Abstract
Protein phosphatase-6 (PP6) is a member of the PPP family of Ser/Thr phosphatases involved in intracellular signaling. PP6 is conserved among all eukaryotes, and genetics in model organisms indicates it has non-redundant functions relative to other PPP phosphatases. PP6 functions in association with conserved SAPS subunits and, in vertebrate species, forms heterotrimers with Ankrd subunits. Multiple studies have demonstrated how PP6 exerts negative control at different steps of nuclear factor kappaB signaling. Expression of PP6 catalytic subunit and the PPP6R1 subunit is especially high in hematopoietic cells and lymphoid tissues. Recent efforts at conditionally knocking out genes for PP6c or PP6R1 (SAPS1) have revealed distinctive effects on development of and signaling in lymphocytes.
Keywords: knockout mice, nuclear factor kappaB, phosphorylation/dephosphorylation, T-cells
Protein phosphatase-6 conservation and multisubunit holoenzymes
Protein phosphatase-6 (PP6) is a member of the PPP family of protein Ser/Thr phosphatases, enzymes with a bimetallic (Fe::Zn) catalytic center that distinguish them in terms of structure and activity from other protein phosphatases [1,2]. PP6 is most closely related in protein sequence (57% identical) to protein phosphatase-2A (PP2A) and to protein phosphatase-4 (PP4). Despite their sequence similarity separate homologues of these phosphatases (PP2A/PP4/PP6) are conserved in eukaryotes, including model organisms Saccharomyces cerevisiae (PPH21/PPH3/SIT4), Schizosaccharomyces pombe (ppa2/pph3/ppe1), Caenorhabditis elegans (LET-92/PPH-4.1/PPH-6) and Drosophila (mts/Pp4-19C/PpV). Human PP6 can complement mutants of either sit4 or ppe1, showing functional homology [3]. Neither PP4 nor PP5 were able to complement the sit4-102 mutation, reinforcing the specificity for PP6 [3]. Preservation of these separate phosphatases during evolution implies that they serve non-redundant functions. Selective knockdown of these individual PPP phosphatases by RNAi in C. elegans produces distinctive phenotypes, supporting this concept [4–6].
In addition to genetics, another approach to discovery of the functions of PPP phosphatases has been to treat intact cells or tissues with selective inhibitors that can enter the cell [7]. Okadaic acid (OA) is a polyketide natural product synthesized by dinoflagellates that accumulates in marine filter feeders such as sponges and shellfish [8]. OA has been isolated from these sources and used as a research tool for almost 30 years [9–11]. Potent in vitro inhibition of the PP2A/PP4/PP6 (type 2A) phosphatases by low nanomolar doses of OA distinguishes these phosphatases from other members of the PPP family [12,13]. Permeation of OA into living cells requires higher doses and longer times, complicating interpretation of results [14]. Nonetheless, there are >5400 articles in the literature (PubMed) that report using OA and the biological effects are almost always attributed to inhibition of PP2A, probably because PP2A is the best known of these type 2A phosphatases and was the first of them to be purified and characterized. Use of reagents such as OA that equally inhibit PP2A/PP4/PP6 has confounded assignment of the biological actions of these individual type 2A phosphatases. There is not much appreciation of PP6 or PP4 as separate PPP phosphatases that are expressed in most cell lines and tissues. In fact, PP6 is the most abundant PPP in HeLa cells, as quantified by mass spectrometry [15]. Some of the reported effects of OA are due to inhibition of PP6, but it is difficult to tell from the experiments which phosphatase is involved.
Most of the PPP family catalytic subunits associate with regulatory subunits to assemble into holoenzymes [1,2]. Thus, by combinatorial association less than a dozen catalytic subunits assemble into hundreds of phosphatase enzymes that are available to counterbalance the actions of ∼400 protein Ser/Thr kinases. The basis for substrate specificity is different for kinases and phosphatases, so there is not a clear correspondence for enzymes that catalyze the opposing reactions at any given phosphosite [16]. Each of the type 2A phosphatases has an exclusive set of regulatory subunits that have been conserved among eukaryotes. In the case of PP6, dedicated subunits were initially discovered in yeast and named SAPs (Sit4-Associated Proteins) [17]. Based on genetic evidence, it was proposed that each of these dimeric holoenzymes (Sit4::SAP) has separate functions [17]. Some years later, three SAP homologues were identified in the human genome [18] and later shown to functionally complement the SAP genes in yeast, demonstrating conservation of subunit–subunit recognition [19]. Curiously, S. pombe, C. elegans and Drosophila each only have a single SAP gene [i.e. Ekc1, ceSAP1(G47G2.5) and dSAP, respectively]. The defining SAPS domain was mapped as the common region of the proteins, and shown to be sufficient for binding the PP6 catalytic subunit [23]. The SAPS domain was modeled as a helical repeat structure resembling p115 golgin, different from the HEAT repeat structure of the A scaffold subunit of PP2A [20]. Frogs, mice and humans have three SAPS, encoded by genes also known as PPP6R1, PPP6R2 and PPP6R3, which comprise a 2R-ohnolog family generated by the two rounds of whole-genome duplication at the origin of the vertebrates [21]. The SAPS are phosphoproteins [22], but how phosphorylation alters their properties or the activity of PP6 is unknown. Another set of PP6 subunits, called Ankrd proteins, were first identified by proteomics using FLAG-SAPS1 [23]. The Ankrd are only expressed in vertebrate organisms, with no corresponding genes found in flies, worms or microorganisms [23]. These proteins (Ankrd 28, 44, 52) associate with SAPS subunits in the C-terminal region outside the SAPS domain and are composed entirely of helical repeat ankyrin domains that presumably mediate protein–protein interactions. Both BRCA1 and DOCK180 have been identified as Ankrd28-binding partners [24–26]. Using proteomics three other groups have independently confirmed the heterotrimeric organization of PP6 in human cells [27–29]. Defining the individual functions of the human PP6 holoenzymes remains one of the challenges for ongoing research. Our approach has been to knock down or delete individual SAPS regulatory subunits [18,30].
