Abstract
The human males absent on the first (MOF)-containing histone acetyltransferase nonspecific lethal (NSL) complex comprises nine subunits including the O-linked N-acetylglucosamine (O-GlcNAc) transferase, isoform 1 (OGT1). However, whether the O-GlcNAc transferase activity of OGT1 controls histone acetyltransferase activity of the NSL complex and whether OGT1 physically interacts with the other NSL complex subunits remain unclear. Here, we demonstrate that OGT1 regulates the activity of the NSL complex by mainly acetylating histone H4 Lys-16, Lys-5, and Lys-8 via O-GlcNAcylation and stabilization of the NSL complex subunit NSL3. Knocking down or overexpressing OGT1 in human cells remarkably affected the global acetylation of histone H4 residues Lys-16, Lys-5, and Lys-8. Because OGT1 is a subunit of the NSL complex, we also investigated the function of OGT1 in this complex. Co-transfection/co-immunoprecipitation experiments combined with in vitro O-GlcNAc transferase assays confirmed that OGT1 specifically binds to and O-GlcNAcylates NSL3. In addition, wheat germ agglutinin affinity purification verified the occurrence of O-GlcNAc modification on NSL3 in cells. Moreover, O-GlcNAcylation of NSL3 by wild-type OGT1 (OGT1-WT) stabilized NSL3. This stabilization was lost after co-transfection of NSL3 with an OGT1 mutant, OGT1C964A, that lacks O-GlcNAc transferase activity. Furthermore, stabilization of NSL3 by OGT1-WT significantly increased the global acetylation levels of H4 Lys-5, Lys-8, and Lys-16 in cells. These results suggest that OGT1 regulates the activity of the NSL complex by stabilizing NSL3.
Keywords: histone acetylase, histone acetylation, O-GlcNAcylation, O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), post-translational modification (PTM), NSL complex
Introduction
O-Linked β-N-acetylglucosamine (O-GlcNAc)3 transferase (OGT) is a ubiquitously expressed and conserved enzyme that catalyzes O-GlcNAc addition to serine (Ser) and threonine (Thr) residues of cytosolic and nuclear proteins (1). Dynamic changes of the post-translational O-GlcNAc modification (O-GlcNAcylation) are controlled by OGT and the glycoside hydrolase O-GlcNAcase (OGA) (2). Numerous studies have confirmed that O-GlcNAcylation is involved in many fundamental cellular processes, including gene transcription (3), cell signaling (4), cell growth (5), and apoptosis (6). Accumulating data reveal that O-GlcNAcylation as “protein code” or “histone code” may provide recognition platforms or executive instructions for subsequent proteins to carry out biological processes (7). For example, O-GlcNAcylation by OGT to the specific repeats region of host cell factor 1 (HCF1) may provide instructions for HCF1 proteolysis (8). In addition, O-GlcNAcylation at histone H2B Ser-112 by OGT stimulates H2B Lys-120 ubiquitination, which further activates gene transcription such as ring finger protein 20 (9, 10), suggesting a complicated correlation between O-GlcNAcylation and the other post-translational modifications.
