Abstract
The increasing use of azole antifungals for the treatment of mucosal and systemic Candida glabrata infections has resulted in the selection and/or emergence of resistant strains. The main mechanisms of azole resistance include alterations in the C. glabrata ERG11 gene (CgERG11), which encodes the azole target enzyme, and upregulation of the CgCDR1 and CgCDR2 genes, which encode efflux pumps. In the present study, we evaluated these molecular mechanisms in 29 unmatched clinical isolates of C. glabrata, of which 20 isolates were resistant and 9 were susceptible dose dependent (S-DD) to fluconazole. These isolates were recovered from separate patients during a 3-year hospital survey for antifungal resistance. Four of the 20 fluconazole-resistant isolates were analyzed together with matched susceptible isolates previously taken from the same patients. Twenty other azole-susceptible clinical C. glabrata isolates were included as controls. MIC data for all the fluconazole-resistant isolates revealed extensive cross-resistance to the other azoles tested, i.e., itraconazole, ketoconazole, and voriconazole. Quantitative real-time PCR analyses showed that CgCDR1 and CgCDR2, alone or in combination, were upregulated at high levels in all but two fluconazole-resistant isolates and, to a lesser extent, in the fluconazole-S-DD isolates. In addition, slight increases in the relative level of expression of CgSNQ2 (which encodes an ATP-binding cassette [ABC] transporter and which has not yet been shown to be associated with azole resistance) were seen in some of the 29 isolates studied. Interestingly, the two fluconazole-resistant isolates expressing normal levels of CgCDR1 and CgCDR2 exhibited increased levels of expression of CgSNQ2. Conversely, sequencing of CgERG11 and analysis of its expression showed no mutation or upregulation in any C. glabrata isolate, suggesting that CgERG11 is not involved in azole resistance. When the isolates were grown in the presence of fluconazole, the profiles of expression of all genes, including CgERG11, were not changed or were only minimally changed in the resistant isolates, whereas marked increases in the levels of gene expression, particularly for CgCDR1 and CgCDR2, were observed in either the fluconazole-susceptible or the fluconazole-S-DD isolates. Finally, known ABC transporter inhibitors, such as FK506, were able to reverse the azole resistance of all the isolates. Together, these results provide evidence that the upregulation of the CgCDR1-, CgCDR2-, and CgSNQ2-encoded efflux pumps might explain the azole resistance in our set of isolates.
Candida glabrata has recently emerged as a significant pathogen in various hospital settings, where it is responsible for an increasing number of systemic infections and candiduria (2, 16). In a recent study, C. glabrata was the second most common non-C. albicans species as a cause of fungemia in the United States and was found to account for 21% of all Candida bloodstream isolates (26). Second only to C. albicans, C. glabrata is also the Candida species most commonly recovered from the oral cavities of human immunodeficiency virus-infected patients (13, 16, 40).
The rise in the number of C. glabrata systemic infections deserves a great deal of concern due to the high mortality rate associated with C. glabrata fungemia and to the propensity of this microorganism to rapidly develop resistance to azole antifungal agents (10, 19). Several studies have revealed that a significant percentage of C. glabrata clinical isolates are resistant to fluconazole (approximately 9%) and itraconazole (37 to 40%) (3, 16, 25). More recently, in a surveillance study conducted by Pfaller et al. (27) to examine the antifungal susceptibilities of Candida species isolated from patients with bloodstream infections stratified by patient age, a trend of decreasing susceptibilities to fluconazole and itraconazole with increasing patient age was observed. In fact, none of the C. glabrata isolates from individuals ≤1 year old were resistant to fluconazole, whereas a higher proportion (5 to 9%) of resistant isolates was found in adult patients. Similarly, among 347 bloodstream, invasive, and colonizing strains of C. glabrata isolated from patients at three urban teaching hospitals in New York City, the overall rates of resistance to fluconazole and itraconazole were 10.7 and 15.2%, respectively (33).
The mechanisms of resistance to azole antifungal agents have been well elucidated in C. albicans and can be mainly categorized as (i) changes in the cell wall or plasma membrane, which lead to impaired azole uptake; (ii) alterations in the affinity of the drug target Erg11p (lanosterol 14α-demethylase) to azoles or in the cellular content of Erg11p due to target site mutation or overexpression of the ERG11 gene; and (iii) the efflux of drugs mediated by membrane transport proteins belonging to the ATP-binding cassette (ABC) transporter family (CDR1 and CDR2) or to the major facilitator superfamily (MDR1 and FLU1). In the last case, the CDR1 and CDR2 genes and the MDR1 gene were shown to be overexpressed in many resistant isolates, and deletion of these genes resulted in hypersensitivity to azoles (34). In addition, compensatory pathways that involve alterations of specific steps in ergosterol biosynthesis have been documented as mechanisms of resistance to the azole and polyene antifungal classes (39).
More recently, increased levels of expression of the ABC transporter genes C. glabrata CDR1 (CgCDR1) and CgCDR2 have been also shown in azole-resistant isolates of C. glabrata (5, 15, 35, 36). Similar to C. albicans, genetic evidence supporting the role of multidrug transporters in the azole resistance of C. glabrata was provided (36). Moreover, Marichal et al. (14) previously showed increased levels of expression of ERG11 in an azole-resistant C. glabrata strain which arose from a chromosomal duplication. In contrast, it has yet to be well explored whether point mutations in the ERG11 gene are also implicated in the resistance of C. glabrata to azoles.
The purpose of the present study was to determine if the molecular mechanisms described above, alone or in combination, were sufficient to explain the phenotype of azole resistance in unmatched clinical C. glabrata isolates obtained from various clinical specimens during a 3-year hospital survey of antifungal resistance or if other (not well-established) mechanisms might correlate with azole resistance. In addition, pairs of susceptible and resistant C. glabrata isolates that had been obtained from the same patient and that had the same genotype were also examined.
MATERIALS AND METHODS
Yeast isolates and growth conditions.
The isolates of C. glabrata included in the present study were from a collection of clinical isolates recovered during an epidemiological survey of antifungal resistance conducted at our institution, a large university hospital in Rome, over a 3-year period (January 2000 through December 2003). They were identified by standard methods (43) and tested for their susceptibilities to amphotericin B, flucytosine, fluconazole, ketoconazole, itraconazole, and voriconazole by the Sensititre Yeast One commercial method (Biomedical Service, Milan, Italy), as recommended by the manufacturer. Twenty-nine isolates for which fluconazole MICs exceeded the established susceptibility breakpoint (MIC ≤ 8 μg/ml) (17, 32) were retested for their antifungal susceptibilities by the NCCLS reference method for confirmation and were then selected for molecular studies (see below). These isolates were recovered from various body sites of 29 separate patients (Table 1). Among the isolates studied, there were 4 fluconazole-susceptible isolates that matched 4 fluconazole-resistant isolates obtained from the same patient, as well as 20 randomly selected clinical isolates of C. glabrata susceptible to azoles (Table 2). In addition, two well-characterized C. glabrata isolates, a susceptible isolate (isolate DSY562 [36]) and a resistant isolate (isolate DSY565 [36]), kindly furnished by Dominique Sanglard, were used as controls. All 53 isolates were kept at −80°C as 20% glycerol stocks and were subcultured, as required, on YEPD (1% yeast extract, 2% peptone, 2% glucose) agar plates at 30°C. For liquid culture, the isolates were grown in YEPD broth at 30°C under constant agitation (240 rpm).
TABLE 1.
