Abstract
Foscarnet is widely used for the treatment of acyclovir-resistant herpesvirus infections, and foscarnet-resistant herpesvirus infections are a serious concern in immunocompromised patients. Twenty-seven single-plaque isolates of herpes simplex virus type 1 (HSV-1) resistant to foscarnet were selected from foscarnet- and acyclovir-sensitive HSV-1 strain TAS by exposure to foscarnet, and the DNA polymerase genes were analyzed. The sensitivities of these mutants to foscarnet, cidofovir, S2242, acyclovir, ganciclovir, and penciclovir were determined. A single amino acid substitution, double amino acid substitutions, and a combination of a single amino acid substitution with a deletion or insertion of amino acid residues in the viral DNA polymerase were demonstrated in 21, 4, and 2 isolates, respectively. Of the 27 isolates, an amino acid substitution of serine for asparagine at amino acid position 724 in the DNA polymerase (724 S-N) was detected in 8 isolates. An amino acid substitution in conserved region II was demonstrated in these eight isolates as well as four other isolates. The mutation in the DNA polymerase responsible for resistance to foscarnet was located between the pre-IV region and conserved region V, especially within conserved region II. All the isolates were sensitive or hypersensitive to cidofovir and ganciclovir. Seven, 5, and 15 of the 27 isolates were also sensitive to S2242, acyclovir, and penciclovir, respectively. Thus, most of the foscarnet-resistant HSV-1 isolates were sensitive or hypersensitive to cidofovir and ganciclovir.
The number of immunodeficient patients, such as those with bone marrow transplants or AIDS, has been increasing. Treatment strategies for herpes simplex virus (HSV) infections in these patients have become a major concern, because immunocompromised patients often experience recurrent or chronic herpesvirus infections. Among the herpesvirus infections, HSV type 1 (HSV-1), HSV-2, and varicella-zoster virus infections are usually treated with acyclovir (ACV). ACV treatment sometimes induces ACV-resistant (ACVr) infections in these patients. ACVr herpesvirus infections are treated with foscarnet (PFA), a pyrophosphate analogue (1, 3, 5, 7, 10-12, 14, 16, 17, 20, 22, 28, 29, 32, 35), and ACVr and/or PFA-resistant (PFAr) herpesvirus infections are also treated with cidofovir (CDV), an acyclic nucleoside analogue (4, 5, 19, 31). These two drugs are commonly prescribed for the treatment of herpesvirus infections.
There have been many reports on PFAr HSV-1 and HSV-2 infections (4, 6, 8-10, 18, 19, 23, 28, 30, 33). Therefore, it is important to study the sensitivities of PFAr HSV-1 strains to other antiviral agents with different mechanisms of action against viral strains and to characterize PFAr viruses genetically. In the present study, we independently selected 27 PFAr HSV-1 isolates to clarify their genotypic characterizations and also determined their sensitivities to other antiherpesvirus drugs. The therapeutic strategies used for the treatment of PFAr HSV-1 infections are also discussed in light of the results of the present study as well as those of previous studies.
MATERIALS AND METHODS
Virus and cells.
An ACV-sensitive and PFA-sensitive HSV-1 strain, strain TAS, isolated from a patient with Wiskott-Aldrich syndrome (25, 26) was used in this study. An African green monkey kidney cell line (Vero cell line; American Type Cell Collection) was used for the selection of PFAr HSV-1 isolates and the plaque reduction assay. Vero cells were grown in Eagle's minimum essential medium (MEM) containing streptomycin and penicillin G and 5% fetal bovine serum (FBS) (MEM-5FBS). Vero cells were cultured in MEM containing the two antibiotics and 2% FBS (MEM-2FBS) when they were used for virus growth.
Antiviral agents.
The antiviral agents used in the present study were selected on the basis of their different mechanisms of action for inhibition of virus replication. The antiviral agents used were PFA, ACV, and ganciclovir (GCV) (Sigma-Aldrich Chemical Company, St. Louis, Mo.); penciclovir (PCV; SmithKline Beecham, West Sussex, United Kingdom); CDV (Gilead Sciences, Foster City, Calif.); and 2-amino-7-[(1,3-dihydroxy-2-propoxy)methyl] purine (S2242; Hoechst, Frankfurt, Germany). PFA is a direct DNA polymerase inhibitor (2). ACV, GCV, and PCV are viral thymidine kinase (TK)-associated antiherpesvirus drugs (2, 34). S2242 possesses anti-HSV-1 and anti-HSV-2 activities, but it displays no TK-associated activity (2, 21). Except for S2242, all the drugs are available in the clinical setting.
Selection of PFAr HSV-1 isolates.