In cells, OGT as a glycosyltransferase interacts with numerous proteins. Besides free OGT, it also forms complexes with certain proteins such as ten-eleven translocation (TET) enzymes, mixed lineage leukemia (MLL)-SET1 methyltransferases, or males absent on the first (MOF) histone acetyltransferase (HAT) to accomplish intricate cellular processes (11–15). TET proteins (including TET1/2/3) regulate gene transcription by catalyzing the conversion of 5-methylcytosine to 5-hydroxymethylcytosine. Research evidence suggests that the coordination of TET2/TET3 and OGT regulates the binding of the SET1-COMPASS histone H3K4 methyltransferase to chromatin, thus further controlling subsequent gene transcription (11). We previously purified and characterized a novel human MOF (hMOF)-containing HAT nonspecific lethal (NSL) complex that was composed of nine subunits including the male-specific lethal (MSL) 1-like protein NSL1, NSL2 (FLJ20436), NSL3 (FLJ10081), HCF1, plant homeodomain-linked finger-containing protein PHF20, O-linked N-acetylglucosamine transferase, isoform 1 (OGT1), WD repeat protein 5, microspherule protein 1 (15), and hMOF as a catalytic subunit of the NSL complex (Fig. 1A, left). Compared with the traditional hMOF-containing MSL complex that plays a key role in fly dosage compensation and is specifically responsible for histone H4 Lys-16 acetylation (H4K16ac) (16), the NSL complex possesses a broader substrate specificity and is able to acetylate H4 residues Lys-16, Lys-5, and Lys-8. Further studies indicate that the NSL complex functions in promoting histone H3 Lys-4 dimethylation activity by MLL-SET complexes in a unidirectional manner (17). Although biochemical purifications combined multidimensional protein identification technology (MudPIT) analysis confirmed that OGT1 is closely associated with the NSL complex (15), there are many questions yet to be resolved such as (i) whether OGT1's O-GlcNAc transferase activity impacts the HAT activity of the NSL complex in cells, (ii) whether OGT1 physically interacts with the other NSL complex subunits, and (iii) whether any NSL complex subunit can be O-GlcNAcylated by OGT1.
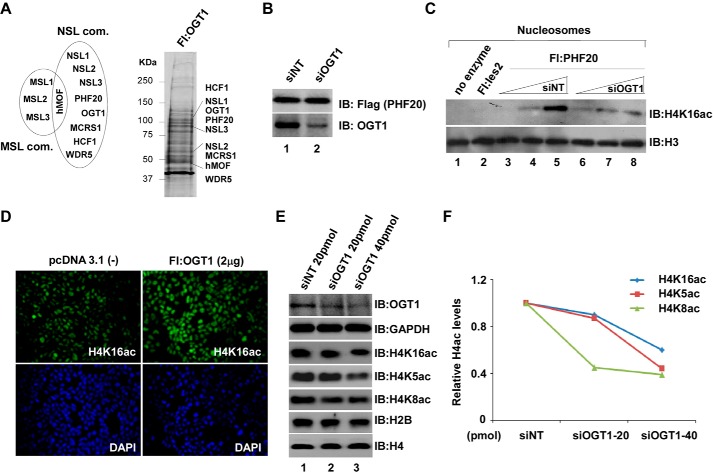
Figure 1.
OGT1 may play a critical role in HAT NSL complex. A, identification of all NSL complex (com) subunits from cells stably expressing FLAG-OGT1. Left, schematic diagram of two distinct hMOF-containing MSL and NSL complexes. Right, anti-FLAG eluates of cells stably overexpressing FLAG-OGT1 were subjected to silver staining. B and C, decline of HAT activity in OGT1 knockdown cells stably expressing FLAG-PHF20. FLAG-PHF20-expressing 293FRT cells were transfected with 20 pmol of OGT1 and non-targeting (NT; as a control) siRNA. 48 h after transfection, cells were harvested and lysed for anti-FLAG IP (B). Anti-FLAG agarose eluates were subjected to HAT assays (C). HAT assays were performed essentially as described (15). Reconstituted nucleosomes were used in HAT assays. IB, immunoblot. D, increase of global H4K16ac in OGT1-overexpressing HeLa cells that were transfected with pcDNA/FLAG-OGT1. 48 h after transfection, immunofluorescence staining was carried out to detect global H4K16ac status. Cell nuclei were stained using Vectashield with DAPI (Vector Laboratories, Inc., catalog number H-1200). E, impact of OGT1 on global acetylation of histone H4 residues Lys-16, Lys-5, and Lys-8. 293T cells were transfected with 20 and 40 pmol of OGT1 and non-targeting siRNA. 48 h later, Western blot analysis was performed using the indicated antibodies. GAPDH, H2B, and H4 are internal controls. F, quantified proteins. Western blot images were quantified using Quantity One software (Bio-Rad). The y axis represents the histone H4 acetylation (H4ac) levels relative to the total H4. Fl, FLAG.
