Abstract
Interactions between the ribosomal exit tunnel and the nascent peptide can affect translation elongation rates. While previous studies have already demonstrated the feasibility of such interactions, little is known about the nature of the stalling peptide sequences and their distribution in the proteome. Here we ask which peptide sequences tend to occupy the tunnel of stalled ribosomes and how they are distributed in the proteome. Using computational analysis of ribosome profiling data from S. cerevisiae, we identified for the first time dozens of short stalling peptide sequences and studied their statistical properties. We found that short peptide sequences associated with ribosome stalling tend significantly to be either over- or underrepresented in the proteome. We then showed that the stalling interactions may occur at different positions along the length of the tunnel, prominently close to the P-site. Our findings throw light on the determinants of nascent peptide-mediated ribosome stalling during translation elongation and support the novel conjecture that mRNA translation affects the proteomic distribution of short peptide sequences.
Keywords: ribosome stalling, nascent peptide, ribosomal exit tunnel, translation, underrepresented, overrepresented
INTRODUCTION
During mRNA translation elongation, the growing polypeptide chain departs from the ribosome through a narrow, negatively charged exit tunnel. The cramped dimensions of the ribosomal exit tunnel preclude tertiary folding of whole large protein domains; however, tertiary/secondary conformations of small segments of the nascent polypeptide as well as folding of small protein domains are usually feasible (Kosolapov and Deutsch 2009; Wilson and Beckmann 2011; Nilsson et al. 2015; Marino et al. 2016). Short sequences within the nascent peptide that chemically/electrostatically interact with ribosomal subunits might interfere with translation via delaying or blocking protein synthesis or via affecting functional properties of the ribosome, such as the A-site of the peptidyl transferase center (Nakatogawa and Ito 2002; Tenson and Ehrenberg 2002; Lawrence et al. 2008; Seidelt et al. 2009; Bhushan et al. 2010; Ramu et al. 2011). Since mRNA translation is the process that consumes most of the cellular energy, genomes are under strong selection to improve this process (Arava et al. 2003; Ingolia et al. 2009; Plotkin and Kudla 2010; Tuller et al. 2010; Gingold and Pilpel 2011). Whereas peptide sequences that block protein synthesis are expected to be selected against (Peil et al. 2013; Woolstenhulme et al. 2013; Navon et al. 2016), peptide sequences that “transiently” halt the ribosome may contribute to cotranslational protein folding or other important nontranslational phenomena, and thus be evolutionarily preferred (Kimchi-Sarfaty et al. 2007; Tsai et al. 2008; Komar 2009; Kramer et al. 2009; Zhang et al. 2009; Ciryam et al. 2013).
Previous studies have shown that the distribution of short peptide sequences in the proteome deviates from what is expected based on the proteomic distribution of their individual amino acids (AA) (Qi et al. 2004; Höhl and Ragan 2007; Tuller et al. 2007). Furthermore, it has been shown that many possible peptide sequences, 5 AA long, are totally absent from all known proteomes although they are “coded” multiple times in noncoding regions (Tuller et al. 2007). In addition, various ribo-seq-based studies in recent years have suggested that the AA content of the nascent peptide may affect translation elongation speed via interactions with the ribosomal exit tunnel (Dana and Tuller 2012; Charneski and Hurst 2013; Artieri and Fraser 2014; Sabi and Tuller 2015). These studies, however, have focused on single amino acids rather than longer peptide sequences.
The scarcity of some peptide sequences from endogenous proteins suggests, among others, that they might have been selected against to improve translation efficiency. Missing peptide sequences are presumably more deleterious to the organism than those that occur in the proteome but are underrepresented relatively to the distribution of their individual amino acids. Alternatively, peptide sequences that are overrepresented in the proteome are expected to contribute to the fitness of the organism. Studying the proteomic distribution of short peptide sequences within proteins may teach us about the nature of missing peptides and the evolutionary reasons for the over- or underrepresentation of certain peptide sequences in the proteome.
In this study, we aim at identifying for the first time the set of short peptide sequences (composed of 2–3 AA) suspected to stall ribosomes during translation elongation. In addition, we aim at understanding the properties of these peptides and whether natural selection acting on endogenous S. cerevisiae genes inhibits or promotes their representation in the proteome. To answer this question, we performed a systems biology study based on multiple data sets from large-scale ribosome profiling experiments in S. cerevisiae (Ingolia et al. 2009; Brar et al. 2012; Gerashchenko et al. 2012; McManus et al. 2014).
RESULTS
Our analyses rely mainly on the comparison of two fundamental features attributed to short peptide sequences: (i) their distribution in the proteome and (ii) their predicted translational-stalling effect inside the ribosomal exit tunnel. The latter is learned from ribosome profiling data, gathered by the ribosome profiling (or ribo-seq) method, which provides high-throughput quantitative measures of the translational status of the entire transcriptome (Ingolia et al. 2009). Ribosome profiling experiments are based on the isolation of ribosome protected fragments that are then sequenced and mapped to the transcriptome yielding ribosomes protected footprints. As slowly decoded codons are covered by ribosomes for a longer time, they tend to create more protected fragments compared to faster codons on the same transcript. Each transcript has a specific read count profile, which represents its unique translational signature. To quantitatively evaluate the translational efficiency distribution along a given transcript while controlling for biases related to sequencing or experimental procedures, we normalized each ribo-seq profile by its corresponding mRNA-seq profile (see details in Materials and Methods).
