ABSTRACT
Deep sequencing has revolutionized our understanding of the bacterial RNA world and has facilitated the identification of 280 small RNAs (sRNAs) in Salmonella. Despite the suspicions that sRNAs may play important roles in Salmonella pathogenesis, the functions of most sRNAs remain unknown. To advance our understanding of RNA biology in Salmonella virulence, we searched for sRNAs required for bacterial invasion into nonphagocytic cells. After screening 75 sRNAs, we discovered that the ablation of InvS caused a significant decrease of Salmonella invasion into epithelial cells. A proteomic analysis showed that InvS modulated the levels of several type III secreted Salmonella proteins. The level of PrgH, a type III secretion apparatus protein, was significantly lower in the absence of InvS, consistent with the known roles of PrgH in effector secretion and bacterial invasion. We discovered that InvS modulates fimZ expression and hence flagellar gene expression and motility. We propose that InvS coordinates the increase of PrgH and decrease in FimZ that promote efficient Salmonella invasion into nonphagocytic cells.
IMPORTANCE Salmonellosis continues to be the most common foodborne infection reported by the CDC in the United States. Central to Salmonella pathogenesis is the ability to invade nonphagocytic cells and to replicate inside host cells. Invasion genes are known to be regulated by protein transcriptional networks, but little is known about the role played by small RNAs (sRNAs) in this process. We have identified a novel sRNA, InvS, that is involved in Salmonella invasion. Our result will likely provide an opportunity to better understand the fundamental question of how Salmonella regulates invasion gene expression and may inform strategies for therapeutic intervention.
KEYWORDS: Salmonella, gene regulation, host cell invasion, noncoding RNA
INTRODUCTION
Salmonella enterica serovar Typhimurium (S. Typhimurium) remains a leading cause of foodborne illness and poses a major public health problem worldwide. Salmonella harbors several pathogenicity islands (SPIs) scattered throughout the chromosome, which comprise functionally distinct virulence genes. Virulent Salmonella strains possess pathogenicity island 1 (SPI-1) and pathogenicity island 2 (SPI-2), encoding two separate type III secretion systems (TTSSs). These TTSSs function to deliver bacterial effectors into the host cell to reprogram host cell functions to promote invasion and intracellular survival, respectively (1). Salmonella TTSSs are composed of more than 20 proteins, including a highly conserved group of integral membrane proteins, a family of cytoplasmic chaperones, and several accessory proteins. The core unit of Salmonella SPI-1 TTSS is the needle complex. The multiring base of the complex is anchored to the bacterial envelope, which is composed of proteins, including InvG, PrgH, and PrgK (2). The filamentous needle is composed of PrgI, which is linked to the base by another substructure, the inner rod. It is known that the deletion of PrgH or PrgK impairs the SPI-1 TTSS assembly and hence effector secretion (3, 4). SPI-1 effectors include SSalmonella invasion protein A (SipA), SipB, SipC, SSalmonella outer protein B (SopB), SopD, SopE, and SopE2. These SPI-1 effectors work in concert to rearrange the host actin cytoskeleton to facilitate Salmonella invasion (5). By contrast, SPI-2 effectors are responsible for Salmonella replication inside phagocytic cells to promote bacterial survival and systemic infection.
The process of Salmonella infection of mammals involves the transition of the bacteria through multiple environmental conditions, from the acidity of the stomach to the low-oxygen environment of the gastrointestinal tract. The pathogen relies on an intricate transcriptional network to stimulate invasion when the pathogen interacts with the nonphagocytic cells associated with the gut wall. The SPI-1 invasion genes are tightly regulated by several SPI-1-encoded classic transcription factors. HilC and HilD are two AraC-like transcriptional regulators which activate HilA expression. In turn, HilA, an OmpR/ToxR family member, directly activates the transcription of several SPI-1 operons involved in effector secretion and bacterial invasion. These operons encode the type III secretory apparatus, secreted effectors, and transcriptional regulators such as InvF, an AraC-like transcriptional regulator (6). InvF activates the expression of SPI-1 Salmonella effector genes from a second HilA-independent promoter (7).
Bacterial sRNAs are small (50 to 250 nucleotides) noncoding RNA molecules. sRNAs usually regulate gene expression through base pairing with a corresponding mRNA target(s) and thereby repress or activate the target genes at the posttranscriptional level. Many trans-acting sRNAs require the Hfq RNA chaperone to form stable base pairing with target mRNAs.
Although rapid progress has been made in the identification of novel Salmonella sRNA transcripts (8–11), the majority of the identified sRNAs are of unknown biological function, and very few sRNAs have been shown to play a role in the regulation of Salmonella virulence (12, 13). IsrM is a pathogenicity island-encoded sRNA that is important for bacterial invasion and intracellular replication inside macrophages (14). We hypothesized that a newly discovered sRNA might control the expression of genes required for Salmonella invasion.
Chao et al. have performed deep sequencing of Hfq-bound transcripts from Salmonella and identified 280 sRNAs (8). The majority of those sRNAs have never been functionally characterized. In this study, we surveyed the role of the previously identified sRNAs for their roles in Salmonella invasion. Using an exhaustive screening approach, we discovered that Salmonella sRNA InvS is essential for Salmonella invasion. Several of the type III effector proteins known to be involved in bacterial invasion were secreted at lower levels in the absence of InvS. InvS also modulates the protein levels of PrgH and FimZ, a type III secretion apparatus protein and a negative regulator that suppresses the expression of Salmonella invasion genes, respectively. We suggest that InvS regulates Salmonella invasion via PrgH and FimZ.
RESULTS
InvS is essential for Salmonella invasion.
To identify S. Typhimurium sRNAs involved in bacterial invasion, we generated chromosomal deletions of 75 sRNA-encoding genes in strain SL1344. The resulting null mutant strains were tested for their ability to invade cultured epithelial cells using the classic gentamicin protection assay (see Table S1 in the supplemental material). The deletion of STnc470 had the biggest impact on the invasion of HeLa cells, with a reduction of approximately 70% compared with that of the wild-type strain (Fig. 1). Accordingly, we renamed STnc470 as InvS. The invasion defect of the ΔinvS mutant was restored when InvS was expressed in trans from a plasmid (Fig. 1), proving that InvS is required for efficient Salmonella invasion.
FIG 1.
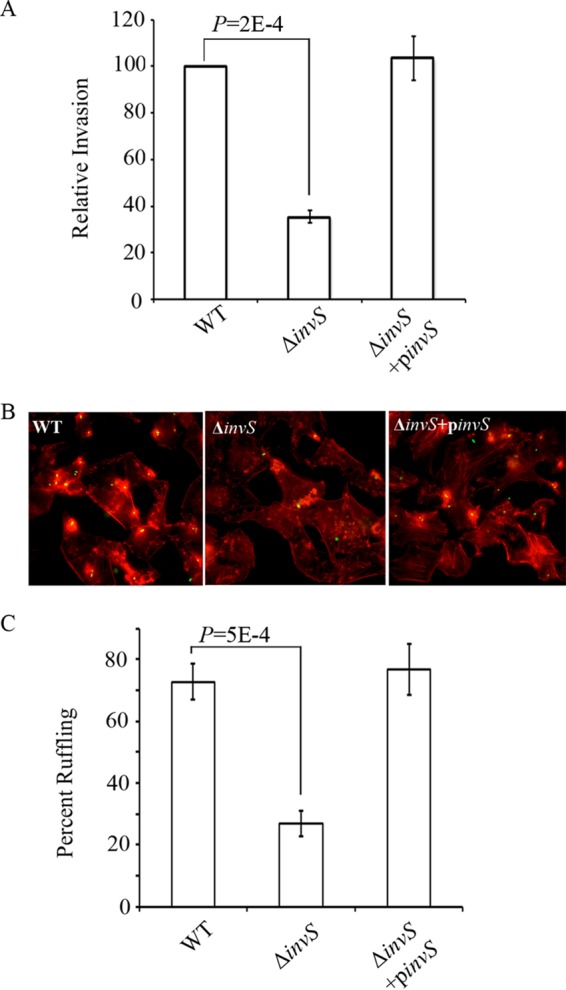
InvS is essential for Salmonella invasion. (A) HeLa cells were infected with Salmonella strains for 15 min at an MOI of 10. Relative bacterial invasion was determined by the gentamicin protection assay as described in Materials and Methods. The invasion rate of the wild-type strain was defined as 100%. The data are the averages from three independent experiments with error bars indicating the standard deviations. (B) HeLa cells were infected with Salmonella for 15 min at an MOI of 10. Actin staining was conducted as described in Materials and Methods to indicate Salmonella-induced ruffling formation. (C) Percentages of infected cells with ruffles were calculated. The data shown were obtained from three independent experiments. Error bars indicate standard deviations. P values were calculated using the Student t test.
