (strain UCE6), Salmonella serovar Muenchen from humans (strain NCPH3633), and Salmonella serovar Typhimurium from pigs isolated on the same farm as the Salmonella serovar Muenchen strain (strain UCE15), were mated with a spontaneous nalidixic acid-resistant derivative of E. coli K-12 strain MG1655 (strain CA32). The antibiotics and concentrations used in the selective plates were as follows: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; tetracycline, 25 μg/ml; and chloramphenicol, 50 μg/ml. The detailed protocol was as described previously (14). After confirmation that the transconjugants were indeed E. coli (pink colonies on MacConkey agar), the plasmid DNA was prepared by the alkaline lysis method, as described previously (30). Restriction analysis was conducted with four restriction enzymes, EcoRI, BamHI, PstI, and HindIII, with a restriction enzyme kit (Roche Diagnostics, Mannheim, Germany), according to the guidelines of the manufacturer. Plasmid DNA fragments were separated on a 0.7% horizontal agarose gel at 20 mA for 18 h.
Southern hybridization.
Southern hybridization for the acetylphosphotransferase gene (aphA1-Iab), which encodes kanamycin resistance, was carried out by the standard protocol (31). Briefly, purified plasmid DNA was restricted with the EcoRI enzyme for 3 h and was then run overnight on 0.7% agarose in 1× TAE (Tris-acetate-EDTA) buffer. The restricted plasmid DNA was denatured in alkaline transfer buffer and transferred onto a positively charged nylon membrane (Roche Diagnostics) by capillary action. The DNA on the membrane was cross-linked by UV irradiation in a Stratalinker apparatus (Stratagene, La Jolla, Calif.). A digoxigenin (DIG)-labeled aphA1-Iab-specific detection probe was generated according to the directions of the manufacturer (Roche Diagnostics). Prehybridization and hybridization of the membrane with the denatured DIG-labeled probe (20 ng/ml of hybridization solution) were done at 50°C. The DIG-labeled probe that bound to the membrane was detected by chemiluminescence with anti-DIG-alkaline phosphatase and substrate after incubation at 37°C for 10 min.
RESULTS
Salmonella isolation and prevalence of Salmonella serovar Muenchen.
Among the total of 1,314 Salmonella isolates collected from porcine fecal samples, 28 (2%) were found to be of Salmonella serovar Muenchen. These isolates were detected from 5 of the 24 farms sampled during the 3-year study period. All the isolates of porcine origin were collected from the southeastern part of North Carolina, the part of the state where pig production is predominant. Twenty-two of the 28 isolates were collected in 1999 from three farms and were all from apparently healthy pigs. Pigs from all three farms also carried MDR Salmonella serovar Typhimurium strains that commonly had R types AxACSSuT and AKSSuT, as reported elsewhere (15). Among the total of 5,520 Salmonella isolates of human origin submitted to NCSPHL from 1998 to 2002, 141 (2.6%) were found to be of Salmonella serovar Muenchen. The incidence rate ranged between 1 and 3% per year. Thirty-nine randomly selected isolates were submitted to our laboratory for further characterization. These were submitted from counties in all four regions of the state: northwest (n = 8), southwest (n = 9), northeast (n = 2), and southeast (n = 20). About half of the human isolates were from the southeast region, where all the porcine isolates also originated. In addition, one human isolate from a bordering county in the state of Virginia was included.
Antimicrobial susceptibilities of porcine and human Salmonella serovar Muenchen isolates.
All Salmonella serovar Muenchen isolates from pigs and humans (n = 67; 28 swine isolates and 39 human isolates) that were collected during the 3-year period were characterized by antimicrobial susceptibility testing. Resistance to 10 of the 12 antimicrobial agents tested was detected, as summarized in Table 2. All isolates were susceptible to ciprofloxacin and amikacin. Thirty-five of the human isolates (89%) were susceptible to all 12 antimicrobials tested. In contrast, only 15% of the porcine isolates were pansusceptible (Table 2). Eight different resistance patterns were detected, among which four were MDR patterns, with resistance patterns closely similar to that of Salmonella serovar Typhimurium phage type DT104 (R type ACSSuT) with or without resistance to additional antimicrobials. Of the 28 isolates of porcine origin, 21 (75%) showed resistance to seven antimicrobial agents (R type ACSSuTAxK), which was similar to the two most common MDR Salmonella serovar Typhimurium R types reported in the same groups of pigs (AxACSSuT and AKSSuT). The porcine MDR isolates were collected from three geographically distinct swine farms that were owned by the same production company. This could be due to multiple factors, including the common management and antimicrobial use practices within this production system or cross contamination among the three farms by farm workers, or the strains might have been persistent and originated from the breeding herd in the production pyramid. One isolate from humans (isolate NCPH3633) showed resistance to 10 of the 12 antimicrobial agents and had the ACSSuTAxKGCfCro R type. This R type is similar to that of MDR strains carrying AmpC previously described predominantly among Salmonella serovar Newport and Typhimurium isolates.