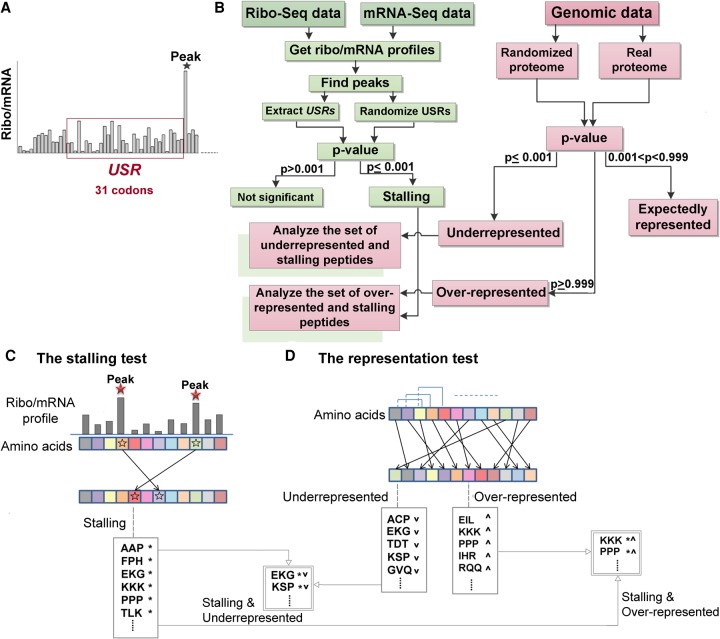
In this study, we focus on ribosome stalling caused by short AA sequences within the nascent peptide. To this end, we first need to identify ribosome stalling positions along each transcript; then, we need to understand which peptide sequence is located inside the tunnel of the stalled ribosome. Here, we defined ribosome stalling (or pausing) positions as peaks in the normalized ribo/mRNA-seq profile, and the peptide sequence inside the tunnel as the AA sequence upstream of the peak (which is equivalent in length to the length of the peptide required to fill the tunnel). We termed this sequence the upstream stalling region (USR) and set its length to 31 codons/AA (Fig. 1A). It is important to emphasize that based on the ribo-seq data, it is impossible to distinguish in large-scale between stalling and transient pausing of ribosomes.
FIGURE 1.
General description of the approach. (A) A ribo/mRNA profile is illustrated. The x-axis represents a codon position. The USR is defined as the AA sequence that encodes the 31 codons upstream of the peak position. (B) The general steps of the approach: Genomic data are analyzed (related blocks/steps are in pink). The pink part to the right describes all the steps related to the genomic data: The AA sequences of all analyzed proteins are randomized and compared to the original sequences in order to identify significantly over- and underrepresented peptide sequences. The green part to the left describes all steps related to the ribo/mRNA-seq data: The USRs are randomized and compared to the original sequences to identify stalling peptide sequences. The two pink boxes shaded in green describe the two steps related to both the genomic and the ribo/mRNA-seq data: The sets of stalling peptide sequences which turned out also to be over- or underrepresented in the proteome are further analyzed and characterized. (C) The stalling test: The codon/AA positions of peaks in the ribo/mRNA profile (corresponding blocks are designated by stars) are randomly changed; accordingly, the randomized versions of the USRs are the AA encoding the codons upstream of the random position (see details in Materials and Methods). Peptide sequences that appear in the real USRs significantly more than in the randomized versions are defined as stalling (designated by asterisks). (D) The representation test: Each protein sequence is permuted so that the original AA content is kept. Underrepresented peptide sequences are defined as those that appear in the randomized protein sequences significantly more than in the real ones (designated by down arrow); overrepresented peptide sequences are defined as those that appear in the real proteins significantly more times than in the randomized versions (designated by up arrow).
The general steps of our approach are described in Figure 1B: In the first test, we perform “the stalling test” and calculate a P-value that seeks to identify short peptide sequences that significantly tend to appear in the USRs compared to random regions of the same length on the same transcript (Fig. 1C). Peptide sequences with P ≤ 0.001 are termed here “stallers” or “stalling peptide sequences.” In the second step, we perform the “representation test” and calculate a P-value that seeks to identify short peptide sequences whose number of occurrences in the proteome significantly deviates from what is expected in a randomized proteome that maintains the “individual” AA distribution of each protein (details in Materials and Methods). Peptide sequences with P ≤ 0.001 are termed here “underrepresented” and those with P ≥ 0.999 are termed “overrepresented” (Fig. 1D). Peptide sequences whose proteomic distribution does not significantly deviate from what is expected based on the independent distribution of their individual AA (0.001 < P < 0.999) are termed here “expectedly represented.”
In this study, we analyzed peptide sequences of 2 and 3 AA. These are termed here “dipeptide and tripeptide sequences,” respectively (see explanation in Materials and Methods regarding the considerations for choosing these lengths).
Strong association between ribosome stalling and proteome composition
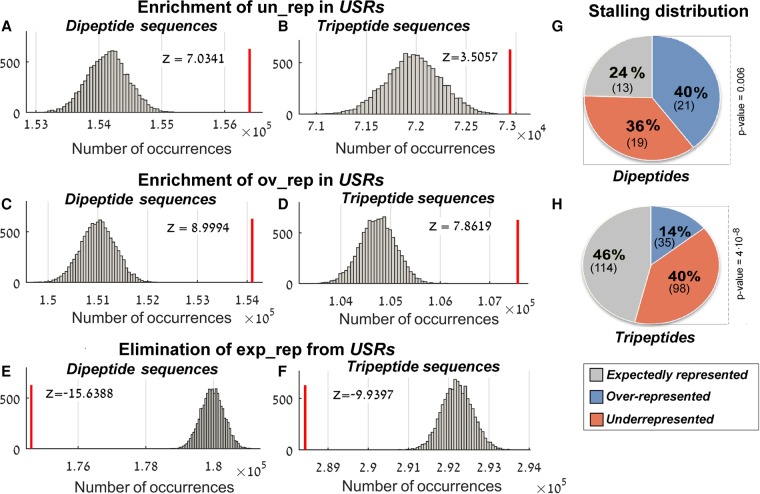
Seeking to determine whether nascent peptide-mediated ribosome stalling plays a role in the evolution of proteins, we analyzed the distribution of short peptide sequences in the USRs. As can be seen in Figure 2, the USRs turned out to be significantly enriched with both over- and underrepresented peptide sequences (P < 10−4, Fig. 2A–D). Alternatively, the expectedly represented peptide sequences turned out to be eliminated from those regions (P < 10−4, Fig. 2E,F).
