Abstract
Background
Pluripotent stem cells, including induced pluripotent stem cells (iPSCs), have the capacity to differentiate towards all three germ layers and have been highlighted as an attractive cell source for the field of regenerative medicine. Thus, stable expression of therapeutic transgenes in iPSCs, as well as thereof derived progeny of hematopoietic lineage, may lay the foundation for innovative cell replacement therapies.
Methods
We have utilized human iPSC lines genetically modified by lentiviral vector technology or targeted integration of reporter genes to evaluate transgene expression during hematopoietic specification and differentiation towards macrophages.
Results
Use of lentiviral vectors equipped with an ubiquitous chromatin opening element (CBX3-UCOE) as well as zinc finger nuclease-mediated targeting of an expression cassette into the human adeno-associated virus integration site 1 (AAVS1) safe harbor resulted in stable transgene expression in iPSCs. When iPSCs were differentiated along the myeloid pathway into macrophages, both strategies yielded sustained transgene expression during the hematopoietic specification process including mature CD14+ and CD11b+ macrophages.
Conclusion
Combination of human iPSC technology with either lentiviral vector technology or designer nuclease-based genome editing allows for the generation of transgenic iPSC-derived macrophages with stable transgene expression which may be useful for novel cell and gene replacement therapies.
Keywords: Gene therapy, Hematopoiesis, Genome editing, Lentivirus, CBX3-UCOE, Macrophages, iPSC
Introduction
Pluripotent stem cells (PSCs) and in particular induced pluripotent stem cells (iPSCs) as first introduced by Kazutoshi Takahashi and Shinya Yamanaka in 2006 [1], hold the promise to revolutionize the field of cell and gene therapies and open new avenues of personalized treatment strategies also for the hematopoietic system. Human iPSCs (hiPSCs) nowadays can be easily generated by a number of reprogramming techniques and from various cell and tissue sources, including peripheral blood-, cord blood- or bone marrow-derived CD34+ hematopoietic stem/progenitor cells (HSPCs). Thereafter, iPSC lines can be differentiated towards the different cell types of all three germ layers, highlighting their attractiveness for the field of regenerative medicine. However, successful iPSC-based gene and cell therapy will rely on the efficient differentiation of iPSCs into the desired cell types. Whereas substantial progress has been made to differentiate hiPSCs into defined mature hematopoietic cells such as granulocytes, macrophages, erythrocytes, megakaryocytes or dendritic cells, lymphoid differentiation or the generation of long-term repopulating hematopoietic stem cells (LTR-HSCs) from human but also murine PSCs is still hampered by the low quality and inefficient quantity of the output cells [2]. The efficient generation of functional mature myeloid cells has been described by several groups utilizing different strategies, which include embryoid body formation, co-culture systems as well as small molecule- or cytokine-driven approaches [3,4,5,6]. In this context, the scalable production of erythrocytes or megakaryocytes/platelets is of particular interest for transfusion medicine [7,8]. Another highly attractive target population for cell-therapeutic strategies is represented by monocyte/macrophages, including tissue-resident macrophages (TRMs).
Following up on recent reports highlighting the self-renewal and repopulating capacity of TRMs as well as their considerable tissue plasticity upon transplantation [9,10], we have recently demonstrated that a single intrapulmonary transplantation of healthy or genetically corrected bone marrow-derived macrophages can substantially improve the disease phenotype of pulmonary alveolar proteinosis in two relevant murine in vivo disease models [11,12]. Although our initial studies used HSPCs derived from murine bone marrow or human cord blood, also iPSC-derived macrophages represent a highly interesting source for cell therapy strategies in diseases related to macrophage dysfunction [13,14]. Here, the embryonic origin of the cells may even serve as an advantage [15], and the transplantation of iPSC-derived macrophages potentially may be superior to the transplantation of adult CD34+ HSC-derived macrophages. Consequently, a combination of hematopoietic cell-based gene therapy applications with the unique potential of iPSCs has been explored for quite some time. However, transcriptional repression or silencing of retroviral transgene expression has long remained as a major hurdle [16]. In particular, the stable genetic modification of PSCs that harbor strong intrinsic silencing mechanisms, presumably as a mode of antiviral defense to protect germ line cells from mutagenesis, has proven a challenge [17,18].
In this context, we here have used lentiviral vectors equipped with an ubiquitous chromatin opening element (CBX3-UCOE) or zinc finger nuclease(ZFN)-mediated targeting of an expression cassette into the human adeno-associated virus integration site 1 (AAVS1) safe harbor in order to provide a tool which allows for stable transgene expression in PSC-derived mature hematopoietic cells. Considering the substantial engraftment potential and tissue plasticity of macrophages including TRMs, derivation of such genetically optimized macrophages may pave the way for the use of iPSC-derived macrophages in a variety of different disease entities.