Sites of isolation, antifungal susceptibilities, and resistance mechanisms for the 33 C. glabrata isolates studied
| Isolate designation | Site of isolation | MIC (μg/ml)a
|
FLC susceptibility categoryb | Gene(s) overexpressed (fold increasec) | |||
|---|---|---|---|---|---|---|---|
| FLC | ITC | KTC | VOR | ||||
| Matched isolates | |||||||
| BPY40 | Blood | 4 | 0.25 | 0.25 | 0.125 | S | |
| BPY41 | Blood | 256 | >16 | 4 | 8 | R | CgCDR1(483.1), CgCDR2 (70.3), CgSNQ2 (11.7) |
| BPY112 | Blood | 2 | 0.125 | 0.125 | 0.125 | S | |
| BPY126 | Blood | 128 | 16 | 4 | 2 | R | CgCDR1 (56.2), CgCDR2 (5.5), CgSNQ2 (18.7) |
| BPY241 | Blood | 4 | 0.25 | 0.125 | 0.125 | S | |
| BPY285 | Blood | 128 | 2 | 2 | 4 | R | CgCDR1 (51.0), CgCDR2 (13.3) |
| BPY449 | Blood | 4 | 0.25 | 0.25 | 0.125 | S | |
| BPY479 | Blood | 256 | 2 | 4 | 2 | R | CgCDR1(132.8), CgCDR2 (45.5) |
| Unmatched isolates | |||||||
| BPY42 | BALd fluid | 128 | >16 | 4 | 4 | R | CgCDR1 (31.9) |
| BPY43 | Urine/catheter | 128 | >16 | 4 | 4 | R | CgCDR1 (100.4) |
| BPY44 | Vagina | 128 | >16 | 4 | 4 | R | CgCDR1 (81.3) |
| BPY45 | Blood | 256 | >16 | 4 | 4 | R | CgCDR1 (130.4) |
| BPY46 | Blood | 128 | >16 | 4 | 4 | R | CgCDR1 (141.0), CgCDR2 (28.4), CgSNQ2 (5.1) |
| BPY47 | Urine | 128 | >16 | 4 | 4 | R | CgCDR1 (90.0), CgCDR2 (6.8) |
| BPY48 | Vagina | 128 | >16 | 2 | 2 | R | CgSNQ2 (25.3) |
| BPY49 | Urine/catheter | 256 | >16 | 4 | 4 | R | CgCDR1 (263.9), CgCDR2 (3.8), CgSNQ2 (5.5) |
| BPY50 | Oral cavity | 256 | 16 | 8 | 4 | R | CgCDR1 (227.1), CgCDR2 (34.7) |
| BPY54 | Sputum | 128 | 16 | 2 | 2 | R | CgCDR1 (18.0), CgSNQ2 (3.7) |
| BPY55 | Vagina | 64 | 4 | 1 | 2 | R | CgSNQ2 (14.7) |
| BPY57 | Urine | 64 | 2 | 1 | 16 | R | CgCDR1 (11.9) |
| BPY59 | Oral cavity | 64 | 2 | 1 | 1 | R | CgCDR2 (31.7), CgSNQ2 (12.7) |
| BPY60 | Decubitus ulcer | 256 | 16 | 4 | 8 | R | CgCDR1 (235.5), CgCDR2 (34.0), CgSNQ2 (20.5) |
| BPY135 | Urine/catheter | 64 | 2 | 1 | 1 | R | CgCDR2 (25.9), CgSNQ2 (9.9) |
| BPY233 | Drainage fluid | 256 | 16 | 4 | 4 | R | CgCDR1 (79.1), CgCDR2 (13.3) |
| BPY51 | Vagina | 16 | 1 | 0.5 | 0.25 | S-DD | CgCDR1 (5.6), CgSNQ2 (2.9) |
| BPY52 | Urine/catheter | 32 | 0.5 | 0.5 | 0.25 | S-DD | CgCDR1 (8.0), CgSNQ2 (4.7) |
| BPY53 | Blood | 16 | 0.5 | 0.5 | 0.25 | S-DD | CgCDR1 (3.8), CgCDR2 (4.8) |
| BPY56 | Sputum | 16 | 0.125 | 0.06 | 0.125 | S-DD | CgCDR1 (4.4) |
| BPY58 | Urine/catheter | 16 | 1 | 0.5 | 1 | S-DD | CgCDR2 (4.7), CgSNQ2 (6.1) |
| BPY150 | Urine/catheter | 32 | 2 | 0.5 | 0.5 | S-DD | CgCDR1 (3.7) |
| BPY174 | Urine/catheter | 16 | 2 | 0.25 | 0.25 | S-DD | CgCDR1 (5.4) |
| BPY221 | Abscess fluid | 16 | 1 | 0.125 | 0.25 | S-DD | CgCDR1 (6.5) |
| BPY270 | Fecese | 16 | 1 | 0.25 | 0.25 | S-DD | CgCDR1 (6.0) |
FLC, fluconazole; ITC, itraconazole; KTC, ketoconazole; VOR, voriconazole. The MICs of these antifungal agents were determined by the broth microdilution method, in accordance with NCCLS document M27-A2 (17).
S, susceptible (MIC, ≤8 μg/ml); S-DD, susceptible dose dependent (MIC, 16 to 32 μg/ml); R, resistant (MIC, ≥64 μg/ml).
Quantification was performed by real-time RT-PCR (see the text for details). The values are averages of four independent experiments and represent increases in the levels of gene expression relative to that of DSY562 (set equal to 1.00).
BAL, bronchoalveolar lavage.
Stool specimen from a patient with an absence of normal bacterial flora.
TABLE 2.
Azole susceptibilities of 20 fluconazole-susceptible C. glabrata isolates according to the levels of expression of resistance-related genes
| Isolate designation | Site of isolation | MIC (μg/ml)a
|
Gene expression (fold increaseb)
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| FLC | ITC | KTC | VOR | CgCDR1 | CgCDR2 | CgSNQ2 | ERG11 | ||
| BPY104 | Vagina | 4 | 0.25 | 0.25 | 0.125 | 0.51 | 1.45 | 1.12 | 0.90 |
| BPY140 | Sputum | 2 | 0.06 | 0.06 | 0.06 | 0.89 | 1.01 | 0.67 | 0.50 |
| BPY164 | Blood | 2 | 0.125 | 0.125 | 0.125 | 0.66 | 1.56 | 1.23 | 0.87 |
| BPY197 | Oral cavity | 4 | 0.5 | 0.125 | 0.125 | 0.54 | 1.86 | 1.12 | 0.98 |
| BPY222 | Urine/catheter | 4 | 0.25 | 0.125 | 0.125 | 1.20 | 0.87 | 1.34 | 0.91 |
| BPY241 | Urine/catheter | 2 | 0.25 | 0.125 | 0.125 | 1.01 | 1.93 | 1.03 | 0.82 |
| BPY324 | Sputum | 4 | 0.25 | 0.25 | 0.125 | 1.11 | 0.47 | 0.92 | 0.67 |
| BPY373 | BALc fluid | 2 | 0.25 | 0.125 | 0.25 | 0.28 | 1.86 | 1.23 | 0.87 |
| BPY376 | Sputum | 1 | 0.125 | 0.125 | 0.06 | 0.33 | 1.80 | 1.05 | 0.71 |
| BPY377 | Oral cavity | 4 | 0.125 | 0.125 | 0.25 | 0.58 | 1.74 | 1.06 | 0.56 |
| BPY388 | BAL fluid | 2 | 0.25 | 0.125 | 0.125 | 1.12 | 1.89 | 0.89 | 0.78 |
| BPY394 | Abscess fluid | 2 | 0.125 | 0.25 | 0.06 | 1.05 | 1.70 | 0.67 | 0.89 |
| BPY455 | Urine/catheter | 4 | 0.5 | 0.25 | 0.25 | 1.52 | 1.43 | 0.78 | 1.12 |
| BPY458 | Blood | 8 | 0.5 | 0.25 | 0.5 | 1.25 | 1.34 | 0.98 | 1.32 |
| BPY480 | Urine/catheter | 4 | 0.25 | 0.25 | 0.125 | 1.32 | 1.65 | 0.89 | 1.02 |
| BPY525 | Vaginal | 2 | 0.125 | 0.25 | 0.125 | 1.56 | 1.43 | 0.71 | 1.09 |
| BPY531 | Blood | 2 | 0.125 | 0.125 | 0.06 | 1.23 | 0.79 | 1.12 | 0.67 |
| BPY544 | Urine/catheter | 4 | 0.25 | 0.125 | 0.06 | 0.67 | 0.34 | 0.98 | 0.70 |
| BPY621 | Blood | 4 | 0.25 | 0.25 | 0.25 | 0.33 | 0.67 | 1.23 | 0.98 |
| BPY640 | Urine/catheter | 4 | 0.5 | 0.125 | 0.25 | 0.54 | 0.87 | 1.12 | 1.54 |
Antifungal susceptibility testing by the NCCLS reference method.
Reference antifungal susceptibility testing of the study isolates was performed by the broth microdilution method described in NCCLS document M27-A2 (17). Powders of the antifungal agents amphotericin B, flucytosine, ketoconazole, fluconazole, voriconazole, and itraconazole were obtained from their respective manufacturers. Briefly, MICs were determined with RPMI 1640 with 2% glucose by use of an inoculum size of 1.5 (±1.0) × 103 cells/ml and incubation at 35°C for 48 h. Quality control was ensured by testing the NCCLS-recommended strains C. krusei ATCC 6258 and C. parapsilosis ATCC 22019 (4). All tests were carried out in duplicate. The interpretive criteria for susceptibility to fluconazole, itraconazole, and flucytosine were those published by Rex et al. (32) and the NCCLS (17) and were as follows: (i) for fluconazole, susceptible, ≤8 μg/ml; susceptible-dose-dependent (S-DD), 16 to 32 μg/ml; and resistant, ≥64 μg/ml; (ii) for itraconazole, susceptible, ≤0.125 μg/ml; S-DD, 0.25 to 0.5 μg/ml; and resistant, ≥1 μg/ml; and (iii) for flucytosine, susceptible, ≤4 μg/ml; intermediate, 8 to 16 μg/ml; and resistant ≥32 μg/ml. Clinical breakpoints for ketoconazole have not yet been proposed, but they are likely to be close to those for itraconazole (44). Likewise, interpretive criteria have not been defined for amphotericin B; however, as suggested by Nguyen et al. (18), isolates for which the amphotericin B MIC was ≤1 μg/ml were defined as susceptible in this study. Although interpretive breakpoints have also not been established for the investigational triazole voriconazole, we have used a susceptibility breakpoint of ≤1 μg/ml, as suggested by Pfaller et al. (26). Assays for susceptibility to fluconazole were also performed in the presence of oligomycin (Sigma, St. Louis, Mo.), verapamil (Sigma), and FK506 (LCLaboratories, Woburn, Mass.). Yeast cells were inoculated in microtiter plate wells containing doubling dilutions of the antifungal agent with fixed concentrations of FK506 (10 μM), oligomycin (50 μM), and verapamil (100 μM) and were incubated to the MIC endpoint, as specified above.
Quantitative real-time RT-PCR.