FIGURE 2.
The distribution of over-, under-, and expectedly represented peptide sequences in the USRs. The red line represents the total number of over/under/expectedly represented peptide sequences in the USRs (designated ov_rep, un_rep, and exp_rep, respectively). The corresponding random distribution is shown in gray. Values in the x-axis are the total number of occurrences in the real/random USRs; the histogram is based on 10,000 randomizations (details in Materials and Methods). Z-scores appear at the top of each sub figure; all corresponding P-values <10−4. (A) Underrepresented dipeptide sequences, (B) underrepresented tripeptide sequences, (C) overrepresented dipeptide sequences, (D) overrepresented tripeptide sequences, (E) expectedly represented dipeptide sequences, and (F) expectedly represented tripeptide sequences. (G,H) Distribution of the number of stalling peptide sequences with respect to proteome representation. The P-value represents the statistical significance of the over- and underrepresented proportion. (G) Dipeptide sequences. (H) Tripeptide sequences.
In addition to the USR composition, the amount of overlap between the group of stalling peptide sequences and the groups of over/underrepresented peptide sequences may also teach us about the relationship between ribosome stalling and the proteome composition. We found that a dominant and statistically significant part of the stalling peptide sequences turned out to be either over- or underrepresented in the proteome; specifically, 40 out of the 53 (76%) stalling dipeptide sequences and 133 out of the 247 (54%) stalling tripeptide sequences have also been identified as over- or underrepresented sequences (Fig. 2G,H).
The list of all dipeptide and tripeptide sequences, their stalling P-values, and their representation P-values are provided in Supplemental Table S1.
All signals shown in Figure 2 remained robust to small changes in the chosen thresholds, such as the threshold value that defines a ribo/mRNA-seq peak and the minimum extent of ribo-seq read counts coverage required to analyze a gene (Supplemental Fig. S1). We also demonstrated that the signal produced by our default parameters is not biased by a subset of peptide sequences that occur in the proteome only a small number of times (Supplemental Fig. S2).
Ranking the stalling effect and proteome representation of short peptide sequences
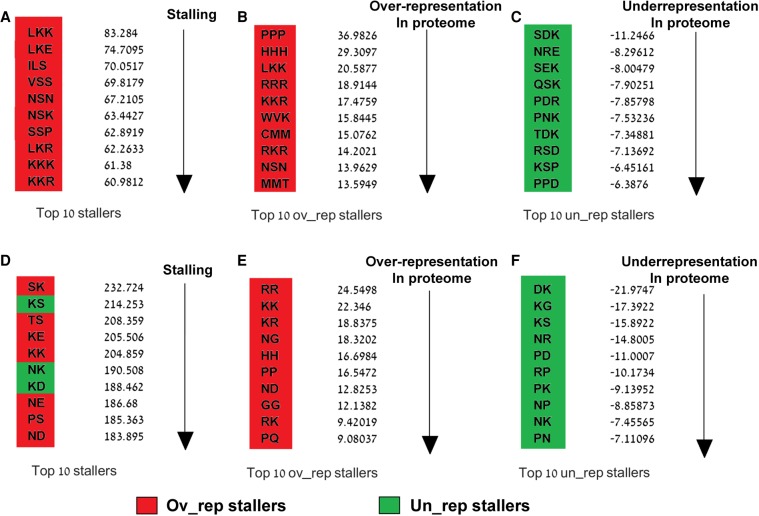
To quantify the extent to which the identified stalling peptide sequences tend to be stalling and over/underrepresented, we ranked them based on a standardized score that compares the real and random USRs/proteome distributions in terms of standard deviations (more details in Materials and Methods). According to the resultant Z-scores, the strongest stallers were the sequences LKK and SK; the most abundant stallers were PPP and RR; and the most underrepresented stallers were SDK and DK (Fig. 3). The top peptide sequences in each category included known stalling peptide sequences in S. cerevisiae such as KKK, KK, and KKR (Charneski and Hurst 2013). Other stallers have also been found in other organisms based on different techniques. For example, the stalling and underrepresented triplet PPD, which was ranked here within the top 10 stalling and underrepresented peptide sequences, has been previously identified as a strong staller in E. coli whose observed frequency of occurrence within E. coli proteins was “smaller” than expected (Peil et al. 2013).
FIGURE 3.
Ranking the stalling motifs. Red and green cells represent over- and underrepresented peptide sequences, respectively. (A,D) Top 10 stalling. Stalling Z-scores based on which peptides were ranked (Materials and Methods) are presented to the right. (B,E) Top 10 overrepresented stalling peptide sequences. Overrepresented Z-scores based on which peptides were ranked (Materials and Methods) are presented to the right. (C,F) Top 10 underrepresented stalling peptide sequences. Underrepresented Z-scores based on which peptides were ranked (Materials and Methods) are presented to the right.
The real and random numbers of occurrences of each peptide sequence in the proteome and in the USRs are presented in Supplemental Figure S3 (for stalling and overrepresented peptides) and Supplemental Figure S4 (for stalling and underrepresented peptides).
