Abstract
Capreomycin, an important drug for the treatment of multidrug-resistant tuberculosis, is a macrocyclic peptide antibiotic produced by Saccharothrix mutabolis subspecies capreolus. The basis of resistance to this drug was investigated by isolating and characterizing capreomycin-resistant strains of Mycobacterium smegmatis and Mycobacterium tuberculosis. Colonies resistant to capreomycin were recovered from a library of transposon-mutagenized M. smegmatis. The transposon insertion site of one mutant was mapped to an open reading frame in the unfinished M. smegmatis genome corresponding to the tlyA gene (Rv1694) in the M. tuberculosis H37Rv genome. In M. smegmatis spontaneous capreomycin-resistant mutants, the tlyA gene was disrupted by one of three different naturally occurring insertion elements. Genomic DNAs from pools of transposon mutants of M. tuberculosis H37Rv were screened by PCR by using primers to the tlyA gene and the transposon to detect mutants with an insertion in the tlyA gene. One capreomycin-resistant mutant was recovered that contained the transposon inserted at base 644 of the tlyA gene. Complementation with the wild-type tlyA gene restored susceptibility to capreomycin in the M. smegmatis and M. tuberculosis tlyA transposon mutants. Mutations were found in the tlyA genes of 28 spontaneous capreomycin-resistant mutants generated from three different M. tuberculosis strains and in the tlyA genes of capreomycin-resistant clinical isolates. In in vitro transcription-translation assays, ribosomes from tlyA mutant but not tlyA+ strains resist capreomycin inhibition of transcription-translation. Therefore, TlyA appears to affect the ribosome, and mutation of tlyA confers capreomycin resistance.
Tuberculosis is the only disease that the World Health Organization has declared a global public heath emergency. Worldwide in 2000, there were about 9 million new cases and 2 million deaths due to tuberculosis (12, 13). The emergence of multidrug-resistant (MDR) tuberculosis has further complicated the treatment and control of the disease (14). Treatment of MDR tuberculosis requires the use of second-line drugs, such as capreomycin, kanamycin, or amikacin (4). In contrast to several antituberculosis drugs, relatively little is known about the mechanism of action or molecular basis of resistance for capreomycin, an important second-line antibiotic (28, 30).
Capreomycin is a macrocyclic peptide antibiotic produced by Saccharothrix mutabolis subspecies capreolus (31, 45) and appears to interfere with translation in mycobacteria (34). Capreomycin can inhibit phenylalanine synthesis in an in vitro translation assay using mycobacterial ribosomes (34). Similar inhibition was seen whether or not ribosomes were preincubated with mRNA, suggesting that capreomycin does not interfere with mRNA binding to the ribosome (34).
Capreomycin is structurally similar to viomycin (26), and many studies show complete cross-resistance between viomycin and capreomycin in Mycobacterium tuberculosis (2, 20, 21, 35, 36). Viomycin has been shown to bind to both the 30S and 50S ribosome subunits (42) and to affect the dissociation of the 70S ribosome of Mycobacterium smegmatis (41). Furthermore, it inhibits ribosomal translocation by arresting peptidyl-tRNA in the ribosomal acceptor site (23). Additionally, changes in the 30S and 50S ribosomal subunits alter binding and confer viomycin resistance in M. smegmatis (8, 22, 33, 40, 41, 43).
Previous reports have linked mutations in the 16S rRNA gene to capreomycin resistance (32, 33); however, these reports are variable and do not completely account for capreomycin resistance in M. tuberculosis. Transposon mutagenesis is a useful tool to search for nonessential genes encoding products that alter a drug target, are involved in drug transport, or convert a prodrug to its active form. To investigate capreomycin resistance in mycobacteria, libraries of transposon mutants from M. smegmatis and M. tuberculosis were utilized. Characterization of transposon and spontaneous M. smegmatis and M. tuberculosis capreomycin-resistant mutants as well as capreomycin-resistant clinical isolates revealed that mutation of the tlyA gene confers capreomycin resistance in mycobacteria.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. smegmatis strain LR222 and M. tuberculosis strains H37Rv, CDC1551, and Beijing F2 and 18 clinical isolates were obtained from the culture collection of the Tuberculosis/Mycobacteriology Branch, Centers for Disease Control and Prevention (CDC). M. tuberculosis cultures were grown in complete Middlebrook 7H9 broth (7H9) (Remel, Lenexa, Kans.) at 37°C or on Middlebrook 7H10 agar (7H10). M. smegmatis was grown in 7H9 broth or on Trypticase soy agar (TSA). In TSA-capreomycin (TSA-Cap; Sigma, St. Louis, Mo.), 7H9-capreomycin (7H9-Cap), 7H10-capreomycin (7H10-Cap), or 7H9-viomycin (7H9-Vio; U.S. Pharmacopeia, Rockville, Md.) medium, each drug was used at a concentration of 10 μg/ml unless stated otherwise. Hygromycin (Invitrogen, Carlsbad, Calif.) was used in the media at a final concentration of 50 μg/ml for mycobacteria when needed. Acetamide (Sigma) was incorporated into media at a final concentration of 0.2% as appropriate. The Escherichia coli EC100 (Epicentre, Madison, Wis.) strain was grown in Luria broth or Luria agar, and hygromycin at a concentration of 200 μg/ml was used when appropriate.
DNA isolation, manipulations, sequencing, PCR, and Southern blotting.
Genomic and plasmid DNAs were purified as previously described (27). All enzymatic reactions were performed as recommended by the manufacturer (Invitrogen, Life Technologies). Oligonucleotide primers were synthesized at the Biotechnology Core Facility, National Center for Infectious Diseases, CDC (sequences available upon request). Amplification reactions (100 μl) were performed as recommended by the manufacturer (PE Applied Biosystems, Foster City, Calif.) in a GeneAmp PCR system 2400 thermocycler (PE Applied Biosystems) by using a three-step cycle of denaturation for 30 s at 96°C, annealing for 30 s at 60 to 68°C (optimized for each primer pair), and extension for 2 min at 72°C. Amplification products were analyzed by electrophoresis through 0.8 to 2.0% agarose-Tris-borate-EDTA gels and visualized by ethidium bromide staining (29). Prior to DNA sequencing, amplicons were purified and concentrated by using a DNA Clean and Concentrator kit (Zymo Research, Orange, Calif.). DNA Sequencing was carried out by using a CEQ Dye Terminator Cycle Sequence Quick Start kit (Beckman Coulter, Fullerton, Calif.) according to the manufacturer's directions. Samples were analyzed on the CEQ8000 sequencer (Beckman). Sequence data were evaluated by using Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html; Tom Hall, Department of Microbiology, North Carolina State University). Southern blot analysis was carried out by using an ECL nucleic acid labeling and detection system (Amersham, Arlington Heights, Ill.) as previously described (27).