InvS is an 89-nucleotide sRNA that was first identified as STnc470 (11). Further characterization has shown that InvS binds Hfq and is derived from the 3′ untranslated region (UTR) of srfN (STM0082) (8). A Northern blot analysis confirmed the size of InvS and that it is cotranscribed with srfN and present as a discrete transcript, consistent with processing of the transcript (Fig. 2).
FIG 2.
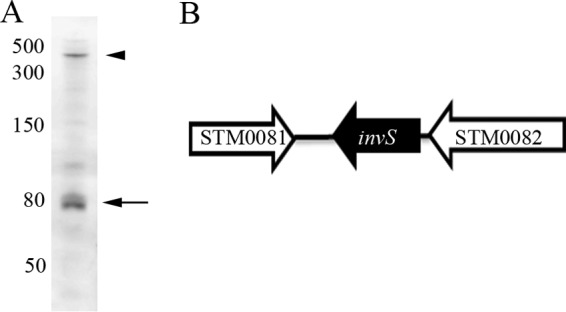
Validation of the InvS transcript. (A) Northern blot analysis identification of the InvS transcript. Comparison with comigrating markers suggests that InvS accumulates as an 89-nt transcript (arrow). Arrowhead, STM0082-STn470 mRNA (400 nt). RNA was isolated from S. Typhimurium 4/74 grown to early stationary phase in LB medium. (Republished from the Proceedings of the National Academy of Sciences [10].) (B) Schematic diagram showing that InvS sRNA is encoded in the 3′ UTR of STM0082.
Overexpression of HilD or InvF rescues the ΔinvS invasion defect.
SPI-1 genes encode proteins involved in the secretion and injection of bacterial effectors into the host cell that promote Salmonella invasion (15, 16). The expression of SPI-1 genes is tightly regulated by a number of transcriptional regulators, including HilD, HilA, and InvF. HilD activates HilA, which in turn upregulates the expression of genes encoding the TTSS, such as proteins encoded by the prg-org and the inv-spa operons (6, 17, 18). The first gene of the inv-spa gene cluster encodes the AraC-like regulator InvF, which activates the expression of genes encoding secreted effectors that are essential for Salmonella invasion, including the sic-sip operon, sopE, and sopB (19).
As a first step toward understanding how InvS facilitates Salmonella invasion, we looked for a role of InvS in the regulation of the transcription factors mentioned above. For this, we tested whether the overexpression of these regulators would rescue the invasion defect of the ΔinvS mutant. HeLa cells were infected with wild-type Salmonella, a ΔinvS null mutant, or the ΔinvS mutant strain expressing one of the following regulators from a plasmid: HilA, HilC, SirA, HilD, or InvF. Invasion rates were assessed using the classic gentamicin protection assay. We found that the overexpression of HilD or InvF restored the invasion defect of the ΔinvS mutant (Fig. 3A) while HilA, HilC, or SirA did not. Our gentamicin protection assay showed plasmids expressing HilA (philA), HilC (philC), and SirA (psirA) were able to restore the invasion deficiency of ΔhilA, ΔhilC, and ΔsirA mutant strains, which indicates that the plasmids are functional (Fig. 3B).
FIG 3.
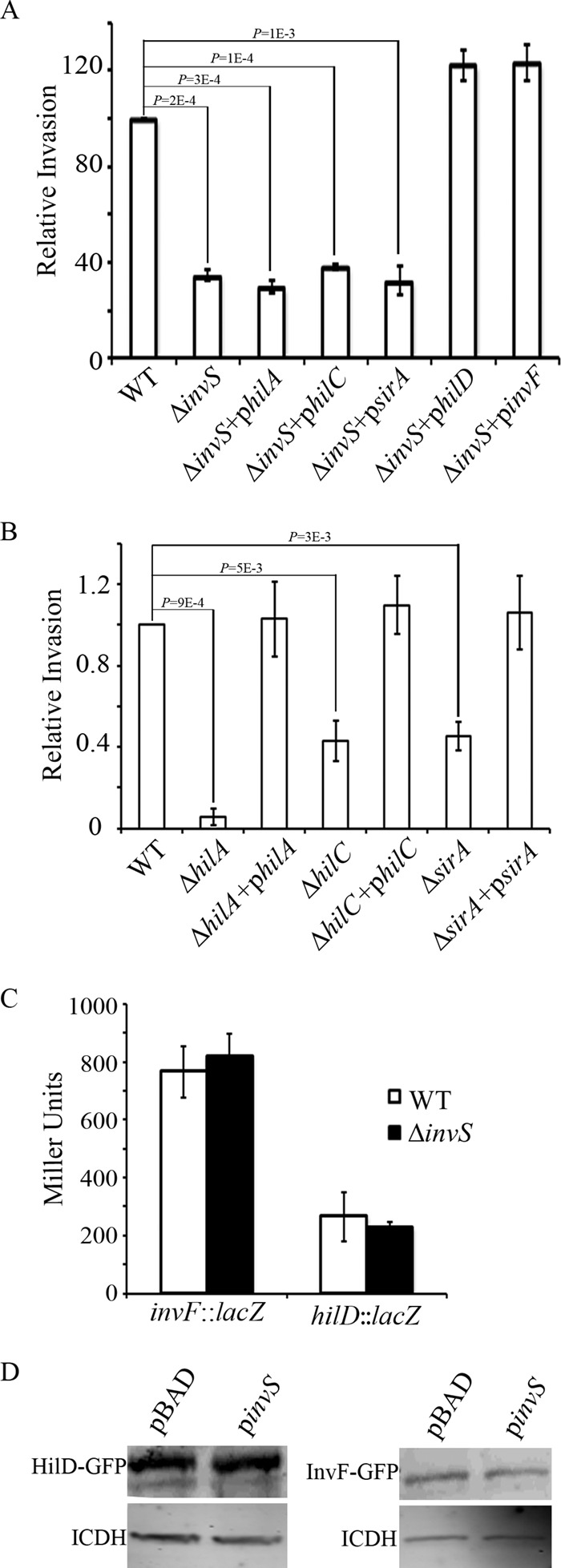
Overexpression of hilD or invF rescues the ΔinvS mutant invasion defect. (A, B) HeLa cells were infected with the indicated Salmonella strains for 15 min at an MOI of 10. Relative bacterial invasion was determined by the gentamicin protection assay. The data shown were obtained from three independent experiments. Error bars indicate standard deviations. P values were calculated using the Student t test. (C) InvS does not change the expression level of hilD or invF. The lacZ reporter gene was placed under transcriptional control of the hilD or invF promoter in either the wild type or the ΔinvS mutant Salmonella strain. β-Galactosidase activity assay was measured as described in Materials and Methods. The data shown were obtained from three independent experiments. Results are presented as the means in Miller units. Error bars indicate standard deviations. (D) InvS does not change the expression of hilD-gfp or invF-gfp. The 5′ UTR along with the full ORFs of HilD and InvF were translationally fused to GFP. Plasmids expressing HilD-GFP or InvF-GFP were cotransformed with plasmids expressing InvS or a vector control as indicated. HilD-GFP or InvF-GFP was detected by Western blotting with polyclonal anti-GFP antibodies. Bacterial isocitrate dehydrogenase (ICDH) was similarly detected using anti-ICDH polyclonal antibodies as the loading control.
It is also reported that HilD is able to activate the transcription of invF from a promoter that is far upstream of its HilA-dependent promoter (6). The loss of HilD results in a more severe effect on invasion than the loss of HilA (6). Using a β-galactosidase fusion, we showed that overexpressing HilD activated invF expression more profoundly than overexpressing HilA. Thus, overexpressing only hilA may not be sufficient to restore the invasion phenotype of the ΔinvS mutant.
We next explored whether InvS regulates the transcription of hilD or invF. A lacZ reporter gene was placed under transcriptional control of the hilD or invF promoter in either the wild type or the ΔinvS mutant Salmonella strain background. The β-galactosidase activities were then monitored under SPI-1-inducing conditions. We found that the expression of lacZ transcribed from the hilD or the invF promoter remained at similar levels in both the wild-type strain and the ΔinvS mutant background (Fig. 3C), suggesting that neither hilD nor invF is regulated by InvS at the transcriptional level. Furthermore, we generated plasmids expressing the HilD-green fluorescent protein (GFP) or the InvF-GFP fusion proteins and monitored their levels in the presence and absence of InvS in E. coli. The HilD-GFP and InvF-GFP levels were not InvS dependent (Fig. 3D). We conclude that hilD and invF are unlikely to be the direct targets of InvS.
