Abstract
Background
The ex vivo generation of human hematopoietic stem cells (HSCs) with long-term repopulating capacity and multi-lineage differentiation potential represents the holy grail of hematopoiesis research. In principle, human induced pluripotent stem cells (hiPSCs) provide the tool for both studying molecular mechanisms of hematopoietic development and the ex vivo production of ‘true’ HSCs for transplantation purposes and lineage-specific cells, e.g. red blood cells, for transfusion purposes. CD43-expressing cells have been reported as the first hematopoietic cells during development, but whether or not these possess multilineage differentiation and long-term engraftment potential is incompletely understood.
Methods
We performed ex vivo generation of hematopoietic cells from hiPSCs using an embryoid body(EB)-based, xeno-product-free differentiation protocol. We investigated the multilineage differentiation potential of different FACS-sorted CD43-expressing cell subsets by colony-forming assays in semisolid media. Further, erythroid differentiation was investigated in more detail using established protocols.
Results
By using CD43, we were able to measure hematopoietic induction efficiency during hiPSC-derived EB differentiation. Further, we determined CD43+ cells as the cell population of origin for in vitro erythropoiesis. Furthermore, colony formation demonstrates that the multipotent hematopoietic stem and progenitor cell fraction is particulary enriched in the CD43hi CD45+ population.
Keywords: Hematopoietic stem cells, Induced pluripotent stem cells, Hematopoiesis, Embryoid body
Introduction
The generation of hematopoietic stem cells (HSCs) from induced pluripotent stem cells is of great interest for both fundamental research questions regarding HSC ontology and potential clinical applications [1,2]. The availability and genetic imbalance between matching donors and recipients as well as risk factors such as graft-versus-host reactions are the major limitations of allogeneic HSC transplantation [3]. The patient-individual generation of HSCs or more mature hematopoietic cells of various lineages for autologous treatment options is therefore an important goal, if not the holy grail within the field of hematopoiesis research [2]. However, hematopoietic differentiation from human embryonic stem cells (hESCs) as well as human induced pluripotent stem cells (hiPSCs) has been shown to be of a primitive rather than a definitive phenotype in multiple studies, especially when performed in absence of the physiological niche [4,5]. Cell lines such as OP-9 may recapitulate the hematopoietic niche and promote definitive hematopoiesis, but the use of xeno-derived cell lines and products is less favorable, if aiming at clinical application [4,6,7].
Following common sense CD34, a 115-kDa transmembrane glycoprotein, firstly described by Civin et al. [8] and cloned by Simmons et al. [9] from a Ka1a library, is a marker widely used to characterize human hematopoietic stem and progenitor cells (HSPCs) and to enumerate HSPC dosages in stem cell transplantation settings [10]. Regardless that its precise mode of action is still unknown, CD34 plays an important role in the interaction of HSPCs and its niche, facilitating mobilization and homing properties. During early embryonic development, however, CD34 expression describes both HSPCs and endothelial progenitor cells (EPCs) within the CD309 (KDR)+ population derived from the embryonic mesoderm [11,12]. Although phenotypical heterogeneity has been reported among adult CD34+ HSPCs, comprising only a small fraction with long-term HSC potential, an efficient marker with similar predictive implications on the potential to reconstitute the adult hematopoietic system lifelong has not fully been defined yet for hiPSC-derived ex vivo hematopoiesis [13].
As a potential candidate, CD43, also known as leukosialin or sialoglycoprotein, has been shown to be expressed on early hematopoietic cells derived from hESCs [4,14]. Comparable to CD34, CD43 is a strongly glycosylated transmembrane sialomucin, expressed on all hematopoietic cells, except mature erythrocytes [15]. Expressed on immature hematopoietic cells, it is downregulated during erythroid differentiation [16]. As a mucin it is believed to function as a repulsive barrier on T cells and myeloid cells, and its downregulation is associated with T-cell activation and migration of myeloid cells from the vasculature [17]. On immature hematopoietic cells, it may prevent adhesion and undesired stimuli to differentiate, but at the same time mediate interaction with selectins to promote migration from the bone marrow niche to the vasculature [18]. Therefore, due to the expression on various hematopoietic cells, CD43 specificity is compromised during hematopoietic differentiation from pluripotent stem cells in vitro. Nonetheless, its expression was detected before onset of CD45 and CD41 expression during hematopoietic differentiation from human pluripotent stem cells using an OP-9 co-culture model [4]. The emergence of CD45 expression correlates with progressive lymphomyeloid commitment as shown by gene expression profiling (e.g. PU1, MPO, IL7RA) [4]. Additionally, early human embryos have been investigated recently regarding early hematopoietic development and determined CD43 expression on the earliest hematopoietic progenitor cells [19]. However, the multi-lineage differentiation potential of CD43 expressing hematopoietic cell subsets, generated in vitro, has not been fully investigated until today.
Here, we generated CD43-expressing cells using a xeno-product- and cell free embryoid body(EB)-based differentiation protocol [20]. We investigated the erythroid differentiation potential of CD43 expressing cells and defined a CD43hi subset as the source of erythroid differentiation from hiPSCs. To address the multilineage differentiation potential we performed colony-forming unit (CFU) assays in semisolid media. Via transcriptional profiling we directly compared CD43hi cells to peripheral blood (PB) and cord blood (CB) derived CD34+ HSPCs. Our data imply multilineage differentiation potential of hiPSC-derived CD43hi cells with a bias towards the erythroid lineage.