Material and Methods
iPSC Cultivation
Human iPSCs were maintained on murine CF-1 feeder cells in ESC medium (knock-out Dulbecco's modified Eagle medium (KO-DMEM) with 20% knock-out serum replacement, 0,1 nmol/l β-mercaptoethanol, 1 mmol/l L-glutamine, 1% non-essential amino acids and 1% penicillin/streptomycin) supplemented with 10-20 ng/ml basic fibroblast growth factor (bFGF; Peprotech, Hamburg, Germany). Passaging was performed every 7-10 days depending on the colony size by using 2 mg/ml collagenase IV (Invitrogen, Carlsbad, CA, USA). Cells were incubated at 37 °C and 5% CO2.
Lentiviral Vector Preparation
SIN-lentiviral vectors were generated according to standard cloning techniques, and lentiviral containing supernatants were produced by transient transfection of the producer cell line HEK293T by calcium phosphate precipitation. Viral supernatants were harvested 24 and 48 h post transfection, filtered, and concentrated by ultracentrifugation for 3 h at 25,000 rpm and 4 °C. Titration was performed on murine SC-1 fibroblasts using serial dilution transduction and flow-cytometric analysis of green fluorescent marker protein(GFP)- or RedStarnuc-positive cells.
Transduction of hiPSCs
For transduction, hiPSCs were dissociated to single cells using TrypLE and subsequently pre-seeded on Matrigel/Geltrex®-coated plates in ESC-medium containing 10 µmol/l Rock Inhibitor Y-27632 (Tocris Bioscience, Bristol, UK) and 10-20 ng/ml bFGF. Cells were transduced with the lentiviral supernatant in the presence of 10 µg/ml protamine sulfate.
Differentiation of hiPSCs
Hematopoietic differentiations of hiPSCs were performed using a previously published protocol [4]. In short, iPSC colonies were disrupted using collagenase IV and used for embryoid body (EB) formation for 5 days in ESC medium containing 10 µmol/l Rock Inhibitor (Y-27632) on an orbital shaker (100 rpm). EBs were selected, manually transferred onto tissue culture plates, and cultivated in differentiation medium I (X-Vivo medium; Stem Cell Technologies, Cologne, Germany) supplemented with 25 ng/ml human IL-3 and 50 ng/ml human M-CSF for macrophage generation. Macrophages generated by myeloid cell-forming complexes (MCFCs) could be harvested from day 10 to day 15 onwards, filtered through a 100 µm mesh and cultivated for 7-10 days for further maturation in differentiation medium II (RPMI1640 medium supplemented with 10% calf serum, 1 mmol/l L-glutamine, 1% penicillin/streptomycin and 50 ng/ml human M-CSF).
Cytospins
Cytospins were prepared with the use of a Shandon cytocentrifuge (Thermo Scientific, Langenselbold, Germany) by spinning of 2 × 105 cells for 10 min at 700 rpm. Slides were stained in May-Grünwald staining solution for 5 min and in 5% Giemsa staining solution for 10 min. Slides were washed intensively, dried until coverage and subsequent analysis using bright-field microscopy.
Flow Cytometry
In brief, cells were harvested using TrypLE and resuspended in phosphate buffered saline (PBS) for analysis. Expression of hematopoietic and macrophage markers was analyzed using a FACSCalibur (Becton & Dickinson, Franklin Lakes, NJ, USA). All antibodies were used as recommended by the manufacturer. Antibodies used: hCD45-PE, hCD11b-APC, hCD14-PE.
Results
Vector Design for Improved Gene Expression in iPSC-Derived Macrophages
In order to prevent silencing of lentiviral transgene expression, a number of epigenetic regulatory elements have been introduced into vector backbones. These regulatory elements can be grouped into two classes: i) elements exhibiting a border or boundary function intended to block the spread of heterochromatin into the integrated expression cassette, e.g. insulators or scaffold/matrix attachment regions (S/MARs), and ii) elements that are able to actively remodel the local chromatin environment and activate transcription such as locus control regions (LCRs) or ubiquitous chromatin opening elements (UCOEs) [19]. UCOEs were initially introduced by Antoniou et al. [20] in 2003 as genomic regions that consist of two divergently transcribed promoters spanning a methylation-free CpG-island and exerting an independent dominant chromatin remodeling function. As the best characterized model, the A2UCOE that is derived from the heterogeneous nuclear ribonucleoprotein A2/B1 (HNRPA2B1)/Chromobox3 (CBX3) housekeeping locus has been shown to mediate stable and reproducible transgene expression even from integrations within centromeric heterochromatin and to efficiently stabilize and enhance gene expression driven by the human cytomegalovirus (hCMV) promoter in mammalian cell lines, when placed upstream of an heterologous promoter [21].