Proteomic analysis of secreted proteins with and without InvS.
Type III secreted effector proteins are known to be involved in promoting Salmonella invasion. An altered secretion of these effectors could potentially affect bacterial invasion. To explore whether InvS affected the levels of the secreted proteins and to identify the potential targets of InvS, we carried out a quantitative proteomic analysis using isobaric tags for relative and absolute quantification (iTRAQ) (20) in the wild-type (WT) and ΔinvS null mutant strains. Proteins were assessed from the pellets to determine total expression levels and from the supernatants to identify secreted amounts. In the bacterial pellets, we calculated the relative protein abundance in ΔinvS versus WT. Since iTRAQ has known issues of underestimating fold changes (21), a threshold P value of ≤0.1 in combination with a minimum of a 1.3-fold change in protein abundance was used (a ΔinvS/WT ratio of <0.7 or >1.3 was considered significant). In the bacterial pellets, we detected more than 200 proteins whose abundances were changed in the ΔinvS mutant (see Table S2). While the majority of these were uncharacterized hypothetical proteins or proteins not known to be related to invasion, we detected a significant decrease in flagellar proteins in the ΔinvS mutant compared with that in the wild-type Salmonella. We also found the level of FlhD was markedly decreased in the ΔinvS mutant compared with that in the wild type. FlhD is a transcriptional regulator that is known to regulate flagellar expression to promote Salmonella invasion. Interestingly, FimZ, a regulator known to facilitate fimbrial protein expression and to repress the expression of flagellar genes by binding to the flhD promoter, was found to be 3.8-fold more abundant in the pellet fraction from the ΔinvS mutant than from the wild type. This is consistent with the increased amount of fimbrial proteins and the decrease of flagellar proteins in pellet fractions from the ΔinvS mutant (Table S2). Flagella have been indicated as essential for efficient bacterial adhesion. It is also reported that flagellum-driven motility forces the bacterium into a “near surface swimming” mode, which promotes Salmonella invasion through “scanning” of the host cell surface (22). In addition, FimZ is known to downregulate Salmonella invasion by activating hilE, which represses the expression of several of the Salmonella invasion genes. Thus, we reasoned that InvS may function to downregulate fimZ to promote Salmonella invasion.
We performed a similar analysis on supernatant fractions, with the exception that peptides were not labeled with iTRAQ due to the challenges of consistently derivatizing secreted proteins of low abundance. The analysis of secreted proteins revealed that the amounts of several Salmonella SPI-1 secreted effectors, including SipA, SopA, SipC, and SopB, in the supernatant fractions of the ΔinvS mutant strain were lower than those of the wild-type bacteria. By contrast, the levels of many other Salmonella effector proteins remain unchanged in the pellet fractions in the ΔinvS mutant strain compared with those of the wild-type bacteria (Table S2). These results indicate that InvS might regulate Salmonella effector secretion. In the bacterial pellets, we failed to detect most of the type III apparatus proteins, which might be due to the low abundances of these proteins in the pellet samples.
InvS regulates Salmonella effector secretion.
Our proteomics data suggested that InvS is involved in Salmonella effector secretion. We sought to examine the expression and secretion of SipA, SipB, and SipC, the three main invasion-related effectors, by Western blotting. Consistent with the proteomics results, the InvS null mutant strain secreted dramatically reduced levels of SipA, SipB, and SipC (Fig. 4A and B). By contrast, the expressions of SipA, SipB, and SipC in the cell-associated fraction were unchanged in both the InvS null mutant strain and the wild type. Taken together, we conclude that InvS is important for the secretion of effector proteins.
FIG 4.
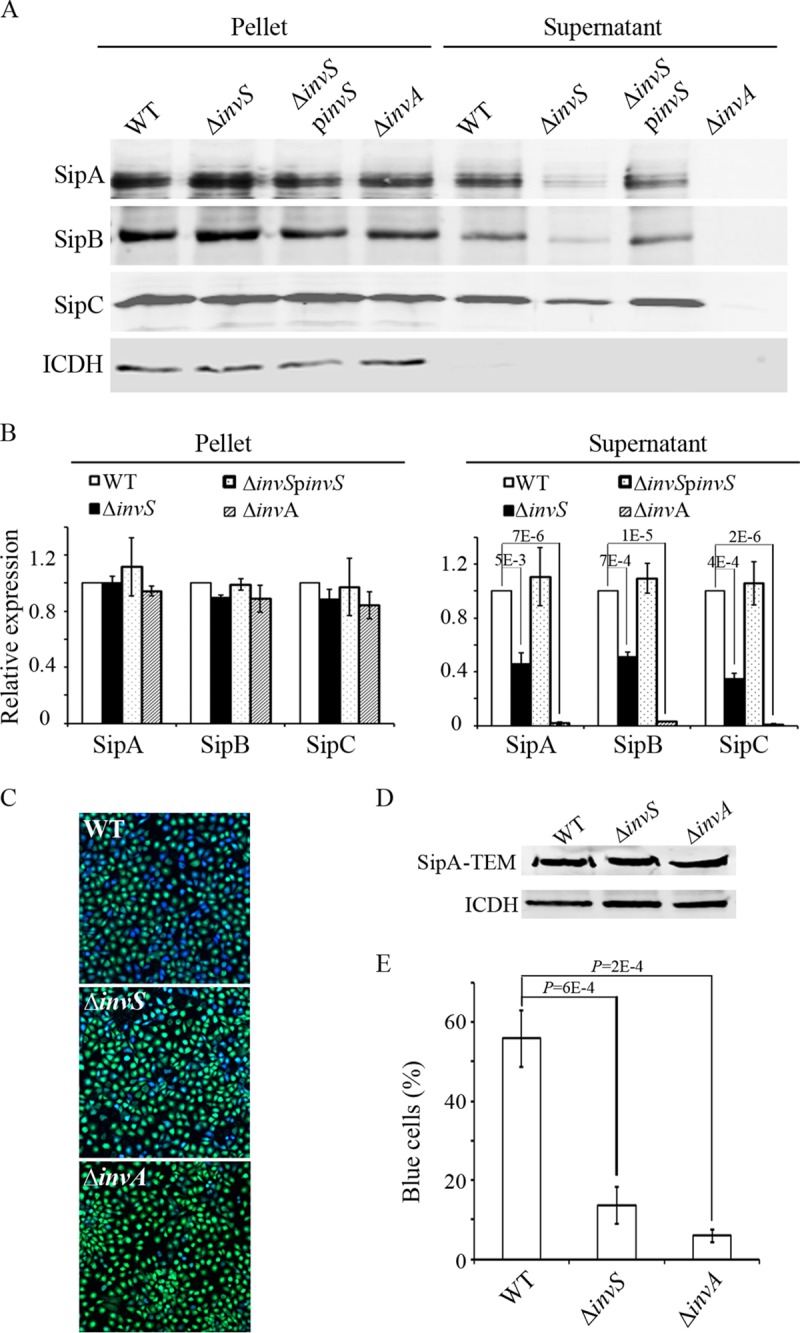
InvS regulates Salmonella effector secretion and translocation. (A) Expression and secretion of invasion-related effectors in Salmonella WT, ΔinvS, ΔinvS/pinvS, and ΔinvA strains. Bacterial strains were grown under SPI-1-inducing conditions and equal amounts of bacterial lysates or culture supernatants were analyzed by Western blotting. (B) Quantification of protein expression in panel A. Protein levels in the WT strain were defined as 1. Values represent relative protein levels after normalization with the expression in the WT. Data are representative of three experiments. (C) HeLa cells were infected with various Salmonella strains carrying a plasmid expressing the SipA-TEM fusion protein. Fifteen minutes after the infection, cells were loaded with CCF4-AM and incubated at room temperature for 2 h. The translocation efficiency was evaluated under a fluorescence microscope. (D) Western blot showing the expression of SipA-TEM in different strains. (E) Quantification of SipA-TEM translocation. Percentages of blue cells were used to measure the translocation efficiency. The data shown were obtained from three independent experiments. Standard deviations are shown. P values were calculated using the Student t test.
To examine if InvS affects Salmonella effector translocation per se, we carried out a β-lactamase-based translocation assay using SipA-TEM1 fusion as a translocation reporter (23). The SipA-TEM1 fusion protein was expressed at similar levels in the wild-type Salmonella and the ΔinvS mutant strain (Fig. 4D). Next, HeLa cells were infected with wild-type Salmonella and the ΔinvS mutant strain expressing SipA-TEM1, and the translocation efficiency was evaluated as previously described (23). As shown in Fig. 4C and E, SipA was translocated at a much lower level from the ΔinvS null mutant than from the wild-type Salmonella. These results support the proteomics data and indicate that InvS is involved in type III effector secretion and translocation during Salmonella infection.