Distribution of PP6 and SAPS subunits in human tissues
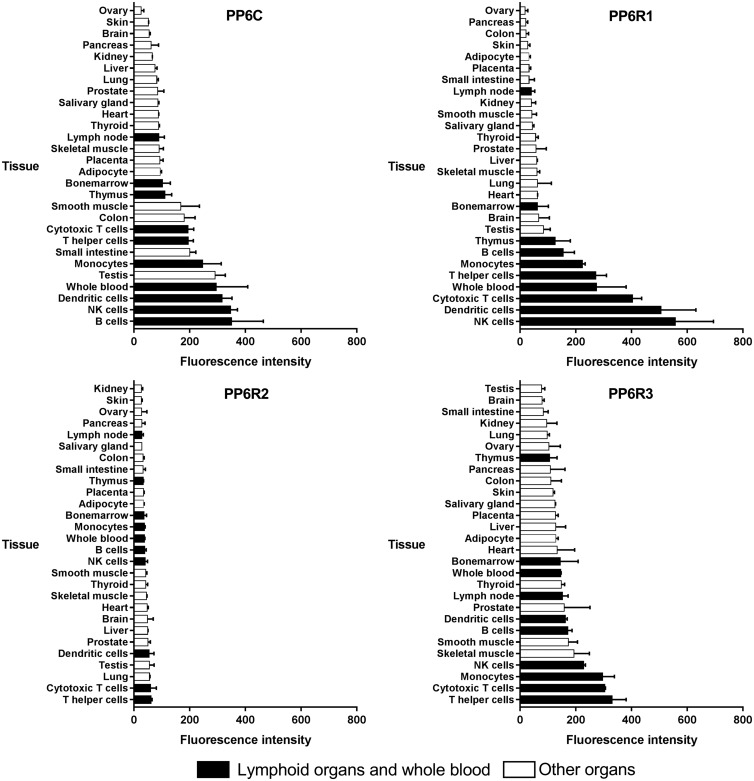
The first survey of expression of PP6c, PPP6R1, PPP6R2 and PPP6R3 genes used northern blot hybridization and a commercial preparation of RNA from 12 human tissues (brain, colon, heart, kidney, liver, lung, muscle, placenta, small intestine, spleen, stomach and testis). The PP6c mRNA was present in all tissues, with highest levels detected in heart, with slightly lower levels in kidney, liver, stomach and brain [18]. All tissues expressed PPP6R1 mRNA at comparable levels (except higher in testis). PPP6R2 mRNA also was detected in all the tissues with the highest levels in testis followed by liver, heart, brain and kidney. Low levels of PP6R3 mRNA were detected in most tissues, except for high levels in heart. There was not an obvious correspondence in relative abundance in different tissues between catalytic subunit and any one of the SAPS [18]. More recent gene microarray results in the BioGPS online database [31,32] reveal a striking distribution pattern, with especially abundant mRNA levels of PP6c, PPP6R1, -R2 and -R3 in immune cells and lymphoid tissues (Figure 1). Some of these cell types were absent from the earlier northern analysis. B lymphocytes, natural killer (NK) cells and dendritic cells have the highest expression of PP6c mRNA, while PP6R1 mRNA is highly abundant in NK cells, dendritic cell and cytotoxic T cells, PP6R2 in T helper cells, cytotoxic T cells and lungs, and PP6R3 in T helper cells, cytotoxic T cells and monocytes. Expression of PP6 and its SAPS subunit genes is by far highest in cells of the hematopoietic system.
Figure 1. mRNA expression in normal human tissues for PP6 subunits: PP6c, PP6R1, PP6R2 and PP6R3.
Protein levels of PP6c and its SAPS regulatory subunits were first measured by western blotting using custom-made anti-peptide antibodies [18]. Highest levels of PP6c protein were found in lung, liver and spleen (a source of lymphocytes), while expression was relatively low in skeletal muscle. These results contrasted with the mRNA distribution of PP6c, suggesting that PP6 protein level is regulated post-transcriptionally, at the level of mRNA translation or protein stability. Lung, bladder, spleen and pancreas contained highest levels of PPP6R1 protein in comparison with other tissue types. Again, similar to PP6c there was discordance between the amounts of PPP6R1 protein and PPP6R1 mRNA levels in different tissue types. Highest levels of PPP6R2 protein were observed in bladder, with lower levels in heart and pancreas, and barely detectable levels in other tissue types. The highest levels of PPP6R3 protein were detected in lung, bladder, spleen and pancreas, in contrast with mRNA expression that was greatest in heart. PP6R1 and PP6R3 proteins had very similar tissue distribution.
