Abstract
DEAD-box proteins are a class of nonprocessive RNA helicases that dynamically modulate the structure of RNA and ribonucleoprotein complexes (RNPs). However, the precise roles of individual members are not well understood. Work from our laboratory revealed that the DEAD-box protein Dbp2 in Saccharomyces cerevisiae is an active RNA helicase in vitro that functions in transcription by promoting mRNP assembly, repressing cryptic transcription initiation, and regulating long noncoding RNA activity. Interestingly, Dbp2 is also linked to glucose sensing and hexose transporter gene expression. DDX5 is the mammalian ortholog of Dbp2 that has been implicated in cancer and metabolic syndrome, suggesting that the role of Dbp2 and DDX5 in glucose metabolic regulation is conserved. Herein, we present a refined biochemical and biological comparison of yeast Dbp2 and human DDX5 enzymes. We find that human DDX5 possesses a 10-fold higher unwinding activity than Dbp2, which is partially due to the presence of a mammalian/avian specific C-terminal extension. Interestingly, ectopic expression of DDX5 rescues the cold sensitivity, cryptic initiation defects, and impaired glucose import in dbp2Δ cells, suggesting functional conservation. Consistently, we show that DDX5 promotes glucose uptake and glycolysis in mouse AML12 hepatocyte cells, suggesting that mammalian DDX5 and S. cerevisiae Dbp2 share conserved roles in cellular metabolism.
Keywords: DEAD-box, RNA, evolution, transcription, glucose metabolism
INTRODUCTION
RNA helicases resolve RNA secondary structures to remodel RNA and ribonucleoprotein complexes (RNPs) in an ATP-dependent manner (Jarmoskaite and Russell 2014). Some RNA helicases use the energy from ATP hydrolysis to translocate on the substrate and processively disrupt RNA duplexes, whereas DEAD-box protein family members do not, due to a lack of processivity (Jarmoskaite and Russell 2014). Instead, this class of enzymes utilizes ATP binding and hydrolysis to catalyze localized changes to RNA structure and/or RNA–protein interactions. This activity translates into a broad array of biochemically distinct roles from acting as RNA chaperones to serving as ATP-regulated platforms for formation of macromolecular complexes (Jarmoskaite and Russell 2014). Members of this class of enzymes share 12 common sequence motifs, which couple ATP binding and hydrolysis to RNA binding and enzyme release, respectively (Cordin et al. 2006). Seemingly subtle differences in the helicase core sequence and structure between family members, such as altered degrees of interdomain communication, have drastic impacts on both thermodynamic interactions and the kinetics of duplex unwinding (Garbelli et al. 2011; Samatanga and Klostermeier 2014). Amino and/or carboxy-terminal extensions can also influence enzymatic activity of individual members through various mechanisms, such as enhanced substrate selection, allosteric regulation, or nucleic acid tethering (Wang et al. 2006; Banroques et al. 2011; Mallam et al. 2011; Rudolph and Klostermeier 2015; Samatanga et al. 2017). Interestingly, 70% of DEAD-box protein genes in yeast are essential and provide nonredundant functions in vivo ranging from ribosome processing to translation and RNA decay (Rocak and Linder 2004). Moreover, orthologous DEAD-box proteins can be identified across species through phylogenetic searches, suggesting that specialized DEAD-box protein roles are conserved (Sharma and Jankowsky 2014). This is somewhat perplexing, however, because DEAD-box proteins lack sequence specificity in vitro (Sengoku et al. 2006; Schutz et al. 2010), leading to questions of how much conservation of function(s) truly exists between orthologous DEAD-box protein enzymes.
The DEAD-box protein Dbp2 is an active ATPase and RNA helicase in Saccharomyces cerevisiae (Cloutier et al. 2012; Ma et al. 2013, 2016). Dbp2 functions in gene regulation by promoting mRNP assembly, repressing cryptic intragenic transcription initiation, and by antagonizing long noncoding RNA (lncRNA) activity, indicative of a central role as a cotranscriptional RNA chaperone (Cloutier et al. 2012, 2013, 2016; Ma et al. 2013). This role appears to be specific for genes linked to energy homeostasis, as deletion of DBP2 results in aberrant expression of glucose metabolic genes in S. cerevisiae (Beck et al. 2014). Moreover, Dbp2 protein localization is responsive to glucose availability (Beck et al. 2014), suggesting that Dbp2 may integrate nutritional status with cellular metabolism.
DDX5 is the human ortholog of Dbp2, which is involved in organismal development and tissue differentiation (Iggo et al. 1991; Stevenson et al. 1998; Kitamura et al. 2001; Abdelhaleem 2005). Whereas DDX5 can unwind RNA duplexes in vitro in end-point assays (Hirling et al. 1989; Rossler et al. 2001; Huang and Liu 2002), refined kinetic and thermodynamic analyses have not been performed to date. Despite this, extensive studies have established that this enzyme functions in multiple aspects of RNA metabolism including pre-mRNA splicing (Zonta et al. 2013), alternative splicing (Kar et al. 2011), microRNA processing (Salzman et al. 2007), and ribosomal RNA processing (Jalal et al. 2007). DDX5 also binds to several transcription factors and acts as a transcriptional cofactor to activate or repress transcription (Fuller-Pace 2013). Overexpression of DDX5 has also been frequently correlated with disease states, especially cancers, with overexpression in breast, colon, prostate cancers, and acute myeloid leukemia (Fuller-Pace 2013; Mazurek et al. 2014). However, the underlying molecular basis for DDX5 in promoting uncontrolled cell growth is not well understood.
Using a combination of biochemical, biophysical, and genetic approaches, we now provide a comprehensive characterization of DDX5 and comparison with Dbp2. Interestingly, we find that the mammalian/avian specific carboxy-terminal extension increases the in vitro unwinding activity of DDX5. Despite this enhanced enzymatic activity, DDX5 fully complements the role of DBP2 in yeast growth, transcriptional fidelity, and glucose import. Finally, we show that DDX5 promotes glucose uptake and glycolysis in mammalian cells, a process that is up-regulated in cancers (Vander Heiden et al. 2009), suggesting a novel entry point for future targeting of cancer-specific metabolism.
RESULTS
DDX5 encodes a mammalian/avian-specific carboxy-terminal extension (CTE)
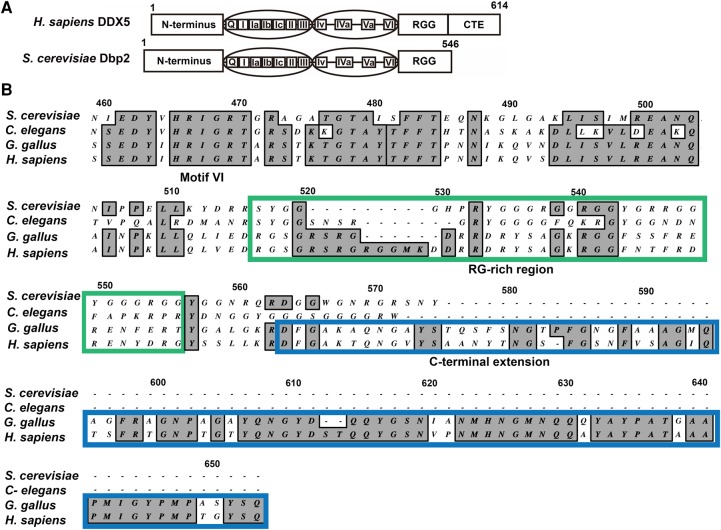
DBP2 was identified as the S. cerevisiae homolog of DDX5 in 1991 through low stringency hybridization screening (Iggo et al. 1991). Human DDX5 and S. cerevisiae Dbp2 share 55% sequence identity and 70% similarity (Fig. 1A). Amino acid alignment of S. cerevisiae Dbp2, C. elegans DDX17, Gallus gallus DDX5, and human DDX5 shows conservation of an arginine and glycine rich (RG-rich or RGG) region in these orthologs (Fig. 1B). RG-rich regions are found in many DEAD-box proteins (Yang and Jankowsky 2005) and are implicated in RNA binding, protein–protein interactions, and/or protein localization (Thandapani et al. 2013). The carboxy termini of G. gallus and human DDX5 share a carboxy-terminal extension (CTE), which is not present in S. cerevisiae or C. elegans, suggesting addition of this accessory domain after divergence of these lineages.
FIGURE 1.
DDX5/Dbp2 orthologs share common DEAD-box core motifs but differ at the carboxy termini. (A) Schematic of human DDX5 and S. cerevisiae Dbp2 proteins. The amino terminus, 12 DEAD-box protein motifs, and RG-rich (or RGG) region are conserved. DDX5 has a carboxy-terminal extension (CTE) that is not conserved with Dbp2. DDX5 and Dbp2 protein sequences have 55% identity and 70% similarity. (B) The CTE is specific to mammalian/avian DDX5. Amino acid alignment of the carboxy terminal ends of Saccharomyces cerevisiae Dbp2 (NP_014287), Caenorhabditis elegans DDX17 (NP_001041134), Gallus gallus DDX5 (NP_990158), and Homo sapiens DDX5 (NP_004387), from motif VI through the CTE. Multiple sequence alignment was performed using the ClustalW alignment tool (Thompson et al. 1994) with 40 open gap penalty in MacVector 11.1.2. Numbers correspond to the alignment, not the amino acid sequences of Dbp2/DDX5. The RG-rich region is highlighted with a green box. The CTE of DDX5 is marked with a blue box.