InvS controls the level of PrgH.
One of the possibilities that might lead to the decreased secretion of a group of type III effectors is the dysfunction of the TTSS apparatus. A GFP-based plasmid assay is available to study sRNA-mediated translational control and to verify potential sRNA targets (24). We used GFP translational fusions to determine whether InvS can modulate the levels of the TTSS apparatus proteins SpaO, InvA, PrgK, and PrgH at the posttranscriptional level. The Salmonella invS null mutant strain was transformed with a PBAD-based sRNA expression vector (with or without InvS) and a constitutive GFP fusion expression vector (pXG30) that carried the 5′ UTR and the full open reading frames (ORFs) of SpaO, InvA, PrgK, and PrgH translationally fused to GFP. The expression of the GFP fusion proteins was examined in the presence or absence of InvS. While the levels of SpaO-GFP, InvA-GFP, and PrgK-GFP remain unchanged with and without InvS, the PrgH-GFP level was decreased in the absence of InvS (Fig. 5A and B). When the PrgH-GFP expression plasmid was introduced into Escherichia coli, no PrgH-GFP was detected by Western blotting (data not shown). The lack of PrgH-GFP expression could be because additional Salmonella factors may be required to maintain a higher level of PrgH-GFP in Salmonella. We also transformed pXG30 expressing PrgH-GFP into the WT and ΔinvS strains and examined the expressions of the fusion proteins by Western blotting. We detected a smaller amount of PrgH-GFP in ΔinvS, which further confirmed that InvS functions to upregulate prgH-gfp expression (Fig. 5C). On the other hand, similar levels of prgH promoter activity were detected in the WT and the ΔinvS mutant strain (Fig. 5D), indicating that InvS may indirectly regulate prgH at the posttranscriptional level. To investigate whether the overexpression of PrgH is able to rescue the InvS-dependent invasion phenotype, we overexpressed PrgH in the ΔinvS mutant strain, and its invasion efficiency was found to be partially restored compared with that of the wild-type Salmonella (Fig. 5E). Overexpressing PrgH in the wild-type strain did not significantly influence invasion levels (Fig. 5E). This result suggests that InvS is required for maintaining PrgH expression and Salmonella invasion. The partial rescue of invasion by the overexpression of prgH in the ΔinvS mutant suggests that InvS may influence additional target genes involved in Salmonella invasion.
FIG 5.
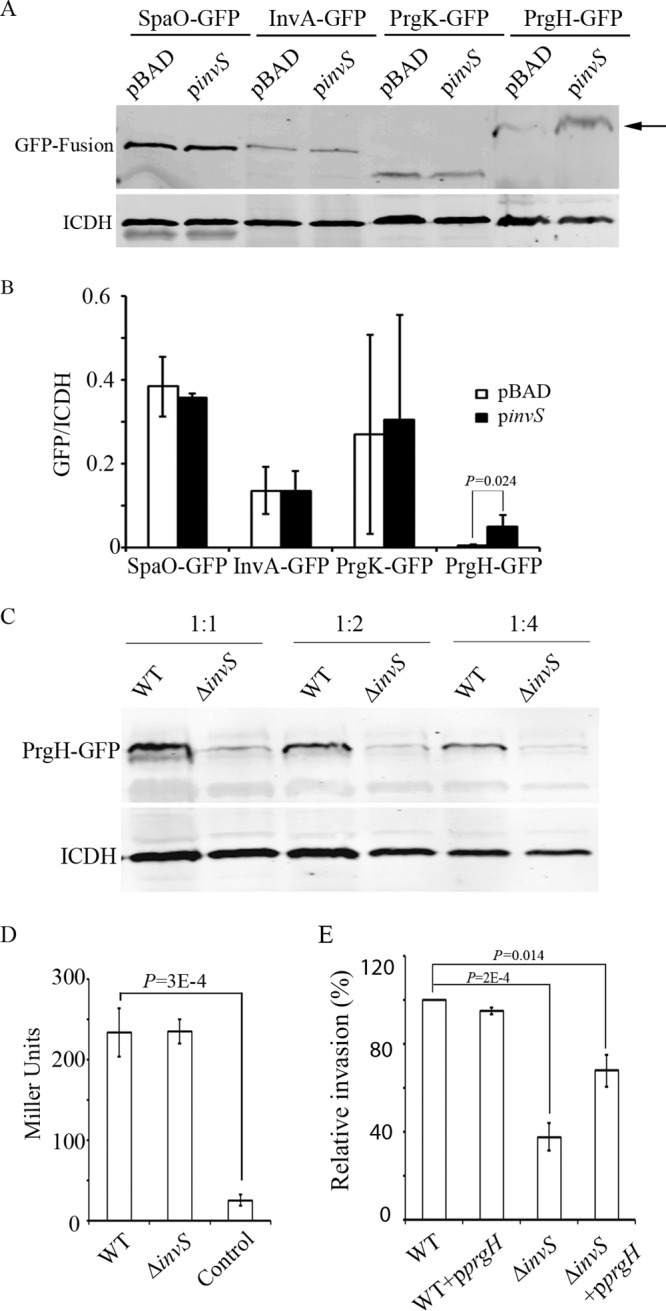
InvS regulates the level of PrgH. (A) InvS upregulates prgH-gfp expression in Salmonella. The 5′ UTR along with the full ORFs of SpaO, InvA, PrgK, and PrgH were translationally fused to GFP. Plasmids expressing the GFP fusion proteins were cotransformed with plasmids expressing InvS or the vector control into the ΔinvS strain. The background strain eliminates the potential effect that can be caused by chromosomal invS. Bacterial isocitrate dehydrogenase (ICDH) was detected using anti-ICDH polyclonal antibodies as the loading control. The levels of GFP fusion proteins were determined by Western blotting with polyclonal anti-GFP antibodies. The arrow indicates the expression of PrgH-GFP. (B) Quantification of GFP fusion protein expression from three independent experiments. Values represent GFP fusion protein expression levels after normalization to the expression of ICDH. The P value was calculated using the Student t test. (C) The expression of prgH-gfp was decreased in the absence of InvS. pXG30-derived PrgH-GFP was transformed into the WT or ΔinvS strain. Western blot of the 2-fold dilution series showing the expression of prgH-gfp. (D) InvS does not regulate the transcriptional level of prgH. A promoterless lacZ gene was placed under the prgH promoter in either the wild-type Salmonella or the ΔinvS mutant strain. β-Galactosidase activity was measured as described in Materials and Methods. The Salmonella WT strain without lacZ was used as the negative control. The data shown were obtained from three independent experiments. Results are presented as the means in Miller units. Error bars indicate standard deviations. (E) Overexpression of prgH partially restores the ΔinvS mutant invasion defect. HeLa cells were infected with Salmonella for 15 min at an MOI of 10. Relative bacterial invasion was determined by the gentamicin protection assay. The data shown were obtained from three independent experiments. The P values were calculated using the Student t test.
InvS regulates the level of FimZ.
Our proteomic analysis showed a higher level of FimZ and lower level of FlhD in the absence of InvS. Next, we used the GFP-based plasmid assay to test if InvS affects the levels of FimZ and FlhD. The 5′ UTR along with the full ORFs of FimZ and FlhD were translationally fused to GFP. The assay was performed in both E. coli and Salmonella ΔinvS. When tested in the ΔinvS background strain, we detected a lesser amount of FimZ-GFP when InvS was coexpressed from a plasmid, pinvS. Interestingly, the difference in the FimZ-GFP levels disappeared when the same plasmids were coexpressed in the E. coli background (Fig. 6A and B). This result suggests that InvS may indirectly repress fimZ expression and that additional cofactors (from Salmonella) may be required for InvS to regulate fimZ expression. The expression of the GFP fusion proteins was also examined in the WT and the ΔinvS mutant strain. We found that the level of FimZ-GFP was higher in the ΔinvS mutant than in the WT strain (Fig. 6C). FimZ is known to negatively regulate flhD expression. It is possible that the increase in FimZ in the ΔinvS background strain led to the decrease of flhD expression. We then performed a gentamicin protection assay to examine if the alteration of the FimZ level is able to work against the effect of InvS and rescue the invasion phenotype of the ΔinvS mutant. We found the double deletion ΔfimZ ΔinvS strain showed an invasion level similar to that of the ΔfimZ strain. The overexpression of FimZ in the wild-type strain resulted in a decrease in the invasion rate (Fig. 7). Western blotting confirmed the decrease of flagella in the ΔinvS strain compared with the wild-type strain (Fig. 8A and B), which is consistent with the result showing that the deletion of invS impaired Salmonella motility (Fig. 8C). The overexpression of FimZ produced a larger amount of FimZ than the invS deletion strain and drastically inhibited flagellar gene expression (Fig. 8). Although the detailed mechanism remains unclear, our results suggest fimZ is an important regulatory component linking InvS and its effects on flagellar expression and Salmonella invasion. InvS facilitates invasion in a fimZ- and flagellum-dependent manner.