Transposon-mutant libraries.
The M. smegmatis LR222 library was constructed by using an EZ::TN Kan Transposome Kit (Epicentre Technologies, Madison, Wis.). Wild-type M. smegmatis electrocompetent cells were electroporated with 1 μl of the transposome as described previously (23); then 1 ml of 7H9 broth was added. After an 18-h incubation at 37°C, aliquots of the cell mixture were spread on 10 TSA plates containing 10 μg of kanamycin (TSA-Kan) per ml. After 72 h at 37°C, 40 colonies were picked for evaluation, and the remaining growth from each plate was resuspended in 7H9 broth with 10 μg of kanamycin (7H9-Kan) per ml and incubated for 18 h at 37°C. The 10 pools were stored at −70°C. The M. tuberculosis H37Rv library was generated in a similar manner except that a total of seven samples of electrocompetent cells were electroporated and outgrowth in 7H9 broth was for 24 to 72 h. The cell mixture from each electroporation was plated on 10 7H10 plates containing 10 μg of kanamycin (7H10-Kan) per ml and held at 37°C for 21 days. Eighteen colonies from each set were picked for evaluation, and those remaining were resuspended in 7H9-Kan broth and incubated 30 days at 37°C. DNA was extracted from a 1.5-ml sample of the culture from each pool. Pools were stored at −70°C. DNA was extracted from the individual colonies selected for evaluation and analyzed by PCR by using primers specific to the Tn903 kanamycin resistance gene (Kanr) in the EZ::TN transposon.
Generation of capreomycin-resistant spontaneous mutants.
Spontaneous capreomycin mutants were generated from M. smegmatis LR222 and pan-susceptible M. tuberculosis strains H37Rv, CDC1551, and Beijing strain F2. All parent strains were susceptible to 10 μg of capreomycin per ml. Portions of a concentrated cell suspension of each strain were spread on each of four plates containing 10 μg of capreomycin (TSA-Cap for M. smegmatis or 7H10-Cap for M. tuberculosis) per ml. Serial 10-fold dilutions of the concentrated cell suspension were plated on media without drug to determine the number of viable cells in the suspension. Once colonies grew on the drug media, isolated colonies were picked and inoculated into 7H9-Cap broth. DNA lysates were made from the cultures and analyzed by PCR and sequencing.
Cloning of mutant genomic DNA into pBluescript.
Genomic DNAs from two capreomycin-resistant M. smegmatis transposon mutants were digested with EcoRI and ligated into an EcoRI restriction site of pBluescript KS(+) (Stratagene, La Jolla, Calif.). The libraries were electroporated into E. coli, and colonies were selected for kanamycin resistance. Plasmid DNAs from three kanamycin-resistant transformants were sequenced by using outward facing primers specific to the Tn903 Kanr gene in the EZ::TN transposon.
Construction of tlyA expressing plasmids.
The open reading frames (ORFs) of the M. smegmatis and M. tuberculosis wild-type tlyA genes were amplified by using primers with BglII restriction sites. The amplicons were purified, digested with BglII, and ligated into the BamHI site of pMF1 (provided by Mark Fisher) which contains the E. coli origin of replication from pUC18, the mycobacteriophage L5 integrase gene and attP site, the Streptomyces hygroscopicus hygromycin resistance selectable marker, and the M. smegmatis acetamidase promoter (M. Fisher, personal communication). This cloning strategy places the tlyA ORF under the control of the acetamidase promoter. The resulting plasmids were designated pMF1-tlyA/sm and pMF1-tlyA/tb for the M. smegmatis tlyA ORF and M. tuberculosis tlyA ORF, respectively.
Susceptibility testing.
Susceptibilities to antituberculosis drugs were determined according to NCCLS guidelines and definitions (24) by using 7H10 agar containing the following: isoniazid (at concentrations of 0.2, 1, and 5 μg/ml; Sigma), rifampin (1 μg/ml; Sigma), ethambutol (5 μg/ml; Sigma), streptomycin (2 and 10 μg/ml; Sigma), rifabutin (2 μg/ml; Pharmacia and Upjohn, Kalamazoo, Mich.), ciprofloxacin (2 μg/ml; Bayer, West Haven, Conn.), kanamycin (5 μg/ml; Sigma), ethionamide (10 μg/ml; Sigma), capreomycin (10 μg/ml; Sigma), p-aminosalicylic acid (2 μg/ml; Sigma), ofloxacin (2 μg/ml; Sigma), or amikacin (4 μg/ml; Sigma). MICs were determined in 7H10 agar by using the following drugs at the indicated concentrations: capreomycin at 10, 20, 40, 80, and 160 μg/ml; viomycin at 10, 20, 40, and 80 μg/ml; kanamycin at 5, 10, 20, 40, and 80 μg/ml; and amikacin at 4, 8, 16, 32, and 64 μg/ml. The MIC was defined as the lowest concentration of drug resulting in complete inhibition of growth or in growth of <1% of the inoculum. MIC controls for spontaneous capreomycin-resistant mutants and clinical isolates consisted of wild-type strains M. tuberculosis H37Rv, CDC1551, and Beijing F2. All control strains were susceptible to 10 μg of capreomycin per ml, which is the critical concentration for the drug as stated by NCCLS (24).
Growth analysis.
Wild-type M. smegmatis bacteria and the tlyA EZ::TN-kan mutant were used separately to inoculate, in triplicate, 7H9 and 7H9-Cap media to a starting optical density at 600 nm (OD600) of 0.2 as measured by an Ultrospec 1100 pro visible spectrophotometer (Biochrom Ltd., Cambridge, England). The cultures were incubated at 37°C for up to 52 h on a tissue culture roller drum. OD600 readings were taken every 4 h. The mean OD600 ± standard deviation (SD) for each time point was calculated.
In vitro transcription-translation.