Material and Methods
Collection of CD34+ HSPCs from PB and CB
Adult PB CD34+ cells were obtained from allogeneic stem cell donations after G-CSF mobilization by apheresis. CB CD34+ cells were collected after normal deliveries. CD34+ cells were further purified by immunomagnetic sorting via MACS (Milteny, Bergisch-Gladbach, Germany), and cells from three donations (adult PB and CB each) were subsequently applied to ex vivo erythropoiesis as biological replicates as previously described [21]. Written informed consent was obtained from all donors; the study was approved by the local ethics committee (Medical University Graz, Austria EK: 27-165 ex 14/15).
Hematopoietic and Erythroid Differentiation of hESCs and hiPSCs
Hematopoietic induction and erythroid differentiation from hESCs (H1, Wicell, WI, USA) and CB CD34-derived hiPSCs has been performed using an EB-based hematopoietic induction protocol followed by a three-stage ex vivo erythropoiesis assay as described earlier (fig. 2A) [20]. The generation of a CB-derived iPSC line by OCT4, SOX2, KLF4, and c-Myc and its characterization has been described previously [20]. CD43+ cells as well as CD43low and CD43hi subpopulations were seeded at a density of 5 × 104 cells/ml upon in vitro erythropoiesis initiation (stage II). Bulk cells from EB dissociation were seeded at a density of 3 × 105 cells/ml; PB and CB HSCs were seeded as 1 × 104 cells/ml.
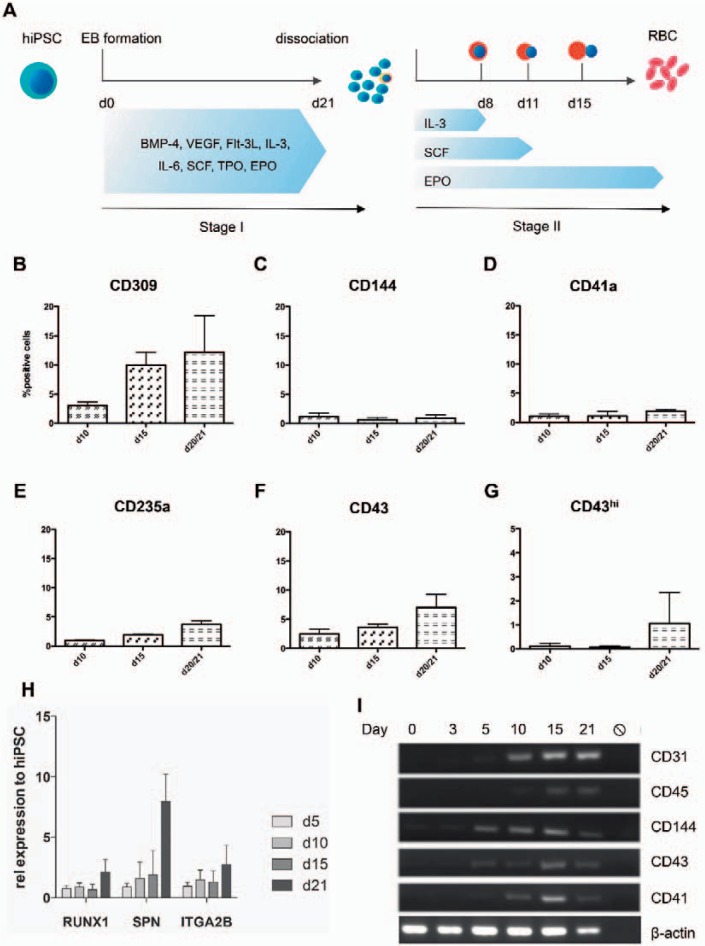
Fig. 2.
Hematopoietic induction during EB differentiation. hiPSCs were dissociated from the MEF layer and allowed to form EBs on day 0 of stage I and subsequently cultured in hematopoietic induction medium for 21 days. After Collagenase B digestion single cells were cultured for in vitro erythropoiesis in a three-stage manner for up to 21 days (A). Expression of surface markers specific for hematopoietic specification (KDR (VEGFR2), CD144, CD41a, CD235a (GPA), CD43) was assessed on EB day 10, 15 and 21 by flow cytometry (B-G). Expression kinetics of RUNX1, CD43 and CD41 was additionally investigated by quantitative RT-PCR and compared to undifferentiated hIPSC (H). CD31, CD45, CD144 (VE-Cadherin), CD43 and CD4 expression was monitored more tightly during EB differentiation using conventional PCR (I). Data were obtained from three independent experiments (n = 3), mean values ± SD are shown.
Sample Collection and RNA Isolation
In order to identify unbiased gene expression profiles across the entire transcriptome between hiPSC-derived CD43+ hematopoietic cells and PB as well as CB HSPCs, samples were collected at day 21 of EB differentiation. Sorting of CD43+ cells was performed after labeling of dissociated EB d21 cells with anti-CD43-APC and anti-CD45-PE on a Becton Dickinson FACS Aria Ilu (Franklin Lakes, NJ, USA). Total RNA was prepared using RNeasy columns (Qiagen, Hilden, Germany) following the manufacturer's instructions.