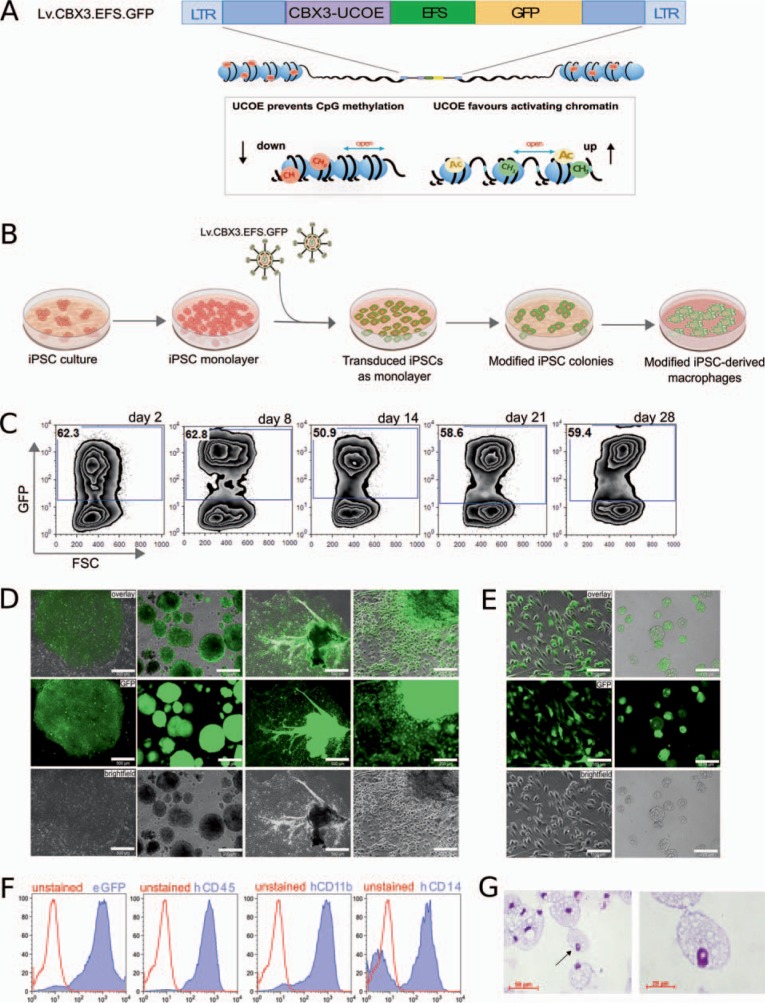
Considering the enormous potential of iPSC-derived hematopoietic cells for autologous gene and cell therapy strategies, but also the pronounced epigenetic repression of transgene expression in these cells, it appeared logical to exploit the anti-silencing activity of UCOEs in the context of iPSC-based gene therapy. The A2UCOE efficiently stabilized transgene expression driven by a truncated elongation factor 1a (EFS) or the viral spleen focus forming virus (SFFV) promoter in murine and human PSCs as well as their differentiated progeny [22,23]. Although very efficient, the reduction of viral titers, as well as the risk of deregulating endogenous gene expression due to its dual promoter activity or the presence of splice signals within the sequence, represented critical problems for the employment of A2UCOE-based vectors. More recently, however, a minimal 0.7 kb CBX3-UCOE (CBX3-UCOE) has been introduced to the field, which retains most if not all of the A2UCOE functionality [24]. To further evaluate the properties of the CBX3-UCOE in hiPSCs, we have designed a third-generation self-inactivating lentiviral vector expressing the GFP from the human EFS promoter. In order to avoid silencing of the GFP transgene, we placed a CBX3-UCOE upstream of the EFS promoter (Lv.CBX3.EFS.GFP) (fig. 1A). Using the Lv.CBX3.EFS.GFP vector, we could efficiently transduce hiPSCs (CD34iPSC11) [22] with approximately 60% transduction efficiency (multiplicity of infection (MOI): 20) and establish a polyclonal transgenic iPSC line (fig. 1B) demonstrating stable transgene expression over 28 days of culture (fig. 1C). Similar results could be obtained for a second iPSC line (CD34iPSC16) [13]. Here lentiviral transduction with a MOI of 50 resulted in >90% stable GFP expression and maintained iPSC morphology (fig. 1D). To proof the anti-silencing effect of the CBX3-UCOE also in the context of differentiation processes and the associated epigenetic repression, we have subjected the transduced iPSC line (CD34iPSC16) to an established hematopoietic differentiation protocol [4] yielding macrophages in the presence of IL-3 and M-CSF. Within the first days of differentiation, transduced iPSCs formed EBs with classical round shape morphology which were positive for GFP as analyzed by fluorescence microscopy (fig. 1D). Following hematopoietic differentiation, EBs successfully attached to culture plates and formed MCFCs within 10-15 days of culture. Of note, MCFCs, as well as adherent endothelial-like stroma cells, remained positive for GFP, indicating stable transgene expression during differentiation (fig. 1D). In addition, MCFCs continuously gave rise to GFP-positive macrophages with classical morphology and a CD45+ CD11b+ CD14+ surface marker profile (fig. 1E-G). Thus, the CBX3-UCOE element efficiently stabilized transgene expression in hematopoietic differentiation cultures of hiPSCs.