In an effort to address the above questions, a series of biochemical and molecular biological experimental approaches were used in this study. As we show below, our results clarify a new collaborative mechanism in which the stability of NSL3 in cells is associated with its O-GlcNAcylation by OGT1, and this modification further impacts the HAT activity of the NSL complex as reflected in changes in global histone H4 acetylation at lysine Lys-5, Lys-8, and Lys-16 in cells.
Results
OGT1 may play a critical role in hMOF-containing NSL complex
The hMOF as a catalytic subunit forms two distinct HAT (NSL and MSL) complexes in cells (Fig. 1A, left). We previously identified a novel hMOF-containing HAT NSL complex that can acetylate histone H4 residues Lys-5, Lys-8, and Lys-16 (15). Interestingly, OGT1 as a subunit of the NSL complex was identified by MudPIT analyses in anti-FLAG eluates of several stably expressed FLAG-tagged subunits of the NSL complex (15). Conversely, all subunits of the NSL complex were also identified by MudPIT in anti-FLAG eluates from cells stably expressing FLAG-OGT1 (Fig. 1A, right), suggesting that OGT1 is stably assembled in the NSL complex. To know the role of OGT1 in the NSL complex, an in vitro HAT assay was first carried out with anti-FLAG eluates from cells stably expressing FLAG-PHF20 (a subunit of the NSL complex) after knocking down OGT1 with siRNA (Fig. 1B). Compared with non-targeting siRNA, decreased H4K16ac on reconstituted nucleosomes in OGT1 knockdown cells was observed (Fig. 1C). To further investigate whether OGT1 affects the global histone H4 acetylation in cells, experiments involving overexpression or knockdown of OGT1 in HeLa or human embryonic kidney (HEK) 293T (293T) cells were performed. As shown in Fig. 1D, a remarkable increase of global H4K16ac by overexpression of OGT1 was visualized with immunofluorescence staining. However, a reduction of global H4K16ac, H4K5ac, and H4K8ac was observed in siOGT1-transfected 293T cells (Fig. 1E), suggesting a role of OGT1 in regulating the global acetylation of histone H4 residues Lys-16, Lys-5, and Lys-8 in cells. Quantified acetylation levels of histone H4 at lysines 5, 8, and 16 are shown in Fig. 1F.
Physical interaction between OGT1 and NSL3 was confirmed by co-transfection/co-immunoprecipitation (co-IP) approaches
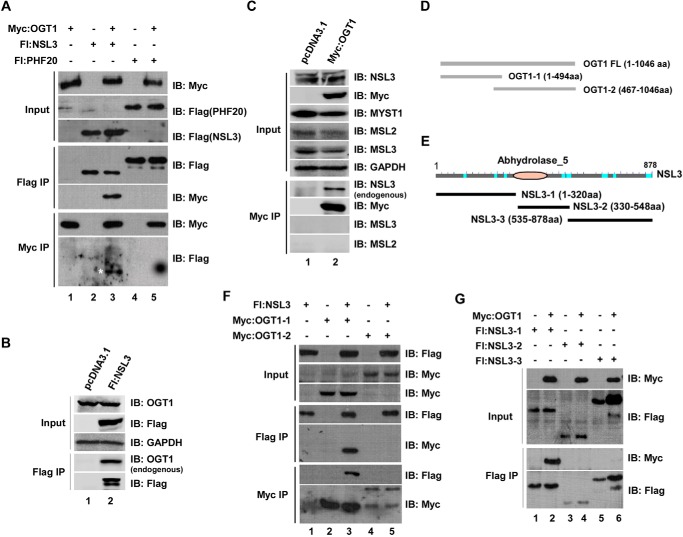
The potential importance of OGT1 in the NSL complex was confirmed by previous experiments. This prompted us to further clarify the structural relationship between OGT1 and the other NSL complex subunits. To do that, co-transfection/co-IP experiments were first designed using Myc-tagged OGT1 as bait. Besides HCF1, which has been reported to interact with OGT1 (18), all other NSL complex subunits were tested. Based on the experimental results, the only direct interaction was observed between OGT1 and NSL3, a specific subunit of the NSL complex (Fig. 2A, lane 3) (other negative data not shown). To verify