Coupled in vitro transcription-translation assays were performed by using S30 extracts obtained from the M. smegmatis wild-type and EZ::TN-kan tlyA mutant. Bacteria were harvested by centrifugation at 6,000 × g for 15 min from 1 liter of 7H9 cultures grown to mid-log phase. The pellets were washed twice with buffer A (10 mM Tris acetate [pH 8.2], 14 mM Mg acetate, 60 mM K acetate, 1 mM dithiothreitol). Pellets were stored at −70°C, subjected to five cycles of freezing and thawing, and then resuspended in 1.82 ml of buffer A per 1 g of pellet (wet weight). The suspensions were passed through a Parr bomb at 2,000 lb/in2 for 45 min on ice to break cells (19). The resulting lysates were treated with 2 μl of 1 M dithiothreitol per 1 ml of lysate and centrifuged twice at 30,000 × g for 30 min to obtain the S30 extracts.
Components of an E. coli S30 Extract System for Linear Templates kit (Promega, Madison, Wis.) were used for the coupled transcription-translation assay. The plasmid pTS73 which contains the luciferase gene expressed from the Tn5 promoter was the DNA template in the assay. The reaction mixture contained 0.08 μg of pTS73/μl reaction mixture, 5 μl of complete amino acid mixture, 20 μl of S30 premixture without amino acids, and 20 μl of M. smegmatis S30 extract. E. coli S30 extract was used as a reagent control. The samples were incubated at 37°C for 2.5 h, and then the reactions were stopped by incubation at 4°C for 20 min. Luciferase activity was measured by using a Luciferase Assay System (Promega) and a TD-20/20 luminometer (Turner Designs, Sunnyvale, Calif.).
RESULTS
Generation of EZ::TN libraries in M. smegmatis and M. tuberculosis.
Electroporation of 1 μl of the EZ::TN Kan transposome into M. smegmatis yielded a total of 5 × 103 kanamycin-resistant colonies that were combined into 10 pools. Each of the 40 kanamycin-resistant colonies studied contained the Tn903 Kanr gene of the EZ::TN transposon in a differently sized fragment (data not shown), suggesting random insertion within the genome. The seven electroporation reactions in M. tuberculosis yielded a total of 6.9 × 103 kanamycin-resistant colonies. A total of 126 colonies were selected for evaluation, and the remaining colonies were combined into 64 pools. Southern blot analysis of 126 mutants revealed that 114 (90%) had an insertion of the EZ::TN transposon into the chromosome and that 12 appeared to be spontaneous Kanr mutants (data not shown). The distribution of the size of the fragments detected with the Tn903 Kanr gene probe (data not shown) suggested that the insertion sites were random.
Isolation of capreomycin-resistant M. smegmatis transposon mutants.
Portions of the library of M. smegmatis EZ::TN transposon mutants were spread on TSA plates containing 10 μg of capreomycin per ml, and resistant colonies were recovered at a frequency of about 1 in 5.5 × 104 bacteria plated. Initially, three capreomycin-resistant mutants were selected, and all three mutants displayed the same 8-kb fragment when EcoRI-digested DNA was hybridized with a probe specific for the Tn903 Kanr gene in the EZ::TN transposon. Genomic DNAs from two of the mutants were cloned into pBluescript and transformed into E. coli, and colonies were selected for kanamycin resistance. Plasmid DNAs from three kanamycin-resistant transformants were sequenced, and the insertion site of the transposon for each was mapped to an ORF in the unfinished M. smegmatis genome sequence, which corresponds to the tlyA gene (Rv1694) of the M. tuberculosis H37Rv sequence (10). One of the EZ::TN-kan capreomycin-resistant mutants was designated P2U and used in further studies.
Isolation of a capreomycin-resistant M. tuberculosis transposon mutant.
DNAs were extracted from each of the 64 pools of M. tuberculosis H37Rv EZ::TN transposon mutants and amplified by using a primer to the tlyA gene and a primer to the aph (Kanr) gene of the EZ::TN transposon. Two of the 64 pools produced PCR products of about 300 bp when a tlyA reverse primer and Tn903 forward primer were used and approximately 700 bp when a tlyA forward primer and Tn903 reverse primer were used. Capreomycin-resistant colonies were obtained from one of the two pools by plating on 7H10 plates containing 10 μg of capreomycin per ml. Amplification and sequencing of DNAs from the capreomycin-resistant colonies confirmed that the EZ::TN transposon was inserted in the tlyA gene and had inserted at base 644 (amino acid Ala). One of these mutants was designated 315-A and utilized for further study. The tlyA transposon mutant 315-A had a MIC for capreomycin of 20 μg/ml.
Complementation of tlyA transposon mutants.
Integrating plasmids were constructed that expressed either the wild-type M. smegmatis or M. tuberculosis tlyA ORF by using the inducible M. smegmatis acetamidase promoter (pMF1-tlyA/sm or pMF1-tlyA/tb, respectively). This promoter is induced approximately eightfold in the presence of 0.2% acetamide (25). The plasmid containing the appropriate tlyA ORF as well as the parent plasmid without the tlyA ORF (pMF1) were electroporated into either the EZ::TN-kan tlyA mutant M. smegmatis P2U or M. tuberculosis 315-A and plated on media containing hygromycin (Hyg agar). Hygromycin-resistant transformants were recovered, grown in 7H9-Hyg broth, and plated on Hyg agar, Hyg-acetamide agar, capreomycin (Cap agar), and Cap-acetamide agar. All strains grew equally well on Hyg agar with and without acetamide (Table 1). The M. smegmatis P2U and M. tuberculosis 315-A transposon mutants containing pMF1 grew on Cap agar and Cap-acetamide agar (Table 1). M. smegmatis P2U with pMF1-tlyA/sm and M. tuberculosis 315-A with pMF1-tlyA/tb grew on Cap agar (uninduced) but not on Cap-acetamide agar (induced) (Table 1), indicating that expression of the integrated wild-type tlyA gene restored capreomycin susceptibility.
TABLE 1.
Complementation of tlyA mutants
| Strain | Plasmid | Growth on 7H10 agara
|
|||
|---|---|---|---|---|---|
| Hygromycin (50 μg/ml)
|
Capreomycin (10 μg/ml)
|
||||
| Uninduced | Induced | Uninduced | Induced | ||
| M. smegmatis tlyA | pMF1 | + | + | + | + |
| M. smegmatis tlyA | pMF1-tlyA/sm | + | + | + | − |
| M. tuberculosis tlyA | pMF1 | + | + | + | + |
| M. tuberculosis tlyA | pMF1-tlyA/tb | + | + | + | − |
Growth was scored after 7 and 28 days for the M. smegmatis and M. tuberculosis strains, respectively, on 7H10 media containing the indicated antibiotic and no acetamide (uninduced) or 0.2% acetamide (induced) to induce expression of the plasmid-encoded tlyA gene. +, visible colonies, −, no growth.