TABLE 2.
Frequency of antimicrobial resistance among porcine and human Salmonella serovar Muenchen isolates
| Strain source | No. (%) of resistant isolates
|
R types (no. of isolates) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | S | Su | T | Ax | Cf | Cro | K | G | ||
| Porcine (n = 28) | 22 (79) | 22 (79) | 21 (75) | 22 (79) | 24 (86) | 22 (79) | 0 (0) | 0 (0) | 21 (75) | 0 (0) | ACSSuTAxK (20), ACSSuTAxKPi (1), ACSuTAx (1), T (2), pansusceptible (4) |
| Human (n = 39) | 1 (3) | 1 (3) | 4 (10) | 2 (5) | 2 (5) | 1 (3) | 1 (3) | 1 (3) | 1 (3) | 1 (3) | Pansusceptible (35), S (2), SSuT (1), ACSSuTAxKGCfCro (1) |
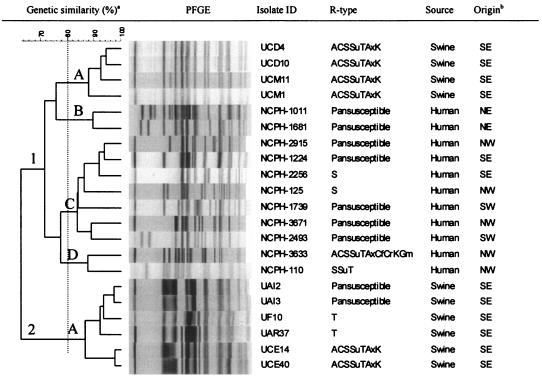
PFGE fingerprinting.
To determine whether there is clonal relationship within and between Salmonella serovar Muenchen isolates of porcine and human origin, particularly the MDR strains, a total of 21 isolates (10 porcine isolates and 11 human isolates) were analyzed by PFGE. PFGE grouped the isolates into two major clusters (clusters 1 and 2) (Fig. 1). Cluster 1 had four subgroups at the 80% genetic similarity threshold. Of the four subgroups in this cluster, three (subgroups 1-B, 1-C, and 1-D) contained isolates exclusively of human origin. The second cluster (cluster 2-A) contained only porcine isolates. Overall, the human and porcine isolates demonstrated unique PFGE patterns. The highly MDR strain from humans (strain NCPH3633), with resistance to 10 antimicrobial agents (R type ACSSuTAxKGCfCro), was clustered with another MDR strain (subgroup 1-D) from humans (strain NCPH110 with R type SSuT). This strain was not clonally related to the MDR strain of porcine origin (R type ACSSuTAxK).
FIG. 1.
PFGE fingerprints of 21 S. enterica serovar Muenchen isolates from swine and humans. a, two major clusters are shown. The vertical dotted line at 80% genetic similarity was considered an arbitrary threshold for grouping of the clusters into subgroups. Clusters 1 (with four subgroups, subgroups 1-A, 1-B, 1-C, and 1-D) and 2 (with one subgroup, subgroup 2-A) are shown. b, origin shows the different regions of North Carolina where the samples originated (NE, northeastern; SE, southeastern; NW, northwestern; SW, southwestern). All the swine isolates originated from the southeastern region of the state, the part of the state where pig production predominates.
Analysis of resistance genes and class 1 integrons.