Fig. 1.
CBX3-UCOE-mediated transgene expression in hiPSC-derived macrophages. A Scheme of the lentiviral vector expressing GFP from the EFS (Lv.CBX3.EFS.GFP) (LTR: long terminal repeat). The lentiviral vector was further equipped with a CBX3-UCOE to prevent epigenetic transgene silencing such as methylation and to favor an open chromatin state. B Experimental scheme of lentiviral-mediated transduction of hiPSCs. Lentiviral particles (Lv.CBX3.EFS.GFP) were used to transduce hiPSC monolayer. After transduction, modified iPSCs were subjected to hematopoietic differentiation to study transgene stability. C Flow-cytometric analysis of GFP in transduced CD34iPSC11 over 28 days of culture. D GFP expression in transduced CD34iPSC16 and during hematopoietic differentiation in EBs and MCFCs (scale bars; column 1 and 3: 500 µm, column 2 and 4: 200 µm). E GFP expression in CD34iPSC16-derived macrophages in cell culture (left column) and unstained cytospin (right column) (scale bars: 100 µm). F Flow-cytometric analysis and G stained cytospins of terminally differentiated macrophages.
ZFN-Mediated Targeting of iPSCs and iPSC-Derived Macrophages
While retroviral-based gene therapy has achieved great successes in hematopoietic stem cell-based gene therapy approaches, even more powerful technologies are available in the context of iPSCs. Here, innovative genome editing strategies that enable either the specific targeting of a safe harbor locus, the correction of a disease-causing mutation by targeted introduction of a ‘superexon’ into the respective endogenous locus, or the directed ‘repair’ of an affected gene by using single-stranded DNA oligonucleotides may be applied. Particularly, the latter two strategies carry the benefit of maintaining the physiological regulation of the targeted gene.
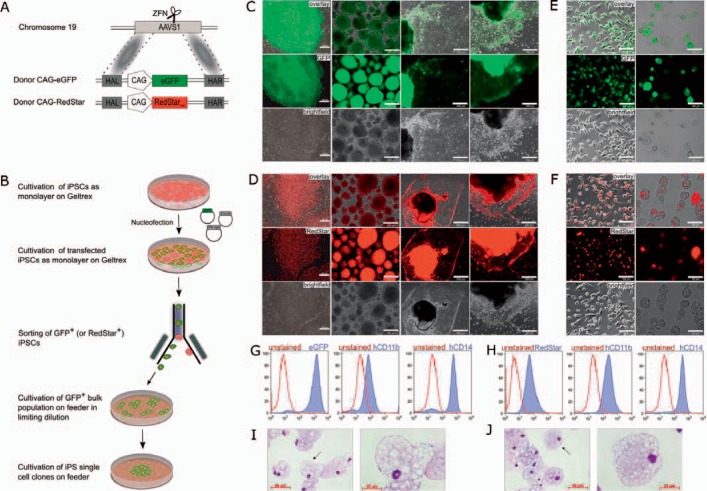
To this end, programmable and highly specific nuclease-based tools have been developed, which include ZFNs, transcription activator-like effector nucleases (TALEN), and the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) protein 9 system [25]. Employing this technology, precise correction or knock-out of desired genes has been demonstrated in multipotent as well as pluripotent stem cells [26,27,28]. However, so far transfer of this technology to primary HSCs has been hampered by a number of problems, foremost the low targeting efficiencies achieved in these cells calling for the application of this technology in more suitable and ideally clonally expandable cell sources, such as iPSCs. Along this line, we have addressed the stability of transgene expression in hiPSC-derived macrophages, which have been genetically modified to constitutively express GFP or RedStarnuc from an exogenous promoter in the AAVS1 safe harbor [29]. As described previously in more detail [29,30], the hiPSC line hCBiPS2 [31] was transfected with AAVS1-specific ZFNs and donor plasmids harboring either GFP or RedStarnuc under the control of the CMV early enhancer element coupled to the chicken β-actin (CAG) promoter (fig. 2A). Following expression of the reporter gene, single cell sorting was performed yielding individual clones positive either for GFP (hCBiPS2_AAVS1eGFPC18) or RedStarnuc (hCBiPS2_AAVS1RedStarC8) (fig. 2B). Extended cultivation of these clones revealed stable transgene expression over time, whereby GFP expression could be detected in the entire cytoplasm, while RedStarnuc expression was restricted to the nucleus (fig. 2C,D). This specific, although expected, pattern is of high interest, as it allows delivering transgene expression in distinct subcellular localization. In order to investigate transgene expression from the AAVS1 locus during hematopoietic differentiation, we subjected both clones to hematopoietic differentiation and evaluated transgene stability in hiPSC-derived macrophages. Both genetically modified iPSC lines formed EBs expressing either GFP or RedStarnuc, thus highlighting stable transgene expression during the formation of the three germ layers. Moreover, stable transgene expression was also observed in MCFCs derived from the EBs upon hematopoietic specification (fig. 2C,D). Of note, expression of GFP or RedStarnuc was also detected in endothelial-like stroma cells. From day 10-15 onwards MCFC-derived macrophages displayed typical morphology on bright-field and cytospin images, which were positive for GFP or RedStarnuc and stained positive for CD11b and CD14 (fig. 2E-J). Of note, expression of RedStarnuc again was only detectable in the nucleus, whereas GFP expression could be detected in the cytoplasm of macrophages (fig. 2E,F). Thus, our data highlight stable and specific transgene expression from the AAVS1 locus during hematopoietic differentiation of hiPSCs towards macrophages (fig. 2E-J).