Characterization of the over- and underrepresented stalling peptide sequences
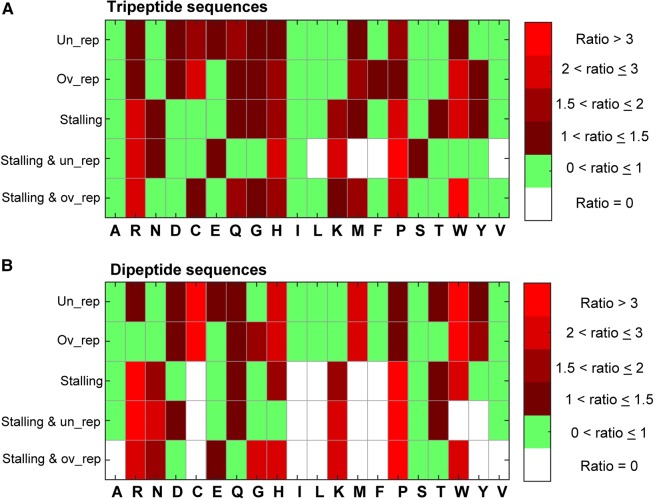
We set out to computationally and statistically characterize the biochemical properties of the stalling peptide sequences that turned out also to be over- or underrepresented in the proteome. To this end, we analyzed the AA composition of these sets. We calculated the ratio between the empirical probability to observe each AA in each set and its probability in the proteome. We also calculated this ratio for other groups including the over- and underrepresented peptide sequences and the group of stalling peptide sequences (Fig. 4). In line with previous studies, proline (P) and arginine (R) were particularly dominant in the stalling groups, with probability to appear in the stalling groups that is two- to fourfold higher than their probability to appear in a certain position in the proteome (Tuller et al. 2011; Charneski and Hurst 2013; Peil et al. 2013; Artieri and Fraser 2014; Sabi and Tuller 2015).
FIGURE 4.
The AA content of different peptide groups. The ratios defined in the color bar represent the ratio between the probability to observe each AA in each group to the probability to observe it in the proteome. White cells correspond to AA that are missing from all peptide sequences in the specified group, green cells correspond to ratios lower than one (meaning that they are more likely to be observed in the proteome than in the specific group), and red cells correspond to ratios higher than one (meaning that they are more likely to be observed in the peptides group than in the proteome). (A) The probability to observe each AA in different groups of tripeptide sequences. (B) The probability to observe each AA in different groups of dipeptide sequences.
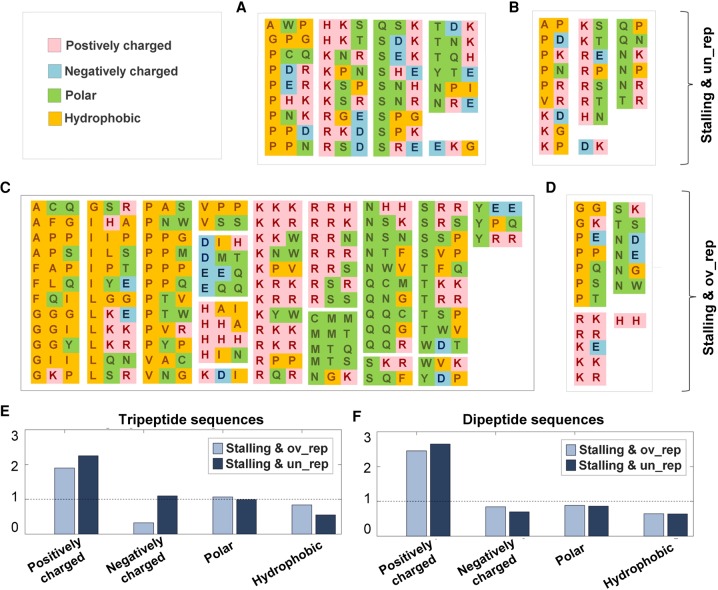
Next, we aimed at understanding whether the over/underrepresented stalling peptide sequences tend to include AA from specific biochemical groups (e.g., positively/negatively charged, hydrophobic, or polar). Interestingly, we observed a significant tendency of the stalling and overrepresented di- and tripeptide sequences to include AA from the same biochemical groups (P < 10−2). We also found that the group of stalling and underrepresented sequences tended to include AA from different biochemical groups (Fig. 5A–D). This tendency, however, turned out to be statistically significant (P < 10−2) only for the dipeptide sequences.
FIGURE 5.
Biochemical groups of AA within over- and underrepresented stalling motifs. (A–D) The AA were colored based on their biochemical classification as appears in the legend at the top left corner. (E,F) The ratio between the probability to observe an amino acid from a specific biochemical group in the set of over/underrepresented stalling peptide sequences (lighter/darker, respectively) and in the proteome. A dashed line is presented for ratios of one.
In addition, it can be seen that while positively charged AA were frequent in both over- and underrepresented stalling sequences (Fig. 5E,F), for tripeptide sequences, negatively charged AA turned out to be more frequent in the set of underrepresented stalling sequences (Fig. 5E).
Finally, we were not able to find evidence that the stalling and over/underrepresented peptides tend to appear in proteins encoded by specific groups of genes in terms of the number of protein–protein interactions, the number of consumed ribosomes, or their function. This analysis supports the conjecture that the results reported here are universal and not related to specific group(s) of genes.