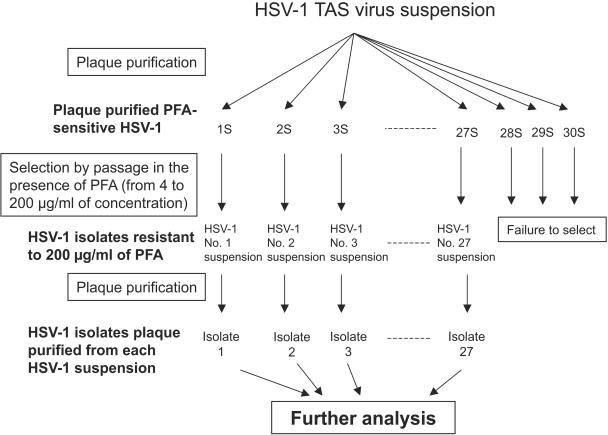
HSV-1 isolates were selected by exposure of wild-type HSV-1 strain TAS to PFA, as summarized in Fig. 1. First, 30 plaque-purified wild-type HSV-1 TAS isolates were obtained. Plaque purification was carried out by selecting viruses that formed plaques in cell culture overlaid with MEM-2FBS with 1% agarose. The plaque purification was performed twice at this step. Next, each of the plaque-purified HSV-1 isolates was propagated independently in Vero cells in the presence of PFA at a concentration of 4 μg/ml, and the virus produced was then inoculated onto cells cultured in MEM-2FBS with PFA at a concentration of 10 μg/ml. The concentration of PFA was gradually increased from 4 to 10, 40, 100, and finally, 200 μg/ml. Plaque purification was carried out for each of the HSV-1 TAS isolates propagated in the presence of PFA at 200 μg/ml in order to obtain a single isolate from each of the virus solutions. Plaque purification was also performed twice at this step. The plaque-purified HSV-1 isolates were first grown in Vero cells seeded into a culture flask (25 cm2), and the cells were then frozen and thawed once to make the working virus solution. Multiple passage of the virus was avoided. The purified HSV-1 isolates were subjected to further analyses of their sensitivities to antiviral agents and genotypic characterization.
FIG. 1.
Schematic representation of the procedures used for the selection of PFAr HSV-1 isolates.
Plaque reduction assay.
HSV-1 TAS and the mutant HSV-1 isolates, which were selected from the infected cells in MEM-2FBS with PFA, were tested for their sensitivities to PFA, CDV, S2242, ACV, GCV, and PCV by plaque reduction assay, as described previously (25-28). The 50% inhibitory concentration (IC50) of each antiviral agent was calculated as the concentration at which the plaque number decreased to half of that in medium without antiviral agents. Five of the purified HSV-1 TAS isolates (isolates 1S to 5S in Fig. 1 and Table 1) were chosen and tested for their sensitivities to the antiviral agents. The IC50s of the antiviral agents tested for the five isolates were determined, and then the log10 values of the IC50s were calculated. The average values and standard deviations (SDs) of the log10 values of the IC50s for isolates 1S to 5S were then calculated. The virus isolates for which the IC50s were higher than 10average + 3 SDs and those for which the IC50s were between 10average + 3 SDs and 10avereage + 2 SDs were defined as highly resistant and moderately resistant, respectively. The mutants for which the IC50 was 10average ± 2 SDs were defined as sensitive to the antiviral agents. Furthermore, the virus isolates for which the IC50 was between 10average − 2 SDs and 10average − 3 SDs and those for which the IC50 was less than10average − 3 SDs were defined as moderately sensitive and highly sensitive, respectively.
TABLE 1.
Sensitivities of PFAr HSV-1 isolates, selected with PFA at a concentration of 200 μg/ml, to PFA, CDV, S2242, ACV, GCV, and PCV and nucleotide and amino acid sequence mutations detected in DNA polymerase gene
| PFAr isolate | Mutation(s) in DNA polymerase
|
IC50 (μg/ml [avg ± SD])a
|
||||||
|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | PFA | CDV | S2242 | ACV | GCV | PCV | |
| 1 | 1481 A-G | 494 N-S | 66 ± 5 | 0.23 ± 0.02 | 2.1 ± 0.1 | 0.7 ± 0.2 | 0.14 ± 0.02 | 0.97 ± 0.09 |
