Abstract
The Australian Ascochyta rabiei (Pass.) Labr. (syn. Phoma rabiei) population has low genotypic diversity with only one mating type detected to date, potentially precluding substantial evolution through recombination. However, a large diversity in aggressiveness exists. In an effort to better understand the risk from selective adaptation to currently used resistance sources and chemical control strategies, the population was examined in detail. For this, a total of 598 isolates were quasi-hierarchically sampled between 2013 and 2015 across all major Australian chickpea growing regions and commonly grown host genotypes. Although a large number of haplotypes were identified (66) through short sequence repeat (SSR) genotyping, overall low gene diversity (Hexp = 0.066) and genotypic diversity (D = 0.57) was detected. Almost 70% of the isolates assessed were of a single dominant haplotype (ARH01). Disease screening on a differential host set, including three commonly deployed resistance sources, revealed distinct aggressiveness among the isolates, with 17% of all isolates identified as highly aggressive. Almost 75% of these were of the ARH01 haplotype. A similar pattern was observed at the host level, with 46% of all isolates collected from the commonly grown host genotype Genesis090 (classified as “resistant” during the term of collection) identified as highly aggressive. Of these, 63% belonged to the ARH01 haplotype. In conclusion, the ARH01 haplotype represents a significant risk to the Australian chickpea industry, being not only widely adapted to the diverse agro-geographical environments of the Australian chickpea growing regions, but also containing a disproportionately large number of aggressive isolates, indicating fitness to survive and replicate on the best resistance sources in the Australian germplasm.
Keywords: chickpea, Ascochyta rabiei, resistance sources, SSR genotype, haplotype and highly aggressive
Introduction
Chickpea (Cicer arietinum L.), is the most widely cultivated legume, grown in over 50 countries across the Indian subcontinent, North Africa, the Middle East, southern Europe, the Americas and Australia. The global production in 2014 was 14 million tons with yields of1 982 kg/ha. The crop is grown in rotation, largely for its high cash return and ability to fix atmospheric nitrogen (Gan et al., 2006). However, significant yield instability remains, largely due to Ascochyta blight caused by the necrotrophic fungal pathogen Ascochyta rabiei (Nene, 1982). The disease causes extensive crop losses globally (Pande et al., 2005), and remains the major biotic constraint to the winter-grown crop in Australia, with all growing regions affected (Bretag et al., 2008). Subsequent inoculum release following increased precipitation over the 2013 to 2016 growing seasons has led to non-manageable epidemics on “resistant” host genotypes (Moore et al., 2016). The recent severity of the disease is likely due to the dispersal of isolates that are highly aggressive, widely adapted and able to survive between the growing seasons in the harsh Australian summer climate.
Ascochyta rabiei is a bipolar heterothallic fungus with one mating type locus and two mating types (Wilson and Kaiser, 1995). Large temporal and spatial variations have been detected within populations from other global regions where both mating types exist (Udupa et al., 1998; Jamil et al., 2000; Peever et al., 2004; Ali et al., 2012). On a global scale, the total gene diversity detected with 19 sequence tag microsatellite primers was estimated to be 0.29 among A. rabiei populations worldwide. Maximum gene diversity was detected among intra-country populations in Canada (0.38), followed by the United States (0.36) and Syria (0.32) (Phan et al., 2003). In other studies, based on different sets of short sequence repeat (SSR) loci, the diversity of the population was estimated to be even higher; 0.55 in Tunisia (Rhaïem et al., 2006), and 0.79 in Iran (Nourollahi et al., 2011). This is in stark contrast to the population diversity observed in Australia, where despite trapping of the putative ascospore in the field (Galloway and MacLeod, 2003), only one mating type has been detected (Barve et al., 2003; Leo et al., 2015). Accordingly, multiple studies have shown a very low gene diversity within the population (ranging from 0.02 to 0.094), consistent with an organism that is reproducing asexually (Phan et al., 2003; Rhaïem et al., 2006; Leo et al., 2015).
The variation in aggressiveness detected within sexually recombinant A. rabiei populations worldwide has led to the erosion of resistant host genotypes (McDonald and Linde, 2002; Peever et al., 2012; Mahiout et al., 2015; Vafaei et al., 2015; Tekin et al., 2017). Although not directly comparable due to a number of factorial differences such as host genotype, isolate and bioassay conditions, several in country studies have identified diversity of aggressiveness within A. rabiei populations. Sets of isolates have been identified that react similarly or differently to a group of host genotypes (Grewal, 1984; Udupa et al., 1998; Jayakumar et al., 2005; Pande et al., 2005; Imtiaz et al., 2015; Vafaei et al., 2015; Baite et al., 2016). Jan and Wiese (1991) reported the presence of 11 “virulent forms” among 39 isolates assessed from the Palouse region of the United States. Navas-Cortés et al. (1998) identified 11 “pathotypes” in India, Pakistan, Spain, and the United States. Next, Jamil et al. (2000) classified 102 isolates from Pakistan into eight virulence forms and 14 “pathotype groups” were identified among 40 Canadian isolates assessed for disease reaction on eight chickpea differential lines (Chongo et al., 2004). Pouralibaba et al. (2008) reported three “pathotype groups” present in north-western Iran, whereas Ghiai et al. (2012) reported 10 “virulent forms” and 16 “pathogenic groups,” respectively, from Iran. Most recently, a new highly virulent “pathotype IV” was reported in Syria and the existence of the four previously identified Syrian pathotypes (Atik et al., 2013) were confirmed (Imtiaz et al., 2015).
In Australia, although it appears that the population is largely clonal based on neutral genetic markers, a broad range of aggressiveness exists (Elliott et al., 2013). Hence the Australian chickpea industry is at risk from selective propagation and dispersal of the fittest and best adapted A. rabiei clones. Indeed, since host resistance is multigenic and partial (Cho et al., 2004), there is a heightened risk of resistance erosion caused by selection and increasing frequency of individual clones, with the ability to overcome singular or multiple defense genes/strategies as well as maintain peak fitness (Andrivon et al., 2007).
Two types of adaptation are recognized in fungal species, generalized adaptation and localized adaptation, both resulting in the production of unique haplotypes with high aggressiveness levels and frequencies, due directly to high survival rates (Leonard, 1977). Elliott et al. (2013) first detected clones with differing levels of aggressiveness within the Australian A. rabiei population and proposed that despite its clonal nature, the population contained a large potential to evolve and adapt to overcome chemical and host resistance management strategies. This proposal was based on a small number of isolates. To better understand and manage this risk a much larger study, encompassing a greater number of isolates from multiple growing regions and host genotypes collected over several growing seasons was required. This rationale is supported by observations over recent seasons of severe disease symptomology on host genotypes widely adopted throughout the Australian growing regions and that, until very recently, were considered “resistant” (in the case of Genesis 090 in the southern Victoria and South Australia regions) or “moderately resistant” (in the case of PBA HatTrick in the northern New South Wales and southern Queensland regions).