Generation of spontaneous capreomycin-resistant mutants.
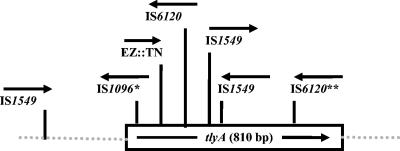
Spontaneous capreomycin-resistant M. smegmatis mutants were recovered at a frequency of 1.2 × 10−5. Amplification and sequencing of the tlyA gene of nine spontaneous M. smegmatis mutants revealed that a naturally occurring M. smegmatis transposon IS6120 (17), IS1549 (27), or IS1096 (9) had inserted into the tlyA gene in each of the nine mutants (Fig. 1). The insertion elements were found in both the forward and reverse direction in relation to the tlyA ORF. Spontaneous capreomycin-resistant mutants of M. tuberculosis were recovered at a frequency of 4.9 × 10−7 and 7.8 × 10−7 for H37Rv and CDC1551, respectively. The frequency of mutants in the Beijing strain F2 could not be determined accurately due to the slow growth of the mutants. Of the mutants studied, 10 of 11 H37Rv mutants, 11 of 11 CDC1551 mutants, and 7 of 8 Beijing strain F2 mutants contained changes within the tlyA gene (Table 2). Among these 28 tlyA mutations, 10 introduced a stop codon, 9 generated a frameshift, and 9 generated a missense mutation (Table 2). For the two isolates that contained wild-type tlyA genes, no mutations were found in the 500 bp upstream of the tlyA ORF. Furthermore, sequence analysis of all 30 M. tuberculosis mutants found that none had mutations in the region of the 16S rRNA gene (rrs) associated with resistance to aminoglycosides (32, 33). Ten M. tuberculosis tlyA spontaneous mutants representing missense, nonsense, and frameshift mutations (Table 2) were evaluated with a panel of 12 antituberculosis drugs including kanamycin and found to be susceptible to all 12 drugs and to have capreomycin MICs of 40 μg/ml (Table 2).
FIG. 1.
Insertion sites in spontaneous capreomycin-resistant M. smegmatis LR222 mutants. The arrows indicate the orientation of the IS1096, IS1549, or IS6120 insertion element in or near the tlyA ORF. *, two mutants with the same insertion element in the same insertion site; **, three separate mutants with the same insertion element in the same insertion site. The location and orientation of the EZ::TN transposon insertion are also noted.
TABLE 2.
tlyA mutations in spontaneous capreomycin-resistant M. tuberculosis mutants
| Straina | Mutation site (base no.) | Nucleotide change | Amino acid change | MIC (μg/ml)b
|
|
|---|---|---|---|---|---|
| CAP | VIO | ||||
| H37Rvc | |||||
| C-201 | 758 | ΔC | Frameshift | 40 | 20 |
| C-202 | 7 | C→T | Arg 3 Stop | 40 | 20 |
| C-203 | 397 | C insertion | Frameshift | 40 | 20 |
| C-206 | 555 | T→G | Phe 185→Leu | ||
| C-210 | 353 | T→C | Leu 118 Pro | 40 | 40 |
| C-211 | 64 | C→T | Gln 22 Stop | ||
| C-212 | 52 | C→T | Arg 18 Stop | ||
| C-213 | 383 | T→A | Val 128 Glu | ||
| C-214 | 26 | ΔC | Frameshift | ||
| C-215 | 200 | C→A | Ala 67 Glu | ||
| C-216 | None | N/A | N/A | ||
| CDC1551c | |||||
| C-302 | 550 | C→T | Gln 184 Stop | ||
| C-307 | 64 | C→T | Gln 22 Stop | ||
| C-308 | 586 | ΔG | Frameshift | 40 | 20 |
| C-309 | 550 | C→A | Gln 184→Lys | ||
| C-310 | 653 | ΔT | Frameshift | ||
| C-311 | 550 | C→T | Gln 184 Stop | ||
| C-312 | 64 | C→T | Gln 22 Stop | 40 | ≤10 |
| C-313 | 310 | ΔG | Frameshift | ||
| C-314 | 205 | A→G | Lys69→Glu | ||
| C-315 | 548 | C→T | Pro183→Leu | 40 | 20 |
| C-316 | 400 | ΔA | Frameshift | ||
| Beijing Fc | |||||
| C-501 | 272 | C→A | Ala91→Glu | 40 | ≤10 |
| C-502 | 712 | G→A | Glu238→Lys | ||
| C-503 | 673-674 | ΔGΔT | Frameshift | ||
| C-504 | 550 | C→T | Gln 184 Stop | 40 | ≤10 |
| C-505 | 477 | ΔG | Frameshift | 40 | ≤10 |
| C-507 | 550 | C→T | Gln 184 Stop | ||
| C-509 | 550 | C→T | Gln 184 Stop | ||
| C-510 | None | N/A | N/A | ||
Mutants were selected on 7H10 containing 10μg of capreomycin/ml.
MICs were determined for a missense, nonsense, and frameshift mutant from each of the three different parent strains.
All parent strains were susceptible to 10 μg of capreomycin/ml and 10 μg of viomycin/ml. CAP, capreomycin; VIO, viomycin; N/A, not applicable.
Mutations in capreomycin-resistant clinical isolates.