Fig. 2.
Stable transgene expression from the AAVS1 locus in hiPSC-derived macrophages. A Scheme of ZFN-mediated targeting of the fluorescence reporter genes GFP or RedStarnuc into the AAVS1 safe harbor locus (HAL, homology arm left; HAR, homology arm right). B Schematic outline of nuclease-mediated genome editing of iPSCs. After nucleofection of iPSCs with suitable ZFNs and donor constructs, GFP- or RedStarnuc-positive cells were sorted to obtain transgenic iPS single cell clones. C GFP expression during hematopoietic differentiation (scale bars; column 1 and 2: 200 µm, column 3 and 4: 500 µm) and E in iPSC-derived macrophages (scale bars: 100 µm). D RedStarnuc expression during hematopoietic differentiation (scale bars; column 1 and 2: 200 µm, column 3 and 4: 500 µm) and F in iPSC-derived macrophages (scale bars; column 1: 100 µm and column 2: 50 µm). G Flow-cytometric analysis of terminally differentiated macrophages derived from iPSCs which either express GFP or H RedStarnuc from the AAVS1 locus. I Cytospins of differentiated macrophages derived from iPSCs which either express GFP or J RedStarnuc.
Discussion
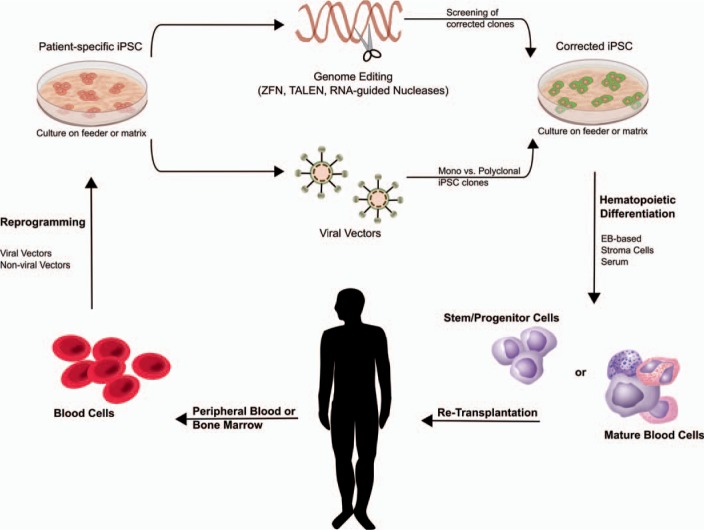
hiPSC-based gene and cell therapy holds great potential for innovative treatment strategies. The concepts most commonly envisioned in this context postulate the generation of customized, patient-derived iPSCs, which, after genetic modification and appropriate differentiation, will be (re)transplanted to the patient as an individualized therapy approach (fig. 3). The efficient generation and subsequent differentiation of iPSCs already has been established for a number of different tissues and organ systems, including cells of the hematopoietic system. However, iPSC-based gene and cell therapy approaches employing patient-specific iPSCs will rely on the efficient genetic modification of the target cells. To this point, we have used genetically modified iPSC lines to address transgene stability during hematopoietic differentiation towards macrophages. While different reports shed light into the mode of action of the full length or minimal UCOE [22,24], the authors have used either lentiviral vectors equipped with a CBX3-UCOE or ZFN-mediated targeting of an expression cassette into the human AAVS1 safe harbor to provide a tool for stable transgene expression in iPSCs and differentiated macrophages.
Fig. 3.
iPSC-based gene and cell therapy. Patient-specific iPSCs can be generated from easily accessible blood samples by overexpression of specific transcription factors using viral or non-viral vectors. Genetic modification can be performed in the pluripotent status, thereby allowing sub-cloning and further safety tests. Thereafter, either gene addition using integrating viral vectors or precise genome editing strategies employing designer nucleases (ZFN, TALEN, CRISPR/Cas9) can be applied. Corrected iPSCs (subclones) can subsequently be differentiated into the disease-affected cell type or stem/progenitor cells, which could then be investigated in vivo.