FIG 6.
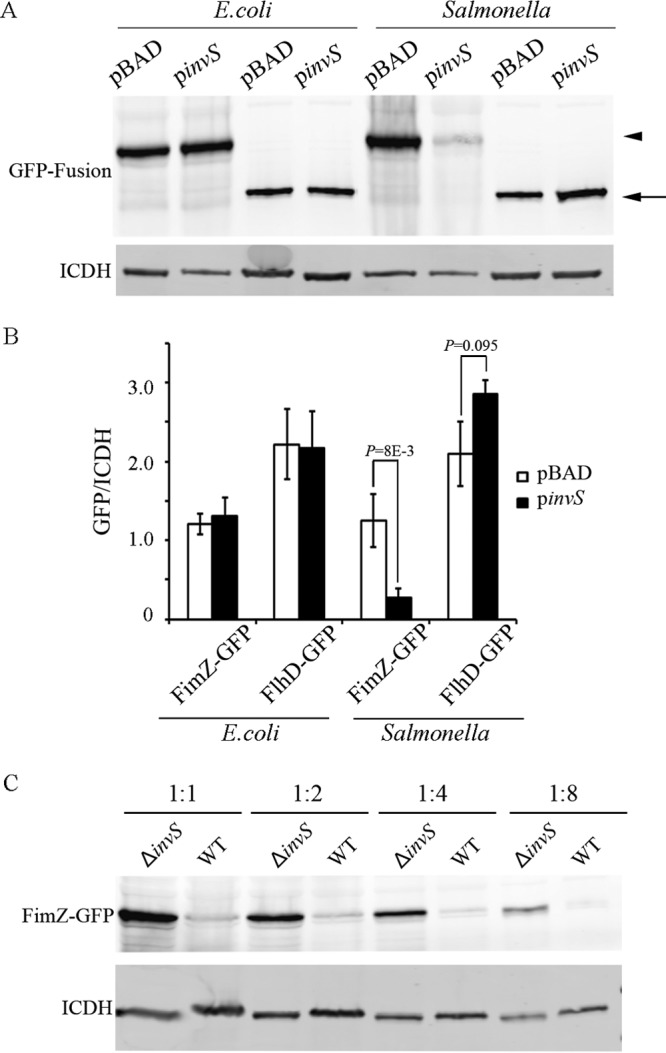
InvS regulates the level of FimZ. (A) InvS downregulates fimZ-gfp expression in Salmonella. The 5′ UTR along with the full ORFs of FimZ and FlhD were translationally fused to GFP. Plasmids derived from pXG30, expressing GFP fusion proteins, were cotransformed with pBAD-derived plasmids expressing InvS or the vector control. Bacterial ICDH was detected using polyclonal anti-ICDH antibodies as the loading control. The levels of GFP fusion proteins were determined by Western blotting with polyclonal anti-GFP antibodies. (B) Quantification of GFP fusion proteins from three independent experiments. Values represent GFP fusion protein levels after normalization with that of ICDH. Data are representative of three experiments. The P values were calculated using the Student t test. (C) The expression of FimZ-GFP was decreased in the presence of InvS. pXG30-derived FimZ-GFP was transformed into the WT or the ΔinvS strain. Western blot of the 2-fold dilution series showing decreases of FimZ-GFP levels in the presence of InvS.
FIG 7.
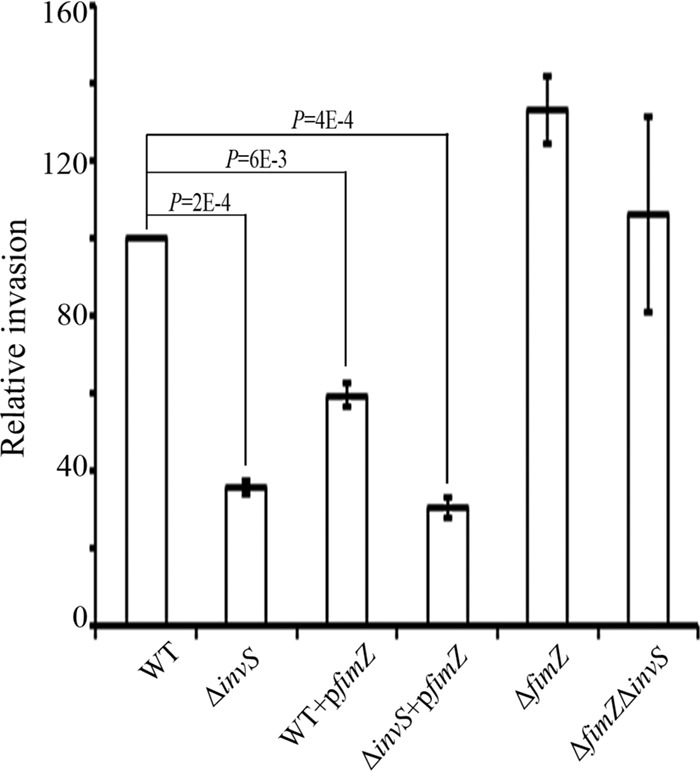
FimZ downregulates Salmonella invasion. HeLa cells were infected with Salmonella for 15 min at an MOI of 10. Relative bacterial invasion was determined by the gentamicin protection assay. The data shown were obtained from three independent experiments. The P values were calculated using the Student t test.
FIG 8.

InvS regulates flagellar expression. (A) InvS upregulates the expression of fliC. The expression of FliC protein was detected by Western blotting with monoclonal anti-FliC antibodies. Bacterial ICDH was detected using anti-ICDH polyclonal antibodies as the loading control. (B) Quantification of FliC expression. FliC expression in the WT strain was defined as 1. Values represent relative protein expression levels after normalization with that in the WT. Data are representative of three experiments. The P values were calculated using the Student t test. (C) Two-microliter samples of Salmonella cultures (optical density at 600 nm, 1.0) were inoculated onto LB plates made up of 0.3% Bacto agar (Difco), and cultures were grown at 37°C. Photos were taken 6 h postinoculation. (D) Halos around the colonies were measured after 6 h of incubation at 37°C. Data are representative of three experiments. The P values were calculated using the Student t test.
DISCUSSION
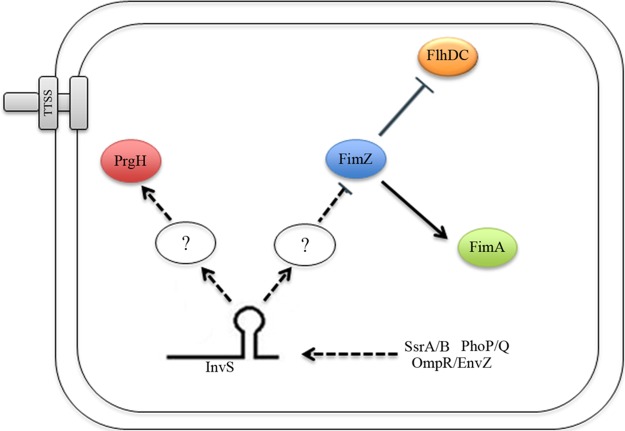
Small RNAs represent a relatively new set of posttranscriptional regulatory molecules that are gaining interest in bacteria. A few bacterial sRNAs are reported to regulate the bacterial stress response and are involved in the regulation of virulence genes. Gong et al. reported that IsrM negatively regulates Salmonella HilE and is essential for Salmonella invasion (14). Ryan et al. have demonstrated that the small RNA DsrA influences the acid tolerance response and virulence of Salmonella (25, 26, 49). Recently, hundreds of novel sRNAs have been identified in Salmonella, but few have been functionally characterized (8, 11, 14, 27). To identify the involvement of these small RNAs in Salmonella virulence, we screened recently identified Salmonella sRNAs for their roles in Salmonella invasion and found that InvS is essential for Salmonella entry into nonphagocytic cells. InvS was originally identified by Hfq coimmunoprecipitation sequencing (Hfq-CoIP-seq) and showed a 2- to 47-fold enrichment under various stress conditions (8, 11). Colgan et al. performed transcriptome sequencing (RNA-seq) to study the differential expression of Salmonella sRNAs. InvS was shown to be positively regulated by two-component regulatory systems, including SsrA/B, PhoP/Q and OmpR/EnvZ (28). The details of the InvS regulatory pathways are not clear. It is known that PhoP/Q regulates both SPI-1 and SPI-2 expression, while SsrA/B and OmpR/EnvZ are able to activate SPI-2 expression (29). It is not known if InvS plays any roles in the cross talk between SPI-1 and SPI-2.