In order to assess risk to currently employed host resistance and chemical control strategies, as well as to better select for resistance longevity, an in depth understanding of the genetic and pathogenic structure of the Australian A. rabiei population is required. Therefore the aims of this study were to (1) assess the genetic structure of the A. rabiei population and any changes in the structure within and between the major chickpea growing regions of Australia and host genotypes sown, and (2) assess the spread and frequency of the most frequently occurring haplotypes containing the most aggressive isolates, to identify those isolates of highest risk to the Australian chickpea industry. Used together, this new knowledge of diversity, haplotype frequency and aggressiveness will enable strategic choice of isolates for application to resistance breeding programs and to assess for sustainability of resistance in newly deployed and soon to be widely adopted host genotypes.
Materials and Methods
Population Structure: SSRs
Isolate Collection and Culturing
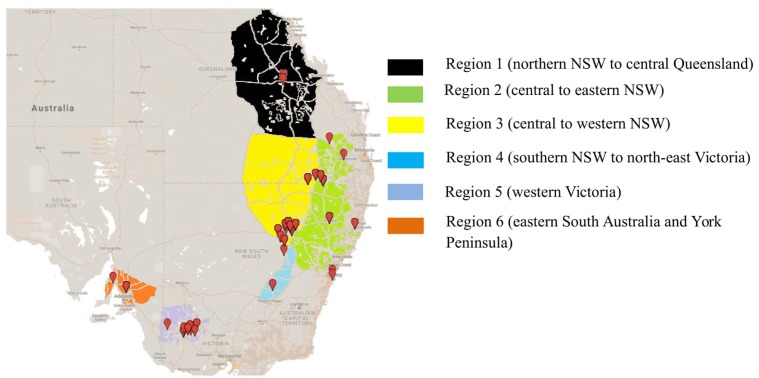
To determine the structure of the Australian A. rabiei population, isolates were collected from commercial chickpea crops and National Variety Trial (NVT) sites during 2013–2015. This was done in a quasi-hierarchical manner, in that wherever possible, infected material was collected from the four corners and one central location within each field. At NVT sites, infected material was collected from as many host genotypes as possible, one sample from each genotype row at each location. For the overall Australian A. rabiei population study, a total of 598 isolates were collected from across the six agro-geographical chickpea growing regions in eastern and southern Australia (Figure 1). The full list of isolates and their available passport data (place of collection, year of collection, and host genotype) is provided in the additional material (Online source 1).
FIGURE 1.
Isolate collection of Ascochyta rabiei from six different agro-geographically classified chickpea growing regions during three consecutive growing seasons (2013–2015).
To assess for selective adaptation on widely grown Australian host genotypes, isolates were intensively collected from Genesis 090 and PBA HatTrick. At the time of study, these were rated as “resistant” and “moderately resistant,” respectively (Pulse Australia, 2009).
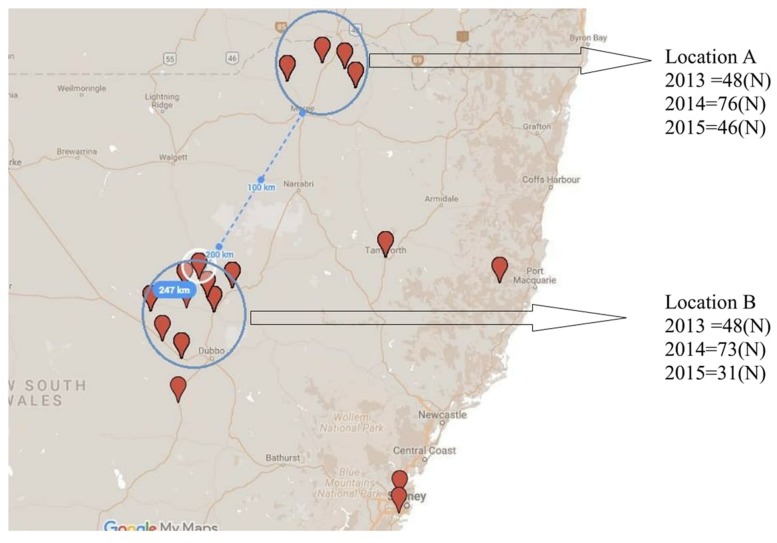
In order to assess for effect of location and track any shift in population structure associated with a single host genotype over time, isolates were collected repeatedly over three consecutive growing seasons (2013, 2014, and 2015) from PBA HatTrick grown in the same two locations (locations A and B), each with a 50 km radius and 247 km apart (Figure 2).
FIGURE 2.
Collection locations and isolate numbers recovered (N) from PBA HatTrick over three consecutive years (2013–2015) for potential association of spatial and temporal effects.
Individual isolates were recovered from pycnidia of only one lesion per infected plant to minimize the likelihood of sampling clones due to short distance dispersal of conidia through rain splash. A single pycnidium per lesion was picked with a sterile needle from an infected chickpea leaf, stem or pod and inoculated into 2 mL of sterile distilled water before streaking onto V8 juice growth agar. Leaf lesions with no visible pycnidia were surface sterilized and placed on V8 juice growth agar. All agar plates were incubated for 14 days at 22 ± 2°C with a 12/12 h near-UV light irradiation (350–400 nm)/dark photoperiod, and resulting cultures were single spored on V8 juice agar media (Elliott et al., 2013).
DNA Extraction and SSR Genotyping
Five hundred and ninety-eight single spored isolates were inoculated separately into 25 mL falcon tubes containing Czapek Dox broth (Difco, Australia) and incubated for 2 weeks at 22 ± 2°C in the dark. Mycelia were then harvested and genomic DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, United States) according to the manufacturer’s instructions.
Seven previously characterized and informative SSR loci were used for determining the genetic structure of the population (Leo et al., 2011). Genotyping was performed using the Multiplex-Ready PCR technique (Hayden et al., 2008), products were separated on a 96 capillary ABI 3730 DNA electrophoresis analyser and allele sizes were analyzed using GeneMapper v4.0 software (Applied Bio-systems) at the Australian Genome Research Facility (AGRF). Allele data was incorporated in population analysis of sizes only relevant to the previously characterized loci repeat polymorphisms.
Molecular Data and Population Structure Analysis
Number of alleles (Na), number of effective alleles (Ne) and Nei’s unbiased gene diversity (Hexp) (Nei, 1978) was used to calculate the genetic diversity was calculated in GenAlex 6.5 (Peakall and Smouse, 2012). The number of multilocus genotypes (MLG), the number of expected MLGs at the smallest sample size based on rarefaction (eMLG), the corrected genotypic diversity index (D), MLG and genotypic evenness (E.5) were calculated using the Poppr package (Kamvar et al., 2014) in R (R Core Team, 2013). Analysis of molecular variance (AMOVA) was performed to examine the variation within and among the above mentioned sub-populations and multilocus analysis was performed to group isolates into haplotypes (online source 1) using GenAlex 6.5. (Peakall and Smouse, 2012).
To visualize the relationships among MLGs in the six sub-populations, SSR data were used to construct a minimum spanning network based on Bruvo’s distance (Bruvo et al., 2004) using the R package Poppr on non-clone-corrected data. The network was visualized using the package igraph (Csardi and Nepusz, 2006). Subsequently, the frequencies of most common haplotype were evaluated separately.