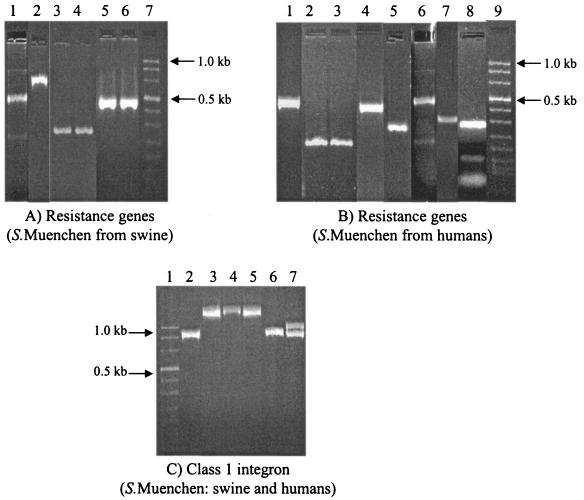
The swine isolates with the ACSSuTAxK R type were similar to Salmonella serovar Typhimurium isolates (R types AxACSSuT and AKSSuT) and predominantly carried resistance genes similar to those carried by Salmonella serovar Typhimurium R type AKSSuT, the aadA2, aphA1-Iab, and tetA(B) genes (Fig. 2). However, none of the β-lactamase genes known to exist in either of the MDR Salmonella serovar Typhimurium strains were detected. The highly MDR strain from humans (strain NCPH3633, R type ACSSuTAxKGCfCro) carried distinctly different genes, including blaTEM, tetA(A), and strA (Fig. 2). None of the Salmonella serovar Muenchen isolates (neither porcine nor human isolates) were found to carry blaCMY-2, blaPSE1, aadA, and tetA(G). All the MDR isolates tested were found to carry class 1 integrons. However, their sizes varied. The swine isolates with R type ACSSuTAxK carried one class 1 integron of 2.0 kb. The human isolate with R type ACSSuTAxKGCfCro carried one integron of 1.0 kb (Fig. 2). By DNA sequencing of the class 1 integron insert, we found that MDR Salmonella serovar Muenchen porcine isolates carried blaOXA-30, in addition to aadA2, on a 2.0-kb class 1 integron. DNA sequencing of the 2.0-kb class 1 integron insert in the swine MDR Salmonella serovar Muenchen isolates revealed blaOXA-30, in addition to the aadA2 gene.
FIG. 2.
PCR amplification of antimicrobial resistance genes and class 1 integron variable region among MDR Salmonella serovar Muenchen isolates from swine and humans. (A) Lanes: 1, sul1; 2, tetA(B); 3 and 4, qacE; 5 and 6, aphA1-Iab; and 7, molecular size marker (0.019 to 1.11 kb). (B) Lanes: 1, tetA(G); 2 and 3, tetA(A) (strains NCPH3633 and NCPH110); 4, strA; 5, qacE; 6, sul1; 7,blaTEM; 8, aadA2; and 9, molecular size marker. (C) Lanes: 1, molecular size marker; 2, strain NCPH3633 (a human Salmonella serovar Muenchen isolate of R type ACSSuTAxKGCfCro), 3, isolate UCE1; 4, isolate UCE3; 5, isolate UCM6 (the isolates in lanes 3, 4, and 5 are swine Salmonella serovar Muenchen isolates of R type ACSSuTAxK); 6, isolate UT30 (Salmonella serovar Typhimurium phage type DT193, swine isolate of R type AKSSuTG); and 7, isolate UW05 (a Salmonella serovar Typhimurium phage type DT104 swine isolate of R type AxACSSuT).
Conjugation assay and plasmid restriction analysis.
To compare the MDR Salmonella serovar Muenchen strains from the two species of hosts, we evaluated the strains for the presence of conjugative plasmids and then compared their sizes and banding patterns using restriction analysis. We also performed the same conjugation and plasmid restriction analysis for porcine isolates of Salmonella serovar Typhimurium in order to compare the plasmid profiles with those of the Salmonella serovar Muenchen isolates. This analysis was conducted with a total of eight isolates, including six porcine Salmonella serovar Typhimurium isolates (isolates UCE15, UCE18, UBC4, UBC5, and UBL6, all of which were R type AKSSuT, and isolate UCM8, R type ACSSuTAx), one porcine Salmonella serovar Muenchen isolate (isolate UCE6, R type ACSSuTAxK), and one human Salmonella serovar Muenchen isolate (isolate NCPH3633, R type ACSSuTAxKGCfCro).
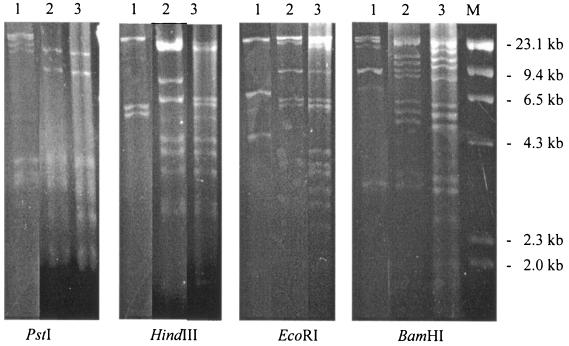
Both the human and the swine strains carried the majority of their resistance genes on self-transferable plasmids. In the MDR isolate from swine of R type ACSSuTAxK (isolate UCE6), the kanamycin and tetracycline resistance markers were readily transferred by conjugation and, thus, were encoded on conjugative or mobilizable plasmids. For the human strain (strain NCPH3633) of R type ACSSuTAxKGCfCro, all four resistance markers for which tests were conducted, including markers for ampicillin, chloramphenicol, kanamycin, and tetracycline resistance, were found to be located on conjugative or mobilizable plasmids; and all resistance markers transferred together. Plasmid restriction analysis of these MDR Salmonella serovar Muenchen isolates with four restriction enzymes, EcoRI, BamHI, PstI, and HindIII, revealed that the plasmid of porcine origin (plasmid pUCE6) had a restriction pattern more closely related to that of a plasmid of an isolate of Salmonella serovar Typhimurium R type AKSSuT (plasmid pUCE15) collected from the same farm at the same time (Fig. 3) than to that of the plasmid from the human serovar Muenchen strain (strain NCPH3633). The plasmids from both porcine Salmonella serovar Muenchen isolates and porcine Salmonella serovar Typhimurium isolates had an estimated size of about 140 kb. The MDR Salmonella serovar Muenchen strain of human origin appeared to have carried a plasmid different from that carried by the MDR porcine Salmonella serovar Muenchen strain, and the restriction patterns were distinctly different in both restriction enzyme analyses (Fig. 3). It also carried a smaller plasmid with an estimated size of 85 kb.