this result, it was further examined whether the endogenous OGT1 or NSL3 protein could be precipitated by overexpressed proteins. As expected, endogenous OGT1 (Fig. 2B, Flag IP/anti-OGT1) was immunoprecipitated by overexpressed FLAG-NSL3. Similarly, the endogenous NSL3 protein (Fig. 2C, Myc IP/anti-NSL3) was specifically immunoprecipitated by overexpressed Myc-OGT1 because no other proteins, such as MSL2 and MSL3, precipitated. Next, to further define the binding region of OGT1 with NSL3, we designed and constructed deletion mutants for OGT1 (Fig. 2D) and NSL3 (Fig. 2E). Co-transfection/co-IP experiments were then carried out. As expected, full-length NSL3 bound to the N-terminal tetratricopeptide repeat (TPR) domain of OGT1 (OGT1-1) (Fig. 2F, lane 3), whereas full-length OGT1 interacted with N-terminal NSL3 (Fig. 2G, Flag IP/anti-Myc).
Figure 2.
Co-transfection/co-IP experiments clarify the physical interaction of OGT1 and NSL3. A, clarification of physical interaction of OGT1 and NSL3 in 293T cells. Cells were co-transfected with the indicated FLAG- or Myc-tagged plasmids. 48 h after transfection, the prepared whole-cell lysate was subjected to co-IP experiments. Anti-FLAG- or anti-Myc-agarose eluates were subjected to SDS-PAGE, and bound proteins were detected by Western blotting with anti-FLAG or anti-Myc antibody. Fl:NSL3, FLAG-NSL3; Fl:PHF20, FLAG-PHF20. B and C, detection of endogenous OGT1 or NSL3 using IP approaches. 293T cells were transfected with Myc-OGT1 or FLAG-NSL3 to perform IP experiments. Immunoprecipitated endogenous NSL3 or OGT1 was then detected by Western blotting using specific antibodies. D and E, a schematic of the deletion mutants of OGT1 (D) and NSL3 (E). F and G, mapping the binding region between OGT1 and NSL3. 293T cells were co-transfected with full-length NSL3 and the indicated deletion mutants of OGT1 (F) or full-length OGT1 and the indicated deletion mutants of NSL3 (G). 48 h later, co-IP experiments were performed with prepared whole-cell lysate. Bound proteins were detected using Western blot analysis with the indicated antibodies. IB, immunoblot.
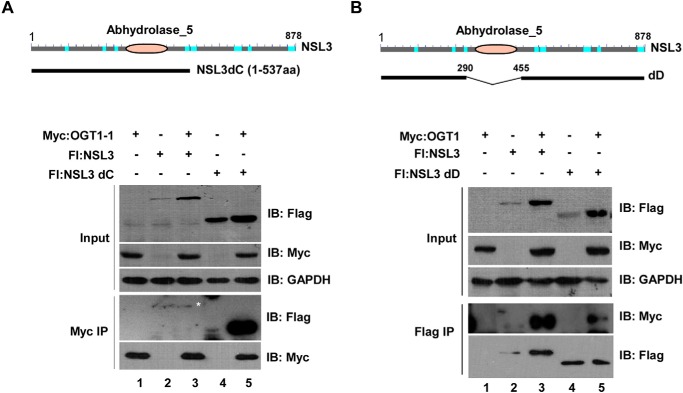
It has been reported that the N terminus of NSL3 contains an abhydrolase domain, but the function of this domain is unclear (19). Thus, to determine whether the abhydrolase domain of NSL3 affects the binding activity between OGT1 and NSL3, we constructed a plasmid that contains full abhydrolase domain region (NSL3dC) (Fig. 3A, upper) and a plasmid lacking the abhydrolase domain (NSLdD) (Fig. 3B, upper). As shown in Fig. 3A (lower panel), compared with full-length NSL3, apparent increased binding activity between NSL3dC and OGT1-1 was observed (Myc IP/anti-FLAG, lane 5). In contrast, the binding activity between OGT1 and NSL3 was significantly decreased by deletion of the abhydrolase domain of NSL3 (Fig. 3B, lower panel, Flag IP/anti-Myc), suggesting the importance of the abhydrolase domain of NSL3 for binding to OGT1.