Expression of PP6c, PPP6R1, -R2 and -R3 proteins was more recently examined by analyzing mass spectrometry data of the human proteome deposited in the Proteomics DB database [33]. Comparing results from microarray and mass spectrometry proteomics demonstrates that the distribution of mRNA is not consistent with protein levels in various tissues. Amounts of PP6c mRNA are lowest in ovary and skin, while protein concentration of PP6c is second and third highest in these tissues (after immune cells and lymphoid organs). The amount of PPP6R1 mRNA is very low in ovary and pancreas, but protein concentration is high in these tissues. PPP6R3 mRNA expression is highest in T helper cells and lowest in brain, while the protein concentration in these two tissues is similar. These observations support the idea that post-transcriptional mechanisms dynamically control expression of PP6 and its SAPS subunits.
Consistent with mRNA expression by microarray, the highest amount of PP6 and SAPS subunits as determined by mass spectrometry appears in immune cells, lymphoid organs and bone marrow. PP6c and PP6R3(SAPS3) protein levels are relatively abundant in bone, NK cells and cytotoxic T cells; PP6R1(SAPS1) — in bone, B cells and NK cells; and PP6R2(SAPS2) — in cytotoxic T cells, B cells and bone. In general, the mRNA distribution pattern provided by microarray and proteomics data from Proteomics DB corresponded well with each other [33]. Abundant expression of PP6 and SAPS subunits in immune cells heralds that its function is probably involved in signaling in these cell types. These results have directed us to study PP6 in lymphocytes.
PP6 opposes activation of nuclear factor kappaB at multiple steps
The signaling pathways that control nuclear factor kappaB (NF-κB) activation have become central to understanding the immune system, starting with discovery of NF-κB 30 years ago [34]. NF-κB is involved in hematopoiesis, thymocyte positive and negative selection, development and activation of innate immune cells, lymphoid organogenesis, TCR and BCR signaling, Ig class switching and cytokine production [35,36]. Despite the predominant role of phosphorylation in activation of the NF-κB pathway, few reports have addressed the role of protein phosphatases. However, there is evidence that PP6 acts to oppose activation of the pathway.
Bouwmeester et al. [37] discovered PP6 in the TNF-α/NF-κB pathway by using tandem affinity purification in a proteomics analysis. Proteins known in the TNF-α/NF-κB pathway, including TNF receptors, TRAFs, a total of 10 kinases, such as MEKK1, MEKK3, NIK, TAK1, IKK, plus NF-κB and the IκB inhibitor subunits, were tagged and stably expressed in TNF-α-responsive HEK293 cells by retrovirus-mediated gene transfer. Proteins associated with tagged components were recovered and identified from peptides by liquid chromatography–tandem mass spectrometry (LC–MS/MS). Out of 680 non-redundant proteins identified, 180 were high-confidence interactors and 80 proteins had not previously been associated with the pathway. PP6c and its putative subunits (KIAA1115, KIAA0685 and KIAA0379, later renamed PPP6R1, PPP6R2 and ANKRD28, respectively) were recovered bound with tagged IκBε but not with tagged IκBα or IκBβ [37]. These results exposed PP6 as an integral component of the TNF-α/NF-κB pathway and support the concept that PP6 is specifically targeted to potential substrates by its regulatory SAPS subunits.
A subsequent study [18] confirmed association of PP6 with IκBε by co-immunoprecipitation with FLAG-tagged PP6R1 (SAPS1) or PP6R2 (SAPS2) ectopically expressed in Cos7 cells. Knockdown of PPP6R1 but not PPP6R3 (as a control) with single siRNAs in HeLa cells resulted in statistically significant reduced levels of IκBε at 30, 60 and 90 min after TNF-α stimulation [18]. The lower IκBε protein levels in the absence of PPP6R1 are attributed to IκBε being more highly phosphorylated and subjected to polyubiquitin-mediated degradation. This conclusion is based on previous reports demonstrating that TNF-α signaling promotes phosphorylation of IκBε and its dissociation from NF-κB, followed by degradation via the proteasome [38,39]. The results implicate PPP6R1 (SAPS1) acting as a partner with PP6 to reduce the phosphorylation of IκBε and thereby limit the activation of NF-κB pathway in response to TNF-α (Figure 2).
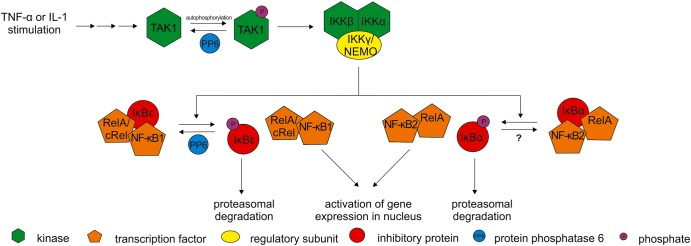
Figure 2. Upon TNF-α or IL-1 stimulation TAK1 becomes activated by autophosphorylation.
TAK1 activates IKK complex that phosphorylates inhibitory proteins (IκBε, IκBα) causing dissociation from transcription factors (RelA, cRel, NF-κB1, NF-κB2). Phosphorylated inhibitory proteins are degraded via the proteasome, and the transcription factors are imported into the nucleus to activate expression of target genes [41]. Inhibitory activity of PP6 on NF-κB signaling can occur in at least two ways: (1) by dephosphorylation and inactivation of TAK1 or (2) by dephosphorylation of the IκBε inhibitory protein.