For quantitative real-time reverse transcription (RT)-PCR analysis, total RNA was extracted from C. glabrata cultures grown to the mid-exponential phase (optical density at 600 nm [OD600], approximately 0.6) with an RNAeasy Protect mini kit (Qiagen, Hilden, Germany), according to the instructions of the manufacturer, by mechanical disruption of the cells with glass beads and an RNase-free DNase treatment step. RNA integrity was assessed by determination of the OD260/OD280 absorption ratio, and the integrity was considered maintained if the ratio was >1.95. Quantitative expression of the CgCDR1, CgCDR2, CgSNQ2, and CgERG11 genes was performed by real-time RT-PCR with an i-Cycler iQ system (Bio-Rad Laboratories, Hercules, Calif.). For the target genes and URA3 reference gene, a primer pair and a Taqman probe, which hybridizes to the region between the primer-specific sequences, were designed with Beacon Designer 2 (version 2.06) software (Premier Biosoft International, Palo Alto, Calif.) and were synthesized by MWG Biotech (Florence, Italy) (Table 3). RT-PCR was performed with a 50-μl volume containing the following reagents: 25 μl of the Platinum Quantitative RT-PCR ThermoScript reaction mixture (Invitrogen Inc., Milan, Italy), 1.5 U of the ThermoScript Plus/Platinum Taq mixture (Invitrogen), each primer pair and the Taqman probe at a concentration of 5 μM, 5 μl of total RNA sample (5, 10, 25, 50, and 100 ng), and distilled water up to the final volume. Samples were subjected to an initial step at 52°C for 45 min for RT; 94°C for 5 min to inactivate the ThermoScript Plus reverse transcriptase and to activate the Platinum Taq polymerase; and 50 cycles, each of which consisted of 15 s at 94°C and 1 min at 59°C. Fluorescence data were collected during the 59°C annealing and extension step and were analyzed with the i-Cycler iQ software. Each reaction was run in quadruplicate. The mean cycle threshold was determined for each transcript and was plotted versus the concentration of input RNA to calculate the slope. The amplification efficiency was then determined for all genes (21, 23). For relative quantification of the target genes, each set of primer pairs and the Taqman probe were used in combination with the primers and probe specific for the URA3 gene in separate reactions. Unless otherwise specified, for each isolate, the fold changes were determined from the mean normalized expression (target gene R0/URA3 R0, where R0 is the starting fluorescence which is proportional to the starting template quantity) relative to the mean normalized expression of DSY562 (susceptible control isolate), as already described (21). A twofold increase in the level of expression of each gene was considered significant.
TABLE 3.
Primers and fluorescent probes used in real-time RT-PCR
| Gene (GenBank accession no.) | Primer or probe | Sequencea | Gene location (5′-3′) |
|---|---|---|---|
| CgCDR1 (AF109723) | CDR1a | TAGCACATCAACTACACGAACGT | 4500-4522 |
| CDR1b | AGAGTGAACATTAAGGATGCCATG | 4647-4670 | |
| CDR1pr | 6FAM-TGCTGCTGCTTCTGCCACCTGGTT-TAMRA | 4621-4644 | |
| CgCDR2 (AF251023) | CDR2a | GTGCTTTATGAAGGCTACCAGATT | 164-187 |
| CDR2b | TCTTAGGACAGAAGTAACCCATCT | 251-274 | |
| CDR2pr | 6FAM-TACCTTTGCGTGCTGGGCGTCACC-TAMRA | 217-240 | |
| CgSNQ2 (AF251022) | SNQ2a | ACCATGTGTTCTGAATCAATCAAT | 360-383 |
| SNQ2b | TCGACATCATTACAATACCAGAAA | 462-485 | |
| SNQ2pr | 6FAM-AACTAATCGCCGCAGGTTGTGACA-TAMRA | 394-317 | |
| ERG11 (L40389) | ERGa | ATTGGTGTCTTGATGGGTGGTC | 928-949 |
| ERGb | TCTTCTTGGACATCTGGTCTTTCA | 1019-1042 | |
| ERGpr | 6FAM-ACTTCCGCTGCTACCTCCGCTTGG-TAMRA | 955-978 | |
| URA3 (L13661) | URAa | GAAAACCAATCTTTGTGCTTCTCT | 168-191 |
| URAb | CATGAGTCTTAAGCAAGCAAATGT | 268-291 | |
| URApr | Texas Red-ACGTCACCACCACCAGCGAATTGT-BHQ2 | 194-217 |
Abbreviations: 6FAM, 6-carboxyfluorescein; TAMRA, 6-carboxy-N,N,N′,N′-tetramethylrhodamine; Texas Red, product from Molecular Probes; BHQ2, Black Hole Quencher 2.
Cloning and sequencing of ERG11 gene.
For amplification of the ERG11 genes from the C. glabrata isolates, primers ERG11fwd (5′-ATGTCCACTGAAAACACTTCTTTGG-3′) and ERG11rev (5′-GTACTTTTGTTCTGGATGTCTCTTTTC-3′) were designed on the basis of positions 561 to 2159 of the CgERG11 sequence (GenBank accession no. LF40389). PCRs were carried out with a 50-μl volume containing 1X Pfx amplification buffer (Invitrogen); dATP, dGTP, dCTP, and dTTP (Roche Diagnostics, Milan, Italy) at a concentration of 200 μM each; primers ERG11a and ERG11b (synthesized by MWG Biotech) at a concentration of 5 μM each; 1 U of Platinum Pfx DNA polymerase (Invitrogen); and 100 ng of genomic DNA. Amplification conditions were as follows: 95°C for 2 min, 95°C for 15 s, 57°C for 1 min, and 68°C for 2 min repeated for 40 cycles. The PCR products were purified and cloned into the Zero Blunt vector system (Invitrogen). The insert DNAs of the recombinant plasmids were sequenced automatically with an ABI Prism 377 sequencer analyzer (Applied Biosystems, Foster City, Calif.). For each clone, the sequences of both strands were analyzed, and multiple-amino-acid alignments were derived by using WinDNASYS software for Windows (version 2.1; Hitachi Software Genetic Systems, San Francisco, Calif.).
Exposure of yeast isolates to fluconazole.
For fluconazole exposure, we followed the protocol described by Niimi et al. (20). Suspensions of C. glabrata cells (OD600, 0.1) freshly prepared in YEPD medium were grown at 30°C to reach an OD600 of 0.3; then, fluconazole was added at a final concentration of 100 μg/ml and the cultures were incubated at 30°C for a further 4 h. Preliminary experiments showed that 4 h was the optimal time for induction (data not shown). In addition, resistant isolates were also exposed to higher concentrations of fluconazole (200 and 400 μg/ml). After the cells were exposed to fluconazole, they were centrifuged at 10,000 × g for 10 min and washed twice with sterile water, and the pellet was used for quantitative RT-PCR analysis (see above). In these experiments, the increases in the levels of gene expression relative to the levels for the same isolates grown in the absence of fluconazole were measured.
Typing of yeast isolates.
Typing analysis of the C. glabrata clinical isolates was performed by a method based on the use of repetitive probes Cg6 and Cg12, as described previously (11). Genomic DNA from each C. glabrata isolate was prepared by a previously described protocol (11). Five micrograms of genomic DNA was cut with EcoRI, and the resulting fragments were electrophoresed in a 0.8% agarose gel and then transferred onto a positively charged nylon membrane (Roche Diagnostics) with a vacuum blotter apparatus (Bio-Rad). The probes were prepared from λ phage DNA by liquid lysate with the QIAgen λ kit (Qiagen) and were labeled with digoxigenin (DIG) by use of a DIG DNA labeling kit (Roche Diagnostics), according to the instructions of the manufacturer. The membranes were prehybridized at 42°C with a buffer containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 2% blocking reagent (Roche Diagnostics), 0.1% N-lauroylsarcosine, and 0.02% sodium dodecyl sulfate. DNA probes were added to the hybridization solution, and the mixture was incubated overnight. The washing and revealing steps were performed with a DIG luminescent detection kit (Roche Diagnostics), as recommended by the supplier. Hybridization patterns were acquired by scanning with Fluor-S MAX 2 (Bio-Rad).
Statistical analysis.
The two-tailed Student t test was used to compare categorical variables. A P value of less than 0.05 was considered statistically significant.
RESULTS
Antifungal susceptibilities of C. glabrata isolates.