| 2 | 1814 C-T | 605 A-V | >200 ± NDb | 0.70 ± 0.10 | 6.7 ± 0.2 | 5.1 ± 1.0 | 0.24 ± 0.03 | 3.2 ± 0.1 |
| 3 | 1814 C-T | 605 A-V | >200 ± ND | 0.70 ± 0.14 | 11 ± 2 | 7.9 ± 0.8 | 0.34 ± 0.07 | 3.1 ± 0.3 |
| 4 | 1814 C-T, 3410 G-A | 605 A-V, silent | >200 ± ND | 1.1 ± 0.5 | 7.6 ± 0.5 | 3.4 ± 2.1 | 0.20 ± 0.01 | 3.6 ± 0.9 |
| 5 | 979 C-T, 1813 G-A | 327 A-T, 605 A-V | >200 ± ND | 1.3 ± 0.3 | 9.9 ± 2.3 | 7.1 ± 1.1 | 0.31 ± 0.02 | 5.7 ± 0.3 |
| 6c | 2104 C-A | 702 L-I | 74 ± 12 | 0.27 ± 0.02 | 1.9 ± 0.3 | 0.69 ± 0.05 | 0.15 ± 0.01 | 1.3 ± 0.1 |
| 7d | 2146 T-C | 716 F-L | >200 ± ND | 0.30 ± 0.03 | 5.9 ± 0.4 | 6.1 ± 0.3 | 0.53 ± 0.07 | 4.2 ± 1.2 |
| 8 | 2156 C-T | 719 A-V | 130 ± 10 | 1.9 ± 0.4 | 4.8 ± 0.7 | 3.9 ± 1.7 | 0.90 ± 0.12 | 5.2 ± 0.4 |
| 9 | 2156 C-T | 719 A-V | 130 ± 4 | 1.7 ± 0.6 | 6.1 ± 0.2 | 4.6 ± 0.4 | 0.75 ± 0.07 | 5.4 ± 0.2 |
| 10 | 2171 G-A | 724 S-N | >200 ± ND | 16 ± 6 | 6.0 ± 0.3 | 7.0 ± 0.5 | 1.7 ± 0.6 | 9.6 ± 3.0 |
| 11 | 2171 G-A, 2747C-T | 724 S-N, 916 A-V | 170 ± 9 | 17 ± 2 | 9.3 ± 1.6 | 2.9 ± 0.3 | 0.72 ± 0.11 | 4.4 ± 0.3 |
| 12 | 2171 G-A | 724 S-N | 150 ± 5 | 12 ± 5 | 6.3 ± 0.4 | 8.2 ± 0.9 | 1.1 ± 0.2 | 6.2 ± 0.5 |
| 13 | 2171 G-A | 724 S-N | >200 ± ND | 16 ± 0.5 | 5.9 ± 0.3 | 9.2 ± 1.8 | 0.92 ± 0.12 | 9.0 ± 0.2 |
| 14 | 2171 G-A | 724 S-N | 150 ± 9 | 12 ± 3 | 6.6 ± 0.2 | 5.9 ± 0.5 | 0.75 ± 0.05 | 6.4 ± 0.4 |
| 15 | 2171 G-A | 724 S-N | 160 ± 7 | 15 ± 2 | 5.2 ± 0.4 | 5.3 ± 0.2 | 0.49 ± 0.03 | 2.9 ± 0.8 |
| 16 | 2171 G-A | 724 S-N | >200 ± ND | 1.4 ± 1.1 | 4.8 ± 0.5 | 3.9 ± 0.6 | 0.39 ± 0.14 | 3.6 ± 0.5 |
| 17 | 2171 G-A | 724 S-N | >200 ± ND | 10 ± 6 | 7.6 ± 0.1 | 13 ± 4 | 1.5 ± 0.1 | 6.4 ± 0.37 |
| 18 | 2332 C-A | 778 L-M | 101 ± 14 | 9.8 ± 1.6 | 3.2 ± 0.8 | 3.0 ± 0.7 | 1.1 ± 0.2 | 5.4 ± 0.80 |
| 19 | 2392 G-A | 798 E-K | >200 ± ND | 0.30 ± 0.02 | 4.6 ± 1.2 | 3.7 ± 0.4 | 0.17 ± 0.02 | 2.6 ± 0.05 |
| 20 | 2392 G-A | 798 E-K | >200 ± ND | 0.28 ± 0.03 | 4.2 ± 1.0 | 3.5 ± 1.2 | 0.20 ± 0.03 | 2.1 ± 0.2 |
| 21 | 2392 G-A | 798 E-K | >200 ± ND | 0.22 ± 0.02 | 6.2 ± 0.4 | 3.7 ± 0.8 | 0.17 ± 0.06 | 2.6 ± 0.3 |
| 22 | 2500 G-T | 834 A-S | 130 ± 14 | 0.33 ± 0.10 | 2.0 ± 0.2 | 1.3 ± 0.2 | 0.14 ± 0.04 | 2.0 ± 0.2 |
| 23 | 2516 C-T | 839 T-I | 130 ± 5 | 2.2 ± 0.4 | 2.3 ± 0.2 | 1.8 ± 0.1 | 0.12 ± 0.02 | 2.0 ± 0.2 |
| 24 | 1697 G-A, 2534C-A | 565 A-G, 842 R-S | 74 ± 12 | 0.27 ± 0.02 | 1.9 ± 0.3 | 0.69 ± 0.06 | 0.15 ± 0.01 | 1.3 ± 0.1 |
| 25 | 2729 C-T | 910 A-V | 140 ± 12 | 1.1 ± 0.5 | 2.5 ± 0.2 | 1.6 ± 0.2 | 0.52 ± 0.05 | 3.2 ± 0.2 |
| 26 | 2872 G-T | 958 V-L | >200 ± ND | 1.1 ± 0.3 | 6.7 ± 1.0 | 6.2 ± 0.1 | 0.49 ± 0.05 | 4.3 ± 1.0 |
| 27 | 2876 G-A | 959 R-H | 82 ± 5 | 0.52 ± 0.05 | 4.0 ± 1.0 | 2.9 ± 0.5 | 0.21 ± 0.03 | 2.2 ± 0.2 |
| S1 | None | None | 16 ± 3 | 1.3 ± 0.4 | 2.2 ± 0.7 | 0.32 ± 0.18 | 0.60 ± 0.14 | 1.8 ± 0.4 |
| S2 | None | None | 38 ± 17 | 34 ± 11 | 3.3 ± 1.1 | 1.5 ± 0.3 | 1.6 ± 0.3 | 3.4 ± 0.8 |
| S3 | None | None | 22 ± 1 | 5.1 ± 0.6 | 1.7 ± 0.3 | 0.38 ± 0.10 | 0.61 ± 0.02 | 1.5 ± 0.2 |
| S4 | None | None | 25 ± 4 | 12 ± 2.2 | 2.2 ± 0.3 | 0.82 ± 0.16 | 1.1 ± 0.2 | 1.8 ± 0.2 |
| S5 | None | None | 21 ± 2 | 4.2 ± 1.1 | 1.8 ± 0.2 | 0.40 ± 0.10 | 0.68 ± 0.02 | 1.0 ± 0.1 |
The averages and SDs of the IC50s of the antiviral compounds for the 27 PFAr HSV-1 isolates and the 5 wild-type HSV-1 isolates were determined from three independent experiments.
ND, not determined.