Using different iPSC lines which express reporter genes either from randomly integrated lentiviral vector expression systems or the safe harbor AAVS1 allowed for the expression of reporter genes in pluripotent cells as well as in thereof derived macrophages, highlighting their suitability to efficiently express therapeutic transgenes in iPSCs.
With regard to retroviral vectors, a 1.5 kb UCOE (A2UCOE) successfully has been exploited in the context of iPSC-based gene therapy approaches, stabilizing transgene expression from various promoter constructs [22,23]. Whereas our initial proof-of-concept studies focused on the expression of fluorescent reporter proteins, the A2UCOE has also been applied efficiently in the context of iPSC-based gene therapy for hereditary pulmonary alveolar proteinosis. Utilizing patient-specific iPSCs, an A2UCOE-equipped lentiviral vector was demonstrated to restore robust and stable expression of the GM-CSF receptor α-chain in hiPSCs and thereof derived macrophages [13]. In order to further optimize the A2UCOE, we recently generated a minimal 0.7 kb CBX3-UCOE [24]. This element allows for the production of high-titer virus preparations, reduces the formation of aberrant transcripts initiated by the dual promoter activity of the A2UCOE, and when investigated in the context of SFFV promoter-driven transgene expression sustains most if not all anti-silencing activity of the A2UCOE element. Moreover, the CBX3-UCOE does not interfere with promoter tissue specificity when combined with a myeloid-specific promoter [24]. To extend this work, we have now used the CBX3-UCOE in combination with the EFS promoter and could prove stable transgene expression in both pluripotent cells and thereof derived macrophages.
As an alternative to lentiviral vector-based gene addition, precise genome editing tools may be employed to either correct a disease-specific mutation or to introduce expression cassettes into a defined genetic locus (fig. 3). Thus, we have employed iPSC lines which have been previously genetically modified by ZFN technology to express GFP and RedStarnuc from a CAG promoter in the AAVS1 locus [29]. In these cells, transgene expression was sustained in the pluripotent state and all along the hematopoietic differentiation pathway towards mature macrophages. Similar results have been described by other groups applying ZFNs in hiPSCs to precisely correct certain gene mutations [32]. In our approach, we have used iPSCs, which were previously modified by ZFNs to express reporter genes from the safe harbor AAVS1. Although this approach relies on the use of exogenous promoter elements similar to the lentiviral approach, precise genome engineering at the AAVS1 locus would minimize the risk of unwanted insertional side effects. Moreover, suitable ZFN pairs can initially be screened for their efficacy and off-targeting specificity, and the designed ZFNs can then in principle be used to incorporate a variety of different expression cassettes at the same AAVS1 site. While we have used a constitutive CAG promoter, this technology can also be applied to tissue-specific promoter elements and may be combined with the use of the CBX3-UCOE to reduce the risk of promoter silencing [22,23]. In addition to ZFNs and TALENs, the use of the RNA-guided DNA endonuclease Streptococcus pyogenes Cas 9 in the CRISPR/Cas9 system has evolved as a promising alternative [25].
Taken together, we demonstrate that lentiviral vectors can be used to introduce genes of interest into iPSCs and that these vectors allow for sustained transgene expression during hematopoietic differentiation when equipped with genetic elements such as the CBX3-UCOE. As an alternative, we show that designer nucleases such as ZFNs can be employed to target expression cassettes to safe harbors, also providing stable transgene expression during hematopoietic differentiation towards macrophages. Showing the feasibility to derive modified iPSC-derived macrophages by both methods, the generation of genetically corrected patient-specific iPSC-derived hematopoietic cells may pave the way for future cell-based treatment strategies.
Disclosure Statement
The authors declare that they have no conflict of interest.
Acknowledgments
The authors thank Theresa Buchegger and Doreen Lüttge (all Hanover Medical School, Hanover, Germany) for technical support. This work was supported by grants from the Else Kröner-Fresenius Foundation (TM: 2013_A24; NL: 2015_A92; MA: 2016_A146), the Deutsche Forschungsgemeinschaft (DFG: Cluster of Excellence REBIRTH; TM Mo886/6-1; NL LA 3680/2-1), the German Center for Lung Research (DZL, BREATH, 82DZL00201 and 82DZL002A1), and Hanover Medical School (NL: ‘Young Academy’ Fellowship award; SM and MA: HiLF fellowship; JK: stipend from the Cluster of Excellence REBIRTH).