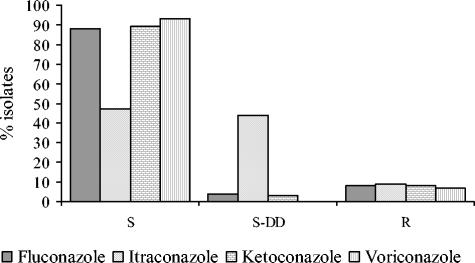
The C. glabrata isolates studied here were obtained during a hospital survey conducted at our institution to monitor the antifungal resistance patterns of Candida sp. infections over a 3-year period. A total of 267 C. glabrata isolates from consecutive infections were screened for antifungal resistance by a routine susceptibility testing method in use in our laboratory and adapted from the NCCLS reference method (8, 24). Figure 1 shows the rates of azole resistance for these C. glabrata isolates. Among the resistant isolates, 7.5, 9.7, 7.5, and 6.7% were resistant to fluconazole, itraconazole, ketoconazole, and voriconazole, respectively, whereas all isolates were found to be susceptible to amphotericin B and flucytosine. The fluconazole MICs for the 29 C. glabrata isolates that originated from different patients hospitalized in intensive care units and oncology wards were superior to the susceptibility breakpoint established for this agent (MIC, ≥8 μg/ml), and the isolates were selected for further phenotypic characterization. These isolates were tested for their susceptibilities to a panel of four azole antifungal agents, including fluconazole, itraconazole, ketoconazole, and voriconazole, by the NCCLS method. The resulting susceptibility profiles confirmed those obtained by the screening method. Overall, according to the interpretive criteria for azole susceptibility mentioned above, the 29 C. glabrata isolates were categorized as follows: 20 were resistant (MICs, ≥64 μg/ml) and 9 were S-DD (MICs, 16 to 32 μg/ml) to fluconazole, 26 were resistant (MICs, ≥1 μg/ml) and 3 were S-DD (MICs, 0.25 to 0.5 μg/ml) or susceptible (MICs, ≤0.125 μg/ml) to itraconazole, and 20 were resistant (MICs, ≥1 μg/ml) and 9 were S-DD (MICs, 0.25 to 0.5 μg/ml) or susceptible (MICs, ≤0.125 μg/ml) to ketoconazole (Table 1). Ultimately, by using an MIC of >1 μg/ml to define resistance to the investigational triazole voriconazole, the 29 isolates included 18 that were resistant and 11 that were susceptible. A detailed analysis of cross-resistance among the four azoles tested showed that 18 isolates were resistant to all azoles, 2 isolates were resistant to three azoles (fluconazole, itraconazole, and ketoconazole), and 6 isolates were resistant to one azole (itraconazole).
FIG. 1.
Distributions of percentages of C. glabrata isolates susceptible to four azole antifungal agents among 267 clinical isolates. S, the isolates are susceptible; S-DD, the isolates are susceptible in a dose-dependent manner; R, the isolates are resistant. The azole susceptibility categories were determined as described in the text.
Genotyping of the 29 isolates showed a very high degree of heterogeneity among them, as 29 unique patterns could be distinguished with probe Cg6 or Cg12, based on differences in the position of one or more bands. Probe Cg6 hybridized to 9 to 16 different bands (from 14 to 4 kb) per isolate, while probe Cg12 hybridized to 7 to 11 different bands (from 14 to 3.5 kb) per isolate. Thus, all isolates were considered epidemiologically unrelated strains (data not shown).
Mechanisms of azole resistance in unmatched isolates.
In previous work (12, 22), sets of matched susceptible and resistant isolates, each one derived from a single strain, were used to investigate the molecular mechanisms underlying the development of azole resistance in Candida species. These isolates, however, are rarely available in clinical settings, so it could be of interest to screen for known resistance mechanisms among randomly collected C. glabrata isolates, as shown by a recent investigation with C. albicans (44).
To determine if the present understanding of C. glabrata resistance mechanisms could account for the resistance phenotypes of unmatched clinical isolates, we analyzed the expression of the resistance-associated and non-resistance-associated C. glabrata genes CgCDR1, CgCDR2, CgSNQ2, and CgERG11 in our collection of 29 C. glabrata isolates. Expression levels were determined by molecular methods that were also applied to matched sets of isolates. Real-time PCR analysis was performed, and for each target gene, the relative amount of the transcript compared to that of the URA3 transcript was determined with total RNA from yeast cells in the logarithmic phase of growth by using a gene-specific fluorescent probe. In addition, a search for point mutations within CgERG11 was performed to assess whether this mechanism was responsible for azole resistance in the isolates studied.
(i) CgCDR1, CgCDR2, and CgSNQ2 expression levels.
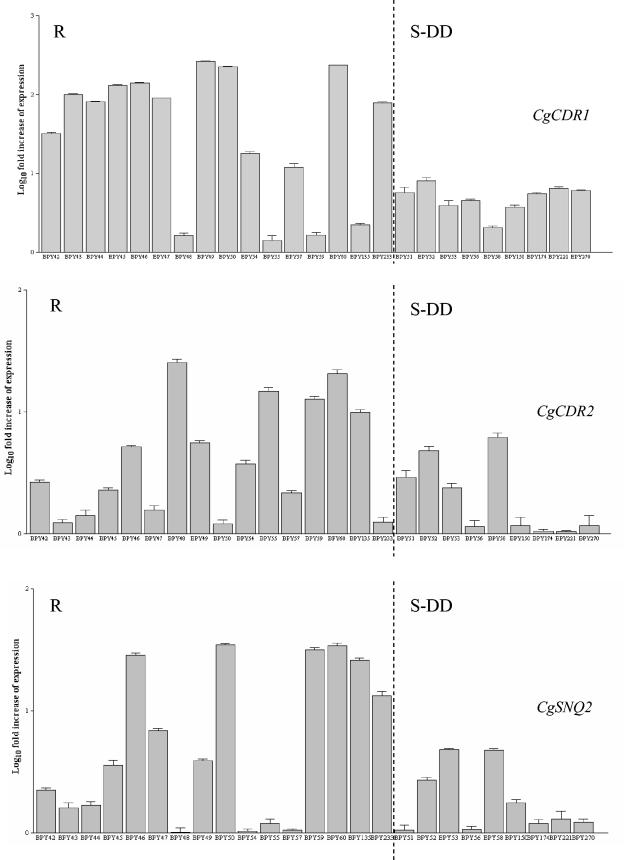
Figure 2 shows the relative quantification of CgCDR1, CgCDR2, and CgSNQ2 (i.e., in reference to that of susceptible control isolate DSY562), expressed as the target gene R0 normalized to the URA3 R0 for 25 of the 29 isolates included in the study (see Materials and Methods for details). The remaining four isolates were analyzed together with their matched isolates, resulting in pairs of resistant and susceptible isolates (see below for results). In Fig. 2, the results for 25 isolates were separated into two groups on the basis of their levels of susceptibility to fluconazole. The results on the left side of Fig. 2 are for resistant isolates (16 isolates), and those on the right side are for S-DD isolates (9 isolates).
FIG. 2.
Expression of CgCDR1, CgCDR2, and CgSNQ2 in the 25 unmatched C. glabrata isolates, as determined by real-time RT-PCR analysis (see the text for details). For each target gene, the relative amount of transcription (i.e., in reference to that by susceptible control isolate DSY562) was compared to that of URA3. The results are reported as the log10 fold increases in gene expression levels. Error bars show standard deviations. The x axis indicates the 25 isolates, arranged as resistant (R) and susceptible dose dependent (S-DD) to fluconazole.
Twelve of the 16 fluconazole-resistant isolates expressed CgCDR1 at higher levels than susceptible control isolate DSY562 did, displaying amounts of CgCDR1 transcript that increased 11.9- to 263.9-fold compared to those detected in DSY562. Of those isolates that upregulated CgCDR1, all were resistant to the other three azoles.
Eight of the 16 fluconazole-resistant isolates expressed CgCDR2 at higher levels than the susceptible control isolate DSY562 did, but a lower relative increase (3.8- to 34.7-fold) than that of CgCDR1 was observed. Six of the eight isolates that upregulated CgCDR2 (isolates BPY46, BPY47, BPY49, BPY50, BPY60, and BPY233) were resistant to the other three azoles, while the remaining two isolates (isolates BPY59 and BPY135) were resistant to two other azoles (ketoconazole and itraconazole).
Eight of the 16 fluconazole-resistant isolates upregulated CgSNQ2, but to a lesser extent than they upregulated CgCDR1 and CgCDR2. The levels of expression in these isolates were 3.7- to 25.3-fold higher than that in susceptible control isolate DSY562. The eight isolates that upregulated CgSNQ2 were resistant to the other three azoles (isolates BPY46, BPY48, BPY49, BPY54, BPY55, and BPY60) or two azoles (isolates BPY59 and BPY135).
Overall, nine isolates concomitantly upregulated more efflux pump genes: CgCDR1, CgCDR2, and CgSNQ2 for three isolates (isolates BPY46, BPY49, and BPY60), CgCDR1 and CgCDR2 for three isolates (isolates BPY47, BPY50, and BPY233), CgCDR2 and CgSNQ2 for two isolates (isolates BPY59 and BPY135), and CgCDR1 and CgSNQ2 for one isolate (isolate BPY54).
Interestingly, of the four isolates that did not upregulate CgCDR1, two isolates (isolates BPY48 and BPY55) showed a relative increase in the level of expression only of CgSNQ2 and were resistant to the four azoles.
The 9 fluconazole S-DD isolates upregulated CgCDR1 or CgCDR2 or upregulated CgSNQ2 (Table 1), but the levels of upregulation were significantly less than those by 16 fluconazole-resistant isolates (3.7- to 8.0-fold for CgCDR1, 4.7- and 4.8-fold for CgCDR2, and 2.9- to 6.1-fold for CgSNQ2).
(ii) CgERG11 upregulation.
In many clinical C. albicans isolates, azole resistance has often been associated with overexpression of the ERG11 gene (12). Likewise, Marichal et al. (14) measured an eightfold increase in ERG11 mRNA levels in an azole-resistant clinical C. glabrata isolate due to amplification of the CgERG11 gene, which in turn resulted from chromosomal duplication. To determine if changes in the levels of expression of CgERG11 could be associated with the resistance phenotype observed in the study isolates, CgERG11 mRNA levels were analyzed (as described above). Beyond expectation, we found that all of the isolates expressed CgERG11 at levels nearly equal to that by susceptible control isolate DSY562.
(iii) CgERG11 sequence analysis.