Activation of the NF-κB pathway is prolonged in keratinocytes derived from keratinocyte-specific Ppp6c conditional knockout mice [40]. The levels of phospho-p65/relA are elevated at 30 and 60 min in knockouts relative to floxed controls. Despite this activation there are no significant differences in protein levels of IκBε or IκBα between knockout and floxed keratinocytes up to 60 min following stimulation by TNF-α or IL-1β. The results suggest that PP6 somehow inhibits activation of the NF-κB pathway in keratinocytes without changes in phosphorylation of IκB. It is possible that keratinocytes have a relatively low level of PPP6R1 that is responsible for targeting IκB. Alternatively, there could be other PP6 substrates in the NF-κB pathway in mouse primary keratinocytes, such as phospho-p65/relA or upstream kinases that support phosphorylation of p65/relA [40].
Other studies have provided evidence that PP6 is involved in negative control of other steps in the NF-κB signaling pathway. TGF-β-activated kinase 1 (TAK1) is a Ser/Thr kinase that undergoes autophosphorylation and activation leading to initiation of NF-κB signaling by phosphorylation of the IκB kinase (IKK) [41]. IL-1β activation of TAK1 involves phosphorylation of Thr187, which is greatly enhanced by treating cells with 100 nM OA. Mass spectrometry proteomics of FLAG-tagged TAK1 complexes revealed association with PP6. Indeed, FLAG-tagged TAK1 and HA-tagged PP6 were co-immunoprecipitated from HEK 293 cells and expression of HA-tagged PP6 reduced phosphorylation of Thr187 in endogenous TAK1 [42]. Knockdown of PP6c by siRNA enhanced IL-1 stimulated phosphorylation of Thr187 and TAK1 and this effect was not mimicked by knockdown of PP2Ac. Other evidence demonstrates that TAB2 protein (TAK1-binding protein 2) recruits PP6 to TAK1 and this involves interaction of PP6c and TAB2 with polyubiquitin chains [43]. Taken together, these results demonstrate specific association of TAK1 and TAB2 with PP6, which dephosphorylates and inactivates this kinase and thereby negatively regulates NF-κB signaling.
The human orthologue of yeast Tip41, also called TIP (type 2A phosphatase-interacting protein), binds human PP6c as well as PP2Ac and PP4c [44]. Overexpression of HA-TIP, T7-TAB1 and FLAG-TAK1 in HEK293T cells resulted in increase in phosphorylation of FLAG-TAK1 and endogenous IKK-β [41]. This could have been due to inhibition of protein phosphatases by the overexpressed HA-TIP; however, it appeared that TIP has unexpected non-canonical functions in this system. Purified GST-TIP directly associates with FLAG-TAK1 and increases its phosphorylation and kinase activity, hence TIP was called TAB4 (as another example of a TAK1-binding protein) [41]. TAB4 associates with the TAK1–TAB1 complex and significantly increases phosphorylation of TAK1 and TAB1 at multiple novel sites, resulting in enhanced activation of TAK1 (Figure 3). The TAB4 kinase-activating function involves association with polyubiquitin chains that is dependent on a Phe-Pro motif at TIP/TAB4 residues 254–255. The ability to activate TAK1 seems to be a separate activity from the TAB4/TIP phosphatase-binding. These reports show that PP6 associates with different protein members of the NF-κB signaling pathway such as Iκ-Bε, TAB2 and TAK1. PP6 clearly is a factor limiting activation of NF-κB, probably through multiple mechanisms.
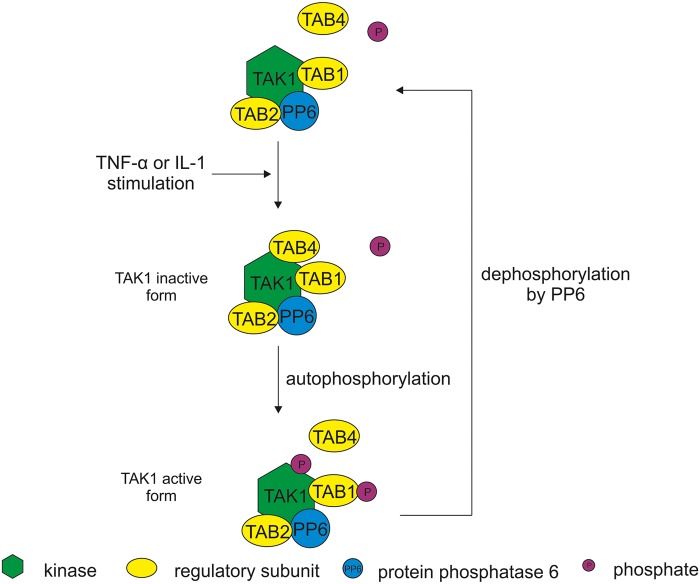
Figure 3. TAB1, TAB2 and PP6c constitutively bind to TAK1 [18,42,43].
TAB2 protein is essential for PP6c binding to TAK1 [43]. Upon TNF-α or IL-1 stimulation, TAB4 (a.k.a. TIP41) associates with inactive (unphosphorylated) form of TAK1. TAB4 binding enhances phosphorylation and activation of the TAK1-TAB1 complex [41]. PP6 inactivates TAK1 by dephosphorylation [42].