References
- 1.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Ackermann M, Liebhaber S, Klusmann JH, Lachmann N. Lost in translation: pluripotent stem cell-derived hematopoiesis. EMBO Mol Med. 2015;7:1388–1402. doi: 10.15252/emmm.201505301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Choi KD, Vodyanik MA, Slukvin II. Generation of mature human myelomonocytic cells through expansion and differentiation of pluripotent stem cell-derived lin-CD34+CD43+CD45+ progenitors. J Clin Invest. 2009;119:2818–2829. doi: 10.1172/JCI38591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lachmann N, Ackermann M, Frenzel E, Liebhaber S, Brennig S, Happle C, Hoffmann D, Klimenkova O, Luttge D, Buchegger T, Kuhnel MP, Schambach A, Janciauskiene S, Figueiredo C, Hansen G, Skokowa J, Moritz T. Large-scale hematopoietic differentiation of human induced pluripotent stem cells provides granulocytes or macrophages for cell replacement therapies. Stem Cell Reports. 2015;4:282–296. doi: 10.1016/j.stemcr.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niwa A, Heike T, Umeda K, Oshima K, Kato I, Sakai H, Suemori H, Nakahata T, Saito MK. A novel serum-free monolayer culture for orderly hematopoietic differentiation of human pluripotent cells via mesodermal progenitors. PLoS One. 2011;6:e22261. doi: 10.1371/journal.pone.0022261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mucci A, Kunkiel J, Suzuki T, Brennig S, Glage S, Kuhnel MP, Ackermann M, Happle C, Kuhn A, Schambach A, Trapnell BC, Hansen G, Moritz T, Lachmann N. Murine iPSC-derived macrophages as a tool for disease modeling of hereditary pulmonary alveolar proteinosis due to Csf2rb deficiency. Stem Cell Reports. 2016;7:292–305. doi: 10.1016/j.stemcr.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng Q, Shabrani N, Thon JN, Huo H, Thiel A, Machlus KR, Kim K, Brooks J, Li F, Luo C, Kimbrel EA, Wang J, Kim K, Italiano J, Cho J, Lu SJ, Lanza R. Scalable generation of universal platelets from human induced pluripotent stem cells. Stem Cell Reports. 2014;3:817–831. doi: 10.1016/j.stemcr.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirose S, Takayama N, Nakamura S, Nagasawa K, Ochi K, Hirata S, Yamazaki S, Yamaguchi T, Otsu M, Sano S, Takahashi N, Sawaguchi A, Ito M, Kato T, Nakauchi H, Eto K. Immortalization of erythroblasts by c-MYC and BCL-XL enables large-scale erythrocyte production from human pluripotent stem cells. Stem Cell Reports. 2013;1:499–508. doi: 10.1016/j.stemcr.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, Greter M, Mortha A, Boyer SW, Forsberg EC, Tanaka M, van Rooijen N, Garcia-Sastre A, Stanley ER, Ginhoux F, Frenette PS, Merad M. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Happle C, Lachmann N, Skuljec J, Wetzke M, Ackermann M, Brennig S, Mucci A, Jirmo AC, Groos S, Mirenska A, Hennig C, Rodt T, Bankstahl JP, Schwerk N, Moritz T, Hansen G. Pulmonary transplantation of macrophage progenitors as effective and long-lasting therapy for hereditary pulmonary alveolar proteinosis. Sci Transl Med. 2014;6:250ra113. doi: 10.1126/scitranslmed.3009750. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Arumugam P, Sakagami T, Lachmann N, Chalk C, Sallese A, Abe S, Trapnell C, Carey B, Moritz T, Malik P, Lutzko C, Wood RE, Trapnell BC. Pulmonary macrophage transplantation therapy. Nature. 2014;514:450–454. doi: 10.1038/nature13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachmann N, Happle C, Ackermann M, Luttge D, Wetzke M, Merkert S, Hetzel M, Kensah G, Jara-Avaca M, Mucci A, Skuljec J, Dittrich AM, Pfaff N, Brennig S, Schambach A, Steinemann D, Gohring G, Cantz T, Martin U, Schwerk N, Hansen G, Moritz T. Gene correction of human induced pluripotent stem cells repairs the cellular phenotype in pulmonary alveolar proteinosis. Am J Respir Crit Care Med. 2014;189:167–182. doi: 10.1164/rccm.201306-1012OC. [DOI] [PubMed] [Google Scholar]
- 14.Suzuki T, Mayhew C, Sallese A, Chalk C, Carey BC, Malik P, Wood RE, Trapnell BC. Use of induced pluripotent stem cells to recapitulate pulmonary alveolar proteinosis pathogenesis. Am J Respir Crit Care Med. 2014;189:183–193. doi: 10.1164/rccm.201306-1039OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchrieser J, James W, Moore MD. Human induced pluripotent stem cell-derived macrophages share ontogeny with MYB-independent tissue-resident macrophages. Stem Cell Reports. 2017;8:334–345. doi: 10.1016/j.stemcr.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Challita PM, Kohn DB. Lack of expression from a retroviral vector after transduction of murine hematopoietic stem cells is associated with methylation in vivo. Proc Natl Acad Sci U S A. 1994;91:2567–2571. doi: 10.1073/pnas.91.7.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teich NM, Weiss RA, Martin GR, Lowy DR. Virus infection of murine teratocarcinoma stem cell lines. Cell. 1977;12:973–982. doi: 10.1016/0092-8674(77)90162-3. [DOI] [PubMed] [Google Scholar]