Point mutations in the ERG11 gene with an effect on the affinity of the enzyme for the azoles have been correlated with azole resistance in C. albicans (22, 37, 44). To elucidate if this mechanism could also be implicated in azole resistance in C. glabrata, the CgERG11 genes of all 29 C. glabrata isolates were cloned and sequenced. All the sequences contained at least one silent nucleotide variation compared to the published sequence of CgERG11 (data not shown). Notably, no variation that led to an amino acid substitution was found in any of the isolates.
Development of azole resistance in sequential isolates.
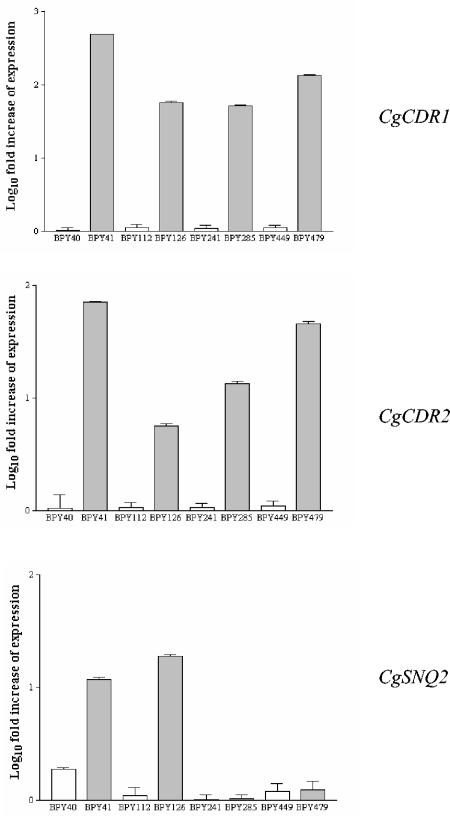
As mentioned above, 25 of the 29 C. glabrata isolates in the present study were unmatched isolates recovered from 25 different patients. The medical records of some of the patients, who were mainly hospitalized in critical care units and oncology wards, were retrospectively reviewed and showed that they were persistently infected with C. glabrata. In fact, we noticed the repeated isolation of a unique strain from the same and/or different body sites, as shown by genotyping analyses (data not shown). In contrast, we documented the in vitro development of azole resistance in 4 of the 29 isolates obtained from separate patients with C. glabrata bloodstream infections. For each patient, the first blood culture isolate (fluconazole susceptible) was recovered before the patient was treated with fluconazole. A second isolate (fluconazole resistant) of the same genotype was obtained from the patient after fluconazole therapy was established (Table 1). For the four patients, the times that elapsed between the time of sampling of the azole-susceptible isolate and the time of sampling of the azole-resistant isolate were 15, 35, 40, and 60 days, respectively. At these times, the patients had received cumulative doses of 6, 10, 12, and 12.2 g of fluconazole, respectively. The four pairs of sequential isolates (isolates BPY40 and BPY41, BPY112 and BPY126, BPY241 and BPY285, and BPY449 and BPY479) were studied as described above. As shown in Table 1, isolates BPY41, BPY126, BPY285, and BPY479 all developed marked resistance to the four azoles tested but expressed different resistance profiles. Interestingly, two isolates (isolates BPY285 and BPY479) showed lower rates of resistance to itraconazole (MICs, 2 μg/ml). When the levels of the CgCDR1, CgCDR2, and CgSNQ2 transcripts were measured, all the resistant isolates had a greater abundance of CgCDR1 and CgCDR2 transcripts than the susceptible isolates, while the amount of CgSNQ2 transcript was increased in only two of the four pairs (isolates BPY40 and BPY41 and isolates BPY111 and BPY126) (Fig. 3).
FIG. 3.
Expression of CgCDR1, CgCDR2, and CgSNQ2 in matched susceptible and resistant isolates from four patients, as determined by real-time RT-PCR analysis (see the text for details). For each target gene, the relative amount of transcription (i.e., in reference to that by susceptible control isolate DSY562) was compared to that of the URA3. The results are reported as the log10 fold increases in gene expression levels. Error bars show standard deviations.
Expression of CgCDR1, CgCDR2, CgSNQ2, and ERG11 in azole-susceptible isolates.
We also evaluated the resistance mechanisms cited above for a set of randomly selected clinical isolates of C. glabrata susceptible to azoles and recovered from patients admitted to our hospital during the study period in order to compare their gene expression with that observed for the 29 isolates (fluconazole resistant and S-DD) studied. Table 2 shows the profiles of expression of the CgCDR1, CgCDR2, and CgSNQ2 efflux pump-encoding genes and ERG11, which encodes the azole target, for the 20 susceptible isolates analyzed. As expected, we found that all isolates expressed basal levels of these genes compared to levels of expression by susceptible control isolate DSY562.
Expression of CgCDR1, CgCDR2, CgSNQ2, and ERG11 in isolates grown in the presence of fluconazole.
In order to evaluate whether the expression profiles observed for our 20 resistant isolates could change after exposure to fluconazole, we performed experiments in which the expression of the CgCDR1, CgCDR2, CgSNQ2, and ERG11 genes by the isolates grown in the presence of fluconazole at a final concentration of 100 μg/ml was analyzed, as described elsewhere (20). The same experiments were also performed with the 24 susceptible isolates, including the 4 isolates of the pairs of sequential isolates, and with the 9 S-DD isolates. The expression levels were compared to those obtained with the same isolates grown in the absence of fluconazole (Tables 4 and 5). Interestingly, the profiles of expression of all target genes, including CgERG11, by the resistant isolates remained unchanged or were only slightly changed, whereas marked increases in the levels of gene expression, particularly of CgCDR1 and CgCDR2, were observed for either the fluconazole-susceptible or -S-DD isolates. Similar results were also obtained for the resistant isolates when the isolates were grown in the presence of higher concentrations of fluconazole (200 and 400 μg/ml) (data not shown).
TABLE 4.
Expression profiles of resistance-related genes for the 24 fluconazole-susceptible C. glabrata isolates grown in the presence of fluconazolea
| Isolate designation | Gene expression (fold increaseb)
|
|||
|---|---|---|---|---|
| CgCDR1 | CgCDR2 | CgSNQ2 | ERG11 | |
| BPY40c | 379.1 | 65.2 | 20.4 | 25.3 |
| BPY104 | 126.4 | 89.3 | 45.3 | 37.1 |
| BPY112c | 79.5 | 14.4 | 21.8 | 26.1 |
| BPY140 | 201.3 | 67.4 | 51.4 | 29.3 |
| BPY164 | 103.2 | 45.2 | 23.1 | 12.4 |
| BPY197 | 75.3 | 32.1 | 12.7 | 25.1 |
| BPY222 | 56.3 | 23.5 | 25.1 | 12.4 |
| BPY241c | 107.3 | 25.6 | 15.3 | 9.5 |
| BPY245 | 120.3 | 34.2 | 21.1 | 18.6 |
| BPY324 | 97.3 | 18.4 | 9.8 | 10.5 |
| BPY373 | 245.2 | 45.2 | 12.4 | 15.1 |
| BPY376 | 252.1 | 76.1 | 15.6 | 20.5 |
| BPY377 | 456.2 | 256.0 | 40.5 | 40.3 |
| BPY388 | 323.6 | 56.4 | 32.1 | 21.4 |
| BPY394 | 129.9 | 39.2 | 23.1 | 17.5 |
| BPY449c | 97.6 | 45.6 | 19.3 | 8.4 |
| BPY455 | 74.4 | 12.5 | 16.2 | 11.5 |
| BPY458 | 126.2 | 32.5 | 28.4 | 14.2 |
| BPY480 | 231.4 | 31.5 | 29.3 | 19.4 |
| BPY525 | 241.3 | 119.3 | 34.5 | 20.3 |
| BPY531 | 187.3 | 56.4 | 12.5 | 21.9 |
| BPY544 | 165.2 | 43.5 | 23.9 | 20.7 |
| BPY621 | 125.4 | 40.2 | 30.4 | 32.8 |
| BPY640 | 98.4 | 32.7 | 21.8 | 19.8 |
Fluconazole was used at a final concentration of 100 μg/ml, as described previously (20).
Quantification was performed as specified in footnote c of Table 1. The values are averages of four independent experiments and represent increases in the levels of gene expression relative to the levels for the same isolates grown in the absence of fluconazole (set equal to 1.00).
These isolates are members of the four pairs of sequential isolates obtained from patients before and after treatment with fluconazole.
TABLE 5.