Pathogenic Population Structure
Plant Material
Four chickpea genotypes with known disease reactions were used as a differential host set to assess isolate aggressiveness (Table 1). ICC3996 is used widely as a resistance source in the Australian chickpea breeding program, and Genesis090 and PBA HatTrick are the most widely grown “resistant” host genotypes in southern and northern regions, respectively. Meanwhile, Kyabra remains a widely grown host genotype in the harsher regions of New South Wales and southern and central regions of Queensland due to high yield and quality but is considered “susceptible” and used as a disease check in NVT sites. Seedlings were grown in 15 cm diameter pots containing commercial grade potting mix. Two replicates were sown for each of the genotype × isolate combinations assessed, with five plants grown per pot/rep. All plants were grown and maintained in the glasshouse facility at 22 ± 5°C under 16 h/8 h day/night photoperiod at The University of Melbourne, Parkville campus, Victoria, Australia.
Table 1.
Differential host genotypes and their known disease response ratings to A. rabiei in Australia.
| Genotype | Resistance level | Reference |
|---|---|---|
| Genesis 090 (kabuli) | R | Pulse Australia, 2017 |
| ICC3996 (desi) | R | Nasir et al., 2000 |
| PBA HatTrick (desi) | MR | Pulse Breeding Australia, 2017 |
| Kyabra (desi) | S | Moore et al., 2015 |
Fungal Materials, Inoculation and Disease Assessment
Two hundred and sixty single spored A. rabiei isolates were selected for phenotyping, representative of the years, regions and host genotype origins within the 2013–2015 collection. This included sub-sets of isolates from targeted regions and genotypes as mentioned in section “Population Structure: SSRs”. Single spored isolates were cultured in V8 juice agar and maintained in the incubator for 14 days at 22 ± 2°C with a 12/12 h near-UV light irradiation (350–400 nm)/dark photoperiod prior to being used in the inoculation bioassay.
Inoculum was prepared as described in Sambasivam et al. (2017) and the mini-dome technique of Chen et al. (2005) was used to initiate disease. The disease severity of each isolate on each of the host genotypes was assessed using the qualitative 1–9 scale of Singh et al. (1981) at 21 days after inoculation (dai) where; scores of 1 or 3 represented a low disease severity; 5 represented a moderate disease severity without significant stem infection, and 7 or 9 represented a high disease severity with stem lesions that would lead to major difficulties in transpiration, photosynthesis and/or breakage.
Highly Aggressive Isolates and Pathogenicity Grouping
Isolates identified as highly aggressive produced a cumulative leaf score of at least 7 on >80% and a stem score of at least 7 on >10% of all of the host plants assessed. Subsequently, this sub-set of isolates were placed into pathogenicity groups based on their ability to cause low, moderate or high disease severity independently on ICC3996, Genesis090 and PBA HatTrick (Table 2).
Table 2.
Criteria used for pathogenicity grouping of the highly aggressive isolates.
| Pathogenicity group | Description |
|---|---|
| 1 | High disease on PBA HatTrick and low disease on Genesis090 and ICC3996 |
| 2 | High disease incidence on PBA HatTrick, moderate disease on Genesis090 and low disease on ICC3996 |
| 3 | High disease on PBA HatTrick, moderate disease on Genesis090 and moderate disease on ICC3996 |
| 4 | High disease on PBA HatTrick, high disease on Genesis090 and moderate disease on ICC3996 |
Highest Risk Isolates to the Australian Chickpea Industry
Isolates of highest risk were identified on the basis of genotype and phenotype data. Accordingly, these belonged to the most frequently detected haplotype and were the most aggressive on the best resistance sources used within the advanced breeding program. Highest risk isolates were also aggressive on the currently deployed “resistant” host genotypes (pathogenicity group 4).
Results
Population Structure: SSRs
Between two and eight alleles were identified for each of the SSR loci across the collection of 598 isolates. The maximum gene diversity (Hexp) at each ranged from 0.020 to 0.183 with an average of 0.066 (Hexp). Locus ArA03T (Hexp = 0.183) was the most informative, followed by ArH05T (Hexp = 0.132) (Table 3).
Table 3.
The informative microsatellite loci used for genotyping the Australian A. rabiei population.
| Locus | Allele size | Number of allele | Number of Allele in regions |
Size of allele | Diversity (Hexp) ±SE | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Region 1 (N = 18) | Region 2 (N = 233) | Region 3 (N = 130) | Region 4 (N = 92) | Region 5 (N = 56) | Region 6 (N = 69) | |||||
| ArA03T | 409–439 | 8 | 5 | 3 | 4 | 2 | 3 | 4 | 409, 412, 415, 421, 424, 427, 430, 433, 439 | 0.183 ± 0.057 |
| ArH05T | 221–257 | 6 | 2 | 4 | 2 | 3 | 2 | 3 | 221, 233, 239, 242, 248, 254 | 0.132 ± 0.035 |
| ArR12D | 185–191 | 3 | 2 | 1 | 2 | 2 | 2 | 2 | 185, 187, 189, 191 | 0.038 ± 0.017 |
| ME14-1-56 | 379–383 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 379, 383 | 0.039 ± 0.039 |
| ME14-1-63 | 313–319 | 3 | 1 | 2 | 1 | 3 | 2 | 1 | 313, 316, 319 | 0.033 ± 0.021 |
| ME14-1-83 | 283–285 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 283, 285 | 0.020 ± 0.019 |
| ME14-1-91 | 339-342 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 333, 339, 342 | 0.021 ± 0.018 |
| Mean (Hexp) = 0.066 ± 0.015 | ||||||||||
N = Number of individuals observed. Hexp = Nei’s unbiased gene diversity (Nei, 1978). SE = Standard error.
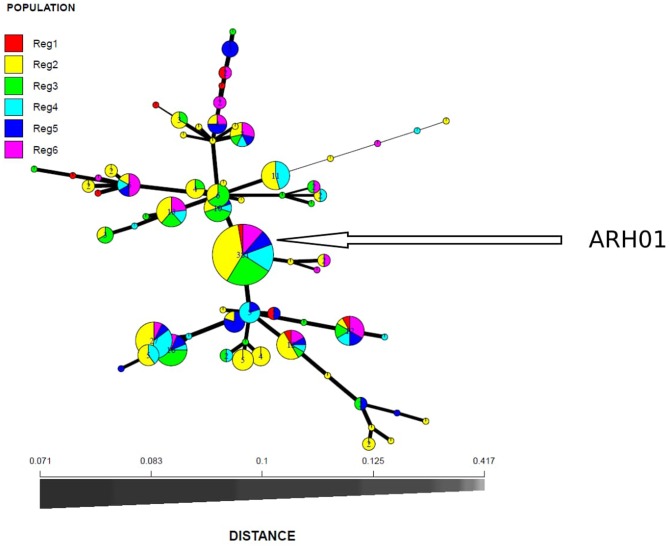
In total, 66 haplotypes were detected, of which 34 were detected just once (n = 1) (Supplementary Table 1). The detection frequency of each haplotype and the genetic relationships among them revealed by the seven SSR loci are presented in Figure 3. The most frequently detected haplotype, ARH01 accounted for 55.35 to 72.30% of the six regional population across all growing seasons (Table 4). In accordance with the overall gene diversity detected, the highest gene diversity was observed in Region 1 (Hexp = 0.161), which contained the most unique and effective alleles (2.28 and 1.20, respectively). Gene diversity in Region 1 was significantly higher than in all other regions (P ≤ 0.001). The eMLG (9.00), E.5 values (0.49), Ne (1.20), and Na (2.28) values were also highest for Region 1, indicating a more diverse population in this region compared to the other analyzed regions. However, the corrected Simpson’s genotypic diversity index (D) did not differ greatly among regions The mean low genotypic diversity (D = 0.57) indicated the consistent, low diversity detected within the entire Australian A. rabiei population (Table 4).