The relationship between tripeptide and dipeptide motifs
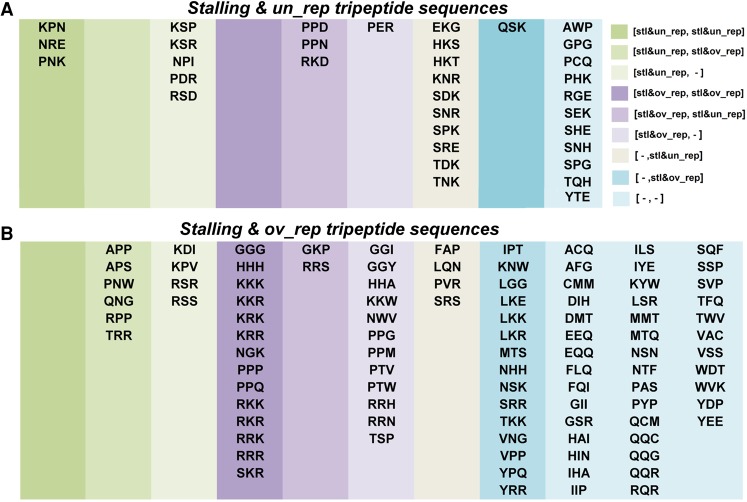
Since each tripeptide sequence is composed of two dipeptide sequences, it is possible that some stalling and over/underrepresentation patterns observed in the tripeptides derive from patterns related only to the dipeptides. Thus, we checked how many over/underrepresented stalling “tripeptide” sequences included over/underrepresented stalling “dipeptide” sequences (Fig. 6). Specifically, out of the 34 stalling and underrepresented tripeptide sequences, 21 (61.7%) included underrepresented stalling dipeptide sequences and five (14.7%) included stalling and overrepresented dipeptide sequences. Out of the 98 overrepresented stalling tripeptide sequences, 16 (16.3%) included underrepresented stalling dipeptide sequences and 49 (50%) included overrepresented stalling dipeptide sequences. Prominently, the group of overrepresented stalling tripeptide sequences that did not include any stalling dipeptide constituted 41.84% of the overrepresented stalling tripeptide sequences. These results demonstrate that the reported tripeptide sequences cannot be trivially related to the dipeptide sequences composing them.
FIGURE 6.
Classification of the tripeptide motifs based on their dipeptide motifs content. The groups of (A) underrepresented stalling tripeptide sequences and (B) overrepresented stalling tripeptide sequences were divided based on the classifications of the two dipeptide sequences they include (where the first dipeptide refers to the first two AA within the tripeptide, and the second dipeptide refers to the last two AA in the tripeptide). The legend at the right corner is written in the following format: [first dipeptide class, second dipeptide class] where three possible classifications were considered for a dipeptide: stalling and underrepresented (designated stl&un_rep), stalling and overrepresented (designated stl&ov_rep), and dipeptides that were neither underrepresented stalling nor overrepresented stalling (designated -). Some of the combinations were not observed, thus some lists are empty.
Stalling positions along the ribosomal exit tunnel
In this study we analyzed stalling sequences of lengths 2 and 3 AA. However, the length of the nascent peptide required to fill the tunnel is approximately 31 AA. This raises the question “where along the length of tunnel do the stalling interactions” tend to occur?
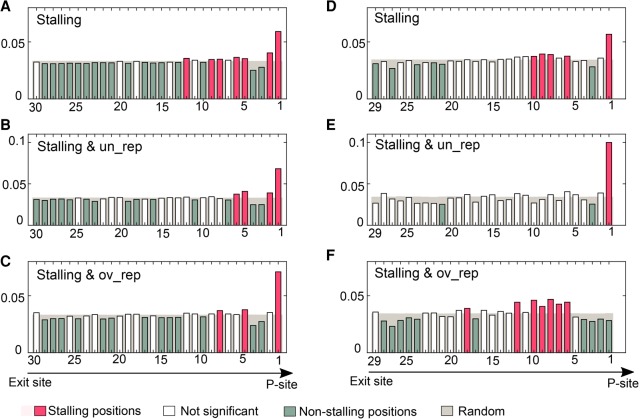
To answer this question, we built a position-specific scoring matrix (PSSM) based on the probabilities of each stalling di- and tripeptide sequence to appear at a specific position along the tunnel. We found out that the USRs are less uniformly distributed than the corresponding random regions with a prominent stalling region close to the P-site (Fig. 7; except for the group of stalling and overrepresented tripeptide sequences). For both the stalling di- and tripeptide sequences, we observed a unique stalling distribution for each sequence. However, whereas the dipeptide sequences exhibited a general stalling tendency close to the P-site, the tripeptide sequences showed different patterns for the over- and underrepresented triplets. In addition to the average pattern observed in Figure 7, we plotted the specific PSSMs of each individual stalling peptide sequence (Supplemental Fig. S5).
FIGURE 7.
The average distribution of stalling peptide sequences along the length of the tunnel. The positions along the tunnel (x-axis) represent the first position the peptide occupies (direction is from exit site to the P-site and as plotted by the arrow below the figure). The height of the bar is the probability to occupy this position (average probability over all peptide sequences defined in the title). Bars corresponding to probabilities in the randomized USRs are shown in gray. Bars are colored red/green if their corresponding probabilities turned out to be significantly (P < 0.05) higher/lower than random, respectively; other bars (P > 0.05) appear in white. (A–C) Dipeptide sequences. (D–F) Tripeptide sequences.
DISCUSSION
Previous studies have separately dealt with the nonuniform distribution of peptides in the proteome (Qi et al. 2004; Höhl and Ragan 2007; Tuller et al. 2007) and the interaction between the translated peptide and the ribosomal exit tunnel (Nakatogawa and Ito 2002; Tenson and Ehrenberg 2002; Kosolapov and Deutsch 2009; Bhushan et al. 2010; Ramu et al. 2011; Tuller et al. 2011; Sabi and Tuller 2015). In this study, we quantitatively demonstrated that these two phenomena are strongly associated: Peptide sequences that tend to occupy the tunnel of stalled ribosomes tend also to be significantly over- or underrepresented in the proteome.
In our proposed workflow, we have taken a novel computational approach of identifying dozens of short peptide sequences predicted to interact with the ribosomal exit tunnel. The fact that many of the stalling tripeptide sequences did not include any stalling dipeptide sequence implies that there may be much more complexity in some of the interactions between the nascent peptide and the ribosomal exit tunnel that involve more than 1–2 AA. Although we have identified a prominent stalling position close to the P-site for most of the stalling peptides, other stalling positions along the tunnel were also observed. Indeed, as the nascent peptide cotranslationally and partially folds inside the tunnel, we do not expect all stalling peptide sequences to occur at the same position. Moreover, biases in the ribo-seq data, including the fact that ribosomes may continue to slowly move even after treatment with cycloheximide, may also affect the stalling position in the tunnel (Kosolapov and Deutsch 2009; Hussmann et al. 2015; Tuller and Zur 2015; Diament and Tuller 2016).