FIG. 3.
Plasmid restriction analysis with four restriction enzymes: PstI, HindIII, EcoRI, and BamHI. Lanes: 1, Salmonella serovar Muenchen (human origin, p3633, R type ACSSuTAxKGCfCro); 2, Salmonella serovar Typhimurium (porcine origin, pUCE15, R type AKSSuT); 3, Salmonella serovar Muenchen (porcine origin, pUCE6, R type ACSSuTAxK); and M, molecular size marker. Relatively more similar plasmid restriction patterns were detected between Salmonella serovar Muenchen and Typhimurium isolates of porcine origin than Salmonella serovar Muenchen isolates of swine and human origin.
Southern hybridization.
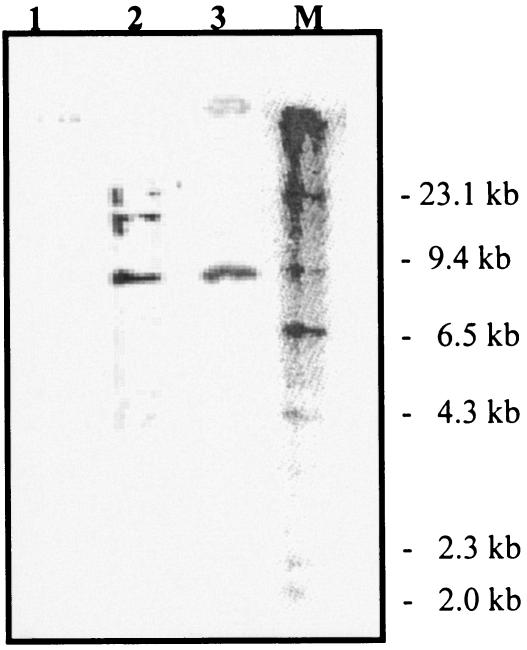
In order to confirm the relatedness of plasmids from the two serovars (serovars Typhimurium and Muenchen) of porcine origin, we performed Southern hybridization using a gene probe that has been known to be common to both MDR strains. As shown in Fig. 4, the DIG-labeled probe specific for the aphA1-Iab gene hybridized at the same location (9.5 kb) in both MDR Salmonella serovar Typhimurium and MDR Salmonella serovar Muenchen strains isolated from the same farm. This finding further supports the close relatedness of the plasmids in isolates of the two serovars.
FIG. 4.
Southern hybridization of three strains: lane 1, Salmonella serovar Muenchen (human origin, p3633, R type ACSSuTAxKGCfCro); lane 2, Salmonella serovar Typhimurium (porcine origin, pUCE15, R type AKSSuT); lane 3, Salmonella serovar Muenchen (porcine origin, pUCE6, R type ACSSuTAxK), and lane M, molecular size marker. A DIG-labeled acetylphosphotransferase gene (aphA1-Iab) was used. Both MDR Salmonella serovar Typhimurium and MDR Salmonella serovar Muenchen isolates of porcine origin had the gene on the same restriction fragment, which was 9.5 kb.
DISCUSSION
Salmonella serovar Muenchen is among the top 10 causes of clinical salmonellosis in humans in the United States. In the present study, 2% of the Salmonella isolates of porcine origin collected during a 3-year epidemiological study in North Carolina were found to be of Salmonella serovar Muenchen. A similar level of prevalence of Salmonella serovar Muenchen among isolates from human salmonellosis cases was also reported in North Carolina during the same period, with an overall prevalence of 2.6% and an incidence rate of 1 to 3% per year. These findings are similar to the prevalence of Salmonella serovar Muenchen isolates among human cases of salmonellosis reported at the national level. According to CDC (10), an approximately 2% prevalence of Salmonella serovar Muenchen is reported at the national level. In the present study, the majority of the isolates of human origin were found to be pansusceptible, whereas 75% of the isolates of porcine origin showed an MDR pattern with an R type of ACSSuTAxK. A high proportion of swine isolates showed MDR phenotypes. However, these isolates were clustered on a few farms (3 of 24 farms sampled) in the southeastern part of the state, the part of the state where pig production predominates. Although these isolates were identified from only a few farms in the present study, they could pose a potential risk for dissemination to other areas and host species, as documented previously (8, 28, 33).