- 18.Hotta A, Ellis J. Retroviral vector silencing during iPS cell induction: an epigenetic beacon that signals distinct pluripotent states. J Cell Biochem. 2008;105:940–948. doi: 10.1002/jcb.21912. [DOI] [PubMed] [Google Scholar]
- 19.Antoniou MN, Skipper KA, Anakok O. Optimizing retroviral gene expression for effective therapies. Hum Gene Ther. 2013;24:363–374. doi: 10.1089/hum.2013.062. [DOI] [PubMed] [Google Scholar]
- 20.Antoniou M, Harland L, Mustoe T, Williams S, Holdstock J, Yague E, Mulcahy T, Griffiths M, Edwards S, Ioannou PA, Mountain A, Crombie R. Transgenes encompassing dual-promoter CpG islands from the human TBP and HNRPA2B1 loci are resistant to heterochromatin-mediated silencing. Genomics. 2003;82:269–279. doi: 10.1016/s0888-7543(03)00107-1. [DOI] [PubMed] [Google Scholar]
- 21.Williams S, Mustoe T, Mulcahy T, Griffiths M, Simpson D, Antoniou M, Irvine A, Mountain A, Crombie R. CpG-island fragments from the HNRPA2B1/CBX3 genomic locus reduce silencing and enhance transgene expression from the hCMV promoter/enhancer in mammalian cells. BMC Biotechnol. 2005;5:17. doi: 10.1186/1472-6750-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackermann M, Lachmann N, Hartung S, Eggenschwiler R, Pfaff N, Happle C, Mucci A, Gohring G, Niemann H, Hansen G, Schambach A, Cantz T, Zweigerdt R, Moritz T. Promoter and lineage independent anti-silencing activity of the A2 ubiquitous chromatin opening element for optimized human pluripotent stem cell-based gene therapy. Biomaterials. 2014;35:1531–1542. doi: 10.1016/j.biomaterials.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 23.Pfaff N, Lachmann N, Ackermann M, Kohlscheen S, Brendel C, Maetzig T, Niemann H, Antoniou MN, Grez M, Schambach A, Cantz T, Moritz T. A ubiquitous chromatin opening element prevents transgene silencing in pluripotent stem cells and their differentiated progeny. Stem Cells. 2013;31:488–499. doi: 10.1002/stem.1316. [DOI] [PubMed] [Google Scholar]
- 24.Muller-Kuller U, Ackermann M, Kolodziej S, Brendel C, Fritsch J, Lachmann N, Kunkel H, Lausen J, Schambach A, Moritz T, Grez M. A minimal ubiquitous chromatin opening element (UCOE) effectively prevents silencing of juxtaposed heterologous promoters by epigenetic remodeling in multipotent and pluripotent stem cells. Nucleic Acids Res. 2015;43:1577–1592. doi: 10.1093/nar/gkv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X, Wang Y, Yan W, Smith C, Ye Z, Wang J, Gao Y, Mendelsohn L, Cheng L. Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point mutation. Stem Cells. 2015;33:1470–1479. doi: 10.1002/stem.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heckl D, Kowalczyk MS, Yudovich D, Belizaire R, Puram RV, McConkey ME, Thielke A, Aster JC, Regev A, Ebert BL . Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nat Biotechnol. 2014;32:941–946. doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cox DB, Platt RJ, Zhang F. Therapeutic genome editing: prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merkert S, Wunderlich S, Bednarski C, Beier J, Haase A, Dreyer AK, Schwanke K, Meyer J, Gohring G, Cathomen T, Martin U. Efficient designer nuclease-based homologous recombination enables direct PCR screening for footprintless targeted human pluripotent stem cells. Stem Cell Reports. 2014;2:107–118. doi: 10.1016/j.stemcr.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merkert S, Martin U. Targeted genome engineering using designer nucleases: state of the art and practical guidance for application in human pluripotent stem cells. Stem Cell Res. 2016;16:377–386. doi: 10.1016/j.scr.2016.02.027. [DOI] [PubMed] [Google Scholar]
- 31.Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C, Zweigerdt R, Gruh I, Meyer J, Wagner S, Maier LS, Han DW, Glage S, Miller K, Fischer P, Scholer HR, Martin U. Generation of induced pluripotent stem cells from human cord blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 32.Connelly JP, Kwon EM, Gao Y, Trivedi NS, Elkahloun AG, Horwitz MS, Cheng L, Liu PP. Targeted correction of RUNX1 mutation in FPD patient-specific induced pluripotent stem cells rescues megakaryopoietic defects. Blood. 2014;124:1926–1930. doi: 10.1182/blood-2014-01-550525. [DOI] [PMC free article] [PubMed] [Google Scholar]