FIGURE 3.
Minimum spanning network based on Bruvo genetic distances representing 66 MLGs observed in six A. rabiei populations from Australia. Node colors represent population membership proportional to the pie size. Node sizes are relatively scaled to logl.75n, where n is the number of samples in the nodes to reduce node overlap. Edge thickness (lines) represent minimum genetic distance between haplotype.
Table 4.
The genetic structure of the population detected within each of the six growing regions.
| Regions | N | MLG | eMLG ± SE | Na ± SE | Ne ± SE | Hexp ± S.E | E.5 | D | % of ARH01 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 18 | 9 | 9 ± 0.00 | 2.28 ± 0.47 | 1.20 ± 0.08 | 0.161 ± 0.05 | 0.498 | 0.555 | 55.55 |
| 2 | 233 | 38 | 6.8 ± 1.73 | 1.85 ± 0.45 | 1.05 ± 0.03 | 0.049 ± 0.03 | 0.269 | 0.555 | 63.09 |
| 3 | 130 | 21 | 5.38 ± 1.52 | 1.85 ± 0.40 | 1.04 ± 0.04 | 0.039 ± 0.03 | 0.312 | 0.592 | 72.30 |
| 4 | 92 | 18 | 6.37 ± 1.49 | 1.85 ± 0.34 | 1.07 ± 0.04 | 0.061 ± 0.03 | 0.382 | 0.592 | 60.86 |
| 5 | 56 | 16 | 7.78 ± 1.49 | 1.71 ± 0.28 | 1.06 ± 0.03 | 0.055 ± 0.02 | 0.393 | 0.554 | 55.35 |
| 6 | 69 | 16 | 6.82 ± 1.50 | 1.85 ± 0.45 | 1.03 ± 0.02 | 0.034 ± 0.01 | 0.362 | 0.652 | 62.31 |
Mean D = 0.57. N = Number of individuals observed. MLG = Number of multilocus genotypes (MLG) observed. eMLG = The number of expected MLG at the smallest sample size based on rarefaction. SE = Standard error. Na = Number of different alleles (Kalinowski, 2005). Ne = Number of effective alleles (Kalinowski, 2005). Hexp = Nei’s unbiased gene diversity (Nei, 1978). E.5 = Genotypic evenness (Pielou, 1975; Ludwig and Reynolds, 1988; Grünwald et al., 2003). D = Corrected Simpson’s Index (Simpson, 1949).
A total of 430 isolates were collected from the two widely adopted and “resistant” or “moderately resistant” host genotypes, Genesis090 and PBA HatTrick. Within these sub-populations, a total of 17 (N = 55) and 47 (N = 373) MLGs were observed on Genesis090 and PBA HatTrick, respectively (Table 5). Regardless of the host genotypes, the most frequently detected haplotype was ARH01, which accounted for 54 and 63% of the isolates detected on Genesis090 and PBA HatTrick, respectively (Table 5). Although more than five times the number of isolates were collected from PBA HatTrick than Genesis090, no significant difference in gene and genotypic diversity measures (P ≤ 0.45) was detected among the isolate groups. Furthermore, genetic diversity were all low (Table 5).
Table 5.
The genetic structure of the A. rabiei population detected on two widely adopted host genotypes.
| Hosts | N | MLG | eMLG ± SE | Na ± SE | Ne ± SE | Hexp ± SE | E.5 | D | % of ARH01 |
|---|---|---|---|---|---|---|---|---|---|
| Genesis090 | 57 | 17 | 4.91 ± 1.32 | 1.85 ± 0.45 | 1.06 ± 0.03 | 0.052 ± 0.02 | 0.37 | 0.662 | 57.89 |
| PBA HatTrick | 373 | 47 | 4.28 ± 1.37 | 2.28 ± 0.42 | 1.05 ± 0.03 | 0.048 ± 0.02 | 0.26 | 0.575 | 64.61 |
N = Number of individuals observed. MLG = Number of multilocus genotypes (MLG) observed. eMLG = The number of expected MLG at the smallest sample size based on rarefaction. SE = Standard error. Na = Number of different alleles (Kalinowski, 2005). Ne = Number of effective alleles (Kalinowski, 2005). Hexp = Nei’s unbiased gene diversity (Nei, 1978). E.5 = Genotypic evenness (Pielou, 1975; Ludwig and Reynolds, 1988; Grünwald et al., 2003). D = Corrected Simpson’s Index (Simpson, 1949).
No Evidence of Temporal or Spacial Population Shift
Although some differences in gene diversity (Hexp) were observed between the 1st and 3rd years of sampling, increasing from 0.024 to 0.053 at Location A, and from 0.024 to 0.059 at Location B, these changes were not significantly different to those detected in the 2nd year of sampling. Also, the frequency of the ARH01 haplotype remained almost static at each independently sampled geographical location, A and B, and across the 3 years of sampling (ranging from 58.62 to 66.67%; Figure 2). Thus, no evidence of either a temporal or spacial population shift in SSR diversity was observed over the period of the study, at the epidemic locations sampled (Table 6).
Table 6.
The genetic structure of the population detected within each of the six growing regions.
| Year | Population locations (N) | MLG | eMLG ± SE | Na ± SE | Ne ± SE | Hexp ± S.E | E.5 | D | % of ARH01 |
|---|---|---|---|---|---|---|---|---|---|
| 2013 | A (48) | 12 | 8.86 ± 1.27 | 1.42 ± 0.29 | 1.02 ± 0.02 | 0.024 ± 0.018 | 0.382 | 0.544 | 66.67 |
| B (48) | 12 | 8.86 ± 1.27 | 1.43 ± 0.29 | 1.02 ± 0.02 | 0.024 ± 0.018 | 0.382 | 0.540 | 66.66 | |
| 2014 | A (72) | 21 | 10.51 ± 1.81 | 1.42 ± 0.29 | 1.03 ± 0.03 | 0.031 ± 0.021 | 0.308 | 0.584 | 63.88 |
| B (72) | 21 | 10.51 ± 1.81 | 1.43 ± 0.29 | 1.03 ± 0.02 | 0.031 ± 0.021 | 0.308 | 0.630 | 63.88 | |
| 2015 | A (42) | 10 | 8.15 ± 1.05 | 1.71 ± 0.47 | 1.06 ± 0.02 | 0.053 ± 0.029 | 0.418 | 0.630 | 66.66 |
| B (29) | 9 | 8.867 ± 9.0 | 1.43 ± 0.20 | 1.06 ± 0.03 | 0.059 ± 0.028 | 0.491 | 0.576 | 58.62 |
N = Number of individuals observed. MLG = Number of multilocus genotypes (MLG) observed. eMLG = The number of expected MLG at the smallest sample size based on rarefaction. SE = Standard error. Na = Number of different alleles (Kalinowski, 2005). Ne = Number of effective alleles (Kalinowski, 2005). Hexp = Nei’s unbiased gene diversity (Nei, 1978). E.5 = Genotypic evenness (Pielou, 1975; Ludwig and Reynolds, 1988; Grünwald et al., 2003). D = Corrected Simpson’s Index (Simpson, 1949).