As reported previously (15), resistant strains were more commonly isolated from the pigs in the production system that used antimicrobials, including β-lactams and drugs of the tetracycline class, more often than from the pigs in another type of large production system. On the basis of the antimicrobial susceptibility patterns, the isolates from humans and swine were different from each other to a large extent. We identified one isolate from a human that was resistant to 10 of the 12 antimicrobials to which isolate resistance was tested. That strain showed resistance to an expanded-spectrum cephalosporin, ceftriaxone, which is the first-line antimicrobial agent used against invasive infections caused by gram-negative organisms. None of the porcine isolates (MDR or pansusceptible) showed resistance to ceftriaxone, although the therapeutic use of ceftiofur, an expanded-spectrum cephalosporin, in swine has been approved since 1995, and ceftiofur was also reportedly used in 16 of the 24 pig groups included in this study (15).
On the basis of PFGE fingerprinting, the human and porcine isolates overall exhibited different PFGE patterns. All five clonally related subgroups (subgroups 1-A to 1-D and 2-A) had isolates derived from a single host (Fig. 1). Cluster 1 predominantly constituted isolates from humans, and one subgroup (1-A) was composed of isolates from swine. The human strain with resistance to 10 of the 12 antimicrobial agents was also grouped with an MDR strain (strain NCPH110) with a different R type, SSuT.
The present findings show that the porcine isolates were not clonally related to Salmonella serovar Muenchen isolates from humans in the study area. The sources of Salmonella isolates from swine and humans were not epidemiologically related, although they originated from similar geographical locations during similar time periods. However, the finding that the isolates from two sources have distinct PFGE patterns is of interest. Salmonella serovars have previously been shown to be clonally disseminated, and often, even epidemiologically unrelated members of the same serovar and phenotype have exhibited similar clonalities on the basis of their PFGE patterns and other approaches to the study of molecular epidemiology (3).
One important finding in the present study is that Salmonella serovar Muenchen isolates of porcine origin had an MDR pattern (R type ACSSuTAxK) similar to those previously reported for Salmonella serovar Typhimurium strains. On all the pig farms where MDR Salmonella serovar Muenchen isolates of R type ACSSuTAxK were identified, we also detected two MDR Salmonella serovar Typhimurium strains of R types AxACSSuT and AKSSuT. These two R types, particularly ACSSuT, have been reported among phage type DT104 isolates that are pandemically disseminated and that are known to cause food-borne illnesses globally (4, 5, 21, 22, 29, 35). R type AKSSuT, which is often associated with phage type DT193, has also been reported at an increasing frequency in recent years by the National Antimicrobial Resistance Monitoring System, in a 2001 CDC report, and in other studies (2, 14, 23, 36). Detailed reports of the phenotypic and genotypic characteristics of the Salmonella serovar Typhimurium strains have been reported elsewhere (14, 15).
We hypothesized that the genetic elements among the MDR Salmonella serovar Muenchen isolates of porcine origin (R type ACSSuTAxK) are related to the two MDR Salmonella serovar Typhimurium strains (R types AxACSSuT and AKSSuT) and could be due to reassortment of the genetic elements for resistance in the two MDR Salmonella serovar Typhimurium strains. In order to prove this hypothesis, we investigated resistance genes, class 1 integrons, and conjugative plasmids and performed plasmid restriction analysis. As reported in Results, the MDR Salmonella serovar Muenchen strain predominantly appeared to have resistance genes similar to those of Salmonella serovar Typhimurium R type AKSSuT. The MDR Salmonella serovar Muenchen strain, however, had a unique β-lactamase gene, blaOXA-30, which was integrated into a class 1 integron. The size of the class 1 integron among the MDR Salmonella serovar Muenchen isolates was 2 kb, while that among the Salmonella serovar Typhimurium isolates of R type AKSSuT is 1 kb, as reported previously (14). The 1-kb difference can be accounted for by the insertion of blaOXA-30, a unique β-lactamase gene (15). Similar to the Salmonella serovar Typhimurium strain of R type AKSSuT, the MDR Salmonella serovar Muenchen strain carried most of its resistance genes on a conjugative plasmid, which was found to be identical to the plasmid of the Salmonella serovar Typhimurium strain, based on PstI restriction analysis.