DDX5 is a more active RNA helicase in vitro than Dbp2
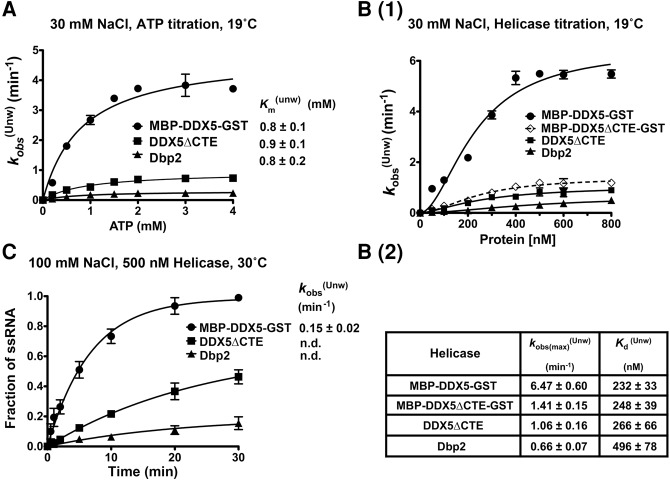
Dbp2 is a bona fide RNA helicase and ATPase in vitro (Cloutier et al. 2012; Ma et al. 2013), with an unwinding rate constant of ∼0.22 min−1 on a 16-bp blunt-end RNA duplex (Ma et al. 2013). This unwinding rate is in the range of other DEAD-box proteins analyzed in vitro (Yang et al. 2007; Mallam et al. 2014). Human DDX5, on the other hand, has only been assayed for unwinding in end-point assays (Hirling et al. 1989; Rossler et al. 2001; Huang and Liu 2002). To directly compare the enzymatic activities of human DDX5 with S. cerevisiae Dbp2 and to determine whether the presence of a CTE impacts these activities, we conducted a series of in vitro assays with recombinant purified DDX5, DDX5ΔCTE, and Dbp2. Note that DDX5 contains an amino-terminal MBP-tag and a carboxy-terminal GST-tag, which were found to be essential for protein solubility and stability, respectively (data not shown). Unwinding assays were performed using a 16-bp blunt-end RNA duplex at “low-salt” (30 mM NaCl) conditions to parallel prior studies of Dbp2 (Ma et al. 2013). We first titrated ATP in the helicase unwinding reactions for MBP-DDX5-GST, DDX5ΔCTE, and Dbp2. This revealed that all three enzymes share similar functional Km’s for ATP (i.e., Km(Unw)) (Fig. 2A), suggesting similar ATP binding affinities. We then titrated in the enzymes in the presence of saturating (2 mM) ATP and found that MBP-DDX5-GST unwinds the RNA duplex faster than Dbp2, as evidenced by a ∼10-fold higher maximum observed unwinding rate (6.47 min−1 versus 0.66 min−1) [Fig. 2B(1)]. In addition, MBP-DDX5-GST has a lower functional Kd of unwinding (i.e., Kd(Unw)) [Fig. 2B(2)], indicating that the RNA substrate is saturated by MBP-DDX5-GST at lower enzyme concentrations as compared to Dbp2. Interestingly, deletion of CTE (DDX5ΔCTE) reduced the unwinding rate of DDX5 by sixfold to 1.06 min−1 without appreciably altering the functional Kd (Fig. 2B). Enhanced unwinding activity of DDX5 when compared with DDX5ΔCTE is not due to the tags, because MBP-DDX5ΔCTE-GST displayed similar unwinding rates to untagged DDX5ΔCTE in vitro (Fig. 2B). This suggests that the CTE itself confers a higher rate of duplex unwinding to DDX5.
FIGURE 2.
The CTE confers higher duplex unwinding activity to human DDX5 over S. cerevisiae Dbp2. (A) MBP-DDX5-GST, DDX5ΔCTE, and Dbp2 have identical functional Km’s for ATP in unwinding assays. RNA unwinding assays were conducted using 300 nM recombinant, purified proteins and the indicated amount of ATP/MgCl2 in the presence of 30 mM NaCl at 19°C. The curves for observed unwinding rates versus ATP were fitted to a binding isotherm: Y = Kobs(max) × X/Km(unw) + X. (B) DDX5 shows higher unwinding activity than DDX5ΔCTE or Dbp2 in “low-salt” conditions. (1) Helicase unwinding assays were conducted at 19°C in the presence of 30 mM NaCl with indicated concentrations of purified recombinant MBP-DDX5-GST, DDX5ΔCTE, Dbp2, or MBP-DDX5ΔCTE-GST. (2) Table of maximum observed unwinding rates and the functional dissociation constants. First, the observed rates (kobs(Unw)) were determined by plotting the fraction of single-stranded (ss) RNA against reaction time and fitting the unwinding curves to the following equation: Y = Ymax × [1 − exp(−kobs(Unw) × X)]. To calculate the maximum observed rates (Kobs(max)(Unw)) and functional dissociation constants (Kd(Unw)), we plotted the observed rates as a function of the protein concentration and fitted the data to a sigmoidal binding isotherm [Y = Kobs(max)(Unw) × Xh/(Kd(Unw)h + Xh)] to generate functional binding curves. The Hill coefficients for these enzymes range from 1.5 to 1.8. (C) DDX5 is an active helicase in “near-physiological salt” conditions. Unwinding assays using 500 nM purified recombinant MBP-DDX5-GST, DDX5ΔCTE, or Dbp2 were performed in the presence of 100 mM NaCl at 30°C. The observed rates were determined as above. n.d., means not determined, when unwinding curves were not convergent. Data above show the mean ± standard deviation (SD) of 2–3 independent replicates.
Given the robust activity of MBP-DDX5-GST in “low-salt” conditions, we tested its unwinding activities in “near-physiological salt” conditions (100 mM NaCl) (Pasic et al. 2001). MBP-DDX5-GST is active (kobs(Unw) ∼0.15 min−1) in the presence of 100 mM NaCl, while DDX5ΔCTE and Dbp2 displayed reduced activities (Fig. 2C). The latter is evidenced by the nonconvergent unwinding curves of DDX5ΔCTE and Dbp2 (Fig. 2C). This indicates that the duplex unwinding activity of DDX5 is more tolerant to high salt. Together, this shows that DDX5 unwinds a blunt-end RNA duplex faster than Dbp2, and that the CTE contributes to higher activity.
DDX5 lacks RNA annealing activity
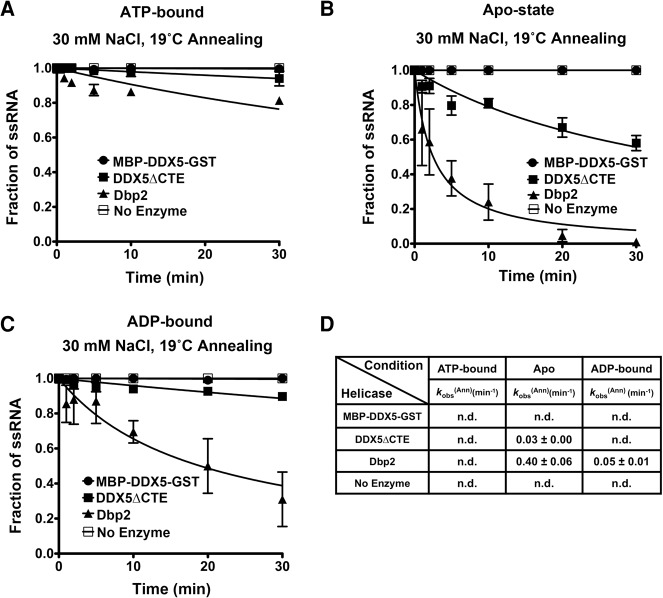
DEAD-box helicases also facilitate duplex annealing, an activity proposed to assist with RNA chaperone activity in vivo (Jarmoskaite and Russell 2014; Ruminski et al. 2016). To know whether DDX5, DDX5ΔCTE, and Dbp2 have different annealing activities, we performed annealing assays with the single-stranded components of the 16-bp duplex. These assays were conducted in the presence of ATP, in the absence of ATP (Apo), or in the presence of ADP. Spontaneous annealing of the substrate was not observed in the absence of enzymes under any conditions (Fig. 3). In agreement with previous studies (Ma et al. 2013), we observed minimal annealing (<10%) in the presence of Dbp2 and ATP (Fig. 3A,D). However, Dbp2 displayed appreciable annealing activity in both the Apo and ADP-bound states with observed annealing rates of ∼0.4 min−1 and ∼0.05 min−1, respectively (Fig. 3B–D). In contrast, MBP-DDX5-GST did not facilitate strand annealing at any nucleotide state (Fig. 3A–D), suggesting that DDX5 lacks annealing activity all together. Removal of the CTE from DDX5 restores some annealing activity in the Apo state (kobs(Ann) ∼0.03 min−1) (Fig. 3B,D), but not to levels equal to Dbp2. This suggests that subtle changes in the core in addition to occurrence of the CTE have altered the biochemical parameters of Dbp2 from fungi to mammals.
FIGURE 3.
DDX5 lacks annealing activity in vitro, irrespective of the nucleotide-binding state, and this is partially due to the CTE. (A) DDX5 lacks annealing activity in the presence of ATP. Duplex annealing assays were conducted as above, but with 0.1 nM radiolabeled 16-nt top strand RNA and 0.1 nM cold complementary RNA in the presence of 30 mM NaCl and 2 mM ATP/MgCl2. (B,C) DDX5 also lacks annealing in the absence of nucleotide or in the presence of ADP, in contrast to Dbp2. Duplex annealing assays using purified recombinant MBP-DDX5-GST, DDX5ΔCTE, Dbp2, or no enzyme, in the absence of ATP (Apo state) (B) or in the presence of 2 mM ADP/MgCl2 (C). (D) Table of the observed annealing rates of DDX5, DDX5ΔCTE, Dbp2, and the no enzyme control in the presence or absence of different nucleotides. The observed annealing rates were determined using the following equation: Y = 1/(1 + kobsAnn × X). n.d., not determined. Data are shown as the mean ± SD of 2–3 independent replicates.