Effects of Ppp6c knockout and Ppp6r1 (SAPS1) knockout on T cells
Ongoing studies by our group and others have focused on actions of PP6 in T lymphocytes. Ppp6c homozygous null mutations are early embryonic lethal [45], necessitating use of conditional knockout approaches to study the role of PP6 in vivo. Ye et al. [46] engineered mice that are homozygous for a LoxP-targeted allele on chromosome 2, which were crossed with Lck-Cre or CD4-Cre mice to drive selective deletion of the gene for PP6 catalytic subunit (PP6c) at early or intermediate stages of thymocyte development, respectively. In an alternative approach, we have produced mice that are homozygous for a LoxP-targeted Ppp6r1 allele (encoding SAPS1) on chromosome 7, which were crossed with Sox2-Cre to delete the SAPS1 regulatory subunit in all adult tissues. The Ppp6r1f/f Sox2-cre mice are viable, fecund and display no difference in longevity compared with Ppp6r1+/+ mice (Brautigan et al., unpublished data).
Immunophenotyping by flow cytometry revealed that loss of PP6c during thymocyte development in Pp6cf/f lck-cre mice significantly impaired the development of α/β-lineage thymocytes. The number of thymocytes in both the CD4+ CD8+ double-positive (DP) as well as CD4+ and CD8+ single-positive (SP) compartments were decreased by 50–60%. The authors noted that PP6c-deficient DP thymocytes were extremely sensitive to antibody cross-linking of the α/β-TCR and subsequently demonstrated that the decreased number of DP and SP thymocytes was due to enhanced sensitivity to negative selection. The number of CD4−/CD8− double-negative (DN) thymocytes in Pp6cf/f lck-cre mice was increased 2-fold. Interestingly, the increased number of DN cells was due to an increase in the number of γ/δ-lineage thymocytes. More detailed analysis revealed significantly increased use of γv1, γv5 and, particularly, γv6 variable region gene segments in PP6c-deficient γδ-thymocytes [46]. Subsequent experiments demonstrated that the increased number of γδ-thymocytes arose from ongoing expansion of fetal-derived γv6/δv1 producing thymocytes rather than increased production of γδ-thymocytes from bone marrow-derived hematopoietic stem cells. Aberrant thymocyte differentiation in Pp6cf/f lck-cre mice led to a dramatic 80–90% reduction in the number of splenic, lymph node and peripheral blood CD4+ and CD8+ T cells in Pp6cf/f lck-cre mice relative to controls.
The small number of CD4+ and CD8+ T cells in the periphery of Pp6cf/f lck-cre mice made it very difficult to study mature CD4+ and CD8+ PP6-deficient T cells. To get around this problem, Ye and colleagues used Pp6cf/f CD4-cre mice [46]. These mice initiate deletion in the DP compartment and were found to contain a larger peripheral CD4+ and CD8+ splenic T cell compartment than Pp6cf/f lck-cre mice, although the number of CD4+ and CD8+ splenic T cells was still reduced by ∼70 and 60%, respectively. Most of the PP6-deficient CD4+ and CD8+ T cells were found to have an activated phenotype. However, both CD4+ and CD8+ T cells had a faster turnover rate and were prone to apoptotic cell death. In addition to disrupted homeostasis of CD4+ and CD8+ T cells, there was a notable increase in the frequency and number of interferon-γ producing CD4+ and CD8+ effector T cells in Pp6cf/f CD4-cre mice as well as a modest increase in the number of IL-4-producing CD4 T cells induced ex vivo by PMA/ionomycin [46]. Thus, loss of PP6c during thymocyte development results in significant defects in thymic output, seemingly as a result of increased negative selection, as well as hyperactivation, increased proliferation and apoptotic cell death in the splenic CD4+ and CD8+ T cell compartments. In addition, PP6c-deficient T cells appear more prone to differentiation and production of cytokines. Importantly, loss of PP6c in CD4+ and CD8+ T cells leads to increased activation of TCR signaling pathways including MAPK, NF-κB and Akt signaling pathways. The consequences of these fascinating phenotypes to immune function remain to be determined. It would be interesting to determine the phenotype of Pp6cf/f mice made PP6-deficient only in mature T cells.
As discussed above, PP6 associates with a set of regulatory subunits called SAPS [17]. To begin to understand how PP6 is tied to different functions in lymphocytes, we have characterized Ppp6r1f/f Sox2-cre mice. These mice do not display defects in T or B cell development and have quantitatively normal T and B cell compartments in the periphery. Thus, SAPS1 does not appear to be involved in directing the activities of PP6 during lymphocyte differentiation nor in immune homeostasis, exposing a clear difference between knockout of PP6c vs. SAPS1. However, ex vivo PMA/ionomycin stimulation of splenocytes from SAPS1-deficient mice resulted in a greater proportion of IL-4-producing CD4+ T cells when compared with corresponding controls. This response was similar in lymphocytes with either PP6c or SAPS1 knockout [46 and unpublished data]. Furthermore, in vitro differentiation assays demonstrate that SAPS1-deficient CD4 T cells favor differentiation into IL-4-producing cells but not differentiation into interferon-γ-producing cells. Interestingly, SAPS1-deficient mice contain 100- to 1000-fold more serum IgE than Ppp6r1f/f or C57BL/6 mice (unpublished data). These results suggest that some signaling events in T cells are specifically targeted by PP6/SAPS1 holoenzymes (i.e. IL-4 production by CD4+ T cells), while other processes are regulated by PP6 holoenzymes not dependent on SAPS1 (i.e. IFNγ production by CD4 and CD8 T cells). We have crossed the Ppp6r1f/f mice to dLck-cre and CD19-cre mice to test for T and B cell-intrinsic aspects of this phenotype.