The reported results have a few important implications: First, they support the novel conjecture that evolutionary selection has shaped the AA composition of endogenous proteins not only by their functionality, but also by the efficiency of their synthesis. This idea has already been suggested in previous studies, however, based on different considerations/mechanisms such as the metabolic cost of amino acids (Akashi and Gojobori 2002). Second, our results may explain the absence of some particular short peptide sequences from various proteomes (Tuller et al. 2007). It is possible that some of the missing peptide sequences induced much stronger stalling interactions with the exit tunnel than those discovered here, and thereby have been eliminated from the proteome by evolutionary selection. One limitation of our approach is that it cannot analyze missing peptides. However, a future study on this topic may be based on synthetic genes that do encode these missing peptides. Third, regardless of the over/underrepresentation in the proteome, our computational method can be generally used to infer short peptide sequences that can lead to ribosome stalling. The fact that some of the stalling peptide sequences identified here have been previously shown to have a stalling effect on ribosomes, demonstrates that our approach can capture relevant peptides. Our method may have important applications in various biomedical fields such as biotechnology, synthetic biology, and specifically proteins and gene expression engineering.
The fact that we specifically referred to sequences that are also over- or underrepresented in the proteome enabled a more reliable detection of stalling peptides in comparison to methods based only on ribo-seq experiments, which tend to include noise and biases (see for example Dana and Tuller 2012; Artieri and Fraser 2014; Diament and Tuller 2016).
The association between stalling and proteome-underrepresentation is intuitive: Evolutionary pressures act to eliminate short peptide sequences that halt the ribosome to improve translation efficiency. The association between stalling and proteome-overrepresentation on the other hand, is less intuitive and can be explained by the fact that translational pausing plays a functional role in cotranslational protein folding, which is known to be fine-tuned by ribosome kinetics (Thanaraj and Argos 1996; Tsai et al. 2008; Komar 2009; Kramer et al. 2009; Zhang et al. 2009; Zhang and Ignatova 2011; Ciryam et al. 2013). Nevertheless, the association between ribosome stalling and proteome-overrepresentation may not be causal. For example, such peptide sequences may have biochemical properties related to other nontranslational processes such as protein functions, interactions, and stability. The same properties may also contribute to the interaction with the exit tunnel. However, as the other processes may be more important to the fitness of the organisms than translation, they are overrepresented in the proteome despite their stalling effect. Indeed, we observed different properties for the over- and underrepresented stalling peptide sequences. The fact that whereas the underrepresented stalling sequences tended to include AA from different biochemical groups and the overrepresented stalling peptide sequences tended to include AA from the same group, may indicate that stretches of the same/different type of AA may contribute (on average) to a better/worse folding and protein-function and thus to an over/underrepresentation, respectively (Senes et al. 2000; Ng and Henikoff 2002; Dobson 2004).
Whether the overrepresented sequences tend to attenuate the stalling effect of the underrepresented sequences, and whether they are conserved across different organisms, are questions deferred to future studies that will involve experimental measurements of the stalling effect of different peptide combinations on translational speed, protein folding, and organismal fitness.
MATERIALS AND METHODS
Coding sequences data
Coding sequences of S. cerevisiae strain s288c were retrieved from the University of California Santa Cruz (UCSC) Genome Browser (https://genome-euro.ucsc.edu/).
Ribosome profiling data
In this study we used a total of 10 data sets of ribosome profiling based on the following experiments in S. cerevisiae: Ingolia et al. (2009), Brar et al. (2012), Mcmanus et al. (2014), and Gerashchenko et al. (2012). The ribosomal footprints were mapped based on Michel et al. (2014) and Diament and Tuller (2016). The specific genomic position assigned to each read represents the location of the ribosomal A-site on the mRNA. In both Michel et al. (2014) and Diament and Tuller (2016), the A-site corresponding to each read was determined by an offset of 15 nucleotides (nt) from the 5′ end of the fragment.
Data filtering and aggregates preparation
Whether it stems from experimental biases or cached translational features, the density of ribosome footprints tends to be significantly elevated at the beginning of the gene (Ingolia et al. 2009, 2011; Tuller et al. 2010). Thus, we have excluded the first 20 codons from all aspects of the analysis described in this study.
The first step of generating the aggregate data set of ribosome profiling was to convert the ribo-seq and mRNA-seq profiles which were mapped with nucleotide resolution into profiles with codon resolution (by averaging read count over the 3 nt of each codon). Then, each ribo-seq and mRNA-seq profile was individually normalized by the total number of read counts in the experiment (excluding the first 20 codons in each profile). Aiming at increasing the statistical power and reliability of the data, we generated two separated aggregates of ribo-seq and mRNA-seq by calculating the average read count at each position of the mRNA over all 10 data sets. In the next step, as suggested in Artieri and Fraser (2014), to account for biases in ribosome profiling we normalized ribo-seq read counts by their corresponding mRNA-seq read counts. Finally, we filtered genes with <70% non-zero read counts in the aggregate of ribo/mRNA. Although our results remained robust also when increasing the coverage threshold up to 90% (Supplemental Fig. S1D,E), we opted for the 70% threshold, as more genes pass this filter so the results are more statistically powerful (Supplemental Fig. S1F).