Gene Expression Analysis by Conventional and qRT-PCR
Complementary DNA synthesis was performed using the High-Capacity cDNA Reverse Transcription Kit following the manufacturer's references, quantitative RT-PCR was performed using Power SYBR Green PCR Master Mix on a 7900HT Fast Real-Time PCR System (all from Thermo Fisher Scientific, Waltham, MA, USA) and specific primers (RUNX1: fw 5´CCGAGAACCTCGAAGACATC´3 rev 3´GTCTGACCCTCATGGCTGT´5, SPN: fw 5´CACTTCAATAACAAGTGACCCTAAGG´3 rev 5´TGGAGGTTGTTGGCTCAGGTA3´ ITGA2B: fw. 5´CTTCTGGAGTGGGTGCTGCT´3 rev. 5´TCTCCCATGGCTGTCCTTGT´3). 18S cDNA was used as a reference gene, adult undifferentiated hiPSCs as a reference sample to determine relative gene expression on day 15 of culture.
Microarray Data Analysis
The microarray (HumanHT-12 4v; Illumina, San Diego, CA, USA) protocol was performed as already described [20]. Gene expression analysis following array normalization with GenomeStudio (Illumina) was done in R. Probe intensities were log2 transformed, and probes with a detection p value below 0.001 were considered for further analysis. A minimum signal of log2(100) was defined as the minimum expression threshold. Heatmaps and dendrograms were calculated with the gplot R-package. Top differentially expressed genes (DEGs) between two groups were determined by the largest fold change between two groups beyond the twofold range.
Genes expressed with variations within the twofold range were considered to be not relevantly up- or downregulated. Statistics of single gene expression data from microarray were plotted and analyzed using GraphPad Prism6 (GraphPad Software, Inc., La Jolla, CA, USA). Gene ontology analysis was performed using DAVID Bioinformatics Resources Version 6.8 software packages (Laboratory of Human Retrovirology and Immunoinformatics, Frederick, MD, USA) [22].
Results
Hematopoietic Induction Efficiency in hiPSC-Derived EBs and Subsequent Erythroid Differentiation
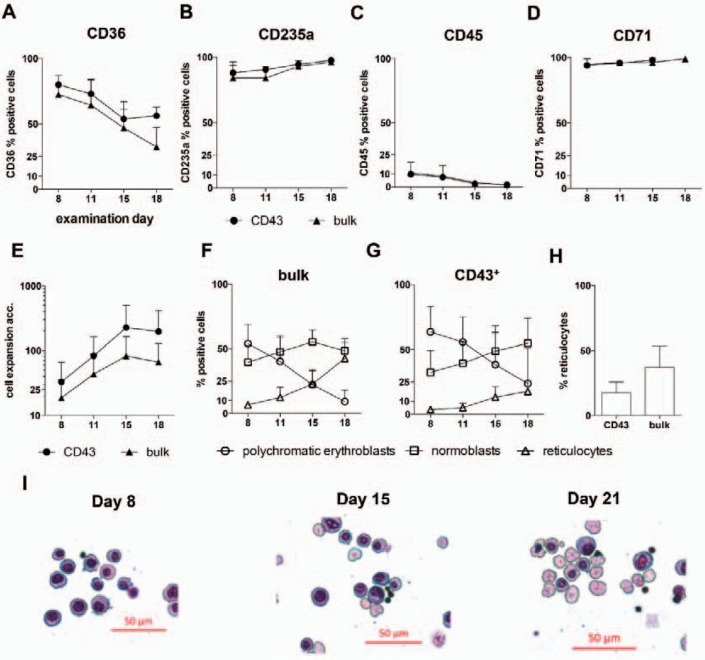
We previously observed that the amount of CD43-expressing cells correlates with the efficiency of the subsequently performed erythroid differentiation [20]. To identify the erythropoiesis-driving cell population in more detail, we performed a fluorescence-activated cell sorting (FACS) approach of different CD43-expressing cell subsets. In order to compare CD43+ with CD43- cells, we initially differentiated CB-derived hiPSCs into hematopoietic cells via EB differentiation as previously described [20]. After 21 days, EBs were dissociated, and single cells were labeled with anti-CD43 APC and anti-CD45 PE flow cytometry antibodies and sorted on a FACS Aria Ilu. We then performed erythroid differentiation and observed that CD43- cells never gave rise to hematopoietic cells in general (data not shown). The percentage of cells staining positive for hematopoietic markers (CD36, CD235a, CD45, CD71) during erythroid differentiation was found similar between CD43+-sorted and bulk cells (fig. 1A-D), while cell expansion was determined similar, yet a bit higher for CD43+ cells (fig. 1E). However, characterization of erythroid cells by cell morphology and neutral benzidine staining revealed a higher number of reticulocytes from bulk cell differentiation compared to CD43+ cells (37.1 ± 14.3% vs. 17.6 ± 7.1%; fig. 1F-H) accompanied by a lower number of polychromatic erythroblasts (7.7 ± 6.7% vs. 23.7 ± 23.8%) on day 15 of in vitro erythropoiesis. These data indicate CD43-expressing cells as the actual cellular origin of in vitro erythropoiesis in our experiments. In addition, these preliminary results suggest the importance of cultivation together with CD43- cells (bulk culture), which probably provide an auxiliary function comparable to the physiological niche-like environment.
Fig. 1.