DDX5 has higher RNA-binding affinity than Dbp2
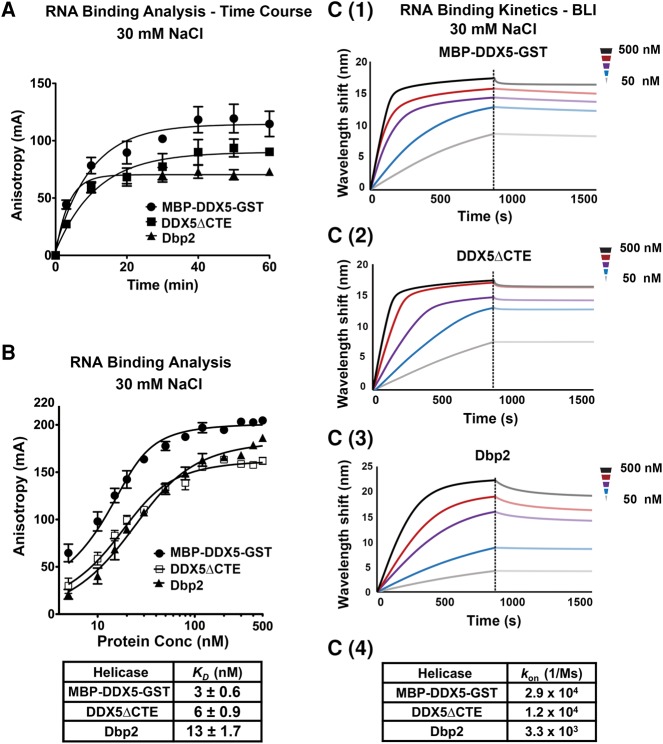
DEAD-box RNA helicases are ATP-dependent RNA-binding proteins (Jarmoskaite and Russell 2014). To further characterize the biochemical parameters of DDX5, DDX5ΔCTE, and Dbp2, we conducted fluorescence anisotropy experiments to measure the RNA-binding affinities using a 6-carboxyfluorescein (6-FAM) labeled 16-nt ssRNA and the nonhydrolyzable ATP analog, AMPPNP. Because DEAD-box proteins release bound RNA upon ATP hydrolysis, AMPPNP “locks” these enzymes onto the RNA to enable measurements of binding affinities (Liu et al. 2014; Samatanga and Klostermeier 2014). Time courses were performed to ensure that the binding reactions reached equilibrium for calculation of dissociation constants (KD’s) (Fig. 4A). Changes in anisotropy were then measured across a range of protein concentrations, revealing that all three proteins bind ssRNA within a similar range, but with full-length DDX5 (MBP-DDX5-GST) binding slightly (approximately fourfold) tighter than Dbp2 (Fig. 4B). This is in line with the approximately twofold higher Kd(Unw) of Dbp2 [Fig. 2B(1–2)]. Similarly, DDX5ΔCTE bound ssRNA with a KD between that of DDX5 and Dbp2 (Fig. 4A), suggesting that the CTE only moderately impacts the RNA-binding affinity of DDX5. We then utilized bio-layer interferometry to analyze the RNA-binding kinetics of MBP-DDX5-GST, DDX5ΔCTE, and Dbp2 by monitoring the association and dissociation of these enzymes to a biotinylated 16-nt ssRNA. Consistent with high ssRNA binding affinities, this revealed faster on rates for full-length DDX5, followed by DDX5ΔCTE, and then Dbp2 (Fig. 4C).
FIGURE 4.
Comparison of RNA-binding affinities and RNA on rates reveals tighter RNA binding by DDX5. (A) RNA-binding reactions reach equilibrium within 60 min. Fluorescence anisotropy assays were conducted using 20 nM indicated proteins, 10 nM 5′-6-FAM labeled 16-nt ssRNA, and AMPPNP. Anisotropy measurements (λex = 495 nm/λem = 520 nm) were taken using a Biotek Synergy 4 plate reader at indicated time points. (B) DDX5, DDX5ΔCTE, and Dbp2 bind ssRNA within a similar range, but with full-length DDX5 binding slightly tighter than DDX5ΔCTE and Dbp2. Fluorescence anisotropy assays were conducted using increasing amounts of MBP-DDX5-GST, DDX5ΔCTE, or Dbp2, 10 nM 16-nt ssRNA, and AMPPNP. Binding reactions were incubated for 60 min before taking anisotropy measurements. Binding data were fitted to the quadratic equation for two site binding: Y = Bmin + (Bmax − Bmin) × ((X + 2[RNA] + KD) − ((X + 2[RNA] + KD)2−8 × X × [RNA])0.5)/4. Data show the mean ± SD of three independent replicates. (C) DDX5 and DDX5ΔCTE show higher on rates than Dbp2 for ssRNA. Bio-layer interferometry (BLI) experiments were conducted to determine the binding kinetics of MBP-DDX5-GST (C1), DDX5ΔCTE (C2), and Dbp2 (C3) on a biotinylated 16-nt ssRNA in the presence of the nonhydrolyzable ATP analog, AMPPNP. Data are shown in background-corrected sensograms depicting the association and dissociation of proteins onto the ssRNA across a range of concentrations. The on-rates of MBP-DDX5-GST, DDX5ΔCTE, and Dbp2 were determined by fitting the real-time binding data with the 2:1 model using the Data Analysis Software (Pall Fortebio) by local fitting (C4). Off rates cannot be determined from this experiment due to the slow disassembly of DEAD-box helicase-AMPPNP–RNA complexes (Liu et al. 2014).
Ectopic expression of human DDX5 complements dbp2Δ growth and transcriptional fidelity defects in S. cerevisiae
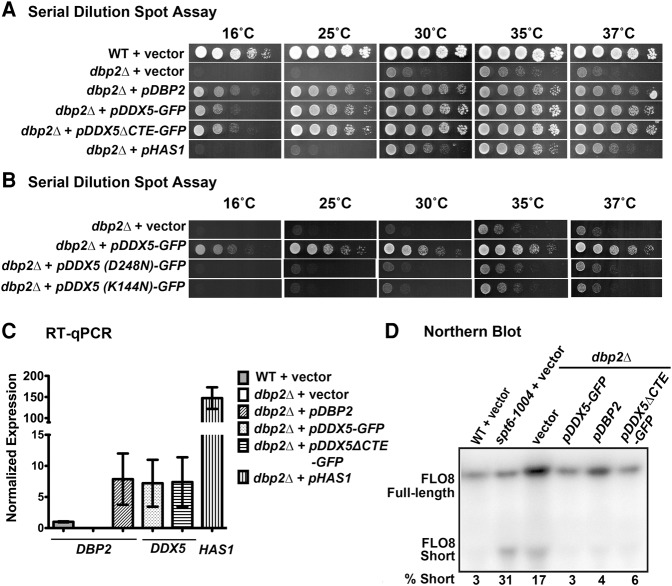
Dbp2 functions in ribosome biogenesis, repression of cryptic transcriptional initiation, and lncRNA-dependent gene regulation in S. cerevisiae (Bond et al. 2001; Cloutier et al. 2012, 2013, 2016). To ask whether DDX5 is functionally equivalent to Dbp2 despite different biochemical parameters, we asked whether ectopic expression of human DDX5 and/or DDX5ΔCTE could rescue the slow growth and cold sensitivity of dbp2Δ cells. To this end, we cloned DDX5 and DDX5ΔCTE into CEN yeast expression vectors under the control of pADH1 promoters, to enable expression at levels near endogenous DBP2. To aid in protein stability (data not shown), we added a carboxy-terminal GFP tag to both DDX5 and DDX5ΔCTE constructs. We then conducted serial dilution analysis of wild-type and dbp2Δ cells harboring either an empty vector or the CEN plasmids with DBP2, DDX5-GFP, or DDX5ΔCTE-GFP (Fig. 5A). We also included dbp2Δ cells harboring a high copy 2μ plasmid expressing HAS1, a DEAD-box protein involved in ribosome biogenesis in S. cerevisiae (Emery et al. 2004), and the sole isolate of a saturating multicopy suppressor screen of dbp2Δ cells. This revealed that DDX5 and DDX5ΔCTE rescue both the slow growth and cold sensitivity defects of dbp2Δ to levels similar to plasmid-based expression of DBP2, whereas HAS1 only partially rescues dbp2Δ cells even when vastly overexpressed (Fig. 5A,C). The expression levels of DDX5 and DDX5ΔCTE are higher than endogenous DBP2 but are similar to plasmid-based expression of DBP2 (Fig. 5C). This shows that DDX5 or DDX5ΔCTE specifically complements dbp2Δ growth defects.
FIGURE 5.
Ectopic expression of human DDX5 or DDX5ΔCTE rescues the growth and cryptic transcription defects of dbp2Δ in S. cerevisiae. (A) Ectopic expression of DDX5-GFP or DDX5ΔCTE-GFP fully rescues dbp2Δ cold sensitivity and slow growth defects. Wild-type (WT) cells transformed with empty vector or dbp2Δ cells transformed with vector only, CEN plasmids expressing DBP2, DDX5-GFP, or DDX5ΔCTE-GFP, or a 2µ HAS1 plasmid were spotted in fivefold serial dilutions on selective media and grown at respective temperatures. HAS1 was isolated from a saturating suppressor screen of dbp2Δ cold sensitivity and was the sole isolate of the screen. HAS1 is a DEAD-box protein gene whose product functions in ribosome biogenesis (Emery et al. 2004). (B) The enzymatic activities of DDX5 are required for growth complementation of dbp2Δ. Serial dilution assays were conducted as above with dbp2Δ cells transformed with vector and CEN-plasmid expressing DDX5-GFP, DDX5(D248N)-GFP, and DDX5(K144N)-GFP. DDX5(K144N) and DDX5(D248N) have mutations in the Walker A and Walker B motifs, respectively, which affect ATP binding and hydrolysis (Cordin et al. 2006), and are shown to be required for the ATPase activities of DDX5 (Zhang et al. 2016). (C) RT-qPCR of DBP2, DDX5, or HAS1 genes in strains above shows similar expression of pDBP2, pDDX5-GFP, and pDDX5ΔCTE-GFP and overexpression of HAS1. Transcript levels were normalized to ACT1 with DBP2 expression in the wild type + vector strain set to one. Data show the mean ± SD of three independent biological replicates. (D) Ectopic expression of DDX5-GFP or DDX5ΔCTE-GFP rescues the cryptic transcription initiation defect in dbp2Δ cells. Northern blotting of endogenous FLO8 transcripts was performed as previously described (Cloutier et al. 2012). Full-length (2.4 kb) and the short, cryptic (0.8 kb) FLO8 transcripts were visualized by autoradiography and quantified by densitometry. (% Short) Percentage of short transcript versus total FLO8 transcripts (Full length + Short).