Isolate 6 possesses not only the described single amino acid substitution (2104 C-A) but also the insertion of 4 amino acid residues (EFDS) between positions 438 and 439 or between positions 442 and 443.
Isolate 7 possessed not only the described single amino acid substitution (716 F-L) but also the deletion of a single amino acid (1) at position 304.
Sequencing.
The nucleotide sequences of the DNA polymerase genes of HSV-1 TAS and the mutant isolates were determined by direct sequencing methods, as described previously (28). The nucleotide sequence of HSV-1 TAS was identical to that of HSV-1 strain R98/0, which was also isolated from the same patient.
Nucleotide sequence accession number.
The nucleotide sequence of HSV-1 TAS has been deposited in the DNA Data Bank of Japan (DDBJ; http://www.ddbj.nig.ac.jp/Welcome.html) under accession number AB070847 (28).
RESULTS
Selection of PFAr HSV-1 isolates.
Thirty HSV-1 isolates were first plaque purified from HSV-1 strain TAS. A plaque-purified HSV-1 isolate, which was grown in MEM-2FBS with 200 μg of PFA/ml, was successfully obtained from each of the 27 series of the 30 parent isolates. All 27 isolates were confirmed to express an intact TK polypeptide and TK activity by Western blotting and a TK activity assay, respectively, as reported previously (24) (data not shown).
Sensitivities to antiviral agents of HSV-1 isolates grown in the presence of PFA.
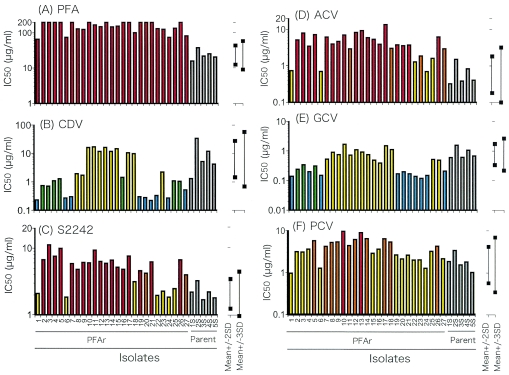
Five plaque-purified isolates of HSV-1 TAS (isolates 1S to 5S) were tested for their sensitivities to the antiviral agents tested. The averages and SDs of the log10 values of the IC50s of the compounds were calculated for each of the five isolates; and the reference values were calculated for determination of sensitivity or resistance to six antiviral agents: PFA, CDV, S2242, ACV, GCV, and PCV (Fig. 2).
FIG. 2.
IC50s of PFA (A), CDV (B), S2242 (C), ACV (D), GCV (E), and PCV (F) for all 27 plaque-purified PFAr isolates that survived in the medium with PFA and those for the 5 plaque-purified wild-type HSV-1 TAS strains (gray bars, isolates 1S to 5S). The last five plaque-purified wild-type HSV-1 TAS strains are considered wild-type HSV-1. The 10average ± 2 SD and 10average ± 3 SD ranges of each compound calculated from the IC50s for plaque-purified isolates (isolates 1S to 5S) are shown. The former are the reference values for the determination of moderately sensitive and moderately resistant, while the latter are those for the determination of highly sensitive and highly resistant. The red, orange, yellow, green, and blue bars indicate highly resistant, moderately resistant, sensitive with normal range, moderately sensitive, and highly sensitive, respectively.
The sensitivity profiles of the isolates selected by exposure to PFA are shown in Table 1 and Fig. 2. All 27 isolates selected after culture in MEM-2FBS with PFA were confirmed to be highly resistant to PFA. Although the virus isolates were selected in MEM-2FBS with PFA at a final concentration of 200 μg/ml, PFA IC50s of less than 200 μg/ml (range, 74 to >200 μg/ml) were demonstrated for 14 isolates. On the other hand, all the PFAr HSV-1 isolates were sensitive to both CDV and GCV. Nine and 10 of the 27 PFAr isolates were highly sensitive to CDV and GCV, respectively (Table 1; Fig. 2). Twenty, 22, and 12 of the 27 PFAr isolates were resistant to S2242, ACV, and PCV, respectively. The patterns of sensitivity of the PFAr HSV-1 isolates to S2242 were similar to those to ACV (Table 1; Fig. 2).
Genotypic characterization of DNA polymerase.
Amino acid changes in the DNA polymerase were demonstrated in all 27 PFAr isolates, as summarized in Table 1; and these changes, along with those of the clinical isolates, are shown in Table 2 (10, 11, 15, 28, 30, 33). A single amino acid substitution, double amino acid substitutions, and a combination of a single amino acid substitution with an amino acid insertion or deletion were demonstrated in 22, 3, and 2 of the 27 PFAr isolates, respectively. The single amino acid (I) deletion detected in the pre-IV region of the DNA polymerase in isolate 7 was at position 304. A unique repeat of nucleotide residues, GAATTCGACAGC GAATTCGAGATG (identical sequences are underlined), was detected between nucleotide positions 1315 and 1338, counted from the initiation codon of the parent virus. The 12 nucleotide residues inserted into the conserved region IV gene, GAATTCGACAGC, which were detected in isolate 6, were identical to those found between nucleotide positions 1315 and 1326, resulting in the appearance of three repeats of this 12-nucleotide sequence, GAATTCGAC(G)AG(T)C(G), as GAATTCGACAGC GAATTCGACAGC GAATTCGAGATG, plus an additional insertion of four amino acid residues (EFDS). It is possible that this insertion occurred between nucleotide positions 1314 and 1315 or between positions 1326 and 1327. Of the 27 PFAr isolates, 8 demonstrated an amino acid substitution of S for N at amino acid position 724 (724 S-N) in the DNA polymerase (Tables 1 and 2). An amino acid substitution in conserved region II was demonstrated in these 8 isolates as well as 4 other isolates (a total of 12 isolates [44%]) (Table 2). The mutations detected in the DNA polymerase were located from the pre-IV region to conserved region V (Table 2).