Phosphorylation of NF-κB signaling proteins is limited by PP6 in thymocytes stimulated by PMA/ionomycin. Phosphorylation of p65/RelA, IKKα/β and Iκ-Bε, but not Iκ-Bα, was enhanced in PP6c-deficient cells relative to floxed controls [46]. Reversed-phase protein array (RPPA) analysis of over 300 proteins and phospho-sites performed with cell extracts prepared from SAPS1-deficient mice revealed that phospho-p65/RelA was the most up-regulated event in naïve splenic CD4+ T cells (unpublished data). However, the level of phospho-TAK1 paradoxically was reduced in PP6-deficient thymocytes, which suggests that increased activation of NF-κB signaling was not simply due to the hyperactivation of TAK1 but possibly involved alternative upstream pathways [46]. Taken together, the findings confirm that PP6 acts together with SAPS1 to limit NF-κB activation in thymocytes as well as splenic CD4+ T cells [46 and unpublished data]. In Table 1, we present a phenotype comparison of Ppp6c knockout and Ppp6r1 (SAPS1) knockout mice models.
Table 1. Phenotype comparison of Ppp6c knockout and Ppp6r1 (SAPS1) knockout mice models [46 and unpublished data].
| PP6c KO | SAPS1 KO | |
|---|---|---|
| Targeted gene | Pppc6 | Ppp6r1 (SAPS1) |
| Chromosome location | Chromosome 2 | Chromosome 7 |
| Cre recombinase | Lck-Cre, CD4-Cre | Sox2-Cre |
| Splenic IL-4+CD4+ T cells (%)1 | Up | Up |
| Splenic CD8+IL-4+ T cells (%)1 | Up | No difference |
| Splenic CD8+INFγ+ T cells (%)1 | Up | No difference |
| Phospho-p65/RelA | Up2 | Up3 |
Following methods were used to measure the parameters: 1 Flow cytometry; 2immunoblot; 3RPPA.
As one approach to better understand PP6 and SAPS1 functions in T cells, we performed gene microarray analysis using naïve CD4+ T cells isolated from spleens of SAPS1 knockout mice and controls. Principal Component Analysis of gene expression changes clearly separated SAPS1 knockout and wild-type C57/B6. Out of more than 25 000 genes, only 242 were significantly up-regulated and 14 down-regulated (cutoff: logFC>|1|, P-value < 0.05). Gene enrichment analysis using the STRING tool [47] revealed that most up-regulated genes were involved in immune system processes, immune response and leukocyte functioning. We are using the changes in gene expression to formulate and answer questions about how deletion of SAPS1 affects T cell functions, as a way to address functions of PP6 in lymphocytes.
Abbreviations
- DN
double-negative
- DP
double-positive
- IKK
IκB kinase
- NF-κB
nuclear factor kappaB
- NK
natural killer
- OA
okadaic acid
- PP2A
protein phosphatase-2A
- PP4
protein phosphatase-4
- PP6
Protein phosphatase-6
- PP6c
PP6 catalytic subunit
- RPPA
reversed-phase protein array
- SAPs
Sit4-associated proteins
- SP
single-positive
- TAK1
TGF-β-activated kinase 1
- TIP
type 2A phosphatase-interacting protein
Funding
The authors were supported by NIH grants [AI-114960, R56 AI-108767, CA-192669] and received generous support from Charles Burnett III.
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Brautigan D.L. (2013) Protein Ser/ Thr phosphatases — the ugly ducklings of cell signalling. FEBS J. 280, 324–325 doi: 10.1111/j.1742-4658.2012.08609.x [DOI] [PubMed] [Google Scholar]
- 2.Shi Y. (2009) Serine/threonine phosphatases: mechanism through structure. Cell 139, 468–484. doi: 10.1016/j.cell.2009.10.006 [DOI] [PubMed] [Google Scholar]
- 3.Bastians H. and Ponstingl H. (1996) The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J. Cell Sci. 109(Pt 12), 2865–2874 PMID: [DOI] [PubMed] [Google Scholar]
- 4.Sumiyoshi E., Sugimoto A. and Yamamoto M. (2002) Protein phosphatase 4 is required for centrosome maturation in mitosis and sperm meiosis in C. elegans. J. Cell Sci. 115(Pt 7), 1403–1410 PMID: [DOI] [PubMed] [Google Scholar]
- 5.Afshar K., Werner M.E., Tse Y.C., Glotzer M. and Gönczy P. (2010) Regulation of cortical contractility and spindle positioning by the protein phosphatase 6 PPH-6 in one-cell stage C. elegans embryos. Development 137, 237–247 doi: 10.1242/dev.042754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kao G., Tuck S., Baillie D. and Sundaram M.V. (2004) C. elegans SUR-6/PR55 cooperates with LET-92/protein phosphatase 2A and promotes Raf activity independently of inhibitory Akt phosphorylation sites. Development 131, 755–765 doi: 10.1242/dev.00987 [DOI] [PubMed] [Google Scholar]