Many classic transcription factors are known to regulate Salmonella invasion by controlling the transcription of invasion-related genes. For example, the transcription of SPI-1 genes can be activated by HilA, HilC, HilD, InvF, and SirA (30). Our results showed that overexpression of HilD and InvF were able to restore the invasion defect of the ΔinvS mutant. So far, there is no evidence to suggest that hilD or invF is a direct target of InvS. Interestingly, we found that the overexpression of HilA failed to rescue the invasion deficiency. This may indicate that InvS is able to regulate invasion in a HilD-dependent but HilA-independent pathway. Singer et al. have demonstrated that HilD directly activates the expression of flagellar genes, while HilA does not affect flagellar gene expression (31); this is consist with our data showing that the deletion of InvS results in a decrease of flagellar expression. Furthermore, it was reported HilD is able to activate the transcription of invF from a promoter that is far upstream of its HilA-dependent promoter. The loss of hilD resulted in a more severe effect on the expression of a subset of SPI-1 genes than the loss of hilA (6). Our data show that overexpressing HilD activates invF expression more profoundly than overexpressing HilA. In addition, it is also possible that additional factors are involved in the InvS-mediated regulation of invasion. InvS may have multiple targets, which might balance out the effect of HilA overexpression. This may explain why overexpressing only hilA is not sufficient to restore the invasion defect of the invS mutant.
We speculated that InvS exerts its function by regulating genes downstream of hilD and invF. These downstream genes may include the Salmonella SPI-1 type III secretion system and type III effectors that are known to play direct roles in Salmonella invasion. To identify the targets of InvS, we noticed that type III effector secretion and translocation are decreased in the absence of InvS. Further analysis revealed that InvS activates the expression of prgH, which is required for the assembly of the type III secretion needle complex. It is known that the deletion of prgH impairs SPI-1 TTSS assembly and effector secretion (3, 4). Consistent with its effect in type III secretion, overexpressing PrgH in the ΔinvS mutant partially rescued the invasion deficiency. It is still unclear how prgH is regulated by InvS. Our results showed similar levels of prgH promoter activity in the WT and the ΔinvS mutant background (Fig. 5D). Thus, it is possible that InvS regulates prgH indirectly or at the protein level. In addition, it is possible that InvS affects additional target genes to regulate Salmonella invasion.
Our proteomic analysis showed higher levels of FimZ in the absence of InvS. FimZ is known as a transcriptional activator responsible for promoting the expression of type I fimbriae and downregulating flagellar synthesis (32). While fimbriae are known to play a role in adhering to infected cells, flagella have been associated with Salmonella motility and invasion. It has been reported that flagellum-driven motility forces the bacterium into a near surface swimming mode, which promotes Salmonella invasion by scanning the host cell surface (22). Our proteomics analysis indicated an increase in fimZ and a decrease in fliC expression in the absence of InvS. The deletion of invS impairs Salmonella motility, suggesting that InvS might function to promote motility to facilitate bacterial invasion. Consistent with this notion, previous reports found that HilD activates the transcription of flagellar genes while HilA does not (31). Our data show that the overexpression of HilD rescued the ΔinvS invasion defect while HilA did not alter the invasion levels. This is in agreement with our data showing that uncontrolled or overexpression of fimZ (in the absence of InvS) leads to a decrease in invasiveness. Taken together, we conclude that InvS coordinates the increase in PrgH and decrease in FimZ leading to more efficient Salmonella invasion (Fig. 9).
FIG 9.
Model for InvS-mediated Salmonella invasion. InvS facilitates Salmonella effector secretion and translocation by positively regulating prgH, which encodes a type III secretion apparatus protein. Furthermore, InvS negatively regulates fimZ, a global regulator that is known to repress Salmonella SPI-1 gene expression by activating HilE. FimZ activates FimA, the major fimbrial unit. FimZ also negatively regulates flagellar synthesis by repressing expression of the master flagellar regulator FlhDC. The regulation of these target mRNAs or proteins by InvS, in turn, promotes Salmonella to invade the host cell. Arrows represent activation while the assertion signs represent inhibition. Dotted lines indicate indirect regulation. The symbol “?” indicates unknown factors or a signaling cascade that may be involved in the pathway.
The exact mechanism by which InvS activates the expression of PrgH and reduces the expression of FimZ is currently unclear. Corcoran et al. have established the GFP-based plasmid assay for the validation of sRNA-mediated target regulation (24). When tested in E. coli, we failed to detect any PrgH-GFP by Western blotting, suggesting that additional factors present in Salmonella might be involved in maintaining the stability of PrgH. Interestingly, our results indicate that InvS downregulates fimZ expression in Salmonella but not in E. coli. The regulation of fimZ and prgH expression is remarkably complex. It is known that prgH is under the regulation of many global regulators, such as HilA, InvF, PhoP, and SirA. Furthermore, Bailey et al. have shown that prgH and other SPI-1 genes are expressed at higher levels in a ramA mutant (33). Previous work showed FimY acts upstream of FimZ to activate the fim operon, while FliZ functions to repress FimZ posttranscriptionally (34). In addition, FimW and FimZ form a coupled feedback loop where they activate their own and each other's expression. Recently, it was reported that the two-component system PhoBR is also capable of inducing fimZ expression (35). Thus, it is possible that additional factors (from Salmonella) are required for InvS to regulate fimZ and prgH expression. Furthermore, Chao et al. showed InvS is associated with Hfq on the basis of results from their coimmunoprecipitation experiments (8). Ansong et al. detected a decrease in FimZ in Δhfq Salmonella (36). In addition, previous studies suggested that cellular RNAs compete for Hfq, and one abundant sRNA can indirectly impact the targets of others by disrupting Hfq-mediated effects (37, 38). It is also possible that InvS indirectly regulates fimZ expression by disrupting the binding of Hfq to fimZ or other RNAs that target fimZ and prgH.
To date, only a fraction of published sRNAs has been functionally characterized, and the roles in bacterial virulence have only been elucidated for a few. We showed that InvS functions to positively regulate prgH expression and negatively regulate fimZ expression, which led to more efficient Salmonella invasion. InvS is highly conserved at the DNA sequence level in all Salmonella enterica serovars, including Typhimurium, Newport, Typhi, Paratyphi, and Enteritidis (39). This pattern of conservation is consistent with the involvement of InvS in SPI-1-mediated invasion throughout the Salmonella enterica species. Our study expands the known sRNA-mediated regulatory network of Salmonella. Additional work on the remaining sRNAs and other regulatory factors will likely describe a coordinated regulatory network revealing the intricate regulation of virulence factors in Salmonella.
MATERIALS AND METHODS
Bacterial strains and mammalian cell lines.
The Salmonella strains used in this study are isogenic derivatives of virulent wild-type (WT) strain SL1344 of Salmonella Typhimurium (40). In-frame chromosomal deletions of genes in Salmonella strains were generated using an allelic-exchange suicide vector pSB890 (41). Briefly, a DNA fragment with the in-frame deletion was cloned into the conjugative suicide vector pSB890. Plasmid constructs were introduced into Salmonella by conjugation and were subsequently integrated into the chromosome by homologous recombination. PCR-generated invS from the Salmonella chromosome was inserted into pBAD via EcoRI and XmaI sites to generate pinvS. Translational gfp fusions were constructed by cloning a PCR insert amplified from the Salmonella chromosome and cloned into pXG30 via NsiI and NheI sites (24). DNA oligomer primers for these PCRs are listed in Table S3 in the supplemental material.
E. coli and Salmonella strains were routinely cultured in Luria-Bertani (LB) broth. Salmonella strains were cultured under SPI-1 TTSS-inducing conditions (LB broth with 0.3 M NaCl) for all of the invasion experiments. Antibiotics were used at the indicated concentrations: ampicillin, 120 μg · ml−1; streptomycin, 25 μg · ml−1; kanamycin, 40 μg · ml−1; and tetracycline, 12 μg · ml−1.
The mammalian cell line HeLa (CCL-2) was purchased from ATCC (Manassas, VA). HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (VWR) supplemented with 10% fetal bovine serum.