TABLE 2.
Mutations in DNA polymerase of the 27 PFAr HSV-1 isolates selected in the present study and the HSV-1 clinical isolates resistant to PFA
| Region in DNA polymerasea
|
Amino acid substitution in DNA polymerase of PFAr HSV-1 isolates
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Region | Amino acid positions | Single amino acid substitution
|
Double amino acid substitution
|
|||||
| Isolates in this study
|
Clinical isolates
|
Isolates in this study
|
||||||
| No. | Description (no. of isolates) | No. | Description (no. of isolates) | Reference | No. | Description (no. of isolates) | ||
| Pre-IV | 1-436 | 0 | 0 | 1 | 327 A-T (isolate 5) (1) | |||
| IV | 437-479 | 0 | 0 | 0 | ||||
| IV-D-region C | 480-530 | 1 | 494 N-S (1) | 0 | 0 | |||
| D-region C | 531-627 | 3 | 605 A-V (3) | 0 | 2 | 565 A-G (isolate 24) (1), 605 A-V (isolate 5) (1) | ||
| D-region C-II | 628-693 | 0 | 0 | 0 | ||||
| II | 694-736 | 11 | 702 L-I (1),b 716 F-L (1),c 719 A-V (2), 724 S-N (7) | 6 | 715 V-G (1), 724 S-N (2), 724 A-T (1), 725 S-G (1), 729 S-N (1) | 10, 28, 30, 33 | 1 | 724 S-N (isolate 11) (1) |
| II-VI | 737-771 | 0 | 0 | 0 | ||||
| VI | 772-791 | 1 | 778 L-M (1) | 2 | 783 L-M (1), 785 D-N (1) | 30 | 0 | |
| VI-III | 792-804 | 3 | 798 E-K (3) | 0 | 0 | |||
| III | 805-845 | 2 | 834 G-T (1), 839 C-T (1) | 1 | 841 G-S (1) | 11 | 1 | 842 R-S (isolate 24) (1) |
| III-I | 846-880 | 0 | 1 | 850 L-I (1) | 30 | 0 | ||
| I | 881-896 | 0 | 0 | 0 | ||||
| I-VII | 897-937 | 1 | 910 A-V (1) | 2 | 912 D-V (1), 920 P-S (1) | 30 | 1 | 916 A-V (isolate 11) (1) |
| VII | 938-946 | 0 | 1 | 941 Y-H (1) | 15 | 0 | ||
| VII-V | 947-952 | 0 | 0 | 0 | ||||
| V | 953-963 | 2 | 958 V-L (1), 959 R-H (1) | 0 | 0 | |||
| Post-V | 964-1235 | 0 | 0 | 0 | ||||
| Total no. of amino acid substitutions | 24 | 13 | 6 | |||||
| Total no. of isolates | 24 | 13 | 3 | |||||
The conserved regions in HSV-1 DNA polymerase are represented by the designations IV, D-region C, II, VI, III, I, VII, and V; and the regions between the conserved regions are represented by the designations pre-IV, IV-D-region C, D-region C-II, II-VI, VI-III, III-I, I-VII, VII-V and post-V. The amino acid positions of these conserved regions for HSV-1 shown here are based on the report by Gilbert et al (13).
The amino acid substitution was that detected in isolate 6 (Table 1).
The amino acid substitution was that detected in isolate 7 (Table 1).
DISCUSSION
There have been many studies of laboratory-derived or clinically isolated PFAr HSV-1 and HSV-2 isolates; however, there have been no studies on the characterization of PFAr HSV-1 with a large panel of PFAr HSV-1 isolates. In the present study, 27 PFAr HSV-1 isolates were independently selected in vitro, and the sensitivities of these viruses to PFA (direct DNA polymerase inhibitor) and viral TK-unassociated antiviral drugs (CDV and S2242) or viral TK-associated antiviral drugs (ACV, GCV, and PCV) were determined. Furthermore, genotypic characterization of the DNA polymerase gene was performed. The findings of the present study are useful for the establishment of treatment strategies for and the diagnosis of PFAr HSV-1 infections. All of the 27 PFAr HSV-1 isolates were sensitive, moderately sensitive, or highly sensitive to both CDV and GCV, while 20 (74%), 22 (81%), and 12 (44%) of the isolates showed decreased sensitivities to S2242, ACV, and PCV, respectively. The mutations were detected from the pre-IV region to conserved region V. It is likely that conserved region II, in which the 724 S-N and 719 A-V mutations were demonstrated, is the hotspot for conferring PFAr resistance. The reported mutations in the HSV-1 DNA polymerase of a total of 13 clinical isolates are summarized in Table 2. Six (46%) of the 13 isolates possessed the mutations responsible for resistance to PFA in conserved region II, and 3 of the 6 isolates possessed a mutation at amino acid position 724. The ratio of the appearance of mutations in conserved region II detected in the present study was similar to the ratio of mutations detected among these clinical isolates.