We found that multiple antimicrobial resistance genes between MDR Salmonella serovar Typhimurium (R type AKSSuT) and Salmonella serovar Muenchen had absolute homology, that the genes were carried on readily transferable conjugative plasmids, and that the plasmid sizes and restriction patterns were similar between the two strains. This is despite the fact that plasmids evolve rapidly and show variable characteristics within a short time span (17, 34). These observations, together with the similar antimicrobial resistance patterns and the identification of these strains from the same geographical locations and the same sampling times, support the argument that the horizontal dissemination of plasmids carrying multiple antimicrobial resistance genes may have occurred between Salmonella serovar Typhimurium and Salmonella serovar Muenchen strains. The potential transfer of resistance genes between closely related organisms such as Salmonella and E. coli has been reported previously (37). Further confirmation of such horizontal dissemination of antimicrobial resistance in experimental and on-farm studies is necessary.
None of the isolates with amoxicillin-clavulanic acid resistance, including the MDR strain of porcine origin, carried blaCMY-2, nor did the human strain of R type ACSSuTAxKGCfCro (strain NCPH3633). Extended-spectrum β-lactamase resistance to amoxcillin-clavulanic acid has previously been shown to be encoded by other genes and by the plasmid-mediated overproduction of blaTEM (commonly referred as inhibitor-resistant TEM [IRT]) (11) and blaPSE-1 (27). Neither of these genes was present among the MDR Salmonella serovar Muenchen isolates of porcine origin. The production of OXA-type enzymes has also been shown to be responsible for amoxicillin-clavulanic acid resistance in E. coli (26). Therefore, we believe that the amoxicillin-clavulanic acid resistance in the porcine strains described in this study may be the result of blaOXA-30 overproduction.
Another interesting finding of the present study was the detection of a highly MDR strain of human origin (strain NCPH3633 of R type ACSSuTAxKGCfCro) with ceftriaxone resistance and an R type that resembles that of MDR isolates carrying AmpC previously reported among Salmonella serovar Newport and Typhimurium strains (1, 6, 16). However, we confirmed that none of the MDR Salmonella serovar Muenchen isolates tested carried blaCMY-2. blaCMY-2 is a member of the plasmid-mediated AmpC-like enzyme family has been shown to be the most common gene associated with ceftriaxone resistance among Salmonella serovars, particularly Salmonella serovars Newport and Typhimurium, in food animals and humans (1, 6, 16). Therefore, we believe that this resistance phenotype might be encoded by other relatively uncommon cephalomycinase (CMY) alleles or may be a result of overexpression of the blaTEM gene carried by this strain (P. L. Winokur, personal communication). The absence of blaCMY-2 also implies that the factors involved in the MDR of this strain are different from the factors involved in the MDR of the epidemic strains carrying AmpC reported among the other serovars. Investigation of the identity of the gene and further characterization are under way.
Even though the samples from swine and humans were not epidemiologically related and were generated on the basis of convenience sampling, the detection of what appears to be a common MDR strain in humans and pigs and the close similarity of the molecular relationship between Salmonella serovars Typhimurium and Muenchen are significant. To our knowledge, this is the first study to show the occurrence of MDR Salmonella serovar Muenchen of porcine origin as well as the occurrence of expanded-spectrum cephalosporin resistance and resistance to multiple other antimicrobials among Salmonella serovar Muenchen isolates from humans and food animals. This study implies that, in addition to the fact that the use of antimicrobials in food animals as well as humans presents a crucial selective pressure that could result in the emergence and persistence of antimicrobial resistance, other factors, such as the existence of distinct animal reservoirs and the specificities of strains for certain hosts, could also play major roles. The MDR strains from human and porcine hosts in this study did not appear to be clonally related, suggesting the presence of another reservoir for MDR isolates, including humans themselves. This could be an important factor for the persistence of such strains in the human population. However, both serovars independently represent a potential public health risk, as food-borne salmonellosis is among the leading means of antimicrobial resistance dissemination between humans and food animals.
Acknowledgments
This work was funded by the North Carolina Agromedicine Institute.
We thank Leslie Wolf (NCSLPH) for providing isolates from humans. We also thank Paul Orndorff for critical review of the manuscript.