Comparison of the erythroid differentiation potential of CD43-expressing cells versus bulk cells. Bulk cells and CD43+ sorted cells from EB day 21 have been subjected to erythroid differentiation (stage II, see fig. 2A) and erythroid surface marker expression was assessed by flow cytometry (A-D). Cumulative cell expansion was a slightly higher in CD43+ cells compared to bulk cells in line with the delayed differentiation (E). Cell morphology of CD43+ and bulk cells differentiation was investigated using cytospins stained with Pappenheim and neutral benzidine (F and G). Bulk cell differentiation revealed a higher number of already enucleated reticulocytes on day 18 of stage II (H). Data were obtained from three independent experiments (n = 3), mean values ± SD are shown. Representative cytospin images from bulk cell differentiation after Pappenheim staining (I).
Profiling of CD43 Expression during EB Formation
To monitor CD43 expression more closely during EB differentiation, we conducted a time course experiment with samples taken on day 5, 10, 15 and 21 of EB maturation, dissociated the EBs, and performed flow cytometry analysis for different markers for early hematopoiesis, such as CD309 (KDR), CD43 (leukosialin), CD41a, CD144 (VE-cadherin) and CD235a (GPA). The percentage of positive cells for each of these markers increased over time during EB differentiation, with CD309+ cells revealing the strongest increase (fig. 2B), followed by CD43 (fig. 2F). CD43+ cells reached an average percentage of 10.5 ± 2.8% on EB day 21 during the experiments conducted for this study (three independent experiments). However, higher numbers of CD43+ cells on EB day 21 have been achieved in other experiments using the same culture conditions (18.8 + 6.8%; 6 independent experiments, fig. 3A). CD41+ and CD235a+ cells reveal a moderate increase with a percentage of <5% of all cells after EB dissociation on day 21 (fig. 2D,E). Further, we investigated gene expression of early hematopoietic markers by both quantitative RT-PCR and conventional PCR and observed early expression of RUNX1, SPN (CD43), and ITGA2B (CD41) during EB differentiation (fig. 2H,I).
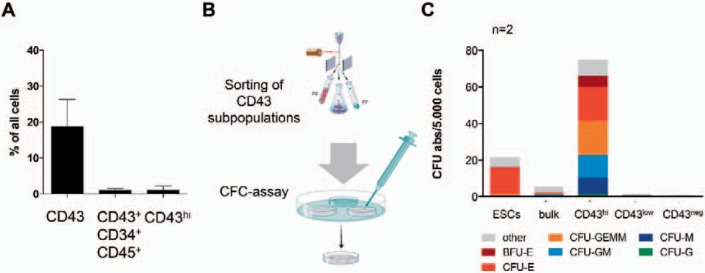
Fig. 3.
Hematopoietic differentiation potential of different CD43 expressing cell subsets. Expression of CD43 and CD34 was assessed by flow cytometry (A; n = 6; mean ± SD). CD43-, CD43low and CD43hi cells were sorted (homogeneity of >98%, data not shown) and subjected to CFC assay in semisolid media (B and C; n = 2). CD43hi cells were applied as 5 × 103 cells/ml, bulk cells from ESCs and hiPSCs, CD43low and CD43- as 3 × 104 cells/ml. Colonies were counted according to their characteristic morphology after 14 days and classified as BFU-E, CFU-E, CFU-M, CFU-G, CFU-GM and CFU-GEMM. Colonies of unknown morphology were classified as other (C).
Impact of the CD43hi Subset for Multilineage Differentiation
As a particular observation, we detected a CD43hi-expressing cell subset appearing as late as day 21 during EB differentiation using flow cytometry. We investigated these cells in more detail, using FACS and transcriptional profiling. CD43hi cells appear during later stages of EB differentiation of hiPSCs and were determined in 1.06 ± 1.29% (range 0.27-2.55%) in the respective analyzed experiments (n = 3) and 2.1 ± 0.47 in other experiments (n = 6) (fig. 3A). All CD43hi cells are CD45+ (low-intermediate) and the majority of these cells co-expresses CD34. CD43low cells are CD45hi/CD34neg. CD43hi, CD43low and CD43neg cell subsets have been sorted in order to investigate their colony-forming potential in a CFC assay in semisolid media supplemented with hematopoietic cytokines (fig. 3B; n = 2). CD43hi CD45low, CD43low CD45+, and CD43- CD45- cell subsets have been sorted in order to investigate their colony-forming potential in a CFC assay in semisolid media supplemented with hematopoietic cytokines (fig. 3B; n = 2). As a reference, hESCs were cultured and subjected to a CFC assay in a similar manner as CB-CD34+ iPSCs (fig. 3C). After 14-21 days, colonies were classified according to their morphology into CFU-M (colony-forming unit monocytes), CFU-GM (colony-forming unit granulocytes, monocytes), CFU-GEMM (immature mixed colonies), BFU-E (burst-forming unit-erythroid), CFU-E (colony-forming unit-erythroid), and hematopoietic colonies of unknown identity, classified as other. While CD43- cells did not possess any hematopoietic colony-forming potential in our experiments, CD43low-expressing cells generated only a few colonies of monocytic origin (4 and 3 CFU-M colonies), possibly indicating their loss of colony-forming potential, in particular their erythroid colony forming potential. The absolute colony number per 5,000 seeded cells was found much higher for CD43hi cells than for unsorted bulk cells, demonstrating the enrichment of hematopoietic progenitors with colony-forming potential in the CD43hi population. Absolute colony numbers are illustrated in figure 3C. CFU-E were found to comprise 24.5% of all CD43hi-derived colonies, compared to 9.1% when bulk cells were applied unsorted and 65.1% CFU-Es from hESCs. Early erythroid colonies (BFU-E) were also found enriched in CD43hi-derived CFC assays (5 and 3%), indicating a high erythroid differentiation potential of CD43hi cells (fig. 3C). Based on these results we suggest, that the CD43hi-expressing population comprises a population of HSPCs with multilineage potential. CD43low cells may represent further differentiated cells, which almost completely lost colony-forming potential and therefore only occasionally give rise to few myeloid colonies.