We recently detected a PFAr HSV-1 infection in a patient with congenital immunodeficiency (28). The isolate was resistant to both PFA and ACV and possessed mutations in both the DNA polymerase and the TK regions that are responsible for PFA resistance and ACV resistance, respectively. Chibo et al. (10) and Stránská et al. (33) also reported on ACVr and PFAr HSV-1 infections in immunocompromised patients and noted that the viruses causing the infections harbored double mutations in the regions encoding DNA polymerase and TK. Infections caused by PFAr HSV-1 isolates that possess mutations in the DNA polymerase region but not in the TK region are also common (30). The present study indicates that CDV can be used for the treatment of most PFAr HSV-1 infections and that GCV can be used for the treatment of infections caused by PFAr HSV-1 without mutations in TK. These results emphasize that the sensitivities of clinical isolates from high-risk patients to the antiviral agents in clinical use should be monitored and that the proper antiviral agents should be selected for the treatment of these patients. If PFAr HSV-1 isolates are sensitive to ACV, PCV, or GCV, these drugs should also be used.
It was revealed that the pattern of sensitivity of the PFAr isolates to ACV was similar to that to S2242. These results strongly suggest that the active forms of ACV and S2242 may inhibit DNA polymerase activity via a similar mechanism.
Double amino acid substitutions were detected in three isolates (isolates 5, 11, and 24). A 605 A-V mutation as well as a 327 A-T mutation was detected in isolate 5, and a 724 S-N mutation as well as a 916 A-V mutation was detected in isolate 11. The 605 A-V and 724 S-N mutations were also demonstrated in isolates 2 to 4 and isolates 10 and 12 to 17, respectively, indicating that these mutations alone can confer resistance to PFA and that it is unknown whether each of the mutations 327 A-T, detected in isolate 5, and 916 A-V, detected in isolate 11, alone can confer resistance to PFA. Furthermore, double amino acid substitutions of 565 A-G and 842 R-S were demonstrated in isolate 24. It is not clear whether each of the mutations detected in isolate 24 independently induce resistance to PFA or whether the combination of two mutations is necessary to confer resistance to PFA.
A combination of a single amino acid substitution and an amino acid deletion (in the pre-IV region) and a combination of a single amino acid substitution and an insertion of 4 amino acid residues (in conserved region IV) were demonstrated in one isolate each. The insertion or deletion of amino acid residues demonstrated in isolate 6 and isolate 7 should be addressed to determine whether they are responsible for PFA resistance. The amino acid substitutions detected in both isolates 6 and 7 were demonstrated in conserved region II. Although further analyses are need, it is likely that the single amino acid substitutions demonstrated in conserved region II can confer resistance to PFA.
As shown in Table 2, mutations in the DNA polymerase-encoding regions of the clinical PFAr isolates responsible for PFA resistance were demonstrated between conserved regions II and VII. However, the mutations responsible for PFA resistance detected in the present study were located between the IV-D-region C and conserved region V (Table 2). Testing of the susceptibilities of clinical isolates to certain antiviral agents, including virus isolation and susceptibility testing, is time-consuming. Therefore, molecular techniques for determination of PFA resistance should be developed to save time while obtaining sufficient information on sensitivity to antiviral agents. For this purpose, the accumulation of data on the nucleotide sequence of the DNA polymerase of wild-type HSV-1 strains and those of PFAr HSV-1 strains is necessary.
In summary, 27 isolates of PFAr HSV-1 were selected in vitro; and sensitivities to six antiviral agents, PFA (direct DNA polymerase inhibitor), CDV and S2242 (drugs independent of viral TK for activation), and ACV, PCV, and GCV (drugs dependent on viral TK for activation), were investigated. Furthermore, genotypic characterization of these PFAr HSV-1 isolates was performed.
Acknowledgments
We thank E. De Clercq, Rega Institute for Medical Research, Katholieke Universiteit Leuven, Leuven, Belgium, for providing us with CDV. We also thank Gerhard Jähane, Hoechst, Frankfurt, Germany, for providing us with compound S2242. We thank M. Ogata, Department of Virology 1, National Institute of Infectious Diseases, Tokyo, Japan, for technical and clerical assistance.