Definition of peaks in ribosomal density
As described in Sabi and Tuller (2015), we defined peaks in ribosomal density (RD) as codon positions along the mRNA for which the normalized read count (ribo/mRNA) is four times higher than the mean ribo/mRNA values of the mRNA excluding the first 20 codons. In addition to the filtering described in the previous section, we only analyzed genes that include peaks; here, a total of 4564 genes. The rationale for choosing a peak cutoff of four was to control the tradeoff between the extent of stalling (which increased with the increase in this cutoff) and the statistical power (which decreased with the increase in the cutoff as less genes are analyzed). The results, however, remain robust also for higher peak thresholds of five, six, and seven (Supplemental Fig. S1).
Randomized proteomes
Of note, 10,000 randomized proteomes were generated by randomly permuting the AA sequence of each protein (excluding the first 20). By performing such permutations, we maintain the original individual AA numbers/distribution of each protein.
The “representation” P-value
Let Nreal,i and Nrandom,i denote the number of occurrences of the ith peptide sequence in the real and randomized proteome, respectively.
The representation P-value of the ith peptide sequence is defined as
This P-value corresponds to the probability that the ith peptide sequence is observed in the real proteome more often than in the randomized one. As we set the significance level to 0.001, peptide sequences for which Prep,i ≤ 0.001 were defined underrepresented and those for which Prep,i ≥ 0.999 were defined overrepresented. All other peptide sequences (0.001 < Prep,i < 0.999) were termed expectedly represented (as their proteomic probability does not significantly deviate from what is expected based on the distribution of their individual AA). The number of over/underrepresented peptide sequences that are expected to be false positives is presented in Supplemental Table S2 and was calculated by applying the representation P-value to a randomized version of the proteome rather than to the real one.
The “representation” Z-score
Let Nreal,i denote the number of occurrences of the ith peptide sequence in the real proteome and let mean(Nrandom,i) and std(Nrandom,i) denote the mean and standard deviations of the number of occurrences of the ith peptide sequences over all 10,000 randomized versions of the proteome. The standardized score of the ith peptide sequence is given by
Z-scorei represents the extent to which the number of occurrences of the ith peptide sequence in the proteome deviates from random in terms of standard deviations. Here, Z-scores ≤ −2 correspond to underrepresented peptides and Z-scores ≥ 2 correspond to overrepresented peptides. The distributions of peptides in the randomized proteomes were verified to be normally distributed using the Kolmogorov–Smirnov test (Massey 1951).
The analyzed peptide length
In this study we analyzed only peptide sequences of lengths 2 and 3 AA. The rationale for choosing these lengths stems from the following two statistical limitations: First, according to the definition of the representation P-value, underrepresented peptides are those that appear in the randomized proteome significantly more times than in the real proteome. However, as peptide sequences become longer, the probability to observe them in the real (and randomized) proteome becomes smaller. While peptide sequences of lengths 2–3 AA were observed in the proteome thousands and hundreds of times, those longer than 3 AA were observed in the proteome only a very few times (Supplemental Table S2). Second, we found that unlike lengths of 2–3 AA, the number of longer stalling sequences that are expected to be false positives was higher than 5% (Supplemental Table S2). Taking these statistical considerations together, we found the length of 3 AA as the maximal length of peptide sequence that allows a statistically sufficient analysis of ribosome-stalling and proteome-representation.
Randomized USRs
Randomized USRs were generated separately for each mRNA by randomly drawing peak positions based on the number of RD peaks in the original profile. Specifically, the randomized version of an mRNA with n peaks would have n random peak positions (Fig. 1C). Accordingly, the randomized USRs of this protein would be the n sequences of 31 AA upstream of each random peak position.
Enrichment of over-, under-, and expectedly represented peptide sequences in the USRs
In order to quantify the extent to which over-, under-, and expectedly represented peptide sequences tend to appear before peaks, we compared their distribution in the real USRs with their distribution in the randomized USRs. Specifically, we summed over all USRs to get the total number of occurrences of the over, under-, and expectedly represented peptide sequences in the real USRs (Nreal). Then, we calculated in the same way their number of occurrences in each of the 10,000 randomized versions of the USRs (Nrandom). Finally, we defined an empirical P-value. For example, for underrepresented peptide sequences, the P-value will be given by
The calculation for the over- and expectedly represented peptide sequences is identical.
The “stalling” P-value
Let Nreal,i and Nrandom,i denote the number of occurrences of the ith peptide sequence in the real and randomized USRs, respectively. The stalling P-value of the ith peptide sequence is defined as
This P-value is corresponding to the probability that the ith peptide sequence is observed in the randomized USRs more often than in the real USRs. Peptides with Pstl,i ≤ 0.001 are termed here stalling peptides. The number of stalling peptide sequences that are expected to be false positives is presented in Supplemental Table S2 and was calculated by applying the stalling P-value on a randomized version of the USR rather than on the real one.
The “stalling” Z-score
Let Nreal,i denote the number of occurrences of the ith peptide sequence in the real USRs, and let mean(Nrandom,i) and std(Nrandom,i) denote the mean and standard deviation of the number of occurrences of the ith peptide sequence over all 10,000 randomized versions of the USRs. The standardized score of the ith peptide sequence is given by
Z-scorei represents the extent to which the number of occurrences of the ith peptide sequence in the USRs deviates from random in terms of standard deviations. Z-scores ≥2 correspond to significantly stalling peptides. Although the random distributions of peptide sequences in the randomized USRs did not turn out to be normal according to the Kolmogorov–Smirnov test (Massey 1951), the stalling Z-score can be used to “visually” estimate the extent to which stalling peptides tend to occupy the exit tunnel of stalled ribosomes.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported in part by a fellowship from the Edmond J. Safra Center for Bioinformatics at Tel-Aviv University and by the Israeli Ministry of Science, Space and Technology.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.059188.116.