CD43hi Cells from hiPSCs May Represent HSPCs with an Advantage towards Erythroid Differentiation
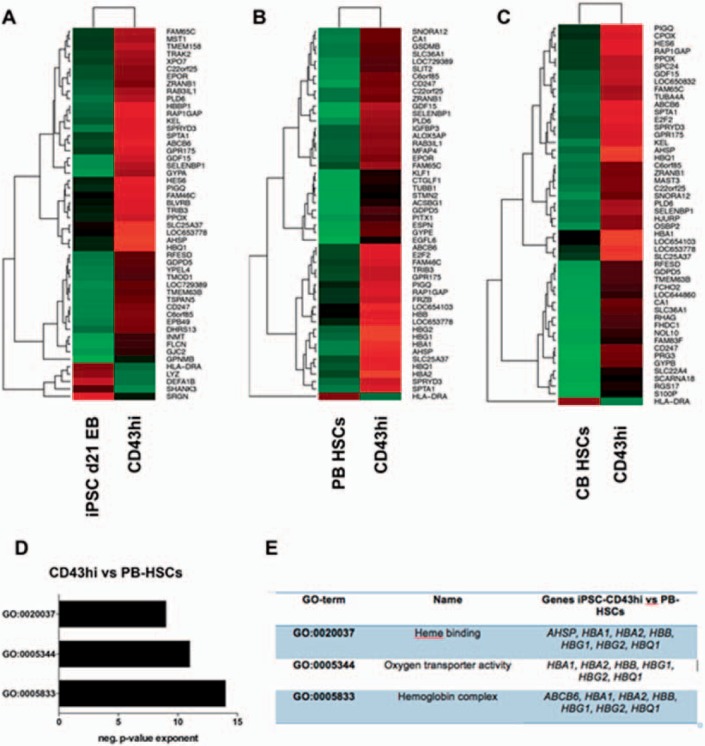
For a more detailed analysis of CD43-expressing cells, we performed transcriptional profiling of CD43hi cells by microarray and compared them to PB and CB CD34+ HSPCs. Correlations between CD43hi cells and CD34+ cells were found low in general (r = 0.607 and 0.635 for PB- and CB-HSCs, respectively; supplementary fig. 1A-C, available at http://content.karger.com/ProdukteDB/produkte.asp?doi=477357), indicating their gene expression profiles are not similar, while the similarities between CB- and PB-HSCs have been found much higher (r = 0.964; our own unpublished data). Further, specific markers for HSPCs were detected low in CD43hi cells by microarray; however, this was also the case for PB- and CB-HSCs (see supplementary fig. 1D, available at http://content.karger.com/ProdukteDB/produkte.asp?doi=477357). Expression levels were here barely detected above the expression threshold of a signal value of 100 arbitrary units. Following this, we performed differential gene expression analysis and found especially erythropoiesis-related genes higher expressed in CD43hi cells than in PB- and CB-HSPCs (fig. 4). Further, these transcripts were enriched in CD43hi cells when compared to hiPSC-derived EB day 21 bulk cells (fig. 4A). Among those genes were transcripts of the α- and β-globin cluster (HBA1, HBQ, HBB, HBG1, HBG2) as well as those for hemoglobin synthesis (AHSP), erythroid membrane proteins, receptors and surface markers (EPOR, GYPA, GYPB, SPTA1) as well as transcription factors (KLF1), as determined by gene ontology analysis using DAVID (fig. 4D,E).
Fig. 4.
Transcriptional profiling of PB- and CB-derived HSCs compared to hiPSC-derived hematopoietic progenitors. CD43hi cells were sorted after 21 days of EB differentiation. Sorted cells (n = 1) as well as HSCs from PB and CB (n = 3, each) were subjected to RNA isolation, cDNA generation and microarray. Differential gene expression was determined by the largest fold change. Top 50 DEGs between CD43hi cells and d21EB bulk cells as well as PB and CB-derived HSCs are shown in heatmaps (A-C). Gene ontology analysis was performed using DAVID. Top 3 GO-terms were determined by significance (negative p value exponent; shown in D). DEG expression analysis revealed strong enrichment for erythroid genes (E).