This study was financially supported by a grant-in-aid (no. 12770416) from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Alvarez-McLeod, A., J. Havlik, and K. E. Drew. 1999. Foscarnet treatment of genital infection due to acyclovir-resistant herpes simplex virus type 2 in a pregnant patient with AIDS: case report. Clin. Infect. Dis. 29:937-938. [DOI] [PubMed] [Google Scholar]
- 2.Andrei, G., R. Snoeck, and E. De Clercq. 1995. Susceptibilities of several drug-resistant herpes simplex virus type 1 strains to alternative antiviral compounds. Antimicrob. Agents Chemother. 39:1632-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bestman-Smith, J., and G. Boivin. 2003. Drug resistance patterns of recombinant herpes simplex virus DNA polymerase mutants generated with a set of overlapping cosmids and plasmids. J. Virol. 77:7820-7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blot, N., P. Schneider, P. Young, C. Janvresse, D. Dehesdin, P. Tron, and J. P. Vannier. 2000. Treatment of an acyclovir and foscarnet-resistant herpes simplex virus infection with cidofovir in a child after an unrelated bone marrow transplant. Bone Marrow Transplant. 26:903-905. [DOI] [PubMed] [Google Scholar]
- 5.Bryant, P., J. Sasadeusz, J. Carapetis, K. Waters, and N. Curtis. 2001. Successful treatment of foscarnet-resistant herpes simplex stomatitis with intravenous cidofovir in a child. Pediatr. Infect. Dis. J. 20:1083-1086. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti, S., D. Pillay, D. Ratcliffe, P. A. Cane, K. E. Collingham, and D. W. Milligan. 2000. Resistance to antiviral drugs in herpes simplex virus infections among allogeneic stem cell transplant recipients: risk factors and prognostic significance. J. Infect. Dis. 181:2055-2058. [DOI] [PubMed] [Google Scholar]
- 7.Chatis, P. A., C. H. Miller, L. E. Schrager, and C. S. Crumpacker. 1989. Successful treatment with foscarnet of an acyclovir-resistant mucocutaneous infection with herpes simplex virus in a patient with acquired immunodeficiency syndrome. N. Engl. J. Med. 320:297-300. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., C. Scieux, V. Garrait, G. Socie, V. Rocha, J. M. Molina, D. Thouvenot, F. Morfin, L. Hocqueloux, L. Garderet, H. Esperou, F. Selimi, A. Devergie, G. Leleu, M. Aymard, F. Morinet, E. Gluckman, and P. Ribaud. 2000. Resistant herpes simplex virus type 1 infection: an emerging concern after allogeneic stem cell transplantation. Clin. Infect. Dis. 31:927-935. [DOI] [PubMed] [Google Scholar]
- 9.Chibo, D., J. Druce, J. Sasadeusz, and C. Birch. 2004. Molecular analysis of clinical isolates of acyclovir resistant herpes simplex virus. Antivir. Res. 61:83-91. [DOI] [PubMed] [Google Scholar]
- 10.Chibo, D., A. Mijch, R. Doherty, and C. Birch. 2002. Novel mutations in the thymidine kinase and DNA polymerase genes of acyclovir and foscarnet resistant herpes simplex viruses infecting an immunocompromised patient. J. Clin. Virol. 25:165-170. [DOI] [PubMed] [Google Scholar]
- 11.Collins, P., B. A. Larder, N. M. Oliver, S. Kemp, I. W. Smith, and G. Darby. 1989. Characterization of a DNA polymerase mutant of herpes simplex virus from a severely immunocompromised patient receiving acyclovir. J. Gen. Virol. 70(Pt 2):375-382. [DOI] [PubMed] [Google Scholar]
- 12.Erlich, K. S., M. A. Jacobson, J. E. Koehler, S. E. Follansbee, D. P. Drennan, L. Gooze, S. Safrin, and J. Mills. 1989. Foscarnet therapy for severe acyclovir-resistant herpes simplex virus type-2 infections in patients with the acquired immunodeficiency syndrome (AIDS). An uncontrolled trial. Ann. Intern. Med. 110:710-713. [DOI] [PubMed] [Google Scholar]
- 13.Gilbert, C., J. Bestman-Smith, and G. Boivin. 2002. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist. Update 5:88-114. [DOI] [PubMed] [Google Scholar]
- 14.Hardy, W. D. 1992. Foscarnet treatment of acyclovir-resistant herpes simplex virus infection in patients with acquired immunodeficiency syndrome: preliminary results of a controlled, randomized, regimen-comparative trial. Am. J. Med. 92:30S-35S. [DOI] [PubMed] [Google Scholar]
- 15.Hwang, C. B., K. L. Ruffner, and D. M. Coen. 1992. A point mutation within a distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J. Virol. 66:1774-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, T. J., and R. Paul. 1995. Disseminated acyclovir-resistant herpes simplex virus type 2 treated successfully with foscarnet. J. Infect. Dis. 171:508-509. [DOI] [PubMed] [Google Scholar]
- 17.Kopp, T., A. Geusau, A. Rieger, and G. Stingl. 2002. Successful treatment of an aciclovir-resistant herpes simplex type 2 infection with cidofovir in an AIDS patient. Br. J. Dermatol. 147:134-138. [DOI] [PubMed] [Google Scholar]
- 18.Langston, A. A., I. Redei, A. M. Caliendo, J. Somani, D. Hutcherson, S. Lonial, S. Bucur, J. Cherry, A. Allen, and E. K. Waller. 2002. Development of drug-resistant herpes simplex virus infection after haploidentical hematopoietic progenitor cell transplantation. Blood 99:1085-1088. [DOI] [PubMed] [Google Scholar]