Eighteen capreomycin-resistant clinical isolates were plated on media containing 10 μg of capreomycin per ml, individual colonies were recovered, and their tlyA and 16S rRNA (rrs) genes were sequenced (Table 3). Four of the isolates, (1, 10, 21, and 22) had mutations in the tlyA gene but not the rrs gene (Table 3). Isolates 10 and 21 had a capreomycin MIC of 80 and 40 μg/ml, respectively, and were kanamycin susceptible. Isolates 1 and 22 had a capreomycin MIC of 80 μg/ml and a kanamycin MIC of 20 μg/ml. Isolate 11 contained a mutation in the tlyA gene and an A1401G mutation in the 16S rRNA gene and had higher MICs for both capreomycin (>160 μg/ml) and kanamycin (>80 μg/ml). Each of the remaining 13 isolates contained an A1401G mutation in the rrs gene and had various levels of capreomycin resistance and high-level kanamycin resistance (MIC of >80 μg/ml) (Table 3); none of the 13 had mutations in the 500 bp upstream of the tlyA ORF. The rrs A1401G mutation in these isolates corresponds to the A1400G mutation in M. tuberculosis and the A1408G mutation in E. coli which have been previously been observed in kanamycin-resistant strains (10, 27, 31, 32). Throughout this report this mutation will be referred to as A1401G in accordance with the revised numbering in the updated M. tuberculosis H37Rv complete genome, accession number NC_000962, National Center for Biotechnology Information (7).
TABLE 3.
Mutations and drug susceptibility in capreomycin-resistant M. tuberculosis clinical isolates
| Clinical isolate no. | Mutation
|
Drug susceptibilitya
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MIC (μg/ml)b
|
INH | RIF | EMB | STR | CIP | ETH | ||||||
| tlyA | rrs | CAP | KAN | VIO | AMK | |||||||
| 1 | C insert in 218L | None | 80 | 20 | 20 | ≤4 | R | R | R | R | S | S |
| 10 | N236K | None | 80 | ≤5 | 20 | ≤4 | R | S | S | Rc | S | S |
| 21 | L150P | None | 40 | ≤5 | ≤10 | ≤4 | R | R | S | R | S | S |
| 22 | ΔA codon 23 | None | 80 | 20 | 40 | ≤4 | S | R | R | R | S | S |
| 11 | R14W | A1401G | >160 | >80 | >80 | >64 | R | R | R | R | S | R |
| 2 | None | A1401G | 80 | >80 | ≤10 | >64 | R | R | R | R | S | S |
| 3 | None | A1401G | 80 | >80 | ≤10 | >64 | R | R | R | R | R | S |
| 4 | None | A1401G | 20 | >80 | ≤10 | >64 | R | R | R | R | S | S |
| 5 | None | A1401G | 20 | >80 | ≤10 | >64 | R | S | S | R | S | S |
| 7 | None | A1401G | 40 | >80 | ≤10 | >64 | R | R | R | R | S | S |
| 9 | None | A1401G | >160 | >80 | 20 | >64 | R | R | R | R | S | R |
| 12 | None | A1401G | >160 | >80 | ≤10 | >64 | R | R | R | R | S | S |
| 13 | None | A1401G | >160 | >80 | 40 | >64 | R | R | R | R | S | S |
| 14 | None | A1401G | >160 | >80 | 40 | >64 | R | S | R | R | S | R |
| 15 | None | A1401G | >160 | >80 | 20 | >64 | R | R | R | R | R | R |
| 16 | None | A1401G | 20 | >80 | ≤10 | >64 | R | R | R | R | S | S |
| 17 | None | A1401G | 40 | >80 | 20 | >64 | R | R | S | R | S | R |
| 18 | None | A1401G | 20 | >80 | 20 | >64 | R | R | R | R | S | R |
R, resistant; S, susceptible; CAP, capreomycin; KAN, kanamycin; VIO, viomycin; AMK, amikacin; INH, isoniazid (5 μg/ml); RIF, rifampin; EMB, ethambutol; STR, streptomycin (10μg/ml); CIP, ciprofloxacin; ETH, ethionamide.
Control strains were susceptible to 10 μg of CAP/ml, 5 μg KAN/ml, 10 μg of VI O/ml, and 4 μg of AMK/ml.
Resistant to 2 μg of streptomycin/ml.
Cross-resistance to viomycin.
M. smegmatis P2U and M. tuberculosis 315-A tlyA transposon mutants were resistant to viomycin at a concentration of 10 μg/ml. Susceptibility to viomycin at a concentration of 10 μg/ml was restored by complementation with pMF1-tlyA/sm or pMF1-tlyA/tb, respectively (the M. smegmatis and M. tuberculosis wild-type parent strains are susceptible to viomycin at a concentration of 10 μg/ml). Among the 10 spontaneous capreomycin-resistant mutants examined, resistance to viomycin was found in 6 of 7 of the tlyA spontaneous mutants obtained from H37Rv and CDC1551 (Table 2). However, the three spontaneous mutants tested that were obtained from the Beijing strain F2 were not resistant to viomycin (Table 2). Viomycin resistance was seen in three (1, 10, and 22) of the four clinical isolates that had only a tlyA mutation (Table 3).
Growth rate of tlyA mutant.
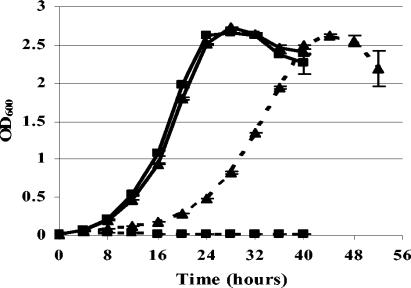
The generation times of M. smegmatis wild-type and tlyA mutant P2U were approximately 3.5 h in 7H9 broth with no drug (Fig. 2). When capreomycin was included in the media, the growth rate of the tlyA mutant P2U bacteria was significantly slower, with a generation time of approximately 6 h (Fig. 2).
FIG. 2.
Comparison of growth in the presence and absence of capreomycin. The growth of tlyA+ capreomycin-susceptible M. smegmatis bacteria (▪) was compared to that of tlyA mutant capreomycin-resistant M. smegmatis bacteria (▴). Each was grown in 7H9 alone (solid line) or 7H9-Cap (10 μg/ml) (dashed line). The experiment under each growth condition was performed in triplicate. The OD600 was determined every 4 h, up to 52 h. Results are presented as the mean OD600 ± SD. Similar results were observed in a second independent experiment.
In vitro transcription-translation assays.