Pathogenic Population Structure and Highest Risk Isolates
Among the 260 isolates assessed, 54 (21%) were highly aggressive and categorized into highly aggressive pathogenicity groups. Of these, 62% belonged to pathogenicity group 1, 2% belonged to pathogenicity group 2, 13% belonged to pathogenicity group 3 and 23% belonged to pathogenicity group 4 (Supplementary Table 2).
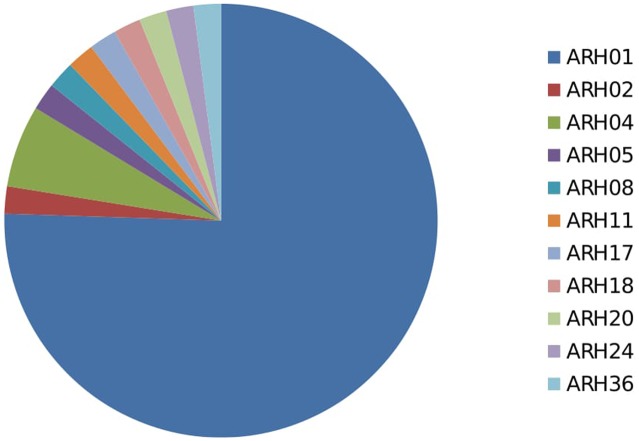
Among the 54 highly aggressive isolates identified, 75% belonged to haplotype ARH01, far more than detected in any other haplotype group, and recovered across all of the growing regions and hosts assessed. Far lower frequencies of the highly aggressive isolates were detected within other haplotype groups (Figure 4; Supplementary Table 3).
FIGURE 4.
Proportion of highly aggressive isolates from different haplotype group.
Discussion
Together with the strategic application of fungicides, chickpea production is reliant on the host containing the optimal combination of A. rabiei resistance alleles for timely recognition and defense initiation. Meanwhile, the pathogen population is under constant selective pressure to alter, to reproduce and spread new versions of itself that are able to survive in new environments, evade detection by the host and potentially overcome early host defenses (Pandey et al., 2016). In a clonal population, this occurs through opportunistic mutation and spread of the fittest and most widely adaptable isolates (Messer et al., 2016). Given the clonal nature of A. rabiei in Australia (Leo et al., 2015), it is unsurprising that we have detected a limited number of haplotypes that occur frequently throughout the chickpea growing regions, independent of host genotype.
Population Structure: SSRs
The overall low genetic diversity found within the comprehensive assessment of the Australian A. rabiei population in this study is a common finding with Leo et al. (2015). Even if compatible mating types were present in the population, it seems likely that the expansion of this population to date has occurred through clonal means, perhaps due to forces suppressing recombination such as an initial imbalance in the mating type ratio of the founder isolates. The life cycles of many fungal species alternate between asexual and sexual multiplication (Hawker, 2016; Laloi et al., 2016) and asexual reproduction is often the major reproductive mechanism during epidemics to quickly increase the frequency of fit individuals (Laloi et al., 2016).
Within the Australian A. rabiei population, the large haplotype group (ARH01) occurs at extremely high frequencies of up to 63% of the entire population within a region. This haplotype likely established during an initial founder effect (Rivas et al., 2004) due to specific fitness characteristics that enabled survival at the point of introduction. The isolates then likely spread to other growing regions through infected seed distribution (Galloway and MacLeod, 2003). Subsequently, isolates that were highly adapted to survive in the range of agro-geographical regions and able to overcome host resistance proliferated through clonal propagation, causing severe disease epidemics when optimal climatic conditions prevailed. The resultant genetic and genotypic diversities observed in the Australian A. rabiei population reflect this founder effect, whereby the establishment and success of the pathogen has occurred through an available niche, provided by the abundance of susceptible host and an optimal environment. The number of founder events that have occurred for A. rabiei in Australia is unknown, but potentially the increasing frequencies of several haplotypes other than ARH01 in the population is an indication of slow evolution of other groups of highly adapted isolates, which should be monitored for increases in highly aggressive isolate frequency.
The genotypic diversity detected among isolates recovered from Genesis090 was not significantly different to that detected among those recovered from PBA HatTrick. Whilst it is likely that the host genotype would contribute to shaping the structure of the pathogen population (Jones and Dangl, 2006; Ley et al., 2006), the number of isolates assessed over the time period in the current study may not have been sufficient to visualize this phenomenon. Given the relatively short period for potential adaptation (<40 years), the clonal Australian population may still be experiencing the original founder effects. More in-depth investigation is required to determine if host factors are contributing to population adaptation. This might be through tracking of specific isolates over time and observations of the host defense responses that are instigated within each of the hosts under investigation. A similar, but smaller scale study was previously conducted by Leo et al. (2016), who found some host-specific differences in defense-related gene expressions. Expression of differential host defense responses to specific isolate populations might also identify factors that impact on survival and reproduction of specific populations and hence inform management strategies within growing regions where particular hosts are grown (Hollomon and Brent, 2009; Bertolini et al., 2012; Ali et al., 2016).
The non-significant differences detected in genotypic diversity over time at both of the locations assessed in New South Wales was unsurprising given the clonal nature of the pathogen and the short period since introduction to Australia. In a similar study, no significant changes were observed over 3 years within a Mycosphaerella graminicola population, DNA fingerprints were used to identify colones produced through asexual reproduction, suggesting and genetic stability of fungus was proposed (Chen et al., 1994).
Although overall seemingly stable and despite being clonal, we cannot ignore the potential for the existing population to change and evolve rapidly in response to an external factor (Messer et al., 2016). Rapid evolution of the wheat pathogen Zymoseptoria tritici was determined to be due to clustering of transposable elements leading to generation of extensive rearrangements and multiple independent gene losses (Hartmann et al., 2017). Rapid population changes may also occur through selective sweeps, potentially linked to host genotypes and/or chemical controls, resulting in “adaptive walk” and genetic shift among a limited number of frequently occurring haplotypes (Orr, 1998; Messer et al., 2016). Evidence of this may become more apparent as the industry adopts new resistant host genotypes such as PBA Seamer, which will need careful monitoring for pathogen population shift (Pulse Australia, 2017).