- 19.LoPresti, A. E., J. F. Levine, G. B. Munk, C. Y. Tai, and D. B. Mendel. 1998. Successful treatment of an acyclovir- and foscarnet-resistant herpes simplex virus type 1 lesion with intravenous cidofovir. Clin. Infect. Dis. 26:512-513. [DOI] [PubMed] [Google Scholar]
- 20.Naik, H. R., N. Siddique, and P. H. Chandrasekar. 1995. Foscarnet therapy for acyclovir-resistant herpes simplex virus 1 infection in allogeneic bone marrow transplant recipients. Clin. Infect. Dis. 21:1514-1515. [DOI] [PubMed] [Google Scholar]
- 21.Neyts, J., J. Balzarini, G. Andrei, Z. Chaoyong, R. Snoeck, A. Zimmermann, T. Mertens, A. Karlsson, and E. De Clercq. 1998. Intracellular metabolism of the N7-substituted acyclic nucleoside analog 2-amino-7-(1,3-dihydroxy-2-propoxymethyl)purine, a potent inhibitor of herpesvirus replication. Mol. Pharmacol. 53:157-165. [DOI] [PubMed] [Google Scholar]
- 22.Safrin, S., T. Assaykeen, S. Follansbee, and J. Mills. 1990. Foscarnet therapy for acyclovir-resistant mucocutaneous herpes simplex virus infection in 26 AIDS patients: preliminary data. J. Infect. Dis. 161:1078-1084. [DOI] [PubMed] [Google Scholar]
- 23.Safrin, S., S. Kemmerly, B. Plotkin, T. Smith, N. Weissbach, D. De Veranez, L. D. Phan, and D. Cohn. 1994. Foscarnet-resistant herpes simplex virus infection in patients with AIDS. J. Infect. Dis. 169:193-196. [DOI] [PubMed] [Google Scholar]
- 24.Saijo, M., T. Suzutani, E. De Clercq, M. Niikura, A. Maeda, S. Morikawa, and I. Kurane. 2002. Genotypic and phenotypic characterization of the thymidine kinase of ACV-resistant HSV-1 derived from an acyclovir-sensitive herpes simplex virus type 1 strain. Antivir. Res. 56:253-262. [DOI] [PubMed] [Google Scholar]
- 25.Saijo, M., T. Suzutani, K. Itoh, Y. Hirano, K. Murono, M. Nagamine, K. Mizuta, M. Niikura, and S. Morikawa. 1999. Nucleotide sequence of thymidine kinase gene of sequential acyclovir-resistant herpes simplex virus type 1 isolates recovered from a child with Wiskott-Aldrich syndrome: evidence for reactivation of acyclovir-resistant herpes simplex virus. J. Med. Virol. 58:387-393. [PubMed] [Google Scholar]
- 26.Saijo, M., T. Suzutani, K. Murono, Y. Hirano, and K. Itoh. 1998. Recurrent aciclovir-resistant herpes simplex in a child with Wiskott-Aldrich syndrome. Br. J. Dermatol. 139:311-314. [DOI] [PubMed] [Google Scholar]
- 27.Saijo, M., T. Suzutani, and I. Yoshida. 1992. Effects of acyclovir, oxetanocin-G, and carbocyclic oxetanocin-G in combinations on the replications of herpes simplex virus type 1 and type 2 in Vero cells. Tohoku J. Exp. Med. 167:57-68. [DOI] [PubMed] [Google Scholar]
- 28.Saijo, M., Y. Yasuda, H. Yabe, S. Kato, T. Suzutani, E. De Clercq, M. Niikura, A. Maeda, I. Kurane, and S. Morikawa. 2002. Bone marrow transplantation in a child with Wiskott-Aldrich syndrome latently infected with acyclovir-resistant (ACVr) herpes simplex virus type 1: emergence of foscarnet-resistant virus originating from the ACVr virus. J. Med. Virol. 68:99-104. [DOI] [PubMed] [Google Scholar]
- 29.Sall, R. K., C. L. Kauffman, and C. S. Levy. 1989. Successful treatment of progressive acyclovir-resistant herpes simplex virus using intravenous foscarnet in a patient with the acquired immunodeficiency syndrome. Arch. Dermatol. 125:1548-1550. [PubMed] [Google Scholar]
- 30.Schmit, I., and G. Boivin. 1999. Characterization of the DNA polymerase and thymidine kinase genes of herpes simplex virus isolates from AIDS patients in whom acyclovir and foscarnet therapy sequentially failed. J. Infect. Dis. 180:487-490. [DOI] [PubMed] [Google Scholar]
- 31.Snoeck, R., G. Andrei, M. Gerard, A. Silverman, A. Hedderman, J. Balzarini, C. Sadzot-Delvaux, G. Tricot, N. Clumeck, and E. De Clercq. 1994. Successful treatment of progressive mucocutaneous infection due to acyclovir- and foscarnet-resistant herpes simplex virus with (S)-1-(3-hydroxy-2-phosphonylmethoxypropyl)cytosine (HPMPC). Clin. Infect. Dis. 18:570-578. [DOI] [PubMed] [Google Scholar]
- 32.Stellbrink, H. J., H. Albrecht, T. Loning, and H. Greten. 1991. Herpes simplex virus type-2 ulcers resistant to acyclovir in an AIDS patient—successful treatment with foscarnet. Klin. Wochenschr. 69:274-278. [DOI] [PubMed] [Google Scholar]
- 33. Stránská, R., A. M. van Loon, R. G. Bredius, M. Polman, E. Nienhuis, M. F. Beersma, A. C. Lankester, and R. Schuurman. 2004. Sequential switching of DNA polymerase and thymidine kinase-mediated HSV-1 drug resistance in an immunocompromised child. Antivir. Ther. 9:97-104. [PubMed] [Google Scholar]
- 34.Suzutani, T., K. Ishioka, E. De Clercq, K. Ishibashi, H. Kaneko, T. Kira, K. Hashimoto, M. Ogasawara, K. Ohtani, N. Wakamiya, and M. Saijo. 2003. Differential mutation patterns in thymidine kinase and DNA polymerase genes of herpes simplex virus type 1 clones passaged in the presence of acyclovir or penciclovir. Antimicrob. Agents Chemother. 47:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Verdonck, L. F., J. J. Cornelissen, J. Smit, J. Lepoutre, G. C. de Gast, A. W. Dekker, and M. Rozenberg-Arska. 1993. Successful foscarnet therapy for acyclovir-resistant mucocutaneous infection with herpes simplex virus in a recipient of allogeneic BMT. Bone Marrow Transplant. 11:177-179. [PubMed] [Google Scholar]