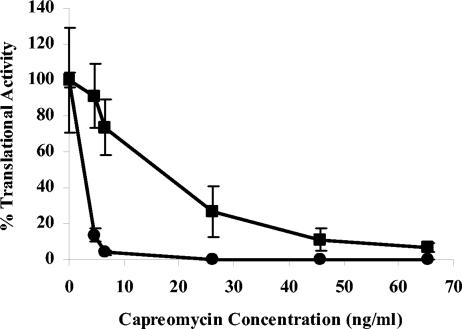
In vitro transcription-translation assays were done by using S30 extracts from M. smegmatis wild-type and tlyA mutant P2U, and transcription-translation activity was measured without capreomycin and in the presence of capreomycin at five concentrations ranging from 4.6 to 65.3 ng/ml (Fig. 3). In the presence of 4.6 ng of capreomycin per ml, transcriptional-translational activity of the wild-type S30 extracts was only 13.6% of the activity seen without capreomycin, whereas the P2U S30 had 91.1% of the activity seen with no drug. A capreomycin concentration of 65.3 ng/ml was required to reduce the activity of the S30 from the tlyA mutant P2U to 5% of the activity seen with no drug.
FIG. 3.
Inhibition of translation by capreomycin. Capreomycin was added to S30 extracts from tlyA+ capreomycin-susceptible M. smegmatis (•) and from tlyA mutant capreomycin-resistant M. smegmatis (▪) at final concentrations of 4.57 to 65.3 ng/ml. The production of luciferase activity in a coupled transcription-translation assay was measured in triplicate samples. Results are presented as the mean percent translational activity ± SD of the activity in the presence of drug to the activity in the absence of drug. Similar results were observed in a second independent experiment.
DISCUSSION
Although transposon mutagenesis cannot be used to identify essential genes, which are often the targets of drugs, transposon mutagenesis can be useful to identify genes whose products may alter the drug target to allow drug binding, may activate the drug, or may be involved in drug transport. In the work presented here, transposon mutagenesis was used to identify a gene, tlyA, that is not essential for survival but plays a role in capreomycin resistance in mycobacteria. Complementation of a capreomycin-resistant tlyA transposon mutant with tlyA expressed from an inducible promoter restored capreomycin susceptibility and confirmed the role of the tlyA gene product in capreomycin resistance.
The frequency of recovering spontaneous tlyA mutations in the M. smegmatis strain was approximately 100-fold higher than that in the M. tuberculosis strains. The tlyA mutations in the M. smegmatis mutants were due to the insertion of naturally occurring transposons into the tlyA ORF. However, the tlyA mutations found in the M. tuberculosis mutants were point mutations. A high level of transposon mobility has been reported in M. smegmatis but is relatively rare in M. tuberculosis (27), which could account for the higher frequency of mutation in M. smegmatis compared to M. tuberculosis.
Capreomycin is structurally similar to viomycin, which has been shown to bind to both the 30S and 50S ribosome subunits (42), to affect the dissociation of the 70S ribosome of M. smegmatis (44), and to inhibit ribosomal translocation by arresting peptidyl-tRNA in the ribosomal acceptor site (23). Furthermore, changes in either the 16S rRNA or 50S ribosomal subunit alters binding and confers resistance to viomycin in M. smegmatis (33, 44). Because capreomycin also interferes with ribosomal function (34) and resistance to it involves the ribosome (33, 42, 44), ribosomal proteins or RNA may be the target of capreomycin, and TlyA may act to alter this target.
To further define the role of TlyA in capreomycin resistance, in vitro transcription-translation assays using S30 extracts containing ribosomes from M. smegmatis wild type and the tlyA mutant were used. The resistance of the mutant extract to inhibition by capreomycin in these assays suggests that resistance is not due to the loss of a transporter for the uptake of capreomycin. Additionally, the dose response suggests that resistance is not due to a loss of function required to activate capreomycin. Rather, the results suggest that the loss of TlyA function has reduced the ability of capreomycin to interact with its target.
The translated product of the tlyA ORF was originally annotated as a hemolysin because extracts of E. coli that express the M. tuberculosis tlyA gene have hemolytic activity (39). More recent in silico data show that the TlyA ORF contains motifs that align well with domains predicted to be involved in rRNA methyltransferase reactions and RNA binding (3, 6), and the ORF displays homology with FtsJ/RrmJ (23S rRNA methylase) of E. coli (6). Furthermore, in silico research by Feder et al. (15) categorized TlyA as an rRNA methyltransferase, and the National Center for Biotechnology Information lists TlyA as a member of COG1189 (clusters of the orthologous groups of proteins), which is a group of predicted rRNA meth-ylases (http://www.ncbi.nlm.nih.gov/genomes/altik.cgi?db= G&gi = 135). Although the possible rRNA methyltransferase activity of TlyA has not been assessed experimentally, it is interesting that 9 of the 12 missense mutations seen in either the M. tuberculosis spontaneous capreomycin-resistant mutants or capreomycin-resistant clinical isolates map at or near the four highly conserved residues of the putative FtsJ/RrmJ catalytic tetrad (K-D-K-E) or in the region implicated in S-adenosylmethionine binding (5, 15, 18).
Given the homology of TlyA to rRNA methyltransferases (15), the reported ability of capreomycin to affect ribosomes (34), and the data presented here indicating that tlyA related capreomycin resistance involves the ribosome, it is possible that TlyA-mediated methylation of rRNA may be required for capreomycin susceptibility. This suggests that ribosomes lacking the methylation would be capreomycin resistant. This is an unusual mechanism, given that resistance to most drugs that interact with ribosomes has been associated with changes in rRNA sequences, acquisition of methylated bases, alterations in ribosomal proteins, or acquisition of activities that inactivate the drug (28, 30, 38). Although unusual, it is not unprecedented as resistance to kasugamycin, an aminoglycoside antibiotic, is due to the loss of a methylase, RsmA (also known as KsgA), which methylates bases 1518 and 1519 of the E. coli ribosomal 16S rRNA (16). Mutation of 16S rRNA at bases 794, 926, and 1519 can also confer kasugamycin resistance (37).
