Abstract
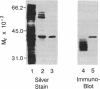
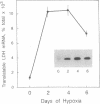
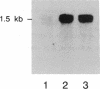
In the roots of barley and other cereals, hypoxia induces a set of five isozymes of L-lactate dehydrogenase [LDH; (S)-lactate:NADH oxidoreductase, EC 1.1.1.27]. Biochemical and genetic data indicate that the five LDH isozymes are tetramers that arise from random association of the products of two Ldh loci. To investigate this system, cDNA clones of LDH were isolated from a lambda gt11 cDNA library derived from hypoxically treated barley roots. The library was screened with antiserum raised against barley LDH purified approximately 3000-fold by an improved three-step procedure. Immunopositive clones were rescreened with a cDNA probe synthesized by the polymerase chain reaction using primers modeled from the amino acid sequences of two tryptic LDH peptides. Two types of LDH clones were found. Nucleotide sequence analysis of one representative insert of each type (respectively, 1305 and 1166 base pairs) revealed open reading frames encoding 10 peptide fragments of LDH. The 1305-base-pair insert included the entire coding region of a 356-residue LDH monomer. The nucleotide sequences of the two LDH cDNAs were 92% identical in the coding region, but highly divergent in the 3' noncoding region, and thus probably correspond to the two postulated Ldh loci. The deduced amino acid sequences of the two barley LDHs were 96% identical to each other and very similar to those from vertebrate and bacterial LDHs. RNA blot hybridization showed a single mRNA band of 1.5 kilobases whose level rose about 8-fold in roots during hypoxic induction, as did the level of translatable LDH message.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Clarke A. R., Wigley D. B., Chia W. N., Barstow D., Atkinson T., Holbrook J. J. Site-directed mutagenesis reveals role of mobile arginine residue in lactate dehydrogenase catalysis. Nature. 1986 Dec 18;324(6098):699–702. doi: 10.1038/324699a0. [DOI] [PubMed] [Google Scholar]
- Dale R. M., Arrow A. A rapid single-stranded cloning, sequencing, insertion, and deletion strategy. Methods Enzymol. 1987;155:204–214. doi: 10.1016/0076-6879(87)55017-0. [DOI] [PubMed] [Google Scholar]
- Eventoff W., Rossmann M. G., Taylor S. S., Torff H. J., Meyer H., Keil W., Kiltz H. H. Structural adaptations of lactate dehydrogenase isozymes. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2677–2681. doi: 10.1073/pnas.74.7.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gerlach W. L., Pryor A. J., Dennis E. S., Ferl R. J., Sachs M. M., Peacock W. J. cDNA cloning and induction of the alcohol dehydrogenase gene (Adh1) of maize. Proc Natl Acad Sci U S A. 1982 May;79(9):2981–2985. doi: 10.1073/pnas.79.9.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. C., Ma Y., Buchbinder B. U., Pyne J. W., Sun S. M., Bliss F. A. Messenger RNA for G1 protein of French bean seeds: Cell-free translation and product characterization. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3196–3200. doi: 10.1073/pnas.75.7.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey H. P., Colbert J. T., Lissemore J. L., Barker R. F., Quail P. H. Molecular cloning of cDNA for Avena phytochrome. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2332–2336. doi: 10.1073/pnas.81.8.2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N. E., Bent A. F., Hanson A. D. Induction of lactate dehydrogenase isozymes by oxygen deficit in barley root tissue. Plant Physiol. 1986 Nov;82(3):658–663. doi: 10.1104/pp.82.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman N. E., Hanson A. D. Purification and properties of hypoxically induced lactate dehydrogenase from barley roots. Plant Physiol. 1986 Nov;82(3):664–670. doi: 10.1104/pp.82.3.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley P. M. Maize pyruvate decarboxylase mRNA is induced anaerobically. Plant Mol Biol. 1989 Aug;13(2):213–222. doi: 10.1007/BF00016139. [DOI] [PubMed] [Google Scholar]
- Kimmel A. R., Berger S. L. Preparation of cDNA and the generation of cDNA libraries: overview. Methods Enzymol. 1987;152:307–316. doi: 10.1016/0076-6879(87)52035-3. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li S. S., Fitch W. M., Pan Y. C., Sharief F. S. Evolutionary relationships of vertebrate lactate dehydrogenase isozymes A4 (muscle), B4 (heart), and C4 (testis). J Biol Chem. 1983 Jun 10;258(11):7029–7032. [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Markert C. L., Shaklee J. B., Whitt G. S. Evolution of a gene. Multiple genes for LDH isozymes provide a model of the evolution of gene structure, function and regulation. Science. 1975 Jul 11;189(4197):102–114. doi: 10.1126/science.1138367. [DOI] [PubMed] [Google Scholar]
- Mierendorf R. C., Percy C., Young R. A. Gene isolation by screening lambda gt11 libraries with antibodies. Methods Enzymol. 1987;152:458–469. doi: 10.1016/0076-6879(87)52054-7. [DOI] [PubMed] [Google Scholar]
- Roberts J. K., Callis J., Jardetzky O., Walbot V., Freeling M. Cytoplasmic acidosis as a determinant of flooding intolerance in plants. Proc Natl Acad Sci U S A. 1984 Oct;81(19):6029–6033. doi: 10.1073/pnas.81.19.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. K., Callis J., Wemmer D., Walbot V., Jardetzky O. Mechanisms of cytoplasmic pH regulation in hypoxic maize root tips and its role in survival under hypoxia. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3379–3383. doi: 10.1073/pnas.81.11.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs M. M., Freeling M., Okimoto R. The anaerobic proteins of maize. Cell. 1980 Jul;20(3):761–767. doi: 10.1016/0092-8674(80)90322-0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter D. B., Sommer S. S. The generation of radiolabeled DNA and RNA probes with polymerase chain reaction. Anal Biochem. 1989 Feb 15;177(1):90–94. doi: 10.1016/0003-2697(89)90019-5. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Walker J. C., Howard E. A., Dennis E. S., Peacock W. J. DNA sequences required for anaerobic expression of the maize alcohol dehydrogenase 1 gene. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6624–6628. doi: 10.1073/pnas.84.19.6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weretilnyk E. A., Hanson A. D. Betaine aldehyde dehydrogenase from spinach leaves: purification, in vitro translation of the mRNA, and regulation by salinity. Arch Biochem Biophys. 1989 May 15;271(1):56–63. doi: 10.1016/0003-9861(89)90255-5. [DOI] [PubMed] [Google Scholar]