Next, we tested the ability of enzymatically deficient DDX5 mutants to rescue dbp2Δ growth defects. ATPase-deficient mutants DDX5(K144N)-GFP and DDX5(D248N)-GFP (Zhang et al. 2016) harboring amino acid substitutions within Motif I (Walker A motif) or Motif II (Walker B motif), respectively, were expressed on CEN plasmids in dbp2Δ cells. The growth of these two strains was compared with dbp2Δ cells transformed with empty vector or wild-type DDX5-GFP in serial dilution spot assays. Neither of the DDX5 mutants rescued the growth defects of dbp2Δ (Fig. 5B), suggesting that the enzymatic activities of DDX5 are required for complementation.
To determine whether ectopic expression of human DDX5 rescues dbp2Δ defects in transcriptional fidelity, we asked if expression of DDX5 and/or DDX5ΔCTE could repress cryptic intragenic initiation at the FLO8 locus (Cloutier et al. 2012). The FLO8 gene is transcribed into a single, full-length transcript (2.4 kb) in wild-type cells (Fig. 5D). In the elongation factor mutant strain spt6-1004 or dbp2Δ cells, however, a short FLO8 transcript (∼0.8 kb) is also transcribed due to usage of a cryptic TATA site within the open reading frame (Kaplan et al. 2003; Cloutier et al. 2012). Consistent with prior studies, we observed an increase in the amount of the short transcript expressed in spt6-1004 or dbp2Δ cells harboring vector alone as compared with wild-type cells (Fig. 5D). In line with our observed growth rescue above, ectopic expression of DBP2, DDX5-GFP, or DDX5ΔCTE-GFP repressed the cryptic transcription in dbp2Δ cells, as evidenced by the decrease in the percentage of short transcript (Fig. 5D). This suggests that DDX5 functions in transcriptional fidelity similar to Dbp2.
DBP2 and DDX5 are required for efficient glucose import in yeast and mammalian cells, respectively
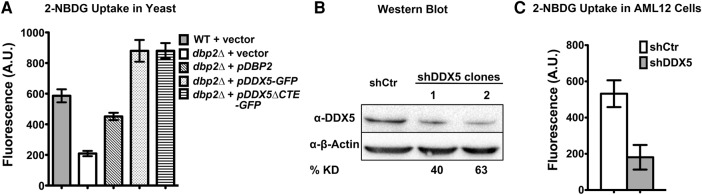
Glucose import is the first and rate-limiting step of glycolysis (Hatanaka 1974), a central pathway in primary metabolism for energy production and homeostasis. In cells lacking DBP2, hexose transporter genes (HXTs) and associated lncRNAs are misregulated, suggesting that Dbp2 may control glucose import (Beck et al. 2014). To test this, we measured the ability of wild-type and dbp2Δ cells to import the fluorescent glucose analog 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) (Zhang et al. 2015). Consistent with HXT gene misregulation, we observed an approximately threefold decrease in 2-NBDG import in dbp2Δ cells when compared with wild type, a defect that was rescued by ectopic expression of DBP2 (Fig. 6A). Next, we asked whether this import defect could be rescued by ectopic expression of human DDX5 or DDX5ΔCTE. Strikingly, expression of DDX5 or DDX5ΔCTE not only rescued the import defect of dbp2Δ cells, but also stimulated import above the levels of wild-type cells (∼50% increase, Fig. 6A). Although we do not know the nature of stimulated glucose import, it is possible that this increase is directly related to the increased helicase activity of the human DDX5 variants when compared with Dbp2.
FIGURE 6.
Both Dbp2 and DDX5 promote glucose import in S. cerevisiae and mammalian cells, respectively. (A) Ectopic expression of DDX5 or DDX5ΔCTE rescues glucose import defects in dbp2Δ cells. Glucose import was determined using 2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose (2-NBDG) uptake assays (Blodgett et al. 2011), which measure the fluorescence intensity of 2-NBDG in cells after lysis. A.U., arbitrary units. Data show the mean ± SD of three independent biological replicates. (B) Western blots show shRNA knockdown levels of DDX5 in mouse hepatocytes (AML12). Western blotting of DDX5 or β-actin was performed from cell lysates from two independent AML12 clones stably expressing a shRNA targeting DDX5 (shDDX5) or a nontargeting control shRNA (shCtr). The percent of knockdown of DDX5 was quantified by densitometry relative to β-actin loading control. (C) DDX5 promotes glucose uptake in AML12 cells. 2-NBDG uptake assays in mouse AML12 cells expressing shDDX5 or shCtr. Cells were incubated at 37°C with fully supplemented culture media containing 100 µM 2-NBDG for 30 min. 2-NBDG uptake was measured as above.
To determine whether DDX5 promotes glucose import in mammalian cells, we knocked down DDX5 in a nontransformed mouse hepatocyte cell line (AML12) using a small hairpin RNA targeting DDX5 mRNA (shDDX5). This resulted in ∼50% knockdown of DDX5 protein levels in two independent clones stably expressing the shDDX5 (Fig. 6B). We then conducted 2-NBDG uptake assays in AML12 cells expressing the nontargeting control shRNA (shCtr) or shDDX5. Interestingly, DDX5 knockdown reduced 2-NBDG import to ∼30% of the wild-type levels (Fig. 6C), suggesting that Dbp2 and DDX5 share conserved roles in promoting glucose import in S. cerevisiae and mammalian cells. This is surprising given the distinct modes of glucose metabolism between fungi and animal cells (see Discussion).
Knockdown of DDX5 decreases glycolysis and increases respiration in mammalian cells
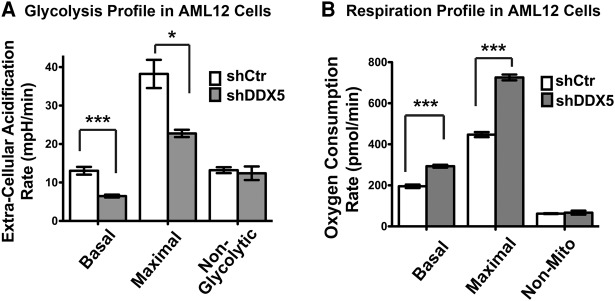
To determine whether DDX5 promotes cellular metabolism, we analyzed the rates of basal glycolysis, maximal glycolytic capacity, and nonglycolytic acidification by measuring extracellular acidification rates (Chen et al. 2016). This reveals that DDX5 knockdown in AML12 cells down-regulates both basal and maximal glycolysis rates, whereas nonglycolytic acidification is not affected (Fig. 7A). Importantly, these changes are comparable to down-regulation of established metabolic regulators in hepatocytes, such as the circadian regulator Bmal1 (Cheng et al. 2015; Jacobi et al. 2015). Next, we measured respiration by determining the oxygen consumption rates at basal and maximal levels, as well as the nonmitochondrial oxygen consumption rate. Interestingly, this reveals that the basal and maximal oxygen consumption rates are up-regulated in cells expressing shDDX5 (Fig. 7B). This up-regulation is presumably due to the need to balance energy production, due to reduced glycolytic activity. Thus, DDX5 promotes glycolysis in mammalian cells, consistent with promoting glucose import as the first step of glycolysis. Taken together, this suggests that both Dbp2 and DDX5 promote glucose metabolism in yeast and mammalian cells.
FIGURE 7.
DDX5 promotes glycolysis in mammalian cells. (A) Metabolic profiles of AML12 mouse hepatocytes with DDX5 knockdown reveals defects in glycolytic capacity. The glycolysis profiles of cells expressing shDDX5 or shCtr were analyzed using the Seahorse XFe24 Analyzer as previously described (Chen et al. 2016). Extracellular acidification rate, which correlates with the rate of glycolysis, was measured following addition of compounds that trigger various stages of glycolysis. The first injection is glucose that induces glycolysis. The second injection is oligomycin, an ATP synthase inhibitor that abolishes mitochondrial ATP production and thus shifts the energy production to glycolysis (Hao et al. 2010), revealing the maximum glycolytic capacity. The final injection is 2-deoxy-d-glucose, which competitively inhibits glucose hexokinase and blocks glycolysis (Brown 1962). The basal, maximal, and nonglycolytic acidification rates were determined using the Seahorse XF Stress Test Report Generator (Seahorse Bioscience). (B) DDX5 knockdown increases respiration in AML12 cells. The respiration profile of AML12 cells was analyzed by measuring the oxygen consumption rates at basal levels and following injection of compounds that target the electron transport chain. Oligomycin decreases basal respiration (Hao et al. 2010). FCCP (carbonyl cyanide-p-(trifluoromethoxy) phenylhydrazone) is an uncoupling agent that dissipates proton gradient (Park et al. 2002); as a result, oxygen is maximally consumed. The third injection is rotenone and antimycin A, which shuts down mitochondrial respiration and enables the calculation of nonmitochondrial respiration. Respiration rates were determined using the Seahorse XF Stress Test Report Generator. All data are shown as the mean ± SD of three independent biological replicates. (*) P-value <0.05, (***) P-value <0.001.
DISCUSSION
DEAD-box RNA helicases are the largest group of RNA helicases in all forms of life (Jarmoskaite and Russell 2014). Despite this, the precise biochemical roles of individual DEAD-box proteins and whether these roles are conserved across organisms are largely unknown. We have provided a comprehensive comparison of the biochemical and biological roles between two DEAD-box helicase orthologs and show that mammalian DDX5 and S. cerevisiae Dbp2 are functionally conserved, despite enzymatic differences. We also provide evidence demonstrating conserved roles for Dbp2/DDX5 in glucose metabolism, revealing a unique biological function for a DEAD-box helicase family branch.
Human DDX5 (or p68) was one of the first members of the DEAD-box helicase family to be shown to have duplex unwinding activity in vitro (Hirling et al. 1989). Despite the ∼30 yr since this study, there has been no kinetic analysis of duplex unwinding by DDX5. Instead, end-point assays using different RNA substrates have provided conflicting reports of processivity and activity for this enzyme (Rossler et al. 2001; Huang and Liu 2002). Our studies provide a kinetic and thermodynamic analysis of DDX5 along with comparison to its yeast counterpart. These studies now show that DDX5 and Dbp2 have subtle differences in enzymatic activities. First, DDX5 is a more active RNA helicase than Dbp2, even at near-physiological salt concentrations. Second, DDX5 lacks annealing activity all together. Finally, the mammalian/avian-specific CTE of DDX5 contributes to these activities.