Expression profiles of resistance-related genes for the 29 fluconazole-resistant and-S-DD C. glabrata isolates grown in the presence of fluconazolea
| Isolate designation | FLC susceptibility categoryb | Genes expression (fold increasec)
|
|||
|---|---|---|---|---|---|
| CgCDR1 | CgCDR2 | CgSNQ2 | ERG11 | ||
| BPY41 | R | 1.68 | 1.07 | 1.86 | 5.86 |
| BPY42 | R | 1.87 | 0.98 | 1.28 | 6.34 |
| BPY43 | R | 1.98 | 1.56 | 0.87 | 4.23 |
| BPY44 | R | 1.01 | 1.23 | 1.34 | 0.98 |
| BPY45 | R | 1.23 | 1.98 | 2.01 | 1.23 |
| BPY46 | R | 0.93 | 0.93 | 1.07 | 0.87 |
| BPY47 | R | 1.43 | 0.98 | 1.34 | 0.98 |
| BPY48 | R | 1.23 | 0.87 | 1.21 | 4.56 |
| BPY49 | R | 1.45 | 1.02 | 0.79 | 1.45 |
| BPY50 | R | 1.76 | 0.89 | 1.07 | 1.78 |
| BPY54 | R | 2.45 | 0.79 | 0.89 | 1.98 |
| BPY55 | R | 2.78 | 1.76 | 3.20 | 7.89 |
| BPY57 | R | 3.03 | 1.23 | 1.67 | 9.56 |
| BPY59 | R | 1.76 | 0.89 | 1.32 | 8.12 |
| BPY60 | R | 0.98 | 0.93 | 1.13 | 1.78 |
| BPY126 | R | 1.23 | 1.12 | 0.89 | 1.42 |
| BPY135 | R | 1.89 | 1.65 | 1.76 | 1.83 |
| BPY233 | R | 2.01 | 0.92 | 1.59 | 1.31 |
| BPY285 | R | 1.76 | 1.16 | 1.33 | 0.99 |
| BPY479 | R | 1.09 | 1.11 | 1.34 | 0.93 |
| BPY51 | S-DD | 82.3 | 8.1 | 18.0 | 25.4 |
| BPY52 | S-DD | 30.9 | 10.5 | 27.3 | 14.4 |
| BPY53 | S-DD | 61.8 | 3.3 | 5.6 | 128.0 |
| BPY56 | S-DD | 43.5 | 21.5 | 4.5 | 30.1 |
| BPY58 | S-DD | 78.9 | 8.3 | 3.8 | 67.8 |
| BPY150 | S-DD | 54.5 | 12.4 | 34.3 | 29.0 |
| BPY174 | S-DD | 29.4 | 10.5 | 9.6 | 34.7 |
| BPY221 | S-DD | 93.2 | 3.5 | 4.6 | 43.6 |
| BPY270 | S-DD | 65.3 | 5.6 | 4.3 | 23.5 |
Fluconazole was used at a final concentration of 100 μg/ml, as described previously (20).
S-DD, susceptible dose dependent (MIC, 16 to 32 μg/ml); R, resistant (MIC, ≥64 μg/ml).
Quantification was performed as specified in footnote c Table 1. The values are averages of four independent experiments and represent increases in the level of gene expression relative to that for the same isolates grown in the absence of fluconazole (set equal to 1.00).
Effects of pump inhibitors on the fluconazole resistance of C. glabrata isolates.
It is known that several compounds are able to reverse the resistance phenotype of yeast strains by inhibiting the drug efflux activity of ABC transporters. Therefore, these compounds are considered potential chemosensitizers for fluconazole-resistant C. glabrata (42). In the present study, we used a functional assay to investigate the effect of the immunosuppressant FK506, the chemosensitizer oligomycin, and the antiarrhythmic drug verapamil on the reversal of fluconazole resistance in our 20 resistant C. glabrata isolates (Table 6). According to the results of other studies (41, 42), FK506 efficiently reverses the fluconazole resistance of all the pump-overexpressing isolates, as shown by the decreased MICs for the isolates. Instead, oligomycin strongly reduced the fluconazole MICs only for the isolates that overexpressed CgCDR2 and/or CgSNQ2, whereas verapamil, a well-known inhibitor of human Mdr1-mediated drug resistance, had no effects on the isolates examined. None of the inhibitors alone had an effect on the growth of the same isolates, nor did they substantially alter the fluconazole MICs for susceptible isolates (data not shown).
TABLE 6.
Effects of ABC transporter inhibitors on the fluconazole resistance of the 20 C. glabrata isolates studied
| Isolate designation | FLC MIC (μg/ml)a
|
|||
|---|---|---|---|---|
| Alone | Plus FK-506 | Plus oligomycin | Plus verapamil | |
| BPY41 | 256 | 32 | 128 | 256 |
| BPY42 | 128 | 16 | 64 | 128 |
| BPY43 | 128 | 16 | 64 | 128 |
| BPY44 | 128 | 8 | 64 | 128 |
| BPY45 | 256 | 8 | 256 | 128 |
| BPY46 | 128 | 16 | 128 | 128 |
| BPY47 | 128 | 16 | 64 | 128 |
| BPY48 | 128 | 0.5 | 1 | 64 |
| BPY49 | 256 | 16 | 128 | 256 |
| BPY50 | 256 | 16 | 256 | 256 |
| BPY54 | 128 | 8 | 64 | 128 |
| BPY55 | 64 | 0.125 | 0.25 | 32 |
| BPY57 | 64 | 2 | 32 | 64 |
| BPY59 | 64 | 0.125 | 0.125 | 32 |
| BPY60 | 256 | 16 | 128 | 256 |
| BPY126 | 128 | 8 | 64 | 128 |
| BPY135 | 64 | 0.25 | 0.5 | 32 |
| BPY233 | 256 | 16 | 128 | 256 |
| BPY285 | 128 | 8 | 64 | 128 |
| BPY479 | 256 | 8 | 256 | 256 |
FLC, fluconazole; the MICs of this antifungal agent were determined by broth microdilution method, in accordance with NCCLS document M27-A2 (17).
DISCUSSION
Although C. glabrata infection is second or third in frequency, after C. albicans, as a difficult-to-treat infection and is associated with a high mortality rate in at-risk hospitalized patients, very little is known to date about the epidemiology, pathogenesis, treatment, and, above all, antifungal resistance of C. glabrata isolates (10). A more recent concern has been the numerous reports describing infections due to C. glabrata with documented in vitro antifungal resistance in compromised patients taking long-term oral antifungal agents (10, 29, 45, 47). Compared to other Candida species, especially C. albicans, the MICs of all azoles for C. glabrata isolates tend to be higher and C. glabrata isolates are intrinsically less susceptible to all antifungal agents, including amphotericin B (10). Although primary in vitro resistance to fluconazole has been reported (31), acquired resistance to azoles is, by far, the most common form of resistance in C. glabrata (45-47) and is most often seen for fluconazole. It is conceivable that the selective pressure of fluconazole, due to the increased use of this antifungal in some hospitals (1, 47), may be responsible for both the emergence of C. glabrata and fluconazole resistance, with the latter being related to the ability of the microorganism to rapidly develop secondary antifungal resistance due to its haploid state (10).
However, the relationship between fluconazole use and the emergence of fluconazole-resistant C. glabrata remains controversial, as recently reported by Pfaller et al. (28), who suggested that factors other than antifungal usage, such as patient age, underlying diseases, and geographic location, likely contribute to the emergence of C. glabrata as cause of bloodstream infections. In this context, Safdar et al. (33) observed a significant discrepancy in antifungal susceptibilities among C. glabrata organisms isolated from hospitals in the same geographic location. Therefore, periodic surveillance programs at individual institutions are critical in determining trends in the distributions and antifungal susceptibilities among Candida species, especially at centers caring for patients at risk.
For this reason, we conducted a survey of antifungal resistance in order to monitor the susceptibility patterns of C. glabrata infections in our institution, a large university hospital in Rome, Italy. The frequency of resistance to fluconazole seen among our isolates during a 3-year study was 7.5%; and although the in vitro resistance of C. glabrata to ketoconazole and itraconazole was believed to be somewhat less common (10), we observed, notably, comparable rates of resistance to these azoles and also to voriconazole. In addition, a careful analysis of the drug resistance profiles of our isolates showed that the majority of them were resistant to multiple azoles, indicating that cross-resistance is a very common feature in azole-resistant C. glabrata isolates, especially in isolates that are capable of expressing multiple mechanisms of resistance, as will be discussed below.
In the present study, we attempted to correlate the resistance phenotypes of 29 clinical C. glabrata isolates from the survey mentioned above to the molecular basis of azole resistance by evaluating the isolates for the different resistance mechanisms. As already reported (34), enhanced drug efflux is an important mechanism of azole resistance in C. glabrata and occurs as a result of the upregulation of multidrug efflux transporter genes, such as CgCDR1 and CgCDR2. Another mechanism of azole resistance in C. glabrata involves increased ERG11-dependent ergosterol synthesis due to an eightfold increase in ERG11 mRNA levels (14). In a recent work, Sanglard et al. (35) explored the role of CgCDR2, which was found to be identical to the PDH1 gene reported by Miyazaki et al. (15), in the phenomenon of azole resistance. They examined the levels of expression of both CgCDR1 and CgCDR2 in an azole-resistant clinical isolate of C. glabrata and showed that while the upregulation of CgCDR1 was significant, CgCDR2 was only moderately expressed in the same strain. On the other hand, when high-frequency azole resistance (HFAR; 2 × 10−4 to 4 × 10−4) was established from an azole-susceptible strain, both CgCDR1 and CgCDR2 were upregulated to high levels. Interestingly, this HFAR was coupled to the loss of mitochondria (35). Likewise, the azole resistance of petite mutants of C. glabrata, obtained by exposure to fluconazole or induced by ethidium bromide (6), was linked to the upregulation of nuclear genes encoding efflux proteins (7).