Because capreomycin is typically used clinically after failure of therapy with first-line drugs (4), capreomycin-resistant clinical isolates are usually resistant to several antituberculosis drugs including kanamycin. Cross-resistance between kanamycin and capreomycin was previously observed in a variable fraction of kanamycin-resistant M. tuberculosis isolates and strains (21, 36) though the molecular mechanism of this resistance was not known. More recent reports show that most isolates with high-level kanamycin resistance have mutations in the 16S rRNA gene, with more than 90% of the mutations being A1401G (designated A1400G in the publications) (11, 28, 32, 33). However, isolates with low-level kanamycin resistance usually do not have changes in the rrs gene, and the molecular basis of resistance is not known (28). Each of the 14 clinical isolates studied that contained the rrs A1401G mutation was also resistant to capreomycin; however, the capreomycin MICs varied from 20 to >160 μg/ml. Based on these data, it is tempting to speculate that the rrs A1401G mutation can confer resistance to both drugs, but it is difficult to explain how a single mutation can yield such a large variation in capreomycin MICs. Also, in a previously published report (1), the presence of the A1401G (A1400G in reference 1) mutation correlated with resistance to 10 μg of capreomycin per ml in only 6 of 12 clinical isolates with this mutation. A caveat with this study is that the susceptibility to capreomycin was determined in one of several submitting laboratories. The fact that all of the clinical isolates in both studies were MDR also limits the ability to draw a conclusion about the A1401G (A1400G) mutation and capreomycin resistance. If the 1401 rrs mutation is truly involved in capreomycin resistance and if TlyA is an rRNA methyltransferase, then a reasonable hypothesis is that capreomycin resistance in mycobacteria is due to the loss of the ability of capreomycin to bind to and inhibit ribosomes because of the lack of methylation of rRNA (tlyA mutation) or changes in the 16S rRNA (rrs mutation).
Overall, the results reported here demonstrate that disruption of the tlyA ORF can confer capreomycin resistance. This was the predominant means of resistance observed in laboratory-generated spontaneous capreomycin-resistant mutants and was also observed in four capreomycin-resistant clinical isolates. However, tlyA or rrs mutations were not found in two of the spontaneous mutants, and only the rrs A1401G mutation was found in 13 of the 18 capreomycin-resistant MDR clinical isolates examined. Further work is needed to elucidate the role of mutations in rrs in capreomycin resistance and to discover the basis of resistance in capreomycin-resistant strains that do not have mutations in tlyA or rrs.
Acknowledgments
This work was supported by CDC funds.
We thank Ray Butler, CDC, for technical assistance with the Parr bomb; Tony Romeo, Emory University, for advice on the in vitro transcription-translation assay and protocol for producing the S30 extracts; Beverly Metchock and David Sikes, CDC, for providing advice and clinical isolates; and Mark Fisher, National Institutes of Health, for plasmid pMF-1.
Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the Public Health Services, or the CDC.
REFERENCES
- 1.Alangaden, G. J., B. N. Kreiswirth, A. Aouad, M. Khetarpal, F. R. Igno, S. L. Moghazeh, E. K. Manavathu, and S. A. Lerner. 1998. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1295-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Algeorge, G., and A. Petre. 1970. Some experimental aspects of cross-resistance between capreomycin and viomycin. Antibiot. Chemother. 16:32-35. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 1999. Novel predicted RNA-binding domains associated with the translation machinery. J. Mol. Evol. 48:291-302. [DOI] [PubMed] [Google Scholar]
- 4.Blumberg, H. M., W. J. Burman, R. E. Chaisson, C. L. Daley, S. C. Etkind, L. N. Friedman, P. Fujiwara, M. Grzemska, P. C. Hopewell, M. D. Iseman, R. M. Jasmer, V. Koppaka, R. I. Menzies, R. J. O'Brien, R. R. Reves, L. B. Reichman, P. M. Simone, J. R. Starke, and A. A. Vernon. 2003. American Thoracic Society/Centers for Disease Control and Prevention/Infectious Diseases Society of America: treatment of tuberculosis. Am. J. Respir. Crit. Care Med. 167:603-662. [DOI] [PubMed] [Google Scholar]
- 5.Bugl, H., E. B. Fauman, B. L. Staker, F. Zheng, S. R. Kushner, M. A. Saper, J. C. Bardwell, and U. Jakob. 2000. RNA methylation under heat shock control. Mol. Cell 6:349-360. [DOI] [PubMed] [Google Scholar]
- 6.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 275:16414-16419. [DOI] [PubMed] [Google Scholar]
- 7.Camus, J. C., M. J. Pryor, C. Medigue, and S. T. Cole. 2002. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology 148:2967-2973. [DOI] [PubMed] [Google Scholar]
- 8.Choi, E. C., M. Misumi, T. Nishimura, N. Tanaka, S. Nomoto, T. Teshima, and T. Shiba. 1979. Viomycin resistance: alterations of either ribosomal subunit affect the binding of the antibiotic to the pair subunit and the entire ribosome becomes resistant to the drug. Biochem. Biophys. Res. Commun. 87:904-910. [DOI] [PubMed] [Google Scholar]
- 9.Cirillo, J. D., R. G. Barletta, B. R. Bloom, and W. R. Jacobs, Jr. 1991. A novel transposon trap for mycobacteria: isolation and characterization of IS1096. J. Bacteriol. 173:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 11.De Stasio, E. A., D. Moazed, H. F. Noller, and A. E. Dahlberg. 1989. Mutations in 16S ribosomal RNA disrupt antibiotic-RNA interactions. EMBO J. 8:1213-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dye, C., M. A. Espinal, C. J. Watt, C. Mbiaga, and B. G. Williams. 2002. Worldwide incidence of multidrug-resistant tuberculosis. J. Infect. Dis. 185:1197-1202. [DOI] [PubMed] [Google Scholar]
- 13.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 14.Farmer, P., and J. Y. Kim. 1998. Community based approaches to the control of multidrug resistant tuberculosis: introducing “DOTS-plus.” BMJ 317:671-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feder, M., J. Pas, L. S. Wyrwicz, and J. M. Bujnicki. 2003. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 302:129-138. [DOI] [PubMed] [Google Scholar]