While we do not know the precise role of the CTE, one possibility is that the CTE alters the conformation of the helicase core and/or RNA substrate, rendering the full-length DDX5 more productive in unwinding cycles. For example, the CTE may help DDX5 adopt a conformation in the Apo and ADP-bound states that is incompatible with stable binding of an A-form RNA duplex, decreasing annealing activity during a round of unwinding. Consistent with this notion, there are major differences in the spatial organizations of the two RecA-like domains among different DEAD-box proteins. For example, the RecA-like domains of eIF4A exhibit an “open” conformation due to a lack of interdomain interactions. (Caruthers et al. 2000), whereas Dhh1 is more compact with extensive interactions between the two domains, even in the absence of nucleotide (Cheng et al. 2005). This results in very different biochemical activities between these two DEAD-box helicases (Dutta et al. 2011). The CTE may also modulate the conformation of the bound RNA. Structural studies of the S. cerevisiae DEAD-box protein Mss116 reveal that the carboxy-terminal extension of Mss116 induces a bend at the 5′ end of a bound RNA (Del Campo and Lambowitz 2009). Such a bend may block strand annealing. Although there are no obvious structural similarities between the CTEs of Mss116 and DDX5, the CTE of Mss116 has α-helical structure (Del Campo and Lambowitz 2009), whereas the CTE of DDX5 is predicted to be unstructured; these differences do not preclude analogous roles for these accessory domains in modulating enzymatic activity.
Despite the differences between yeast Dbp2 and human DDX5 in vitro, ectopic expression of either DDX5 or DDX5ΔCTE fully complements dbp2Δ defects including slow growth, cold sensitivity, cryptic transcription, and impaired glucose import. This suggests that DDX5 may function in transcription through mechanisms similar to Dbp2, including regulating lncRNA activities or facilitating mRNP assembly. In fact, DDX5 has been linked to lncRNA-regulated gene expression in several recent studies (Huang et al. 2015; Wongtrakoongate et al. 2015; Zhang et al. 2016). Moreover, the DDX5 ortholog in Drosophila melanogaster facilitates RNA release from the chromatin and nuclear export, two processes requiring efficient mRNP assembly (Buszczak and Spradling 2006). These roles are not mutually exclusive as both lncRNAs and mRNAs are transcribed, processed, and packaged by similar cellular machineries in vivo (Quinn and Chang 2016).
On the surface, the ability of DDX5 to fully rescue dbp2Δ is surprising because S. cerevisiae and humans diverged about 1 billion years ago (Douzery et al. 2004). Dbp2 works in concert with the protein cofactor Yra1 in vivo (Cloutier et al. 2012; Ma et al. 2013), an interaction that seems to be conserved in mammalian cells (Zonta et al. 2013). This suggests that certain protein–protein and/or protein–RNA interfaces may be preserved during the course of evolution. Consistently, a large-scale screen reveals that 200 human genes out of 424 tested complement their yeast orthologs in growth analyses (Kachroo et al. 2015). Eight DEAD-box protein genes were included in this screen, among which the ribosome biogenesis factors FAL1 and DBP6 (Kressler et al. 1997, 1998) were complemented by their human orthologs EIF4A3 and DDX51. In addition, DDX3X rescues the growth defects of a Schizosaccharomyces pombe strain lacking its ortholog ded1 (Epling et al. 2015), which functions in translation initiation (Iost et al. 1999; Senissar et al. 2014). This suggests that orthologous complementation may be a feature of DEAD-box protein genes.
Our studies show that both Dbp2 and DDX5 promote glycolysis in yeast and mammalian cells, respectively, indicating that both of these enzymes regulate common mRNAs and/or lncRNAs linked to cellular metabolism. This is further supported by the fact that genes involved in glycolysis are specifically misregulated upon DDX5 knockdown in acute myeloid leukemia cells (Mazurek et al. 2014) and documentation of a DDX5 polymorphism (S480A) linked to metabolic syndrome (Guo et al. 2010). The conserved role in glucose metabolism is surprising, since fungi and mammalian cells have distinct modes of glucose metabolism. After glucose is catabolized to pyruvate, yeast cells undergo ethanol fermentation to convert pyruvate to ethanol and CO2. This allows yeast to use glycolysis and fermentation as their only energy production source when glucose is present (Barnett 2003). In contrast, most mammalian cells use respiration to convert pyruvate to acetyl-CoA, which enters the Krebs cycle and feeds electrons to oxidative phosphorylation (Berg et al. 2002). Although the fate of pyruvate is different between yeast and mammalian cells, the upstream steps of glycolysis are conserved. For example, both require sugar transporters to import glucose into the cell, the first and rate-limiting step of glucose metabolism (Hatanaka 1974; Ozcan and Johnston 1999).
Both fungi and mammals encode numerous high and low affinity glucose transporters whose expression and activity are controlled by the extracellular concentrations of specific sugars (Ozcan and Johnston 1999; Mueckler and Thorens 2013). Our studies show that both Dbp2 and DDX5 promote glucose uptake, consistent with a specific role in promoting the first step of glycolysis. This is also consistent with prior studies showing misregulation of hexose transporter genes (HXTs) and associated lncRNAs in the absence of DBP2 in S. cerevisiae (Beck et al. 2014). It is possible that DDX5 regulates the analogous glucose transporters (GLUTs) similarly in mammalian cells, genes that are also up-regulated in cancers (Macheda et al. 2005). Cancer cells frequently acquire glucose dependence due to high levels of glycolytic activity (i.e., Warburg effect) (Vander Heiden et al. 2009). This “cancer cell metabolism” fulfills the need for massive macromolecule synthesis of fast-growing cells while reducing mitochondria-dependent apoptosis (Vander Heiden et al. 2009). Thus, inhibition of DDX5 may be a novel way to target cancer metabolism in the future.
MATERIALS AND METHODS
Plasmids and oligonucleotides are listed in Table 1 and Table 2. Yeast strains are listed in Table 3. Quantitative (q) PCR primers are listed in Table 4.
TABLE 1.
Yeast and bacterial plasmids
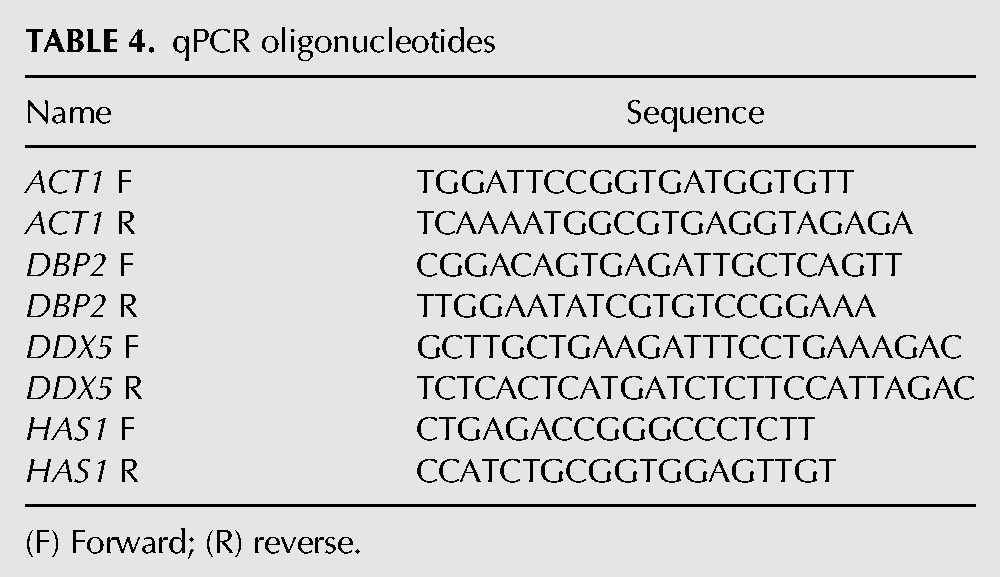
TABLE 2.
PCR oligonucleotides
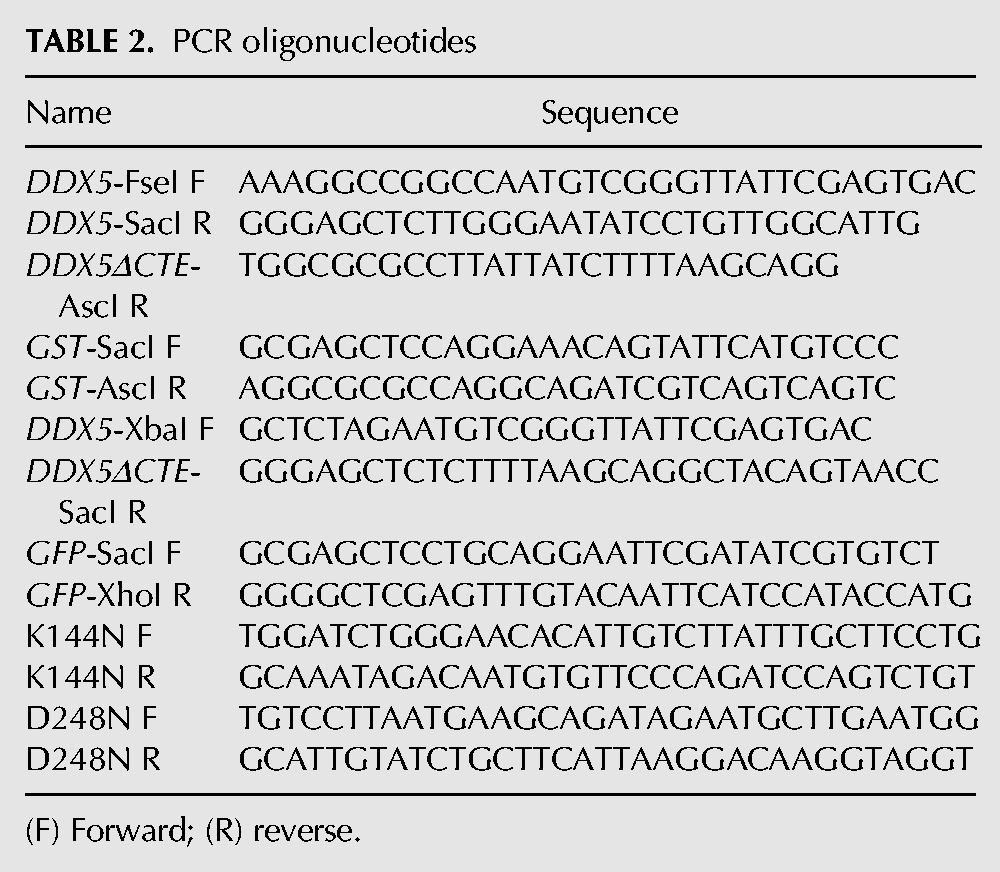
TABLE 3.