The Pathogenic Population Structure
Evaluating A. rabiei populations on a set of differentials with different levels of resistance is useful for monitoring aggressiveness changes and to identify the most aggressive isolates. These isolates are required for selective breeding and, potentially, disease management strategies, particularly if differential factors underpinning aggressiveness are able to be dissected. Despite the SSR clonal composition, a similar wide diversity in aggressiveness was detected within the Australian population as previously detected elsewhere (Iqbal et al., 2004; Benzohra et al., 2011; Atik et al., 2013; Mahiout et al., 2015). Highly aggressive isolates were able to cause differential disease severities across a host set including two of the most widely adopted cultivars that underpin the Australian chickpea industry, Genesis090 and PBA HAtTrick. Genesis090 was introduced from ICARDA, Syria, where it was tested as FLIP94-090C, while PBA HatTrick is a cross of cv. Jimbour and the resistant Iranian landrace ICC14903 (Pulse Australia, 2017). However, these host genotypes have in recent years experienced an increase in susceptibility to A. rabiei (Moore et al., 2016), in keeping with our observation of increased frequencies of highly aggressive isolates within the population.
Increased aggressiveness within the Australian A. rabiei population was highlighted by the recent first observation of pycnidia formation on ICC3996, one of the resistance pillars of the chickpea breeding program, in field trials (Moore et al., 2016). Consequently, erosion of resistance in host genotypes that contain alleles from this source is highly likely as the adapted highly aggressive isolates spread and most likely become more frequent. Other sources of resistance will be necessary in the immediate future to underpin the chickpea breeding programs in Australia. Such material should be selected based on ability to resist the diversity of the pathogen population since different genetic mechanisms are likely to be controlling aggressiveness between the different pathogenicity groups (Hamid and Strange, 2000). Although the most destructive isolates detected in this study were of Pathogenicity Group 4, we must remember that this classification was based on disease severity on ICC3996. It is highly likely that differential reactions would occur on other resistance sources and that these should be more fully characterized for their own disease reactions to the representative pathogen population before being used within the breeding program.
Conclusion
Within the adapted clonal groups detected in this study, we can surmise that isolates were selected by their ability to overcome resistance within the widely adopted “resistant” host genotypes such as Genesis090 and PBA HatTrick. This would help to explain the occurrence of a greater frequency of highly aggressive isolates within the ARH01 haplotype group, creating “super isolates” of the very highest pathogenicity ranking able to survive in many locations and on a wide range of host genotypes. These isolates represent the very highest risk to the Australian chickpea industry. However, several factors must be considered when selecting accessions that appear “resistant” to these: (1) This study suggests rapid changes in aggressiveness of the pathogen population, (2) only one mating type of A. rabiei has been detected in Australia but, if both mating types were present, the sexual reproductive cycle may quickly become active to create a recombinant pool population, and (3) isolate sampling and testing is limited by time and resources leading to the possibility of missing some important aggressive isolates. To extend stability of resistance, our growers must maintain their best practice in farming systems including growing clean seed, a minimum of 3-year rotations, effective distances between chickpea crops and fungicide spray regimes. Meanwhile, further studies are required to better understand the genetics of resistance in order to develop host genotypes with different resistance gene combinations, to potentially reduce selective adaptation of the pathogen.
Author Contributions
YM assessed the molecular structure of the A. rabiei population and constructed the manuscript. PS assessed the pathogenic structure of the A. rabiei population and drafted the manuscript. JD, KH, KM, CL, SK, JB, and AL contributed to analyses and manuscript production. RF supervised the research and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. The research was funded by the Grains Research and Development Cooperation within project UM00052.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01029/full#supplementary-material
References
- Ali H., Alam S., Attanayake R., Rahman M., Chen W. (2012). Population structure and mating type distribution of the chickpea blight pathogen Ascochyta rabiei from Pakistan and the United States. J. Plant Pathol. 94 99–108. [Google Scholar]
- Ali S., Soubeyrand S., Gladieux P., Giraud T., Leconte M., Gautier A., et al. (2016). CLONCASE: estimation of sex frequency and effective population size by clonemate resampling in partially clonal organisms. Mol. Ecol. Resour. 16 845–861. 10.1111/1755-0998.12511 [DOI] [PubMed] [Google Scholar]
- Andrivon D., Pilet F., Montarry J., Hafidi M., Corbière R., Achbani E. H., et al. (2007). Adaptation of Phytophthora infestans to partial resistance in potato: evidence from French and Moroccan populations. Phytopathology 97 338–343. 10.1094/PHYTO-97-3-0338 [DOI] [PubMed] [Google Scholar]
- Atik O., Ahmed S., Abang M. M., Imtiaz M., Hamwieh A., Baum M., et al. (2013). Pathogenic and genetic diversity of Didymella rabiei affecting chickpea in Syria. Crop Prot. 46 70–79. 10.1016/j.cropro.2012.12.012 [DOI] [Google Scholar]
- Baite M. S., Dubey S., Singh B. (2016). Morphological variability in the Indian isolates of Ascochyta rabiei causing blight in chickpea and evaluation of chickpea cultivars. Indian J. Plant Prot. 44 74–82. [Google Scholar]
- Barve M., Arie T., Salimath S., Muehlbauer F., Peever T. (2003). Cloning and characterization of the mating type MAT locus from Ascochyta rabiei teleomorph Didymella rabiei and a MAT phylogeny of legume-associated Ascochyta spp. Fungal Genet. Biol. 39 151–167. 10.1016/S1087-1845(03)00015-X [DOI] [PubMed] [Google Scholar]
- Benzohra I. E., Bendahmane B. S., Labdi M., Benkada M. Y. (2011). Identification of pathotypes and physiological races in Ascochyta rabiei (Pass.) Labr., The Agent of Ascochyta Blight in Chickpea (Cicer arietinum) in Algeria. World Appl. Sci. J. 15 978–984. [Google Scholar]