- 7.Swingle M., Ni L. and Honkanen R.E. (2007) Small-molecule inhibitors of ser/thr protein phosphatases: specificity, use and common forms of abuse. Methods Mol. Biol. 365, 23–38 doi: 10.1385/1-59745-267-X:23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickey R.W., Bobzin S.C., Faulkner D.J., Bencsath F.A. and Andrzejewski D. (1990) Identification of okadaic acid from a Caribbean dinoflagellate, Prorocentrum concavum. Toxicon 28, 371–377 doi: 10.1016/0041-0101(90)90074-H [DOI] [PubMed] [Google Scholar]
- 9.Cohen P., Holmes C.F.B. and Tsukitani Y. (1990) Okadaic acid: a new probe for the study of cellular regulation. Trends Biochem. Sci. 15, 98–102 doi: 10.1016/0968-0004(90)90192-E [DOI] [PubMed] [Google Scholar]
- 10.Dounay A.B. and Forsyth C.J. (2002) Okadaic acid: the archetypal serine/threonine protein phosphatase inhibitor. Curr. Med. Chem. 9, 1939–1980 doi: 10.2174/0929867023368791 [DOI] [PubMed] [Google Scholar]
- 11.Valdiglesias V., Prego-Faraldo M.V., Pásaro E., Méndez J. and Laffon B. (2013) Okadaic acid: more than a diarrheic toxin. Mar. Drugs 11, 4328–4349 doi: 10.3390/md11114328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honkanen R.E. and Golden T (2002) Regulators of serine/threonine protein phosphatases at the dawn of a clinical era? Curr. Med. Chem. 9, 2055–2075. doi: 10.2174/0929867023368836 [DOI] [PubMed] [Google Scholar]
- 13.Prickett T.D. and Brautigan D.L. (2006) The α4 regulatory subunit exerts opposing allosteric effects on protein phosphatases PP6 and PP2A. J. Biol. Chem. 281, 30503–30511 doi: 10.1074/jbc.M601054200 [DOI] [PubMed] [Google Scholar]
- 14.Favre B., Turowski P. and Hemmings B.A. (1997) Differential inhibition and posttranslational modification of protein phosphatase 1 and 2A in MCF7 cells treated with calyculin-A, okadaic acid, and tautomycin. J. Biol. Chem. 272, 13856–13863 doi: 10.1074/jbc.272.21.13856 [DOI] [PubMed] [Google Scholar]
- 15.Nagaraj N., Wisniewski J.R., Geiger T., Cox J., Kircher M., Kelso J. et al. (2011) Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 7, 548 doi: 10.1038/msb.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sparks J.W. and Brautigan D.L. (1986) Molecular basis for substrate specificity of protein kinases and phosphatases. Int. J. Biochem. 18, 497–504 doi: 10.1016/0020-711X(86)90159-X [DOI] [PubMed] [Google Scholar]
- 17.Luke M.M., Della Seta F., Di Como C.J., Sugimoto H., Kobayashi R. and Arndt K.T. (1996) The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol. Cell. Biol. 16, 2744–2755 doi: 10.1128/MCB.16.6.2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefansson B. and Brautigan D.L. (2006) Protein phosphatase 6 subunit with conserved Sit4-associated protein domain targets IκBε. J. Biol. Chem. 281, 22624–22634 doi: 10.1074/jbc.M601772200 [DOI] [PubMed] [Google Scholar]
- 19.Morales-Johansson H., Puria R., Brautigan D.L. and Cardenas M.E. (2009) Human protein phosphatase PP6 regulatory subunits provide Sit4-dependent and rapamycin-sensitive sap function in Saccharomyces cerevisiae. PLoS ONE 4, e6331 doi: 10.1371/journal.pone.0006331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guergnon J., Derewenda U., Edelson J.R. and Brautigan D.L. (2009) Mapping of protein phosphatase-6 association with its SAPS domain regulatory subunit using a model of helical repeats. BMC Biochem. 10, 24 doi: 10.1186/1471-2091-10-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh P.P., Arora J., Isambert H. and Christos A. (2015) Identification of ohnolog genes originating from whole genome duplication in early vertebrates, based on synteny comparison across multiple genomes. PLoS Comput. Biol. 11, e1004394 doi: 10.1371/journal.pcbi.1004394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. www.phosphosite.org under SAPS1, SAPS2, SAPS3.