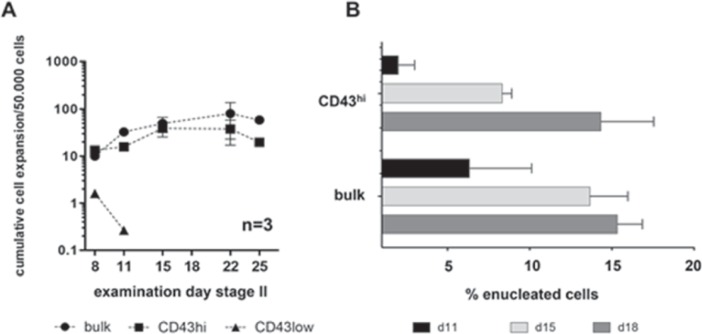
To investigate the erythroid differentiation potential of different CD43 cell subpopulations, we performed in vitro differentiation of bulk cells as well as of CD43hi, CD43low and CD43- sorted cells as described earlier in this and our previous work [20]. CD43- cells did not proliferate during the first 8 days, most cells died, and the cell cultures were terminated prematurely. CD43low cells gave rise to few hematopoietic cells, yet with growth arrest (fig. 5A), and skewed differentiation towards cells with myeloid morphology (data not shown). CD43hi cells, however, displayed similar cell expansion of 20- to 60-fold compared to bulk cell differentiation when seeded at an initial density of 50,000 cells/ml after sorting on day 0 of stage II. Moreover CD43hi cells seemed to enucleate at an almost similar rate (up to 15%) compared to bulk cells until day 18 of culture (fig. 5B) pointing at terminal differentiation as expected for erythroid generated from hiPSC sources.
Fig. 5.
Proliferation and enucleation behavior of CD43hi vs. CD43low cells. CD43hi and CD43low sorted cells were subjected to erythroid differentiation (stage II). CD43hi cells were seeded at an initial density of 5 × 104 cells/ml, CD43low and bulk cells at 3 × 105 cells/ml. Cumulative expansion was calculated as per 5 × 104 cells (A). Enucleation was determined on d11, 15 and 18 of stage II differentiation as % enucleated cells of all erythroid cells (B).
Discussion
Our study confirms the efficient generation of HSPCs from hiPSCs using an EB- based, xeno-product-free differentiation protocol [20]. Additionally, we defined CD43hi CD45low cells as HSPCs with multilineage differentiation potential with a bias towards erythroid differentiation.
Hematopoietic differentiation from hiPSCs enables the investigation of early development and promises to be a source of cell-based therapies in the future [5,23]. Besides the generation of distinct hematopoietic lineages, the generation of HSCs from hiPSCs is a central purpose of biomedical research and transfusion medicine in particular [24,25]. Although significant efforts were done, true HSCs with long-term engraftment potential cannot be expanded indefinitely in vitro until now [26,27]. In contrast, undifferentiated hiPSCs can theoretically be expanded indefinitely in vitro while maintaining their pluripotency for all three germ layers; they can be stored, thawed, and differentiated into the desired tissue afterwards as well as bioengineered orgene-modified on demand. A patient-specific generation of hiPSCs-derived HSPCs would offer both the potential for autologous transplantation omitting graft-versus-host immunogenicity [8,29] and autologous cell/tissue replacement without the risk of rejection, paving the way for ultimate personalized medicine [30,31,32]. However, long-term engrafting HSCs from hiPSCs have to date only been generated by way of teratoma formation on the basis of murine feeder cell layers such as OP-9 cells or the mouse as a host organism [6,33]
CD43 has been described previously as a pan-hematopoietic marker for hESC-derived hematopoiesis [4]. However CD43+ CD45+ CD34+ cells have been described as myeloid-skewed multipotent hematopoietic cells when expanded with GM-CSF [14]. In contrast to this, we report here the emergence of CD43hi CD45 low CD34+/- cells possessing enhanced erythroid differentiation potential.
As determined by flow cytometry, the fraction of cells in our cultures expressing hematopoietic surface markers increases over time during EB differentiation (stage I) until day 21. Flow cytometry-activated cell sorting experiments additionally lead to the conclusion, that a subset of CD43hi-expressing cells might possess multilineage differentiation potential in vitro, as revealed by CFC assay of CD43hi cells after EB dissociation. This is in line with a recent report by Ivanovs et al. [34], reporting CD43-expressing cells derived from early human embryos after elective medical termination of pregnancy and transplanted into preconditioned adult NOD.Cg-Prkdcscid Il2rgtm1Wjl/Sz mice. These cells were subsequently able to reconstitute the hematopoietic system of the mice with multilineage differentiation potential. Based on this and our own findings, we suggest that CD43 is a useful surface marker for the purification of HSPCs from hESC and hiPSC in vitro differentiation. However, CD43hi expression may mark a heterogeneous hematopoietic cell population, lineage-specifically skewed by the growth factors and further ingredients of the differentiation media used. During our experiments arising HSPCs might be primed towards erythroid differentiation immediately after emergence by the cytokine cocktail used during stage I.
Using microarray analysis, we were able to confirm the enrichment in erythroid progenitors among CD43hi cells compared to d21 EB bulk cells as well as CB- and PB-derived HSCs. We differentially compared global gene expression patterns of hiPSC-derived CD43hi cells to PB- and CB-HSPCs and found CD43hi cells strongly enriched for expression of erythroid gene as additionally determined be gene ontology analysis. Membrane and cytoskeleton proteins and transcription factors specific for erythroid cells as well as globin genes and genes involved in heme biosynthesis were found highly, if not exclusively, expressed in CD43hi cells compared to PB- and CB-HSPCs.
During terminal erythroid differentiation of adult PB-HSPCs, CD43 is downregulated, however initially maintained on high levels until day 11 of culturing. At this time point most cells represent the stage of polychromatic erythroblasts. Therefore, high CD43 expression during lineage commitment and erythroid specification might not allow the use of CD43 alone as a marker for a pure HSPC population. With our study we confirmed that by using CD43 expression as a marker, we additionally isolate cells that are committed to the erythroid lineage.