Fluorescent F-actin staining.
An F-actin staining assay was conducted as described previously (42). HeLa cells were infected with Salmonella at a multiplicity of infection (MOI) of 10 unless indicated otherwise. Infected cells were fixed with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 15 min and permeabilized with 0.2% Triton X-100 in PBS. Salmonella was stained using rabbit anti-Salmonella O-antigen group B (Difco), and then visualized with Alexa Fluor 488 (Invitrogen). F-actin was visualized by staining with Texas Red-conjugated phalloidin (Molecular Probes, Eugene, OR).
Gentamicin protection assay.
Salmonella infection of HeLa cells was conducted as previously described (43). Briefly, Salmonella cells were cultured to an optical density at 600 nm (OD600) of 1.0 in LB broth with 0.3 M NaCl at 37°C. Bacteria were then added to HeLa cells at an MOI of 10 and incubated for 15 min at 37°C in 5% CO2. After the infection, cells were washed twice with PBS to remove extracellular bacteria and were incubated further in DMEM containing 10% fetal bovine serum and 16 μg of gentamicin per ml. At different time points after gentamicin treatment, infected cells were washed three times in PBS and lysed with 1% Triton X-100 and 0.1% sodium dodecyl sulfate (SDS). Cell lysates were then serially diluted and plated on selective medium.
GFP-based two-plasmid assay.
Bacteria were transformed with a PBAD-based sRNA expression vector (with or without invS) and a expression vector (pXG30) that constitutively expresses corresponding GFP fusion proteins. Double transformants were grown overnight in LB medium containing appropriate antibiotics at 37°C, followed by subculture (1:200 dilution) until OD600 of 1.0 in LB broth containing 0.2% l-arabinose for the induction of invS expression. Western blotting was performed using polyclonal anti-GFP antibodies to monitor the expression of GPF fusion proteins (24).
Protein translocation assay.
Salmonella strains expressing the β-lactamase fusions were used to infect monolayers of HeLa cells seeded in 96-well plates at an MOI of 20. Fifteen minutes after the infection, CCF4-AM (Invitrogen, Carlsbad, CA) was added to the wells. CCF4-AM enters cells and is cleaved by intracellular esterase, leading to the accumulation of CCF4. CCF4, emitting green fluorescence, is a β-lactamase substrate and emits blue fluorescence upon cleavage. After incubating with CCF4-AM for 2 h at room temperature, infected cells were examined under a fluorescence microscope to quantify the numbers of green and blue cells. Experiments were performed in triplicates. Approximately 300 cells were counted in each sample.
β-Galactosidase assay.
Salmonella carrying LacZ fusions were grown at 37°C overnight, followed by subculture in an SPI-1-inducing condition until the OD600 reached 1.0. The β-galactosidase activity was measured according to standard protocols (44).
RNA isolation and Northern hybridization.
RNA isolation and Northern hybridization experiments were performed as previously described (11, 45). Briefly, RNA was prepared by hot phenol extraction, followed by DNase I treatment. Five to 10 micrograms of total RNA was denatured for 5 min at 95°C in RNA loading buffer (95% formamide, 0.1% xylene cyanol, 0.1% bromophenol blue, and 10 mM EDTA), separated on polyacrylamide gels, and transferred onto Hybond-XL membranes (GE Healthcare). The 5′-end γ-32P-labeled oligonucleotides (Fermentas) were hybridized to membranes overnight at 42°C, and then washed with 5× saline-sodium citrate buffer (SSC) with 0.1% SDS, 1× SSC with 0.1%SDS, and 0.5× SSC with 0.1% SDS for 15 min each. Signals were visualized using a phosphorimager (Typhoon FLA 7000; GE Healthcare). The probes used are listed in Table S3.
Protein digestion, isobaric labeling, and peptide fractionation.
WT and ΔinvS Salmonella strains were cultured under SPI-1-inducing conditions to an OD600 of 1.0 and were centrifuged to separate the supernatant and pellet. Salmonella cells were lysed by vortexing with silica beads in 50 mM NH4HCO3 buffer, while supernatant proteins were obtained by precipitation with trichloroacetic acid. Cell lysates and supernatant proteins were then denatured in 8 M urea prepared in 50 mM NH4HCO3 containing 5 mM dithiothreitol for 30 min at 37°C. Samples were then alkylated by adding 400 mM iodoacetamide to a final concentration of 10 mM and incubating for 30 min at room temperature protected from light. The reaction was diluted 8-fold with 50 mM NH4HCO3 and incubated for 4 h at 37°C with trypsin at an enzyme/protein ratio of 1/50 (m/m). Samples were desalted with C18 SPE cartridges (Discovery C18, 1 ml, 50 mg; Sulpelco) as previously described (36). Peptides derived from cell lysates were labeled with 4-plex isobaric tags for relative and absolute quantification (iTRAQ) reagent (Applied Biosystems) according to the manufacturer's recommendations and were fractionated by high-pH reverse-phase liquid chromatography as previously described (46), while peptides derived from the supernatant fraction were left unlabeled and unfractionated. Briefly, peptides were loaded into a C18 column (Eclipse XDB C18, 5 μm, 4.6 by 150 mm; Agilent Technologies) connected to a high-performance liquid chromatograph (Waters 1525 binary HPLC pump) and eluted at 0.5 ml/min with the following gradient: 0 to 5% solvent B (solvent A, 10 mM ammonium formate [FA]; solvent B, 10 mM FA in 90% acetonitrile [ACN]) in 10 min, 5 to 35% solvent B in 60 min, 35 to 70% solvent B in 15 min, and holding at 70% for 10 min. Peptides were collected into 60 fractions, further concatenated into 15 fractions, and dried in a vacuum centrifuge. The supernatant was left unfractionated but was subjected to two steps of clean-up with C18 reverse-phase and strong cation exchange (SCX) cartridges to eliminate small molecule contamination (36).
Quantitative proteomic analysis.
Peptides were dissolved in 0.1% formic acid and loaded into a C18 trap column (200 μm by 0.5 mm, ChromXP C18-CL, 3 μm, 120 Å; Eksigent) connected to a nanoHPLC system (Ekspert nanoLC 400; Eksigent). The separation was performed in a capillary C18 column (75 μm by 15 cm, ChromXP C18-CL, 3 μm, 120 Å) at 200 nl/min with the following gradient: 1 min in 5% solvent B (solvent A, 0.1% FA; solvent B, 80% ACN:0.1% FA), 5 to 35% solvent B in 60 min, 35 to 80% solvent B in 1 min, 6 min in 80% solvent B, 80 to 5% solvent B in 1 min, and hold in 5% for 11 min. Eluting peptides were directly analyzed in an electrospray ionization mass spectrometer (5600 TripleTOF; AB Sciex). Full mass spectrometry spectra were collected in the range of 400 to 2000 m/z, and the top 20 most intense parent ions were submitted to fragmentation for 100 ms each using rolling-collision energy.
The identification and quantification of peptides were performed with Paragon software as part of the ProteinPilot package (AB Sciex) by searching tandem mass spectra against Salmonella enterica serovar Typhimurium SL1344 sequences downloaded from Uniprot KnowledgeBase on 11 November 11 2014. Database searches were performed considering trypsin digestion, cysteine residue alkylation with iodoacetamide, and biological modifications asfactors. Peptides were filtered with a confidence score above 95, which resulted in a false-discovery rate of ∼1.3% in protein level. The iTRAQ channel intensities were extracted using ProteinPilot and intensities from different peptide-spectrum matches and peptides from the same protein were summed together. Sample load was then normalized by total channel intensity and significance was tested by analysis of variance (ANOVA) using InfernoRDN (formerly, DAnTE) (47). For the label-free supernatant samples, peak areas were extracted with Skyline (48) before being normalized by linear regression and central tendency, and were tested by ANOVA using InfernoRDN.
Accession number(s).
The raw proteomic data were deposited in the Proteomics Identifications (PRIDE) public repository under accession numbers PXD003589 and PXD003590.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Bindley Bioscience Center and Mark Hall for access to their instrumentation.
This project was partially funded by the Indiana Clinical and Translation Science Institute.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00824-16.