Yeast strains
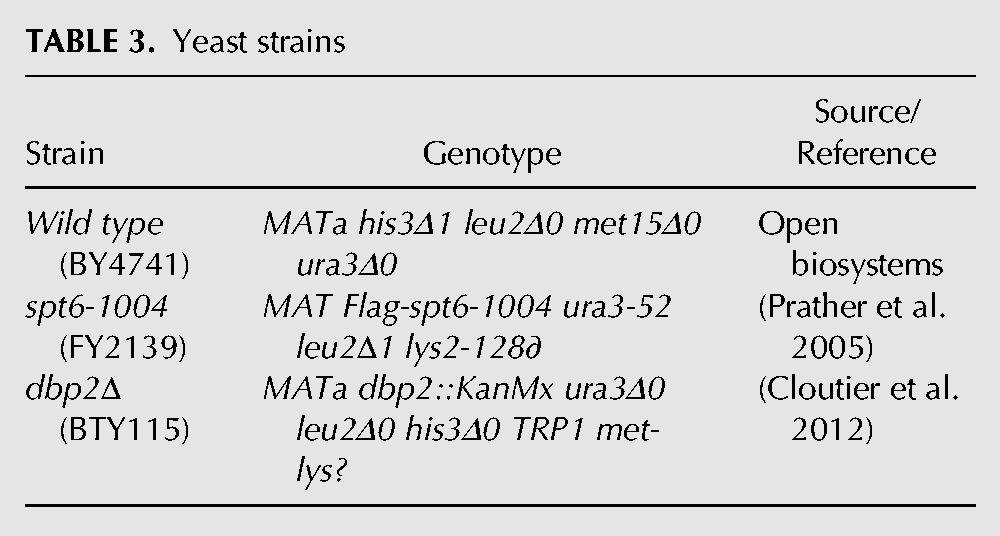
TABLE 4.
qPCR oligonucleotides
Plasmids and cloning
The fully sequenced human DDX5 (Gene ID: 1655) cDNA was obtained from the Mammalian Gene Collection (MGC) (Dharmacon, clone ID: 3528578); DDX5 was subcloned using PCR primers with a 5′ FseI site (DDX5-FseI F) and a 3′ SacI site (DDX5-SacI R) leaving out the stop codon. GST was subcloned from the pGEX-5X-3 vector using GST-SacI F and GST-AscI R primers, with the reverse primer adding in two extra stop codons. The resulting DDX5 and GST coding sequences were subcloned in place of DBP2 in the pMAL-TEV-DBP2 (Ma et al. 2013) to construct pMAL-TEV-DDX5-GST. Expression of human DDX5 in E. coli without the carboxy-terminal tag resulted in a truncated DDX5 lacking the last 90 amino acids, a region that coincides with the CTE (data not shown). We used this to inform us on construction of the DDX5ΔCTE, which was then constructed using PCR primers DDX5-FseI F and DDX5ΔCTE-AscI R (or DDX5ΔCTE-SacI R) to correspond to the truncated protein.
To construct yeast expression plasmids, DDX5 and DDX5ΔCTE were subcloned into DBP2-PL-ADH-p415 using bacterial expression vectors as templates and primers DDX5-XbaI F and DDX5-SacI R, or DDX5-XbaI F and DDX5ΔCTE-SacI R. GFP was then subcloned from pCP-MS2-GFP using primers GFP-SacI F and GFP-XhoI R. Site-directed mutagenesis of p415-DDX5-GFP was conducted using mutation primer pairs K144N F and K144N R, or D248N F and D248N R.
Recombinant protein expression and purification
pMAL-TEV-DBP2 was expressed and Dbp2 was purified as previously described (Ma et al. 2013). Expression of pMAL-TEV-DDX5-GST or pMAL-TEV-DDX5ΔCTE-GST in Rosetta E. coli (DE3) cells (Novagen) was induced using 0.2 mM IPTG at 16°C overnight. Cells were lysed in buffer containing 50 mM HEPES (pH 7.5) and 50 mM NaCl. MBP-DDX5-GST was purified from the soluble lysate using glutathione resin (GE healthcare) followed by cation-exchange chromatography using SP sepharose (Sigma-Aldrich), and eluted with elution buffer 1 (50 mM Tris–HCl [pH 8.0], 300 mM NaCl, 20% glycerol). Full-length DDX5 is not soluble when expressed with an amino-terminal His tag, and undergoes proteolysis without a carboxy-terminal tag (data not shown). MBP-DDX5ΔCTE-GST was purified the same way but eluted with elution buffer 2 (50 mM Tris-HCl [pH 8.0], 500 mM NaCl, 20% glycerol). pMAL-TEV-DDX5ΔCTE was expressed and purified as above, except that amylose resin (NEB) was used for affinity chromatography. The MBP-tag was cleaved from MBP-DDX5ΔCTE by overnight digestion with Tev protease (Life Technologies) at 16°C. DDX5ΔCTE was then purified using SP sepharose and eluted with elution buffer 2. The protein concentrations were measured using Bradford Assays according to the manufacturer's instructions (Thermo Scientific, 1856209). All proteins were stored at −80°C until use without freeze and thaw cycles.
Helicase unwinding assays
The unwinding assays of purified recombinant MBP-DDX5-GST, MBP-DDX5ΔCTE-GST, DDX5ΔCTE, and Dbp2 were conducted as described using the same 16-bp blunt-end RNA duplex (Ma et al. 2013). The top strand (5′-AGCACCGUAAAGACGC-3′) (IDT) of the duplex was 5′ radiolabeled and annealed to an unlabeled bottom strand (5′-GCGUCUUUACGGUGCU-3′) (IDT) (Jankowsky and Putnam 2010). Reactions were conducted in 60 µL mixtures containing 40 mM Tris-HCl (pH 8.0), 2.5 mM MgCl2, 2 mM DTT, 120 U Superase-in (Life Technologies), 0.1 nM RNA duplex. “Low-salt” condition contains 30 mM NaCl and indicated concentrations of protein. “Near-physiological salt” condition contains 100 mM NaCl and 500 nM protein. The reactions were preincubated at either 19°C (low salt) or 30°C (near-physiological) for 10 min prior to addition of 2 mM or indicated concentrations of ATP/MgCl2. Aliquots were taken at the indicated time points and resolved on 10% native polyacrylamide gels for phosphor-imaging. To determine the observed unwinding rates, the fractions of single-stranded RNAs at each time point were quantified and fitted to the following equation: Y = Ymax × (1 − exp(−kobs(Unw) × X)). In the enzyme titration experiment, Kobs(Unw) determined as above were plotted as a function of the enzyme concentration and fitted using the sigmoidal equation: Y = Kobs(max)(Unw) × Xh/(Kd(Unw)h + Xh) to yield the functional binding curves. The ATP titration curves were fitted to the following equation: Y = Kobs(max) × X/Km(unw) + X.
Annealing assays
RNA annealing assays were conducted similarly to helicase unwinding assays, as described previously (Ma et al. 2013). Briefly, the RNA duplex used in the unwinding assay was denatured at 95°C for 2 min to generate ssRNA substrates for annealing assays. Annealing assays were performed in the “low-salt” condition above with no nucleotide, 2 mM ATP/MgCl2 or 2 mM ADP/MgCl2 as indicated. The observed annealing rates were determined using the following equation: Y = 1/(1 + kobs(Ann) × X).
Fluorescence anisotropy assays
Fluorescence anisotropy assays were performed in 40 μL reactions containing 40 mM Tris–HCl (pH 8.0), 2.5 mM MgCl2, 2 mM DTT, 40 U Superase-in (Life Technologies), 30 mM NaCl, 2 mM AMPPNP, 10 nM 16-nt fluorescent RNA oligonucleotide (5′-6-FAM-AGCACCGUAAAGACGC-3′) (IDT), and varying amounts of purified proteins. The reaction mixtures were incubated at room temperature for 60 min in black, half-area 96-well microplates (Corning) to reach equilibrium. The anisotropy signals of 6-FAM (λex = 495 nm and λem = 520 nm) were measured using a Biotek Synergy 4 plate reader, and fitted to the quadratic equation for two analytes:1 ligand binding model using Prism: Y = Bmin + (Bmax − Bmin) × ((X + 2[RNA] + KD) − ((X + 2[RNA] + KD)2 − 8 × X × [RNA])0.5)/4.
Bio-layer interferometry
The on rates of purified recombinant MBP-DDX5-GST, DDX5ΔCTE, and Dbp2 were analyzed using an Octet Red384 instrument (Pall Fortebio) at 30°C. The High Precision Streptavidin Biosensors (Pall ForeBio) were equilibrated in the reaction buffer containing 40 mM Tris–HCl (pH 8.0), 2.5 mM MgCl2, 2 mM DTT, 40 U Superase-in, 2 mM AMPPNP, and 30 mM NaCl for 60 sec, loaded with a 16-nt biotinylated ssRNA (5′-biotin-AGCACCGUAAAGACGC-3′) for 120 sec, and equilibrated again in reaction buffer for 120 sec. The association and dissociation steps were performed by dipping the loaded sensors in wells containing increasing amounts of purified proteins for 800 sec, and then transferring them to wells containing reaction buffer for 800 sec. The binding kinetics were calculated using the ForteBio Data Analysis software (Pall Fortebio). The on rates were determined by locally fitting the association data to a 2:1 (2 analytes:1 ligand) model.