Previous studies aimed at obtaining an understanding of the molecular mechanisms of resistance in C. glabrata have been carried out by analyzing serial isolates from the same patient with decreased susceptibilities to azoles, in other words, matched sets of isolates that represent the susceptible and resistant versions of the same strain (5, 30, 36). Those studies were limited in that they examined a small number of clinical isolates (30, 36) or by the fact that these isolates originated from a single patient population (5). In this context, we believed that a thorough investigation at the molecular level of the azole resistance in C. glabrata should result from the analysis of a large number of unmatched clinical isolates that had different levels of azole resistance, such as those recovered in our hospital, and that were thus representative of various clinical settings. A similar investigation has previously been conducted by White et al. (44), who analyzed a collection of 36 unmatched clinical isolates of C. albicans for the known molecular mechanisms of resistance by standard methods. Remarkably, these analyses underlined the fact that the mechanisms of resistance identified in matched sets of susceptible and resistant isolates are not sufficient to explain resistance in arbitrarily selected isolates and that additional mechanisms have yet to be discovered.
As already mentioned, the set of C. glabrata isolates used in the present study was analyzed for the well-characterized molecular mechanisms of azole resistance. First, we examined the expression of CgCDR1 and CgCDR2 in our fluconazole-resistant isolates and observed that most isolates upregulated both genes and that the high levels of transcripts correlated with their high-level resistance to fluconazole and other azoles, including voriconazole. When we compared the levels of expression of CgCDR1 with those of CgCDR2, we noticed that CgCDR1 upregulation was always clearly manifested in isolates that concomitantly expressed both genes, while CgCDR2 was expressed at moderate levels. These findings add support to the idea that CgCDR1 is more closely associated with azole resistance than CgCDR2 (36). The importance of CgCDR1 and CgCDR2 in participating in azole resistance in C. glabrata was strengthened by the fact that both genes appeared to be upregulated at low levels in isolates that exhibited a fluconazole S-DD phenotype. Nevertheless, no upregulation of CgCDR1 or CgCDR2 was observed in two resistant isolates. For these isolates, it was likely that not-yet-characterized multidrug efflux transporters might be involved.
The gene for the CgSNQ2 pump, recently identified by Sanglard et al. (35) to be similar to a portion of an ABC transporter gene, SNQ2, from Saccharomyces cerevisiae, has not previously been associated with resistance. Some evidence by Sanglard et al. (35) showed that expression of CgSNQ2 was little affected by azole resistance in clinical and HFAR mutant strains of C. glabrata. In our study, a slight upregulation of the CgSNQ2 gene could be detected in part in either fluconazole-resistant or -S-DD isolates. More interestingly, CgSNQ2 accounted for the azole resistance of the four isolates that did not upregulate CgCDR1, alone or in combination with CgCDR2. In the two isolates mentioned above, in which no appearance of upregulation of CgCDR1 and CgCDR2 was found, only CgSNQ2 was upregulated, so relatively large amounts of CgSNQ2 transcripts were detected, and this feature correlated with their phenotype of resistance to multiple azoles. These data support in part the concept that ABC transporter genes with high degrees of similarity, such as CgCDR1 and CgCDR2, can differ in their abilities to confer azole resistance, as already documented for the homologues CDR1 and CDR2 in C. albicans (34). On the other hand, genes such as CgSNQ2, for which little involvement in the acquisition of resistance to azole antifungals has been demonstrated, may play a determinant role in this process. This finding needs to be confirmed by further investigation.
Second, in light of the fact that in C. albicans clinical isolates decreased susceptibilities to multiple azole derivatives were mainly associated with the upregulation of CDR genes (i.e., CDR1 and CDR2), as well as with the presence of specific point mutations in the ERG11 gene, we examined the role of ERG11 in determining the azole resistance phenotypes of our isolates. Confirming previous results (7), sequencing of CgERG11 and analysis of its expression performed with our isolates showed no alteration or overproduction, favoring the hypothesis that CgERG11 is not involved in the azole resistance of C. glabrata. However, one of the two fluconazole-resistant isolates of C. glabrata from a patient with oropharyngeal candidiasis studied by Redding et al. (30) showed the upregulation not only of CgCDR1 and CgCDR2 but also of CgERG11. In contrast, the other strain showed no upregulation of the three genes, thus indicating that the development of resistance to fluconazole by C. glabrata is a highly varied process involving multiple molecular mechanisms.
To complement the RNA analysis of azole-resistant clinical isolates, we analyzed the expression of the target genes in 20 randomly selected azole-susceptible clinical isolates of C. glabrata. As expected, the results provided evidence that the levels of expression of CgCDR1, CgCDR2, CgSNQ2, and CgERG11 in these isolates were always low. On the other hand, we evaluated whether the expression profiling in our 20 resistant isolates was influenced by exposure to fluconazole. Interestingly, we demonstrated that the upregulation of efflux pump-encoding genes was unchanged or was only minimally changed, suggesting that some genetic alteration that perhaps affects a regulatory gene(s) occurred, thereby resulting in unaltered resistance profiles. In support of this, when cultures of 24 susceptible isolates, including 4 pretreatment isolates matched to their corresponding resistant isolates, were treated with fluconazole, we found that all three ABC transporter genes, as well as the ERG11 gene, were markedly upregulated (Table 4). Finally, FK506, which was previously reported to be an inhibitor of ABC pumps (9), reversed the resistance phenotype in our isolates, probably by impairing the drug efflux enzyme activity, although other ABC pump-independent mechanisms, such as an interaction with the calcineurin pathway, could be involved (38).
In summary, the results obtained in the present study emphasize the role of the upregulation of the ABC efflux transporters CgCDR1, CgCDR2, and CgSNQ2 as a major mechanism of azole resistance in C. glabrata. Although this mechanism has not been sufficient to explain the increased level of resistance to fluconazole seen in isolates from 1 of the 20 patients studied by Bennett et al. (5), it proved to be exclusive in our collection of isolates. The multifactorial nature of azole resistance in C. glabrata must not be disregarded, and more comprehensive studies dealing with other possible underlying resistance mechanisms are warranted.
Acknowledgments
This work was supported by a grant from the Istituto Superiore di Sanità (National Program of Research on AIDS).
We thank Dominique Sanglard (Institute of Microbiology, CHUV, Lausanne, Switzerland) for helpful suggestions and critical reading of manuscript and David Soll for providing the Cg6 and Cg12 probes. We are greatly indebted to Marcello Porretta and Marilena La Sorda for excellent technical assistance.
REFERENCES
- 1.Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Baddley, J. W., A. M. Smith, S. A. Moser, and P. G. Pappas. 2001. Trends in frequency and susceptibilities of Candida glabrata bloodstream isolates at a university hospital. Diagn. Microbiol. Infect. Dis. 39:199-201. [DOI] [PubMed] [Google Scholar]
- 3.Barchiesi, F., L. Falconi Di Francesco, D. Arzeni, F. Caselli, D. Gallo, and G. Scalise. 1999. Electrophoretic karyotyping and triazole susceptibility of Candida glabrata clinical isolates. Eur. J. Clin. Microbiol. Infect. Dis. 18:184-187. [DOI] [PubMed] [Google Scholar]
- 4.Barry, A. L., M. A. Pfaller, S. D. Brown, A. Espinel-Ingroff, M. A. Ghannoum, C. Knapp, R. P. Rennie, J. H. Rex, and M. G. Rinaldi. 2000. Quality control limits for broth microdilution susceptibility tests of ten antifungal agents. J. Clin. Microbiol. 38:3457-3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett, J. E., K. Izumikawa, and K. A. Marr. 2004. Mechanism of increased fluconazole resistance in Candida glabrata during prophylaxis. Antimicrob. Agents Chemother. 48:1773-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun, S., C. Aubry, O. Lima, R. Filmon, T. Berges, D. Chabasse, and J. P. Bouchara. 2003. Relationships between respiration and susceptibility to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 47:847-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brun, S., T. Berges, P. Poupard, C. Vauzelle-Moreau, G. Renier, D. Chabasse, and J. P. Bouchara. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48:1788-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davey, K. G., A. Szekely, E. M. Johnson, and D. W. Warnock. 1998. Comparison of a new commercial colorimetric microdilution method with a standard method for in-vitro susceptibility testing of Candida spp. and Cryptococcus neoformans. J. Antimicrob. Chemother. 42:439-444. [DOI] [PubMed] [Google Scholar]
- 9.Egner, R., B. E. Bauer, and K. Kuchler. 2000. The transmembrane domain 10 of the yeast Pdr5p ABC antifungal efflux pump determines both substrate specificity and inhibitor susceptibility. Mol. Microbiol. 35:1255-1263. [DOI] [PubMed] [Google Scholar]