REFERENCES
- 1.Allen, K. J., and C. Poppe. 2002. Occurrence and characterization of resistance to extended-spectrum cephalosporins mediated by beta-lactamase CMY-2 in Salmonella isolated from food-producing animals in Canada. Can. J. Vet. Res. 66:137-144. [PMC free article] [PubMed] [Google Scholar]
- 2.Baggesen, D. L., and F. M. Aarestrup. 1998. Characterisation of recently emerged multiple antibiotic-resistant Salmonella enterica serovar Typhimurium DT104 and other multiresistant phage types from Danish pig herds. Vet. Rec. 143:95-97. [DOI] [PubMed] [Google Scholar]
- 3.Beltran, P., J. M. Musser, R. Helmuth, J. J. Farmer III, W. M. Frerichs, I. K. Wachsmuth, K. Ferris, A. C. McWhorter, J. G. Wells, and A. Cravioto. 1988. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc. Natl. Acad. Sci. USA 85:7753-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besser, T. E., C. C. Gay, J. M. Gay, D. D. Hancock, D. Rice, L. C. Pritchett, and E. D. Erickson. 1997. Salmonellosis associated with S. Typhimurium DT104 in the USA Vet. Rec. 140:75. [PubMed] [Google Scholar]
- 5.Briggs, C. E., and P. M. Fratamico. 1999. Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43:846-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli, A., F. Tosini, W. P. Giles, M. E. Rupp, S. H. Hinrichs, F. J. Angulo, T. J. Barrett, and P. D. Fey. 2002. Characterization of plasmids carrying CMY-2 from expanded-spectrum cephalosporin-resistant Salmonella strains isolated in the United States between 1996 and 1998. Antimicrob. Agents Chemother. 46:1269-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carlson, S. A., L. F. Bolton, C. E. Briggs, H. S. Hurd, V. K. Sharma, P. J. Fedorka-Cray, and B. D. Jones. 1999. Detection of multiresistant Salmonella typhimurium DT104 using multiplex and fluorogenic PCR. Mol. Cell. Probes 13:213-222. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 1999. Outbreak of Salmonella serotype Muenchen infections associated with unpasteurized orange juice—United States and Canada, June 1999. Morb. Mortal. Wkly. Rep. 48:582-585. [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2000. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: a manual. National Center for Infectious Diseases, Centers for Disease Control and Prevention, Atlanta, Ga.
- 10.Centers for Disease Control and Prevention, Public Health Laboratory Information Systems. 2001. Salmonella: annual report. [Online.] http://www.cdc.gov/ncidod/dbmd/phlisdata/salmtab/2001/SalmonellaAnnualSummary2001.pdf. Accessed 15 September 2004.
- 11.Espinasse, F., R. Gheorghiu, A. Poiata, R. Labia, and M. H. Nicolas-Chanoine. 1997. Reduced susceptibility to co-amoxiclav in Escherichia coli, Salmonella typhimurium and Klebsiella pneumoniae isolated in Romania between 1985 and 1993. J. Antimicrob. Chemother. 39:103-106. [DOI] [PubMed] [Google Scholar]
- 12.Frana, T. S., S. A. Carlson, and R. W. Griffith. 2001. Relative distribution and conservation of genes encoding aminoglycoside-modifying enzymes in Salmonella enterica serotype Typhimurium phage type DT104. Appl. Environ. Microbiol. 67:445-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Funk, J. A., P. R. Davies, and W. A. Gebreyes. 2001. Risk factors associated with Salmonella enterica prevalence in three-site production systems in North Carolina, USA. Berl. Munch. Tierarzt. Wochenschr. 114:335-338. [PubMed] [Google Scholar]
- 14.Gebreyes, W. A., and C. Altier. 2002. Molecular characterization of multidrug resistant Salmonella enterica subsp. enterica serovar Typhimurium isolates from swine. J. Clin. Microbiol. 40:2813-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebreyes, W. A., S. Thakur, P. R. Davies, J. A. Funk, and C. Altier. 2004. Trends in antimicrobial resistance, phage types and integrons among Salmonella serotypes from pigs, 1997-2000. J. Antimicrob. Chemother. 53:997-1003. [DOI] [PubMed] [Google Scholar]
- 16.Gupta, A., J. Fontana, C. Crowe, B. Bolstorff, A. Stout, S. Van Duyne, M. P. Hoekstra, J. M. Whichard, T. J. Barrett, F. J. Angulo, and The National Antimicrobial Resistance Monitoring System PulseNet Working Group. 2003. Emergence of multidrug-resistant Salmonella enterica serotype Newport infections resistant to expanded-spectrum cephalosporins in the United States. J. Infect. Dis. 188:1707-1716. [DOI] [PubMed] [Google Scholar]