When we first performed in vitro erythropoiesis of CD43+ cells and compared these to the differentiation of unsorted bulk cells after EB dissociation, we found a comparable differentiation potential between the two groups. However, we observed lower enucleation rates, as reflected in a lower number of reticulocytes on day 18 of culture, and a higher number of polychromatic erythroblasts on the same day. This indicates an more immature differentiation status probably due to decelerated maturation and enucleation arrest. For the differentiation of CD43hi cells, we observed a similar proliferation and enucleation behavior compared to unsorted bulk cell differentiation. However, data from three independent experiments might not deliver a complete description of their differentiation potential as hiPSC-derived hematopoiesis is known to be rather heterogeneous. The recapitulation of the complex spatiotemporal condition during hematopoietic development is challenging and might be hampered by spontaneous differentiation during EB culturing, leading to different outcomes and results. Additionally, the differentiation of sorted hiPSC-derived hematopoietic cells may deprive them from important interactions with accessory cells that are provided by the CD43- compartment and represent a component of the physiological niche. Consequently, this phenomenon might lead to decreased terminal maturation and influence cell proliferation as well as enucleation. Further, cell integrity after enucleation might be hampered by the lack of niche interactions and by causing premature cell death misleading to a more immature picture of the cell culture.
Although our data would possibly benefit from more statistical power provided by additional experiments, we believe that our preliminary results contribute significantly to the current understanding of hiPSC-derived early and late hematopoiesis. Further, the generation of an erythroid-skewed hematopoietic progenitor cell might be of great relevance to transfusion medicine and the ex vivo generation of red blood cells. More detailed studies would provide insights in the existence of further hematopoiesis-derived cell subsets and in the molecular mechanisms underlying their cell type-specific developmental pathways. This gained knowledge might allow for the invention of novel cellular therapies in the field of transfusion medicine.
Disclosure Statement
The authors declare no conflicting interests.
Supplementary Material
Supplementary data
Acknowledgments
The authors thank Dr. Christoph Bartenhagen for helping with microarray data analysis and Tasnim Arroum for performing qRT-PCR experiments.
References
- 1.Daniel MG, Pereira C-F, Lemischka IR, Moore KA. Making a hematopoietic stem cell. Trends Cell Biol. 2016;26:202–214. doi: 10.1016/j.tcb.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahlster L, Daley GQ. Progress towards generation of human haematopoietic stem cells. Nat Cell Biol. 2016;18:1111–1117. doi: 10.1038/ncb3419. [DOI] [PubMed] [Google Scholar]
- 3.Barker CC, Butzner JD, Anderson RA, Brant R, Sauve RS. Incidence, survival and risk factors for the development of veno-occlusive disease in pediatric hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2003;32:79–87. doi: 10.1038/sj.bmt.1704069. [DOI] [PubMed] [Google Scholar]
- 4.Vodyanik MA, Thomson JA, Slukvin II. Leukosialin (CD43) defines hematopoietic progenitors in human embryonic stem cell differentiation cultures. Blood. 2006;108:2095–2105. doi: 10.1182/blood-2006-02-003327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditadi A, Sturgeon CM, Keller G. A view of human haematopoietic development from the Petri dish. Nat Rev Mol Cell Biol. 2017;18:56–67. doi: 10.1038/nrm.2016.127. [DOI] [PubMed] [Google Scholar]
- 6.Amabile G. In vivo generation of transplantable human hematopoietic cells from induced pluripotent stem cells. Blood. 2013;121:1–3. doi: 10.1182/blood-2012-06-434407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ichii M, Frank MB, Iozzo RV, Kincade PW. The canonical Wnt pathway shapes niches supportive of hematopoietic stem/progenitor cells. Blood. 2012;119:1683–1692. doi: 10.1182/blood-2011-07-369199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Civin CI, Strauss LC, Brovall C, Fackler MJ, Schwartz JF, Shaper JH. Antigenic analysis of hematopoiesis. III. A hematopoietic progenitor cell surface antigen defined by a monoclonal antibody raised against KG- 1a cells. J Immunol. 1984;133:157–165. [PubMed] [Google Scholar]
- 9.Simmons DL, Satterthwaite AB, Tenen DG, Seed B. Molecular cloning of a cDNA encoding CD34, a sialomucin of human hematopoietic stem cells. J Immunol. 1992;148:267–271. [PubMed] [Google Scholar]
- 10.Sidney LE, Branch MJ, Dunphy SE, Dua HS, Hopkinson A. Concise review: evidence for CD34 as a common marker for diverse progenitors. Stem Cells. 2014;32:1380–1389. doi: 10.1002/stem.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortés F, Debacker C, Péault B, Labastie MC. Differential expression of KDR/VEGFR-2 and CD34 during mesoderm development of the early human embryo. Mech Dev. 1999;83:161–164. doi: 10.1016/s0925-4773(99)00030-1. [DOI] [PubMed] [Google Scholar]