Multicopy suppressor screening of dbp2Δ growth defects
HAS1 was isolated by screening a 2μ S. cerevisiae genomic library (AB320, ATCC 37323) (Kaelin et al. 1992) for genes that suppress the slow growth of dbp2Δ at 25°C. This resulted in six suppressors. Rescuing plasmids were isolated from colonies and subjected to sequencing using primers YEp13 F (5′-TGCTCGCTTCGCTACTTGGA-3′) and YEp13 R (5′-ATACCCACGCCGAAACAAGC-3′). Plasmids containing HAS1 were isolated from five of the six plasmids, suggesting saturation of the screen.
Yeast growth analysis
Serial dilution assays were conducted as previously described (Cloutier et al. 2012). Yeast strains were grown in SC-LEU+2% glucose to mid-log phase and spotted in fivefold serial dilutions onto SC-LEU+2% glucose agar plates, and incubated at 16°C, 25°C, 30°C, 35°C, or 37°C until wild-type cells were fully grown.
RT-qPCR analysis
RT-qPCR analysis was conducted as previously described (Cloutier et al. 2012). Transcript levels were normalized to ACT1 transcript levels, and the relative expression level of DBP2 in WT + vector strain was arbitrarily set to one. The qPCR primers used are listed in Table 4.
Northern blot
Yeast strains were grown in SC-LEU+2% glucose to mid-log phase, and RNA was isolated using acid–phenol extraction. Thirty micrograms of total RNA was resolved on a 1.2% formaldehyde–agarose gel and transferred to a nylon membrane (Invitrogen) overnight. Northern blotting was conducted as previously described (Cloutier et al. 2012). The targeted transcripts were visualized using PhosphoImager (GE healthcare) and quantified by densitometry using ImageQuant (GE healthcare).
Mammalian cell culture and shRNA knockdown
Mouse hepatocyte AML12 cells were grown in DMEM/F12 media (Gibco) supplemented with 10% FBS, 1× Penicillin–Streptomycin (Life Technologies), 0.4× Insulin–Transferrin–Selenium–Ethanolamine (ITS-X) (Life Technologies). To knock down the mouse Ddx5 (Gene ID: 13207) gene expression, the GIPZ DDX5 shRNA (Dharmacon) was transfected into AML12 cells using the Lipofectamine 2000 (Thermo Fisher) according to the manufacturer's instructions. Stable expression of shDDX5 in AML12 cells was maintained by 4 µg/mL puromycin (Thermo Fisher). Western blotting analyses were conducted as previously described (Zhang et al. 2016) to confirm knockdown using a DDX5 (05-850, Millipore) and β-actin (A5441, Sigma-Aldrich) primary antibodies.
2-NBDG uptake assay in yeast and AML12 cells
2-NBDG [2-(N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)-2-deoxyglucose, Thermo Fisher #N13195] uptake assays were conducted as previously described (Blodgett et al. 2011; Zhang et al. 2015) with some modifications. Yeast strains were grown to log phase in 25-mL cultures and shifted to YP+0.1% glucose+100 µM 2-NBDG for 1 h at 30°C. After 2-NBDG uptake, cells were washed with 1× Tris-EDTA (TE) buffer, and lysed using zymolyase and sonication in 200 µL lysis buffer containing 20 mM Tris (pH 7.4), 1% sodium deoxycholate, and 40 mM KCl (Blodgett et al. 2011). 2-NBDG levels were determined by fluorescence of 2-NBDG (λex = 465 nm, λem = 540 nm) in the resulting cell lysate after normalization to cell density. AML12 cells were plated at 4 × 105 cells per well in a six-well culture the day prior to the experiment. Cells were washed twice in PBS and incubated with the uptake media (growth media+100 µM 2-NBDG) for 30 min at 37°C in a standard CO2 incubator. Media were aspirated and cells were trypsin digested and counted. Cells were then collected by centrifugation and lysed by sonication with 400 µL lysis buffer containing 20 mM Tris (pH 7.4), 1% sodium deoxycholate, and 40 mM KCl. Fluorescence intensity of 2-NBDG in the cell lysate was measured (λex = 465 nm, λem = 540 nm), and normalized to cell count. Data show the mean ± SD of three independent biological replicates.
Glycolysis and respiration analysis
AML12 cells were seeded in a 24-well microplate (Seahorse Bioscience) at 20,000 cells per well and incubated in a standard 37°C CO2 incubator overnight. Prior to the experiment, cells were washed twice with Assay Media (Seahorse Bioscience) and transferred to a 37°C non-CO2 incubator with 575 µL Assay Media/well for 1 h. The glycolysis profile was analyzed by measuring the extracellular acidification rates at basal level and following injections of various compounds that trigger different states of glycolysis using the Glycolysis Test Kit (Seahorse Bioscience). The first injection is glucose to induce glycolysis under basal conditions. The second injection is oligomycin, an ATP synthase inhibitor that shifts the energy production to glycolysis and reveals glycolytic capacity (Hao et al. 2010). The final injection is 2-deoxy-d-glucose, a glucose analog that competitively inhibits glucose hexokinase and shuts down glycolysis (Brown 1962). The mitochondrial profile was analyzed by measuring the oxygen consumption rates at basal level and after injections of drugs that each target a component of the electron transfer chain using the Mito Stress Test Kit (Seahorse Bioscience). First, oligomycin inhibits ATP synthase and decreases basal oxygen consumption, showing that oxygen consumption measured in this experiment is coupled with ATP synthesis. FCCP [carbonyl cyanide-p-(trifluoromethoxy) phenylhydrazone] is an uncoupling agent that allows unlimited electron flow through the electron transfer chain (Park et al. 2002) and reveals maximal oxygen consumption. The third injection is a mix of rotenone, a complex I inhibitor, and antimycin A, a complex III inhibitor. This combination shuts down mitochondrial respiration and enables the measurement of nonmitochondrial respiration. The metabolic rates at different stages of glycolysis and mitochondrial respiration were determined using the Seahorse XF Stress Test Report Generator (Seahorse Bioscience).
ACKNOWLEDGMENTS
We thank the Andrisani laboratory (Purdue University) for providing the AML12 cell line and Xin Liu from the Fierke laboratory (University of Michigan) and the Briggs laboratory (Purdue University) for assistance with fluorescence anisotropy assays. We also thank members from the Tran laboratory for helpful discussions. This work was supported by National Institutes of Health R01GM097332 to E.J.T., P30CA023168 for core facilities at the Purdue University Center for Cancer Research, and an American Heart Association Predoctoral Fellowship 16PRE31170005 to Z.X.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.060335.116.
REFERENCES
- Abdelhaleem M. 2005. RNA helicases: regulators of differentiation. Clin Biochem 38: 499–503. [DOI] [PubMed] [Google Scholar]
- Banroques J, Cordin O, Doere M, Linder P, Tanner NK. 2008. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol 28: 3359–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banroques J, Cordin O, Doere M, Linder P, Tanner NK. 2011. Analyses of the functional regions of DEAD-box RNA “helicases” with deletion and chimera constructs tested in vivo and in vitro. J Mol Biol 413: 451–472. [DOI] [PubMed] [Google Scholar]
- Barnett JA. 2003. A history of research on yeasts 5: the fermentation pathway. Yeast 20: 509–543. [DOI] [PubMed] [Google Scholar]
- Beck ZT, Cloutier SC, Schipma MJ, Petell CJ, Ma WK, Tran EJ. 2014. Regulation of glucose-dependent gene expression by the RNA helicase Dbp2 in Saccharomyces cerevisiae. Genetics 198: 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. 2002. The glycolytic pathway is tightly controlled. In Biochemistry, 5th ed, Section 16.2 WH Freeman, New York. [Google Scholar]
- Blodgett AB, Kothinti RK, Kamyshko I, Petering DH, Kumar S, Tabatabai NM. 2011. A fluorescence method for measurement of glucose transport in kidney cells. Diabetes Technol Ther 13: 743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond AT, Mangus DA, He F, Jacobson A. 2001. Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol Cell Biol 21: 7366–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. 1962. Effects of 2-deoxyglucose on carbohydrate metabolism: review of the literature and studies in the rat. Metabolism 11: 1098–1112. [PubMed] [Google Scholar]
- Buszczak M, Spradling AC. 2006. The Drosophila P68 RNA helicase regulates transcriptional deactivation by promoting RNA release from chromatin. Genes Dev 20: 977–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB. 2000. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci 97: 13080–13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Ahmad N, Liu X. 2016. Combining p53 stabilizers with metformin induces synergistic apoptosis through regulation of energy metabolism in castration-resistant prostate cancer. Cell Cycle 15: 840–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Coller J, Parker R, Song H. 2005. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA 11: 1258–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y-S, Seibert O, Kloting N, Dietrich A, Strassburger K, Fernandez-Veledo S, Vendrell JJ, Zorzano A, Bluher M, Herzig S, et al. 2015. PPP2R5C couples hepatic glucose and lipid homeostasis. PLoS Genet 11: e1005561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110: 119–122. [DOI] [PubMed] [Google Scholar]
- Cloutier SC, Ma WK, Nguyen LT, Tran EJ. 2012. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J Biol Chem 287: 26155–26166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Petell CJ, Tran EJ. 2013. Long noncoding RNAs promote transcriptional poising of inducible genes. PLoS Biol 11: e1001715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Al Husini N, Dhoondia Z, Ansari A, Pascuzzi PE, Tran EJ. 2016. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol Cell 61: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. 2006. The DEAD-box protein family of RNA helicases. Gene 367: 17–37. [DOI] [PubMed] [Google Scholar]
- Del Campo M, Lambowitz AM. 2009. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell 35: 598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douzery EJP, Snell EA, Bapteste E, Delsuc F, Philippe H. 2004. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci 101: 15386–15291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta A, Zheng S, Jain D, Cameron CE, Reese JC. 2011. Intermolecular interactions within the abundant DEAD-box protein Dhh1 regulate its activity in vivo. J Biol Chem 286: 27454–27470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery B, de la Cruz J, Rocak S, Deloche O, Linder P. 2004. Has1p, a member of the DEAD-box family, is required for 40S ribosomal subunit biogenesis in Saccharomyces cerevisiae. Mol Microbiol 52: 141–158. [DOI] [PubMed] [Google Scholar]