- Bertolini L., Feltrin C., Gaudencio Neto S., Martins L., Tavares K., Rodrigues V., et al. (2012). Animal cloning: survival of the fittest. Ciência Animal 22 82–105. [Google Scholar]
- Bretag T., MacLeod W., Kimber R., Moore K., Knights E., Davidson J. (2008). Management of ascochyta blight in chickpeas in Australia. Australas. Plant Pathol. 37 486–497. 10.1071/AP08046 [DOI] [Google Scholar]
- Bruvo R., Michiels N. K., D’SOUZA T. G., Schulenburg H. (2004). A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Mol. Ecol. 13 2101–2106. 10.1111/j.1365-294X.2004.02209.x [DOI] [PubMed] [Google Scholar]
- Chen R. S., Boeger J., McDonald B. (1994). Genetic stability in a population of a plant pathogenic fungus over time. Mol. Ecol. 3 209–218. 10.1111/j.1365-294X.1994.tb00054.x [DOI] [Google Scholar]
- Chen W. M., James E. K., Chou J. H., Sheu S. Y., Yang S. Z., Sprent J. I. (2005). B-rhizobia from mimosa pigra, a newly discovered invasive plant in taiwan. New Phytol. 168 661–675. 10.1111/j.1469-8137.2005.01533.x [DOI] [PubMed] [Google Scholar]
- Cho S., Chen W., Muehlbauer F. J. (2004). Pathotype-specific genetic factors in chickpea (Cicer arietinum L.) for quantitative resistance to ascochyta blight. Theor. Appl. Genet. 109 733–739. 10.1007/s00122-004-1693-x [DOI] [PubMed] [Google Scholar]
- Chongo G., Gossen B., Buchwaldt L., Adhikari T., Rimmer S. (2004). Genetic diversity of Ascochyta rabiei in Canada. Plant Dis. 88 4–10. 10.1094/PDIS.2004.88.1.4 [DOI] [PubMed] [Google Scholar]
- Csardi G., Nepusz T. (2006). The igraph software package for complex network research. InterJournal Complex Syst. 1695 1–9. [Google Scholar]
- Elliott V. L., Taylor P. W., Ford R. (2013). Changes in foliar host reaction to Ascochyta rabiei with plant maturity. J. Agric. Sci. 5 29 10.5539/jas.v5n7p29 [DOI] [Google Scholar]
- Galloway J., MacLeod W. (2003). Didymella rabiei, the teleomorph of Ascochyta rabiei, found on chickpea stubble in Western Australia. Australas. Plant Pathol. 32 127–128. 10.1071/AP02076 [DOI] [Google Scholar]
- Gan Y. T., Siddique K. H. M., MacLeod W. J., Jayakumar P. (2006). Management options for minimizing the damage by ascochyta blight (Ascochyta rabiei) in chickpea (Cicer arietinum L.). Field Crops Res. 97 121–134. 10.1016/j.fcr.2005.10.002 [DOI] [Google Scholar]
- Ghiai S., Razavi M., Shahriyari D. (2012). Study on pathogenic and molecular variability in some isolates of Ascochyta rabiei causal agent of Ascochyta blight of chickpea in Iran. Appl. Entomol. Phytopathol. 79 199–218. [Google Scholar]
- Grewal J. (1984). “Evidence of physiologic races in Ascochyta rabiei of chickpea,” in Proceedings of the Workshop on Ascochyta Blight, and Wintering. (of) Chickpeas, eds Saxena M. C., Singh K. B. (Aleppo: ICARDA; ), 55–65. [Google Scholar]
- Grünwald N. J., Goodwin S. B., Milgroom M. G., Fry W. E. (2003). Analysis of genotypic diversity data for populations of microorganisms. Phytopathology 93 738–746. 10.1094/PHYTO.2003.93.6.738 [DOI] [PubMed] [Google Scholar]
- Hamid K., Strange R. N. (2000). Phytotoxicity of solanapyrones A and B produced by the chickpea pathogen Ascochyta rabiei (Pass.) Labr. and the apparent metabolism of solanapyrone A by chickpea tissues. Physiol. Mol. Plant Pathol. 56 235–244. 10.1006/pmpp.2000.0272 [DOI] [Google Scholar]
- Hartmann F. E., Sánchez-Vallet A., McDonald B. A., Croll D. (2017). A fungal wheat pathogen evolved host specialization by extensive chromosomal rearrangements. ISME J. 11 1189–1204. 10.1038/ismej.2016.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawker L. E. (2016). The Physiology of Reproduction in Fungi. Cambridge: Cambridge University Press. [Google Scholar]
- Hayden M., Nguyen T., Waterman A., McMichael G., Chalmers K. (2008). Application of multiplex-ready PCR for fluorescence-based SSR genotyping in barley and wheat. Mol. Breed. 21 271–281. 10.1007/s11032-007-9127-5 [DOI] [Google Scholar]
- Hollomon D. W., Brent K. J. (2009). Combating plant diseases the Darwin connection. Pest. Manag. Sci. 65 1156–1163. 10.1002/ps.1845 [DOI] [PubMed] [Google Scholar]
- Imtiaz M., Abang M., Malhotra R., Ahmed S., Bayaa B., Udupa S., et al. (2015). Pathotype IV, a new and highly virulent pathotype of Didymella rabiei, causing Ascochyta blight in chickpea in Syria. Arch. Phytopathol. Plant Prot. 48 921–930. [DOI] [PubMed] [Google Scholar]
- Iqbal S., Ghafoor A., Ayub N., Ahmad Z. (2004). Pathogenic diversity in Ascochyta rabiei isolates collected from Pakistan. Pak. J. Bot. 36 429–438. [Google Scholar]
- Jamil F., Sarwar N., Sarwar M., Khan J., Geistlinger J., Kahl G. (2000). Genetic and pathogenic diversity within Ascochyta rabiei (Pass.) Lab. populations in Pakistan causing blight of chickpea (Cicer arietinum L.). Physiol. Mol. Plant Pathol. 57 243–254. 10.1006/pmpp.2000.0303 [DOI] [Google Scholar]
- Jan H., Wiese M. (1991). Virulence forms of Ascochyta rabiei affecting chickpea in the Palouse. Plant Dis. 75 904–906. 10.1094/PD-75-0904 [DOI] [Google Scholar]
- Jayakumar P., Gan Y., Gossen B., Warkentin T., Banniza S. (2005). Ascochyta blight of chickpea: infection and host resistance mechanisms. Can. J. Plant Pathol. 27 499–509. 10.1080/07060660509507251 [DOI] [Google Scholar]
- Jones J. D., Dangl J. L. (2006). The plant immune system. Nature 444 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kalinowski S. T. (2005). HP-rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 5 187–189. 10.1111/j.1471-8286.2004.00845.x [DOI] [Google Scholar]
- Kamvar Z. N., Tabima J. F., Grünwald N. J. (2014). Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2:e281 10.7717/peerj.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi G., Montarry J., Guibert M., Andrivon D., Michot D., Le May C. (2016). Aggressiveness changes in populations of Didymella pinodes over winter and spring pea cropping seasons. Appl. Environ. Microbiol. 82 4330–4339. 10.1128/AEM.00480-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo A. E., Ford R., Linde C. C. (2015). Genetic homogeneity of a recently introduced pathogen of chickpea, Ascochyta rabiei, to Australia. Biol. Invasion. 17 609–623. 10.1007/s10530-014-0752-8 [DOI] [Google Scholar]
- Leo A. E., Ford R., Linde C. C., Shah R. M., Oliver R., Taylor P. W., et al. (2011). Characterization of fifteen newly developed microsatellite loci for the chickpea fungal pathogen Ascochyta rabiei. Mol. Ecol. Resour. 11 418–421.21429157 [Google Scholar]
- Leo A. E., Linde C. C., Ford R. (2016). Defence gene expression profiling to Ascochyta rabiei aggressiveness in chickpea. Theor. Appl. Genet. 129 1333–1345. 10.1007/s00122-016-2706-2 [DOI] [PubMed] [Google Scholar]
- Leonard K. (1977). Selection pressures and plant pathogens. Ann. N. Y. Acad. Sci. 287 207–222. 10.1111/j.1749-6632.1977.tb34240.x [DOI] [Google Scholar]