- 12.Ditadi A, Sturgeon CM. Directed differentiation of definitive hemogenic endothelium and hematopoietic progenitors from human pluripotent stem cells. Methods. 2016;101:65–72. doi: 10.1016/j.ymeth.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Spangrude G, Heimfeld S, Weissman I. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 14.Choi KD, Vodyanik MA, Slukvin II. Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+ CD43+ CD45+ progenitors. J Clin Invest. 2009;119:2818–2829. doi: 10.1172/JCI38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remold-O'Donnell E, Zimmerman C, Kenney D, Rosen FS. Expression on blood cells of sialophorin, the surface glycoprotein that is defective in Wiskott-Aldrich syndrome. Blood. 1987;70:104–109. [PubMed] [Google Scholar]
- 16.Carlsson SR, Fukuda M. Isolation and characterization of leukosialin, a major sialoglycoprotein on human leukocytes. J Biol Chem. 1986;261:12779–12786. [PubMed] [Google Scholar]
- 17.Cruz-Muñoz ME, Salas-Vidal E, Salaiza-Suazo N, Becker I, Pedraza-Alva G, Rosenstein Y. The CD43 coreceptor molecule recruits the zeta-chain as part of its signaling pathway. J Immunol. 2003;171:1901–1908. doi: 10.4049/jimmunol.171.4.1901. [DOI] [PubMed] [Google Scholar]
- 18.Anzai N, Gotoh A, Shibayama H, Broxmeyer HE. Modulation of integrin function in hematopoietic progenitor cells by CD43 engagement: possible involvement of protein tyrosine kinase and phospholipase C-gamma. Blood. 1999;93:3317–3326. [PubMed] [Google Scholar]
- 19.Rybtsov S, Batsivari A, Bilotkach K, Paruzina D, Senserrich J, Nerushev O, Medvinsky A. Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43(-) embryonic precursor. Stem Cell Reports. 2014;3:489–501. doi: 10.1016/j.stemcr.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorn I, Klich K, Arauzo-Bravo MJ, Radstaak M, Santourlidis S, Ghanjati F, Radke TF, Psathaki OE, Hargus G, Kramer J, Einhaus M, Kim JB, Kögler G, Wernet P, Schöler HR, Schlenke P, Zaehres H. Erythroid differentiation of human induced pluripotent stem cells is independent of donor cell type of origin. Haematologica. 2015;100:32–41. doi: 10.3324/haematol.2014.108068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Betz J, Dorn I, Kouzel IU, Bauwens A, Meisen I, Kemper B, Bielaszewska M, Mormann M, Weymann L, Sibrowski W, Karch H, Schlenke P, Müthing J. Shiga toxin of enterohaemorrhagic Escherichia coli directly injures developing human erythrocytes. Cell Microbiol. 2016;18:1339–1148. doi: 10.1111/cmi.12592. [DOI] [PubMed] [Google Scholar]
- 22.Huang DW, Lempicki RA, Sherman BT. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 23.Paes BCMF, Moço PD, Pereira CG, Porto GS, de Sousa Russo EM, Reis LCJ, Covas DT, Picanço-Castro V. Ten years of iPSC: clinical potential and advances in vitro hematopoietic differentiation. Cell Biol Toxicol. 2017;33:233–250. doi: 10.1007/s10565-016-9377-2. [DOI] [PubMed] [Google Scholar]
- 24.Vo LT, Daley GQ. De novo generation of HSCs from somatic and pluripotent stem cell sources. Blood. 2015;125:2641–2648. doi: 10.1182/blood-2014-10-570234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Focosi D, Amabile G, Di Ruscio A, Quaranta P, Tenen DG, Pistello M. Induced pluripotent stem cells in hematology: current and future applications. Blood Cancer J. 2014;4:e211. doi: 10.1038/bcj.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Park B, Yoo KH, Kim C. Hematopoietic stem cell expansion and generation: the ways to make a breakthrough. Blood Res. 2015;50:194–203. doi: 10.5045/br.2015.50.4.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster JA, Stupnikov MR, Ma G, Liao W, Lai R, Ma Y, Aguila JR. Expansion of hematopoietic stem cells for transplantation: current perspectives. Exp Hematol Oncol. 2012;1:12. doi: 10.1186/2162-3619-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Araki R, Uda M, Hoki Y, Sunayama M, Nakamura M, Ando S, Sugiura M, Ideno H, Shimada A, Nifuji A, Abe M. Negligible immunogenicity of terminally differentiated cells derived from induced pluripotent or embryonic stem cells. Nature. 2013;494:100–104. doi: 10.1038/nature11807. [DOI] [PubMed] [Google Scholar]
- 29.Zhao T, Zhang Z, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira LMR, Mostajo-Radji MA. How induced pluripotent stem cells are redefining personalized medicine. Gene. 2013;520:1–6. doi: 10.1016/j.gene.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 31.Robinton DA, Daley GQ. The promise of induced pluripotent stem cells in research and therapy. Nature. 2012;481:295–305. doi: 10.1038/nature10761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chun YS, Byun K, Lee B. Induced pluripotent stem cells and personalized medicine: current progress and future perspectives. Anat Cell Biol. 2011;44:245255. doi: 10.5115/acb.2011.44.4.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki N, Yamazaki S, Yamaguchi T, Okabe M, Masaki H, Takaki S, Otsu M, Nakauchi H. Generation of engraftable hematopoietic stem cells from induced pluripotent stem cells by way of teratoma formation. Mol Ther. 2013;21:1424–1431. doi: 10.1038/mt.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanovs A, Rybtsov S, Anderson RA, Turner ML, Medvinsky A. Identification of the niche and phenotype of the first human hematopoietic stem cells. Stem Cell Reports. 2014;2:449–456. doi: 10.1016/j.stemcr.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data