REFERENCES
- 1.Agbor TA, McCormick BA. 2011. Salmonella effectors: important players modulating host cell function during infection. Cell Microbiol 13:1858–1869. doi: 10.1111/j.1462-5822.2011.01701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galán JE, Unger VM. 2004. Structural insights into the assembly of the type III secretion needle complex. Science 306:1040–1042. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sukhan A, Kubori T, Wilson J, Galán JE. 2001. Genetic analysis of assembly of the Salmonella enterica serovar Typhimurium type III secretion-associated needle complex. J Bacteriol 183:1159–1167. doi: 10.1128/JB.183.4.1159-1167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kimbrough TG, Miller SI. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc Natl Acad Sci U S A 97:11008–11013. doi: 10.1073/pnas.200209497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou D, Mooseker MS, Galán JE. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092–2095. doi: 10.1126/science.283.5410.2092. [DOI] [PubMed] [Google Scholar]
- 6.Akbar S, Schechter LM, Lostroh CP, Lee CA. 2003. AraC/XylS family members, HilD and HilC, directly activate virulence gene expression independently of HilA in Salmonella typhimurium. Mol Microbiol 47:715–728. doi: 10.1046/j.1365-2958.2003.03322.x. [DOI] [PubMed] [Google Scholar]
- 7.Rakeman JL, Bonifield HR, Miller SI. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J Bacteriol 181:3096–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kröger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, Hinton JC. 2013. An infection-relevant transcriptomic compendium for Salmonella enterica serovar Typhimurium. Cell Host Microbe 14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 10.Kröger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, Chao Y, Sittka A, Hebrard M, Handler K, Colgan A, Leekitcharoenphon P, Langridge GC, Lohan AJ, Loftus B, Lucchini S, Ussery DW, Dorman CJ, Thomson NR, Vogel J, Hinton JC. 2012. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4:e1000163. doi: 10.1371/journal.pgen.1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barquist L, Vogel J. 2015. Accelerating discovery and functional analysis of Small RNAs with new technologies. Annu Rev Genet 49:367–394. doi: 10.1146/annurev-genet-112414-054804. [DOI] [PubMed] [Google Scholar]
- 13.Hebrard M, Kröger C, Srikumar S, Colgan A, Handler K, Hinton J. 2012. sRNAs and the virulence of Salmonella enterica serovar Typhimurium. RNA Biol 9:437–445. doi: 10.4161/rna.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong H, Vu GP, Bai Y, Chan E, Wu R, Yang E, Liu F, Lu S. 2011. A Salmonella small non-coding RNA facilitates bacterial invasion and intracellular replication by modulating the expression of virulence factors. PLoS Pathog 7:e1002120. doi: 10.1371/journal.ppat.1002120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galán JE, Curtiss R 3rd. 1989. Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A 86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galan JE, Zhou D. 2000. Striking a balance: modulation of the actin cytoskeleton by Salmonella. Proc Natl Acad Sci U S A 97:8754–8761. doi: 10.1073/pnas.97.16.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darwin KH, Miller VL. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol 181:4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ellermeier CD, Ellermeier JR, Slauch JM. 2005. HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57:691–705. doi: 10.1111/j.1365-2958.2005.04737.x. [DOI] [PubMed] [Google Scholar]
- 19.Darwin KH, Miller VL. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol Microbiol 35:949–960. doi: 10.1046/j.1365-2958.2000.01772.x. [DOI] [PubMed] [Google Scholar]
- 20.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K. 2013. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ow SY, Salim M, Noirel J, Evans C, Rehman I, Wright PC. 2009. iTRAQ underestimation in simple and complex mixtures: “the good, the bad and the ugly.” J Proteome Res 8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 22.Misselwitz B, Barrett N, Kreibich S, Vonaesch P, Andritschke D, Rout S, Weidner K, Sormaz M, Songhet P, Horvath P, Chabria M, Vogel V, Spori DM, Jenny P, Hardt WD. 2012. Near surface swimming of Salmonella Typhimurium explains target-site selection and cooperative invasion. PLoS Pathog 8:e1002810. doi: 10.1371/journal.ppat.1002810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dewoody R, Merritt PM, Houppert AS, Marketon MM. 2011. YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol Microbiol 79:1445–1461. doi: 10.1111/j.1365-2958.2011.07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JC, Vogel J. 2012. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol 84:428–445. doi: 10.1111/j.1365-2958.2012.08031.x. [DOI] [PubMed] [Google Scholar]
- 25.Chao Y, Vogel J. 2010. The role of Hfq in bacterial pathogens. Curr Opin Microbiol 13:24–33. doi: 10.1016/j.mib.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 26.Sittka A, Pfeiffer V, Tedin K, Vogel J. 2007. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padalon-Brauch G, Hershberg R, Elgrably-Weiss M, Baruch K, Rosenshine I, Margalit H, Altuvia S. 2008. Small RNAs encoded within genetic islands of Salmonella typhimurium show host-induced expression and role in virulence. Nucleic Acids Res 36:1913–1927. doi: 10.1093/nar/gkn050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colgan AM, Kroger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, Hokamp K, Hinton JC. 2016. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet 12:e1006258. doi: 10.1371/journal.pgen.1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu X, Hensel M. 2010. Systematic analysis of the SsrAB virulon of Salmonella enterica. Infect Immun 78:49–58. doi: 10.1128/IAI.00931-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Golubeva YA, Sadik AY, Ellermeier JR, Slauch JM. 2012. Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190:79–90. doi: 10.1534/genetics.111.132779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer HM, Kuhne C, Deditius JA, Hughes KT, Erhardt M. 2014. The Salmonella Spi1 virulence regulatory protein HilD directly activates transcription of the flagellar master operon flhDC. J Bacteriol 196:1448–1457. doi: 10.1128/JB.01438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clegg S, Hughes KT. 2002. FimZ is a molecular link between sticking and swimming in Salmonella enterica serovar Typhimurium. J Bacteriol 184:1209–1213. doi: 10.1128/jb.184.4.1209-1213.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J, Piddock LJ. 2010. RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol 192:1607–1616. doi: 10.1128/JB.01517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saini S, Slauch JM, Aldridge PD, Rao CV. 2010. Role of cross talk in regulating the dynamic expression of the flagellar Salmonella pathogenicity island 1 and type 1 fimbrial genes. J Bacteriol 192:5767–5777. doi: 10.1128/JB.00624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baxter MA, Jones BD. 2015. Two-component regulators control hilA expression by controlling fimZ and hilE expression within Salmonella enterica serovar Typhimurium. Infect Immun 83:978–985. doi: 10.1128/IAI.02506-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. 2009. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One 4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. 2009. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 39.Srikumar S, Kroger C, Hebrard M, Colgan A, Owen SV, Sivasankaran SK, Cameron AD, Hokamp K, Hinton JC. 2015. RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella Typhimurium. PLoS Pathog 11:e1005262. doi: 10.1371/journal.ppat.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 41.Dai S, Zhou D. 2004. Secretion and function of Salmonella SPI-2 effector SseF require its chaperone, SscB. J Bacteriol 186:5078–5086. doi: 10.1128/JB.186.15.5078-5086.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen LM, Hobbie S, Galán JE. 1996. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science 274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- 43.Li D, Wang X, Wang L, Zhou D. 2013. The actin-polymerizing activity of SipA is not essential for Salmonella enterica serovar Typhimurium-induced mucosal inflammation. Infect Immun 81:1541–1549. doi: 10.1128/IAI.00337-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nutt JD, Pillai SD, Woodward CL, Sternes KL, Zabala-Diaz IB, Kwon YM, Ricke SC. 2003. Use of a Salmonella typhimurium hilA fusion strain to assess effects of environmental fresh water sources on virulence gene expression. Water Res 37:3319–3326. doi: 10.1016/S0043-1354(03)00244-6. [DOI] [PubMed] [Google Scholar]
- 45.Pfeiffer V, Sittka A, Tomer R, Tedin K, Brinkmann V, Vogel J. 2007. A small non-coding RNA of the invasion gene island (SPI-1) represses outer membrane protein synthesis from the Salmonella core genome. Mol Microbiol 66:1174–1191. doi: 10.1111/j.1365-2958.2007.05991.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, Yang F, Gritsenko MA, Wang Y, Clauss T, Liu T, Shen Y, Monroe ME, Lopez-Ferrer D, Reno T, Moore RJ, Klemke RL, Camp DG II, Smith RD. 2011. Reversed-phase chromatography with multiple fraction concatenation strategy for proteome profiling of human MCF10A cells. Proteomics 11:2019–2026. doi: 10.1002/pmic.201000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Polpitiya AD, Qian WJ, Jaitly N, Petyuk VA, Adkins JN, Camp DG II, Anderson GA, Smith RD. 2008. DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics 24:1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. 2010. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryan D, Ojha UK, Jaiswal S, Padhi C, Suar M. 2016. The small RNA DsrA influences the acid response and virulence of Salmonella enterica serovar Typhimurium. Front Microbiol doi: 10.3389/fmicb.2016.00599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.