- 17.Kron, M. A., D. M. Shlaes, C. Currie-McCumber, and A. A. Medeiros. 1987. Molecular epidemiology of OHIO-1 beta-lactamase. Antimicrob. Agents Chemother. 31:2007-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai-King, N. G., M. R. Mulvey, I. Martin, G. A. Peters, and W. Johnson. 1999. Genetic characterization of antimicrobial resistance in Canadian isolates of Salmonella serovar Typhimurium DT104. Antimicrob. Agents Chemother. 43:3018-3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Madsen, L., F. M. Aarestrup, and J. E. Olsen. 2000. Characterisation of streptomycin resistance determinants in Danish isolates of Salmonella Typhimurium. Vet. Microbiol. 75:73-82. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita, S., S. Yamada, M. Inaba, J. Kusunoki, Y. Kudoh, and M. Ohashi. 1992. Serovar distribution and drug resistance of Salmonella isolated from imported and domestic cases in 1980-1989 in Tokyo. Kansenshogaku Zasshi 66:327-339. [DOI] [PubMed] [Google Scholar]
- 21.Metzer, E., V. Agmon, N. Andoren, and D. Cohen. 1998. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium phage-type DT104 among Salmonellae causing enteritis in Israel. Epidemiol. Infect. 121:555-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molbak, K., D. L. Baggesen, F. M. Aaerestrup, J. M. Ebbessen, J. Engberg, K. Frydendahl, P. Gerner-Smidt, A. M. Petersen, and H. C. Wegener. 1999. An outbreak of multidrug-resistant Salmonella enterica serotype Typhimurium DT104. N. Engl. J. Med. 341:1420-1425. [DOI] [PubMed] [Google Scholar]
- 23.National Antimicrobial Resistance Monitoring System, Centers for Disease Control and Prevention. 2003. Annual report, 2001 [Online.] http://www.cdc.gov/narms/annual/2001/annual_01.htm. Accessed 15 September 2004.
- 24.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial disc and dilution susceptibility tests for bacteria isolated from animals; approved standard, 2nd ed., M31-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 25.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing; 12th informational supplement, M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 26.Nicolas-Chanoine, M. H. 1997. Inhibitor-resistant beta-lactamases. J. Antimicrob. Chemother. 40:1-3. [DOI] [PubMed] [Google Scholar]
- 27.Poirel, L., M. Guibert, S. Bellais, T. Naas, and P. Nordmann. 1999. Integron- and carbenicillinase-mediated reduced susceptibility to amoxicillin-clavulanic acid in isolates of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 from French patients. Antimicrob. Agents Chemother. 43:1098-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Proctor, M. E., M. Hamacher, M. L. Tortorello, J. R. Archer, and J. P. Davis. 2001. Multistate outbreak of Salmonella serovar Muenchen infections associated with alfalfa sprouts grown from seeds pretreated with calcium hypochlorite. J. Clin. Microbiol. 39:3461-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribot, E. M., R. K. Wierzba, F. J. Angulo, and T. J. Barrett. 2002. Salmonella enterica serotype Typhimurium DT104 isolated from humans, United States, 1985, 1990, and 1995. Emerg. Infect. Dis. 8:387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Preparation of plasmid DNA by alkaline lysis with SDS: minipreparation, p. 1.32-1.34. In Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1998. Characterization of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 160:37-41. [DOI] [PubMed] [Google Scholar]
- 32.Silverstolpe, L., J. Kjelland, U. Plazikow, and G. Vahlne. 1961. An epidemic among infants caused by Salmonella Muenchen. J. Appl. Bacteriol. 24:134-196. [Google Scholar]
- 33.Taylor, D. N., I. K. Wachsmuth, Y. H. Shangkuan, E. V. Schmidt, T. J. Barrett, J. S. Schrader, C. S. Scherach, H. B. McGee, R. A. Feldman, and D. J. Brenner. 1982. Salmonellosis associated with marijuana: a multistate outbreak traced by plasmid fingerprinting. N. Engl. J. Med. 306:1249-1253. [DOI] [PubMed] [Google Scholar]
- 34.Thal, L. A., J. W. Chow, J. E. Patterson, M. B. Perri, S. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Molecular characterization of highly gentamicin-resistant Enterococcus faecalis isolates lacking high-level streptomycin resistance. Antimicrob. Agents Chemother. 37:134-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Threlfall, E. J., J. A. Frost, L. R. Ward, and B. Rowe. 1996. Increasing spectrum of resistance in multiresistant Salmonella Typhimurium. Lancet 347:1053-1054. [DOI] [PubMed] [Google Scholar]
- 36.Threlfall, E. J., C. J. Teale, R. H. Davies, L. R. Ward, J. A. Skinner, A. Graham, C. Cassar, and K. Speed. 2003. A comparison of antimicrobial susceptibilities in nontyphoidal salmonellas from humans and food animals in England and Wales in 2000. Microb. Drug Resist 9:183-189. [DOI] [PubMed] [Google Scholar]
- 37.Winokur, P. L., D. L. Vonstein, L. J. Hoffman, E. K. Uhlenhopp, and G. V. Doern. 2001. Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45:2716-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]