- Epling LB, Grace CR, Lowe BR, Partridge JF, Enemark EJ. 2015. Cancer-associated mutants of RNA helicase DDX3X are defective in RNA-stimulated ATP hydrolysis. J Mol Biol 427: 1779–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller-Pace FV. 2013. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta 1829: 756–763. [DOI] [PubMed] [Google Scholar]
- Garbelli A, Beermann S, Di Cicco G, Dietrich U, Maga G. 2011. A motif unique to the human DEAD-box protein DDX3 is important for nucleic acid binding, ATP hydrolysis, RNA/DNA unwinding and HIV-1 replication. PLoS One 6: e19810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hong F, Loke J, Yea S, Lim CL, Lee U, Mann DA, Walsh MJ, Sninsky JJ, Friedman SL. 2010. A DDX5 S480A polymorphism is associated with increased transcription of fibrogenic genes in hepatic stellate cells. J Biol Chem 285: 5428–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim L, Zipor G, Aronov S, Gerst JE. 2007. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods 4: 409–412. [DOI] [PubMed] [Google Scholar]
- Hao W, Chang C-PB, Tsao C-C, Xu J. 2010. Oligomycin-induced bioenergetic adaptation in cancer cells with heterogeneous bioenergetic organization. J Biol Chem 285: 12647–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatanaka M. 1974. Transport of sugars in tumor cell membranes. Biochim Biophys Acta 355: 77–104. [DOI] [PubMed] [Google Scholar]
- Hirling H, Scheffner M, Restle T, Stahl H. 1989. RNA helicase activity associated with the human p68 protein. Nature 339: 562–564. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu Z-R. 2002. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem 277: 12810–12815. [DOI] [PubMed] [Google Scholar]
- Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo F, et al. 2015. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature 528: 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Iggo RD, Jamieson DJ, MacNeill SA, Southgate J, McPheat J, Lane DP. 1991. P68 RNA helicase: identification of a nucleolar form and cloning of related genes containing a conserved intron in yeasts. Mol Cell Biol 11: 1326–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iost I, Dreyfus M, Linder P. 1999. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem 274: 17677–17683. [DOI] [PubMed] [Google Scholar]
- Jacobi D, Liu S, Burkewitz K, Kory N, Knudsen NH, Alexander RK, Unluturk U, Li X, Kong X, Hyde AL, et al. 2015. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metab 22: 709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal C, Uhlmann-Schiffler H, Stahl H. 2007. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res 35: 3590–3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Putnam A. 2010. Duplex unwinding with DEAD-box proteins. Methods Mol Biol 587: 245–264. [DOI] [PubMed] [Google Scholar]
- Jarmoskaite I, Russell R. 2014. RNA helicase proteins as chaperones and remodelers. Annu Rev Biochem 83: 697–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, Marcotte EM. 2015. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348: 921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin WGJ, Krek W, Sellers WR, DeCaprio JA, Ajchenbaum F, Fuchs CS, Chittenden T, Li Y, Farnham PJ, Blanar MA. 1992. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70: 351–364. [DOI] [PubMed] [Google Scholar]
- Kaplan CD, Laprade L, Winston F. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301: 1096–1099. [DOI] [PubMed] [Google Scholar]
- Kar A, Fushimi K, Zhou X, Ray P, Shi C, Chen X, Liu Z, Chen S, Wu JY. 2011. RNA helicase p68 (DDX5) regulates tau exon 10 splicing by modulating a stem-loop structure at the 5′ splice site. Mol Cell Biol 31: 1812–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A, Nishizuka M, Tominaga K, Tsuchiya T, Nishihara T, Imagawa M. 2001. Expression of p68 RNA helicase is closely related to the early stage of adipocyte differentiation of mouse 3T3-L1 cells. Biochem Biophys Res Commun 287: 435–439. [DOI] [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P. 1997. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 17: 7283–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, de la Cruz J, Rojo M, Linder P. 1998. Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S-ribosomal-subunit assembly in Saccharomyces cerevisiae. Mol Cell Biol 18: 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam AA, Jankowsky E. 2014. DEAD-box helicases form nucleotide-dependent, long-lived complexes with RNA. Biochemistry 53: 423–433. [DOI] [PubMed] [Google Scholar]
- Ma WK, Cloutier SC, Tran EJ. 2013. The DEAD-box protein Dbp2 functions with the RNA-binding protein Yra1 to promote mRNP assembly. J Mol Biol 425: 3824–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma WK, Paudel BP, Xing Z, Sabath IG, Rueda D, Tran EJ. 2016. Recruitment, duplex unwinding and protein-mediated inhibition of the dead-box RNA helicase Dbp2 at actively transcribed chromatin. J Mol Biol 428: 1091–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macheda ML, Rogers S, Best JD. 2005. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol 202: 654–662. [DOI] [PubMed] [Google Scholar]
- Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM. 2011. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci 108: 12254–12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallam AL, Sidote DJ, Lambowitz AM. 2014. Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase. Elife 3: e04630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek A, Park Y, Miething C, Wilkinson JE, Gillis J, Lowe SW, Vakoc CR, Stillman B. 2014. Acquired dependence of acute myeloid leukemia on the DEAD-box RNA helicase DDX5. Cell Rep 7: 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueckler M, Thorens B. 2013. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med 34: 121–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. 1999. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev 63: 554–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park KS, Jo I, Pak K, Bae SW, Rhim H, Suh SH, Park J, Zhu H, So I, Kim KW. 2002. FCCP depolarizes plasma membrane potential by activating proton and Na+ currents in bovine aortic endothelial cells. Pflugers Arch 443: 344–352. [DOI] [PubMed] [Google Scholar]
- Pasic L, Sepcic K, Turk T, Macek P, Poklar N. 2001. Characterization of parazoanthoxanthin A binding to a series of natural and synthetic host DNA duplexes. Arch Biochem Biophys 393: 132–142. [DOI] [PubMed] [Google Scholar]
- Prather D, Krogan NJ, Emili A, Greenblatt JF, Winston F. 2005. Identification and characterization of Elf1, a conserved transcription elongation factor in Saccharomyces cerevisiae. Mol Cell Biol 25: 10122–10135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JJ, Chang HY. 2016. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet 17: 47–62. [DOI] [PubMed] [Google Scholar]
- Rocak S, Linder P. 2004. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 5: 232–241. [DOI] [PubMed] [Google Scholar]
- Rossler OG, Straka A, Stahl H. 2001. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res 29: 2088–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Klostermeier D. 2015. When core competence is not enough: functional interplay of the DEAD-box helicase core with ancillary domains and auxiliary factors in RNA binding and unwinding. Biol Chem 396: 849–865. [DOI] [PubMed] [Google Scholar]
- Ruminski DJ, Watson PY, Mahen EM, Fedor MJ. 2016. A DEAD-box RNA helicase promotes thermodynamic equilibration of kinetically trapped RNA structures in vivo. RNA 22: 416–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman DW, Shubert-Coleman J, Furneaux H. 2007. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem 282: 32773–32779. [DOI] [PubMed] [Google Scholar]
- Samatanga B, Klostermeier D. 2014. DEAD-box RNA helicase domains exhibit a continuum between complete functional independence and high thermodynamic coupling in nucleotide and RNA duplex recognition. Nucleic Acids Res 42: 10644–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samatanga B, Andreou AZ, Klostermeier D. 2017. Allosteric regulation of helicase core activities of the DEAD-box helicase YxiN by RNA binding to its RNA recognition motif. Nucleic Acids Res 45: 1994–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutz P, Karlberg T, van den Berg S, Collins R, Lehtio L, Hogbom M, Holmberg-Schiavone L, Tempel W, Park H-W, Hammarstrom M, et al. 2010. Comparative structural analysis of human DEAD-box RNA helicases. PLoS One 5: e12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. 2006. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell 125: 287–300. [DOI] [PubMed] [Google Scholar]
- Senissar M, Le Saux A, Belgareh-Touze N, Adam C, Banroques J, Tanner NK. 2014. The DEAD-box helicase Ded1 from yeast is an mRNP cap-associated protein that shuttles between the cytoplasm and nucleus. Nucleic Acids Res 42: 10005–10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma D, Jankowsky E. 2014. The Ded1/DDX3 subfamily of DEAD-box RNA helicases. Crit Rev Biochem Mol Biol 49: 343–360. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson RJ, Hamilton SJ, MacCallum DE, Hall PA, Fuller-Pace FV. 1998. Expression of the ‘dead box’ RNA helicase p68 is developmentally and growth regulated and correlates with organ differentiation/maturation in the fetus. J Pathol 184: 351–359. [DOI] [PubMed] [Google Scholar]
- Thandapani P, O'Connor TR, Bailey TL, Richard S. 2013. Defining the RGG/RG motif. Mol Cell 50: 613–623. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. 2006. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA 12: 959–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongtrakoongate P, Riddick G, Fucharoen S, Felsenfeld G. 2015. Association of the long non-coding RNA steroid receptor RNA activator (SRA) with TrxG and PRC2 complexes. PLoS Genet 11: e1005615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E. 2005. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry 44: 13591–13601. [DOI] [PubMed] [Google Scholar]
- Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. 2007. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell 28: 253–263. [DOI] [PubMed] [Google Scholar]
- Zhang W, Cao Y, Gong J, Bao X, Chen G, Liu W. 2015. Identification of residues important for substrate uptake in a glucose transporter from the filamentous fungus Trichoderma reesei. Sci Rep 5: 13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Xing Z, Mani SKK, Bancel B, Durantel D, Zoulim F, Tran EJ, Merle P, Andrisani O. 2016. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology 64: 1033–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonta E, Bittencourt D, Samaan S, Germann S, Dutertre M, Auboeuf D. 2013. The RNA helicase DDX5/p68 is a key factor promoting c-fos expression at different levels from transcription to mRNA export. Nucleic Acids Res 41: 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]