- Ley R. E., Peterson D. A., Gordon J. I. (2006). Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124 837–848. 10.1016/j.cell.2006.02.017 [DOI] [PubMed] [Google Scholar]
- Ludwig J. A., Reynolds J. F. (1988). Statistical Ecology: A Primer in Methods and Computing. Hoboken NJ: John Wiley & Sons. [Google Scholar]
- Mahiout D., Bendahmane B. S., Benkada M. Y., Rickauer M. (2015). Physiological Characterisation of Ascochyta rabiei (Pass.) Lab. Isolated from diseased chickpea Fields in Six Regions of Northwestern Algeria. Am. Eur. J. Agric. Environ. Sci. 15 1136–1146. [Google Scholar]
- McDonald B. A., Linde C. (2002). The population genetics of plant pathogens and breeding strategies for durable resistance. Euphytica 124 163–180. 10.1023/A:1015678432355 [DOI] [Google Scholar]
- Messer P. W., Ellner S. P., Hairston N. G. (2016). Can population genetics adapt to rapid evolution? Trends Genet. 32 408–418. 10.1016/j.tig.2016.04.005 [DOI] [PubMed] [Google Scholar]
- Moore K., Hobson K., Dron S., Harden S., Bithell P., Sambasivam R., et al. (2016). Available at. https://grdc.com.au/Research-and-Development/GRDC-Update-Papers/2016/03/Chickpea-Ascochyta-latest-research-on-variability-and-implications-for-management. [Google Scholar]
- Moore K., Ryley M., Cumming G., Jenkins L. (2015). Available at. http://www.pulseaus.com.au/growing-pulses/bmp/chickpea/ascochyta-blight [Google Scholar]
- Nasir M., Bretag T., Kaiser W., Meredith K., Brouwer J. (2000). Screening chickpea germplasm for Ascochyta blight resistance. Australas. Plant Pathol. 29 102–107. 10.1071/AP00018 [DOI] [Google Scholar]
- Navas-Cortés J., Perez-Artes E., Jimenez-Diaz R., Llobell A., Bainbridge B., Heale J. (1998). Mating type, pathotype and RAPDs analysis in Didymella rabiei, the agent of ascochyta blight of chickpea. Phytoparasitica 26 199–212. 10.1007/BF02981435 [DOI] [Google Scholar]
- Nei M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene Y. (1982). A review of Ascochyta blight of chickpea. Int. J. Pest Manage. 28 61–70. 10.1080/09670878209370675 [DOI] [Google Scholar]
- Nourollahi K., Javannikkhah M., Naghavi M. R., Lichtenzveig J., Okhovat S. M., Oliver R. P., et al. (2011). Genetic diversity and population structure of Ascochyta rabiei from the western Iranian Ilam and Kermanshah provinces using MAT and SSR markers. Mycol. Prog. 10 1–7. 10.1007/s11557-010-0668-3 [DOI] [Google Scholar]
- Orr H. A. (1998). The population genetics of adaptation: the distribution of factors fixed during adaptive evolution. Evolution 52 935–949. 10.1111/j.1558-5646.1998.tb01823.x [DOI] [PubMed] [Google Scholar]
- Pande S., Siddique K., Kishore G., Bayaa B., Gaur P., Gowda C., et al. (2005). Ascochyta blight of chickpea (Cicer arietinum L.): a review of biology, pathogenicity, and disease management. Crop Pasture Sci. 56 317–332. 10.1016/j.biotechadv.2008.06.001 [DOI] [Google Scholar]
- Pandey D., Rajendran S. R. C. K., Gaur M., Sajeesh P., Kumar A. (2016). Plant defense signaling and responses against necrotrophic fungal pathogens. J. Plant Growth Regul. 35 1159–1174. 10.1007/s00344-016-9600-7 [DOI] [Google Scholar]
- Peakall R., Smouse P. E. (2012). GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research an update. Bioinformatics 28 2537–2539. 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peever T., Chen W., Abdo Z., Kaiser W. (2012). Genetics of virulence in Ascochyta rabiei. Plant Pathol. 61 754–760. 10.1111/j.1365-3059.2011.02566.x [DOI] [Google Scholar]
- Peever T., Salimath S., Su G., Kaiser W., Muehlbauer F. (2004). Historical and contemporary multilocus population structure of Ascochyta rabiei (teleomorph: Didymella rabiei) in the Pacific Northwest of the United States. Mol. Ecol. 13 291–309. 10.1046/j.1365-294X.2003.02059.x [DOI] [PubMed] [Google Scholar]
- Phan H., Ford R., Taylor P. (2003). Population structure of Ascochyta rabiei in Australia based on STMS fingerprints. Fungal Divers. 13 111–129. [Google Scholar]
- Pielou E. C. (1975). Ecological Diversity. New York, NY: Wiley. [Google Scholar]
- Pouralibaba H., Mahmoudi F., Keshavarz K., Nour E. K. (2008). Identification of pathotypes of Didymella rabiei causing agent of chickpea blight disease, in different parts of iran using trap nursery. J. Plant Pathol. 44 170–175. [Google Scholar]
- Pulse Australia (2009). Available at: http://www.pulseaus.com.au/storage/app/media/crops/pulses/2016 [Google Scholar]
- Pulse Australia (2017). Available at: http://www.pulseaus.com.au/growing-pulses/bmp/chickpea [Google Scholar]
- Pulse Breeding Australia (2017). Available at: https://grdc.com.au/research/trials,-programs-and-initiatives/pba/link3.aspx [Google Scholar]
- R Core Team (2013). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. [Google Scholar]
- Rhaïem A., Chérif M., Harrabi M., Strange R. (2006). First report of Didymella rabiei on chickpea debris in Tunisia. Tunis. J. Plant Prot. 1 13–18. [Google Scholar]
- Rivas G. G., Zapater M. F., Abadie C., Carlier J. (2004). Founder effects and stochastic dispersal at the continental scale of the fungal pathogen of bananas Mycosphaerella fijiensis. Mol. Ecol. 13 471–482. 10.1046/j.1365-294X.2003.02043.x [DOI] [PubMed] [Google Scholar]
- Sambasivam P., Taylor P., Ford R. (2017). Pathogenic variation and virulence related responses of Ascochyta lentis on lentil. Eur. J. Plant Pathol. 147 265–277. 10.1007/s10658-016-0999-2 [DOI] [Google Scholar]
- Simpson E. (1949). Measurement of diversity. Nature 163:688 10.1038/163688a0 [DOI] [Google Scholar]
- Singh K. B., Hawtan G. C., Nane Y. L., Reddy M. V. (1981). Resistance to chickpea against Ascochyta blight. Plant Dis. 65 586–587. 10.1094/PD-65-586 [DOI] [Google Scholar]
- Tekin M., Sari D., Catal M., Ikten C., Smykal P., Penmetsa R. V., et al. (2017). Eco-geographic distribution of Cicer isauricum PH Davis and threats to the species. Genet. Resour. Crop Evol. 78 1–11. [Google Scholar]
- Udupa S., Weigand F., Saxena M., Kahl G. (1998). Genotyping with RAPD and microsatellite markers resolves pathotype diversity in the ascochyta blight pathogen of chickpea. Theor. Appl. Genet. 97 299–307. 10.1007/s001220050899 [DOI] [Google Scholar]
- Vafaei S. H., Rezaee S., Moghadam A. A., Zamanizadeh H. R. (2015). Virulence diversity of Ascochyta rabiei the causal agent of Ascochyta blight of chickpea in the western provinces of Iran. Arch. Phytopathol. Plant Prot. 48 921–930. 10.1080/03235408.2016.1143600 [DOI] [Google Scholar]
- Wilson A. D., Kaiser W. (1995). Cytology and genetics of sexual incompatibility in Didymella rabiei. Mycologia 87 795–804. 10.2307/3760856 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.