REFERENCES
- Akashi H, Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc Natl Acad Sci 99: 3695–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci 100: 3889–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artieri CG, Fraser HB. 2014. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res 24: 2011–2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan S, Gartmann M, Halic M, Armache JP, Jarasch A, Mielke T, Berninghausen O, Wilson DN, Beckmann R. 2010. [α]-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat Struct Mol Biol 17: 313–317. [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. 2012. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charneski CA, Hurst LD. 2013. Positively charged residues are the major determinants of ribosomal velocity. PLoS Biol 11: e1001508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciryam P, Morimoto RI, Vendruscolo M, Dobson CM, O'Brien EP. 2013. In vivo translation rates can substantially delay the co-translational folding of the E. coli cytosolic proteome. Biophys J 104: 578a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana A, Tuller T. 2012. Determinants of translation elongation speed and ribosomal profiling biases in mouse embryonic stem cells. PLoS Comput Biol 8: e1002755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diament A, Tuller T. 2016. Estimation of ribosome profiling performance and reproducibility at various levels of resolution. Biol Direct 11: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM. 2004. Principles of protein folding, misfolding and aggregation. Semin Cell Dev Biol 15: 3–16. [DOI] [PubMed] [Google Scholar]
- Gerashchenko MV, Lobanov AV, Gladyshev VN. 2012. Genome-wide ribosome profiling reveals complex translational regulation in response to oxidative stress. Proc Natl Acad Sci 109: 17394–17399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Pilpel Y. 2011. Determinants of translation efficiency and accuracy. Mol Syst Biol 7: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhl M, Ragan MA. 2007. Is multiple-sequence alignment required for accurate inference of phylogeny? Syst Biol 56: 206–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. 2015. Understanding biases in ribosome profiling experiments reveals signatures of translation dynamics in yeast. PLoS Genet 11: e1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. 2007. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 315: 525–528. [DOI] [PubMed] [Google Scholar]
- Komar AA. 2009. A pause for thought along the co-translational folding pathway. Trends Biochem Sci 34: 16–24. [DOI] [PubMed] [Google Scholar]
- Kosolapov A, Deutsch C. 2009. Tertiary interactions within the ribosomal exit tunnel. Nat Struct Mol Biol 16: 405–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Boehringer D, Ban N, Bukau B. 2009. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16: 589–597. [DOI] [PubMed] [Google Scholar]
- Lawrence MG, Lindahl L, Zengel JM. 2008. Effects on translation pausing of alterations in protein and RNA components of the ribosome exit tunnel. J Bacteriol 190: 5862–5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino J, von Heijne G, Beckmann R. 2016. Small protein domains fold inside the ribosome exit tunnel. FEBS Lett 590: 655–660. [DOI] [PubMed] [Google Scholar]
- Massey FJ Jr. 1951. The Kolmogorov-Smirnov test for goodness of fit. J Am Stat Assoc 46: 68–78. [Google Scholar]
- McManus CJ, May GE, Spealman P, Shteyman A. 2014. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res 24: 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel AM, Fox G, M Kiran A, De Bo C, O'Connor PB, Heaphy SM, Mullan JP, Donohue CA, Higgins DG, Baranov PV. 2014. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res 42: D859–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell 108: 629–636. [DOI] [PubMed] [Google Scholar]
- Navon SP, Kornberg G, Chen J, Schwartzman T, Tsai A, Puglisi EV, Puglisi JD, Adir N. 2016. Amino acid sequence repertoire of the bacterial proteome and the occurrence of untranslatable sequences. Proc Natl Acad Sci 113: 7166–7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. 2002. Accounting for human polymorphisms predicted to affect protein function. Genome Res 12: 436–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson OB, Hedman R, Marino J, Wickles S, Bischoff L, Johansson M, Müller-Lucks A, Trovato F, Puglisi JD, O'Brien EP, et al. 2015. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep 12: 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peil L, Starosta AL, Lassak J, Atkinson GC, Virumäe K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. 2013. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci 110: 15265–15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. 2010. Synonymous but not the same: the causes and consequences of codon bias. Nat Rev Genet 12: 32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi J, Wang B, Hao B-I. 2004. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J Mol Evol 58: 1–11. [DOI] [PubMed] [Google Scholar]
- Ramu H, Vázquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, Micura R, Mankin AS. 2011. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell 41: 321–330. [DOI] [PubMed] [Google Scholar]
- Sabi R, Tuller T. 2015. A comparative genomics study on the effect of individual amino acids on ribosome stalling. BMC Genomics 16: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, et al. 2009. Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326: 1412–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senes A, Gerstein M, Engelman DM. 2000. Statistical analysis of amino acid patterns in transmembrane helices: the GxxxG motif occurs frequently and in association with β-branched residues at neighboring positions. J Mol Biol 296: 921–936. [DOI] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell 108: 591–594. [DOI] [PubMed] [Google Scholar]
- Thanaraj TA, Argos P. 1996. Ribosome-mediated translational pause and protein domain organization. Protein Science 5: 1594–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CJ, Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM, Nussinov R. 2008. Synonymous mutations and ribosome stalling can lead to altered folding pathways and distinct minima. J Mol Biol 383: 281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Zur H. 2015. Multiple roles of the coding sequence 5′ end in gene expression regulation. Nucleic Acids Res 43: 13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Chor B, Nelson N. 2007. Forbidden penta-peptides. Protein Science 16: 2251–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. 2010. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell 141: 344–354. [DOI] [PubMed] [Google Scholar]
- Tuller T, Veksler-Lublinsky I, Gazit N, Kupiec M, Ruppin E, Ziv-Ukelson M. 2011. Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol 12: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson DN, Beckmann R. 2011. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol 21: 274–282. [DOI] [PubMed] [Google Scholar]
- Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. 2013. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci 110: E878–E887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z. 2011. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr Opin Struct Biol 21: 25–31. [DOI] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. 2009. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 16: 274–280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.