- 23.Stefansson B., Ohama T., Daugherty A.E. and Brautigan D.L. (2008) Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 47, 1442–1451 doi: 10.1021/bi7022877 [DOI] [PubMed] [Google Scholar]
- 24.Vincent A., Berthel E., Dacheux E., Magnard C. and Venezia N.L.D. (2016) BRCA1 affects protein phosphatase 6 signalling through its interaction with ANKRD28. Biochem. J. 473, 949–960 doi: 10.1042/BJ20150797 [DOI] [PubMed] [Google Scholar]
- 25.Tachibana M., Kiyokawa E., Hara S., Iemura S.-I., Natsume T., Manabe T. et al. (2009) Ankyrin repeat domain 28 (ANKRD28), a novel binding partner of DOCK180, promotes cell migration by regulating focal adhesion formation. Exp. Cell Res. 315, 863–876 doi: 10.1016/j.yexcr.2008.12.005 [DOI] [PubMed] [Google Scholar]
- 26.Kiyokawa E. and Matsuda M. (2009) Regulation of focal adhesion and cell migration by ANKRD28-DOCK180 interaction. Cell Adh. Migr. 3, 281–284 doi: 10.4161/cam.3.3.8857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng K., Bastos R.N., Barr F.A. and Gruneberg U. (2010) Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 191, 1315–1332 doi: 10.1083/jcb.201008106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rusin S.F., Schlosser K.A., Adamo M.E. and Kettenbach A.N. (2015) Quantitative phosphoproteomics reveals new roles for the protein phosphatase PP6 in mitotic cells. Sci. Signal 8, rs12 doi: 10.1126/scisignal.aab3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Couzens A.L., Knight J.D.R., Kean M.J., Teo G., Weiss A., Dunham W.H. et al. (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase-phosphatase interactions. Sci. Signal 6, rs15 doi: 10.1126/scisignal.2004712 [DOI] [PubMed] [Google Scholar]
- 30.Mi J., Dziegielewski J., Bolesta E., Brautigan D.L., Larner J.M. and Blagosklonny M.V. (2009) Activation of DNA-PK by ionizing radiation is mediated by protein phosphatase 6. PLoS ONE 4, e4395 doi: 10.1371/journal.pone.0004395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C., Jin X., Tsueng G., Afrasiabi C. and Su A.I. (2016) BioGPS: building your own mash-up of gene annotations and expression profiles. Nucleic Acids Res. 44, D313–D316 doi: 10.1093/nar/gkv1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su A.I., Wiltshire T., Batalov S., Lapp H., Ching K.A., Block D. et al. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl Acad. Sci. 101, 6062–6067 doi: 10.1073/pnas.0400782101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M. et al. (2014) Mass-spectrometry-based draft of the human proteome. Nature 509, 582–587 doi: 10.1038/nature13319 [DOI] [PubMed] [Google Scholar]
- 34.Sen R. and Baltimore D. (1986) Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell 46, 705–716 doi: 10.1016/0092-8674(86)90346-6 [DOI] [PubMed] [Google Scholar]
- 35.Gerondakis S. and Siebenlist U. (2010) Roles of the NF-κB pathway in lymphocyte development and function. Cold Spring Harbor Perspect. Biol. 2, a000182 doi: 10.1101/cshperspect.a000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hayden M.S., West A.P. and Ghosh S. (2006) NF-κB and the immune response. Oncogene 25, 6758–6780 doi: 10.1038/sj.onc.1209943 [DOI] [PubMed] [Google Scholar]
- 37.Bouwmeester T., Bauch A., Ruffner H., Angrand P.-O., Bergamini G., Croughton K. et al. (2004) A physical and functional map of the human TNF-α/NF-κB signal transduction pathway. Nat. Cell Biol. 6, 97–105 doi: 10.1038/ncb1086 [DOI] [PubMed] [Google Scholar]
- 38.Li Q. and Verma I.M. (2002) NF-κB regulation in the immune system. Nat. Rev. Immunol. 2, 725–734 doi: 10.1038/nri910 [DOI] [PubMed] [Google Scholar]
- 39.Moynagh P.N. (2005) The NF-κB pathway. J. Cell Sci. 118, 4589–4592 doi: 10.1242/jcs.02579 [DOI] [PubMed] [Google Scholar]
- 40.Hayashi K., Momoi Y., Tanuma N., Kishimoto A., Ogoh H., Kato H. et al. (2015) Abrogation of protein phosphatase 6 promotes skin carcinogenesis induced by DMBA. Oncogene 34, 4647–4655 doi: 10.1038/onc.2014.398 [DOI] [PubMed] [Google Scholar]
- 41.Prickett T.D., Ninomiya-Tsuji J., Broglie P., Muratore-Schroeder T.L., Shabanowitz J., Hunt D.F. et al. (2008) TAB4 stimulates TAK1–TAB1 phosphorylation and binds polyubiquitin to direct signaling to NF-κB. J. Biol. Chem. 283, 19245–19254 doi: 10.1074/jbc.M800943200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kajino T., Ren H., Iemura S.-i., Natsume T., Stefansson B., Brautigan D.L. et al. (2006) Protein phosphatase 6 down-regulates TAK1 kinase activation in the IL-1 signaling pathway. J. Biol. Chem. 281, 39891–39896 doi: 10.1074/jbc.M608155200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Broglie P., Matsumoto K., Akira S., Brautigan D.L. and Ninomiya-Tsuji J. (2010) Transforming growth factor β-activated kinase 1 (TAK1) kinase adaptor, TAK1-binding protein 2, plays dual roles in TAK1 signaling by recruiting both an activator and an inhibitor of TAK1 kinase in tumor necrosis factor signaling pathway. J. Biol. Chem. 285, 2333–2339 doi: 10.1074/jbc.M109.090522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McConnell J.L., Gomez R.J., McCorvey L.R.A., Law B.K. and Wadzinski B.E. (2007) Identification of a PP2A-interacting protein that functions as a negative regulator of phosphatase activity in the ATM/ATR signaling pathway. Oncogene 26, 6021–6030 doi: 10.1038/sj.onc.1210406 [DOI] [PubMed] [Google Scholar]
- 45.Ogoh H., Tanuma N., Matsui Y., Hayakawa N., Inagaki A., Sumiyoshi M. et al. (2016) The protein phosphatase 6 catalytic subunit (Ppp6c) is indispensable for proper post-implantation embryogenesis. Mech. Dev. 139, 1–9 doi: 10.1016/j.mod.2016.02.001 [DOI] [PubMed] [Google Scholar]
- 46.Ye J., Shi H., Shen Y., Peng C., Liu Y., Li C. et al. (2015) PP6 controls T cell development and homeostasis by negatively regulating distal TCR signaling. J. Immunol. 194, 1654–1664 doi: 10.4049/jimmunol.1401692 [DOI] [PubMed] [Google Scholar]
- 47.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J. et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–D452 doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]