- 16.Formenoy, L. J., P. R. Cunningham, K. Nurse, C. W. Pleij, and J. Ofengand. 1994. Methylation of the conserved A1518-A1519 in Escherichia coli 16S ribosomal RNA by the ksgA methyltransferase is influenced by methylations around the similarly conserved U1512. G1523 base pair in the 3′ terminal hairpin. Biochimie 76:1123-1128. [DOI] [PubMed] [Google Scholar]
- 17.Guilhot, C., B. Gicquel, J. Davies, and C. Martin. 1992. Isolation and analysis of IS6120, a new insertion sequence from Mycobacterium smegmatis. Mol. Microbiol. 6:107-113. [DOI] [PubMed] [Google Scholar]
- 18.Hager, J., B. L. Staker, H. Bugl, and U. Jakob. 2002. Active site in RrmJ, a heat shock-induced methyltransferase. J. Biol. Chem. 277:41978-41986. [DOI] [PubMed] [Google Scholar]
- 19.Kikuta-Oshima, L. C., F. D. Quinn, W. R. Butler, T. M. Shinnick, and C. H. King. 1995. Isolation of RNA from Mycobacterium tuberculosis using a nitrogen decompression chamber. BioTechniques 18:987-990. [PubMed] [Google Scholar]
- 20.Koseki, Y., and S. Okamoto. 1963. Studies on cross-resistance between capreomycin and certain other anti-mycobacterial agents. Jpn. J. Med. Sci. Biol. 16:31-38. [DOI] [PubMed] [Google Scholar]
- 21.McClatchy, J. K., W. Kanes, P. T. Davidson, and T. S. Moulding. 1977. Cross-resistance in M. tuberculosis to kanamycin, capreomycin and viomycin. Tubercle 58:29-34. [DOI] [PubMed] [Google Scholar]
- 22.Mizuguchi, Y., K. Suga, and T. Yamada. 1979. Interaction between 30 S ribosomal components in a viomycin-resistant mutant of Mycobacterium smegmatis. Microbiol. Immunol. 23:595-604. [DOI] [PubMed] [Google Scholar]
- 23.Modolell, J., and D. Vazquez. 1977. The inhibition of ribosomal translocation by viomycin. Eur. J. Biochem. 81:491-497. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical laboratory Standards. 2003. Susceptibility testing of Mycobacteria, Norcardiae, and other aerobic actinomycetes. Approved standard M24-A. National Committee for Clinical Laboratory Standards, Wayne, Pa. [PubMed]
- 25.Parish, T., E. Mahenthiralingam, P. Draper, E. O. Davis, and M. J. Colston. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143:2267-2276. [DOI] [PubMed] [Google Scholar]
- 26.Paunescu, E., M. Stoinescu, C. Zaharescu, and E. Dragusanu. 1970. Some correlations between chemical structure and mode of action of tuberculostatica. Researches on capreomycin and isoxyl. Antibiot. Chemother. 16:10-16. [DOI] [PubMed] [Google Scholar]
- 27.Plikaytis, B. B., J. T. Crawford, and T. M. Shinnick. 1998. IS1549 from Mycobacterium smegmatis forms long direct repeats upon insertion. J. Bacteriol. 180:1037-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sander, P., A. Meier, and E. C. Bottger. 1996. Ribosomal drug resistance in mycobacteria. Res. Microbiol. 147:59-67. [DOI] [PubMed] [Google Scholar]
- 31.Sutton, W. B., R. S. Gordee, W. E. Wick, and L. Stanfield. 1966. In vitro and in vivo laboratory studies on the antituberculous activity of capreomycin. Ann. N. Y. Acad. Sci. 135:947-959. [DOI] [PubMed] [Google Scholar]
- 32.Suzuki, Y., C. Katsukawa, A. Tamaru, C. Abe, M. Makino, Y. Mizuguchi, and H. Taniguchi. 1998. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J. Clin. Microbiol. 36:1220-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taniguchi, H., B. Chang, C. Abe, Y. Nikaido, Y. Mizuguchi, and S. I. Yoshida. 1997. Molecular analysis of kanamycin and viomycin resistance in Mycobacterium smegmatis by use of the conjugation system. J. Bacteriol. 179:4795-4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trnka, L., and D. W. Smith. 1970. Proteosynthetic activity of isolated ribosomes of mycobacteria and its alteration by rifampicin and related tuberculostatic drugs. Antibiot. Chemother. 16:369-379. [DOI] [PubMed] [Google Scholar]
- 35.Tsukamura, M. 1969. Cross-resistance relationships between capreomycin, kanamycin, and viomycin resistances in tubercle bacilli from patients. Am. Rev. Respir. Dis. 99:780-782. [DOI] [PubMed] [Google Scholar]
- 36.Tsukamura, M., and S. Mizuno. 1975. Cross-resistance relationships among the aminoglucoside antibiotics in Mycobacterium tuberculosis. J. Gen. Microbiol. 88:269-274. [DOI] [PubMed] [Google Scholar]
- 37.Vila-Sanjurjo, A., and A. E. Dahlberg. 2001. Mutational analysis of the conserved bases C1402 and A1500 in the center of the decoding domain of Escherichia coli 16 S rRNA reveals an important tertiary interaction. J. Mol. Biol. 308:457-463. [DOI] [PubMed] [Google Scholar]
- 38.Walsh, C. 2000. Molecular mechanisms that confer antibacterial drug resistance. Nature 406:775-781. [DOI] [PubMed] [Google Scholar]
- 39.Wren, B. W., R. A. Stabler, S. S. Das, P. D. Butcher, J. A. Mangan, J. D. Clarke, N. Casali, T. Parish, and N. G. Stoker. 1998. Characterization of a haemolysin from Mycobacterium tuberculosis with homology to a virulence factor of Serpulina hyodysenteriae. Microbiology 144:1205-1211. [DOI] [PubMed] [Google Scholar]
- 40.Yamada, T., K. Masuda, K. Shoji, and M. Hori. 1972. Analysis of ribosomes from viomycin-sensitive and -resistant strains of Mycobacterium smegmatis. J. Bacteriol. 112:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamada, T., Y. Mizugichi, K. H. Nierhaus, and H. G. Wittmann. 1978. Resistance to viomycin conferred by RNA of either ribosomal subunit. Nature 275:460-461. [DOI] [PubMed] [Google Scholar]
- 42.Yamada, T., Y. Mizuguchi, and K. Suga. 1976. Localization of co-resistance to streptomycin, kanamycin, capreomycin, and tuberactinomycin in core particles derived from ribosomes of viomycin-resistant Mycobacterium smegmatis. J. Antibiot. (Tokyo) 29:1124-1126. [DOI] [PubMed] [Google Scholar]
- 43.Yamada, T., A. Nagata, Y. Ono, Y. Suzuki, and T. Yamanouchi. 1985. Alteration of ribosomes and RNA polymerase in drug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 27:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, T., and K. H. Nierhaus. 1978. Viomycin favours the formation of 70S ribosome couples. Mol. Gen. Genet. 161:261-265. [DOI] [PubMed] [Google Scholar]
- 45.Zierski, M. 1969. Capreomycin and other drugs in the treatment of pulmonary tuberculosis. Tubercle 50(Suppl.):37-39. [PubMed] [Google Scholar]