- 10.Fidel, P. L., Jr., J. A. Vazquez, and J. D. Sobel. 1999. Candida glabrata: review of epidemiology, pathogenesis, and clinical disease with comparison to C. albicans. Clin. Microbiol. Rev. 12:80-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockhart, S. R., S. Joly, C. Pujol, J. D. Sobel, M. A. Pfaller, and D. R. Soll. 1997. Development and verification of fingerprinting probes for Candida glabrata. Microbiology 143:3733-3746. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Ribot, J. L., R. K. McAtee, L. N. Lee, W. R. Kirkpatrick, T. C. White, D. Sanglard, and T. F. Patterson. 1998. Distinct patterns of gene expression associated with development of fluconazole resistance in serial Candida albicans isolates from human immunodeficiency virus-infected patients with oropharyngeal candidiasis. Antimicrob. Agents Chemother. 42:2932-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maenza, J. R., W. G. Merz, M. J. Romagnoli, J. C. Keruly, R. D. Moore, and J. E. Gallant. 1997. Infection due to fluconazole-resistant Candida in patients with AIDS: prevalence and microbiology. Clin. Infect. Dis. 24:28-34. [DOI] [PubMed] [Google Scholar]
- 14.Marichal, P., H. Vanden Bossche, F. C. Odds, G. Nobels, D. W. Warnock, V. Timmerman, C. Van Broeckhoven, S. Fay, and P. Mose-Larsen. 1997. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob. Agents Chemother. 41:2229-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyazaki, H., Y. Miyazaki, A. Geber, T. Parkinson, C. Hitchcock, D. J. Falconer, D. J. Ward, K. Marsden, and J. E. Bennett. 1998. Fluconazole resistance associated with drug efflux and increased transcription of a drug transporter gene, PDH1, in Candida glabrata. Antimicrob. Agents Chemother. 42:1695-1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran, G. P., D. J. Sullivan, and D. C. Coleman. 2002. Emergence of non-Candida albicans Candida species as pathogens, p. 37-53. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 17.National Committee for Clinical Laboratory Standards. 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts. Approved standard M27-A2. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Nguyen, M. H., C. J. Clancy, V. L. Yu, Y. C. Yu, A. J. Morris, D. R. Snydman, D. A. Sutton, and M. G. Rinaldi. 1998. Do in vitro susceptibility data predict the microbiologic response to amphotericin B? Results of a prospective study of patients with Candida fungemia. J. Infect. Dis. 177:425-430. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen, M. H., J. E. Peacock, A. J. Morris, D. C. Tanner, M. L. Nguyen, D. R. Snydman, M. M. Wagener, M. G. Rinaldi, and V. L. Yu. 1996. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am. J. Med. 100:617-623. [DOI] [PubMed] [Google Scholar]
- 20.Niimi, M., Y. Nagai, K. Niimi, S. Wada, R. D. Cannon, Y. Uehara, and B. C. Monk. 2002. Identification of two proteins induced by exposure of the pathogenic fungus Candida glabrata to fluconazole. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 782:245-252. [DOI] [PubMed] [Google Scholar]
- 21.Peirson, S. N., J. N. Butler, and R. G. Foster. 2003. Experimental validation of novel and conventional approaches to quantitative real-time PCR data analysis. Nucleic Acids Res. 31:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perea, S., J. L. Lopez-Ribot, W. R. Kirkpatrick, R. K. McAtee, R. A. Santillan, M. Martinez, D. Calabrese, D. Sanglard, and T. F. Patterson. 2001. Prevalence of molecular mechanisms of resistance to azole antifungal agents in Candida albicans strains displaying high-level fluconazole resistance isolated from human immunodeficiency virus-infected patients. Antimicrob. Agents Chemother. 45:2676-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller, M. A., S. Arikan, M. Lozano-Chiu, Y. Chen, S. Coffman, S. A. Messer, R. Rennie, C. Sand, T. Heffner, J. H. Rex, J. Wang, and N. Yamane. 1998. Clinical evaluation of the ASTY colorimetric microdilution panel for antifungal susceptibility testing. J. Clin. Microbiol. 36:2609-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfaller, M. A., R. N. Jones, G. V. Doern, H. S. Sader, R. J. Hollis, S. A. Messer, and The SENTRY Participant Group. 1998. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibilities of isolates collected in 1997 in the United States, Canada, and South America for the SENTRY Program. J. Clin. Microbiol. 36:1886-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfaller, M. A., D. J. Diekema, R. N. Jones, H. S. Sader, A. C. Fluit, R. J. Hollis, S. A. Messer, and The SENTRY Participant Group. 2001. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and in vitro susceptibilities to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY antimicrobial surveillance program. J. Clin. Microbiol. 39:3254-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfaller, M. A., D. J. Diekema, R. N. Jones, S. A. Messer, R. J. Hollis, and the SENTRY Participants Group. 2002. Trends in antifungal susceptibility of Candida spp. isolated from pediatric and adult patients with bloodstream infections: SENTRY Antimicrobial Surveillance Program, 1997 to 2000. J. Clin. Microbiol. 40:852-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfaller, M. A., S. A. Messer, L. Boyken, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2003. Variation in susceptibility of bloodstream isolates of Candida glabrata to fluconazole according to patient age and geographic location. J. Clin. Microbiol. 41:2176-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Redding, S. W., W. R. Kirkpatrick, B. J. Coco, L. Sadkowski, A. W. Fothergill, M. G. Rinaldi, T. Y. Eng, and T. F. Patterson. 2002. Candida glabrata oropharyngeal candidiasis in patients receiving radiation treatment for head and neck cancer. J. Clin. Microbiol. 40:1879-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redding, S. W., W. R. Kirkpatrick, S. Saville, B. J. Coco, W. White, A. Fothergill, M. Rinaldi, T. Eng, T. F. Patterson, and J. Lopez-Ribot. 2003. Multiple patterns of resistance to fluconazole in Candida glabrata isolates from a patient with oropharyngeal candidiasis receiving head and neck radiation. J. Clin. Microbiol. 41:619-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rex, J. H., M. A. Pfaller, A. L. Barry, P. W. Nelson, C. D. Webb, the NIAID Mycoses Study Group, and the Candidemia Study Group. 1995. Antifungal susceptibility testing of isolates from a randomized, multicenter trial of fluconazole versus amphotericin B as treatment of nonneutropenic patients with candidemia. Antimicrob. Agents Chemother. 39:40-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rex, J. H., M. A. Pfaller, J. N. Galgiani, M. S. Bartlett, A. Espinel-Ingroff, M. A. Ghannoum, M. Lancaster, F. C. Odds, M. G. Rinaldi, T. J. Walsh, A. L. Barry, et al. 1997. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and Candida infections. Clin. Infect. Dis. 24:235-247. [DOI] [PubMed] [Google Scholar]
- 33.Safdar, A., V. Chaturvedi, B. S. Koll, D. H. Larone, D. S. Perlin, and D. Armstrong. 2002. Prospective, multicenter surveillance study of Candida glabrata: fluconazole and itraconazole susceptibility profiles in bloodstream, invasive, and colonizing strains and differences between isolates from three urban teaching hospitals in New York City (Candida Susceptibility Trends Study, 1998 to 1999). Antimicrob. Agents Chemother. 46:3268-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanglard, D., and J. Bille. 2002. Current understanding of the modes of action and resistance mechanisms to conventional and emerging antifungal agents for treatment of Candida infections, p. 349-383. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, D.C.
- 35.Sanglard, D., F. Ischer, and J. Bille. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanglard, D., F. Ischer, D. Calabrese, P. A. Majcherczyk, and J. Bille. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanglard, D., F. Ischer, L. Koymans, and J. Bille. 1998. Amino acid substitutions in the cytochrome P-450 lanosterol 14α-demethylase (CYP51A1) from azole-resistant Candida albicans clinical isolates contribute to resistance to azole antifungal agents. Antimicrob. Agents Chemother. 42:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanglard, D., F. Ischer, O. Marchetti, J. Entenza, and J. Bille. 2003. Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol. Microbiol. 48:959-976. [DOI] [PubMed] [Google Scholar]
- 39.Sanglard, D., F. Ischer, T. Parkinson, D. Falconer, and J. Bille. 2003. Candida albicans mutations in the ergosterol biosynthetic pathway and resistance to several antifungal agents. Antimicrob. Agents Chemother. 47:2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schoofs, A. G., F. C. Odds, R. Colebunders, M. Ieven, and H. Goossens. 1998. Cross-sectional study of oral Candida carriage in a human immunodeficiency virus (HIV)-seropositive population: predisposing factors, epidemiology and antifungal susceptibility. Mycoses 41:203-211. [DOI] [PubMed] [Google Scholar]
- 41.Schuetzer-Muehlbauer, M., B. Willinger, R. Egner, G. Ecker, and K. Kuchler. 2003. Reversal of antifungal resistance mediated by ABC efflux pumps from Candida albicans functionally expressed in yeast. Int. J. Antimicrob. Agents 22:291-300. [DOI] [PubMed] [Google Scholar]
- 42.Wada, S., M. Niimi, K. Niimi, A. R. Holmes, B. C. Monk, R. D. Cannon, and Y. Uehara. 2002. Candida glabrata ATP-binding cassette transporters Cdr1p and Pdh1p expressed in a Saccharomyces cerevisiae strain deficient in membrane transporters show phosphorylation-dependent pumping properties. J. Biol. Chem. 277:46809-46821. [DOI] [PubMed] [Google Scholar]
- 43.Warren, N. G., and K. C. Hazen. 1999. Candida, Cryptococcus, and other yeasts of medical importance, p. 1184-1199. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 44.White, T. C., S. Holleman, F. Dy, L. F. Mirels, and D. A. Stevens. 2002. Resistance mechanisms in clinical isolates of Candida albicans. Antimicrob. Agents Chemother. 46:1704-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wingard, J. R. 1994. Infections due to resistant Candida species in patients with cancer who are receiving chemotherapy. Clin. Infect. Dis. 19:S49-S53. [DOI] [PubMed] [Google Scholar]
- 46.Wingard, J. R. 1995. Importance of Candida species other than C. albicans as pathogens in oncology patients. Clin. Infect. Dis. 20:115-125. [DOI] [PubMed] [Google Scholar]
- 47.Wingard, J. R., W. G. Merz, M. G. Rinaldi, C. B. Miller, J. E. Karp, and R. Saral. 1993. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob. Agents Chemother. 37:1847-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]