Figure 3.
Abhydrolase domain of NSL3 is required for its binding activity to OGT1. A, enhancement of binding activity between abhydrolase domain-containing N-terminal NSL3 and N-terminal OGT1-1. 293T cells were co-transfected with N-terminal OGT1–1 and full-length NSL3 or a deletion mutant of NSL3 (NSL3dC) (upper panel), and bound proteins were detected using Western blotting with the indicated antibodies. B, reduction of binding activity between NSL3 without the abhydrolase domain (NSLdD) and OGT1. Similarly, 293T cells were co-transfected with OGT1 and abhydrolase domain-deleted NSL3 (upper panel), and bound proteins were measured by Western blotting with anti-FLAG or anti-Myc antibody. IB, immunoblot; Fl, FLAG.
O-GlcNAcylation of NSL3 by OGT1 was confirmed by in vitro and in vivo approaches
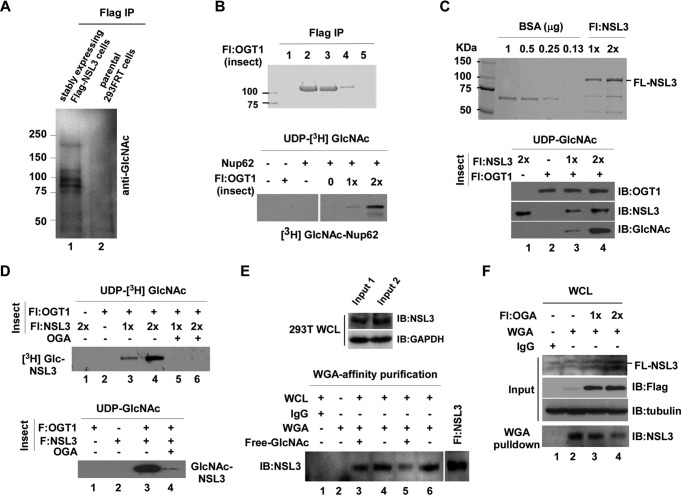
The physical interaction between OGT1 and NSL3 raises the possibility that NSL3 may be a substrate of OGT1. To confirm our speculation, we first tested whether there were O-GlcNAc-modified proteins in anti-FLAG eluates from cells stably expressing FLAG-tagged NSL3. As shown in Fig. 4A, compared with parental 293FRT cells, multi-O-GlcNAc-modified signals were detected in the FLAG-NSL3 lane, suggesting the existence of O-GlcNAcylated NSL complex subunits, possibly including NSL3 and HCF1 (as has been reported). To further address this result, insect cell-expressed and purified FLAG-OGT1 (Fig. 4B, upper) and FLAG-NSL3 (Fig. 4C, upper) were used in an in vitro O-GlcNAc transferase assay. We first tested the enzyme activity of purified OGT1 by examining the O-GlcNAcylation of Nup62 (a known substrate of OGT1) by OGT1. After confirming the glycosylation activity of purified OGT1 (Fig. 4B, lower), the impact of OGT1 on recombinant NSL3 in an in vitro O-GlcNAc transferase assay was measured. As shown in Fig. 4C (lower, lanes 3 and 4), O-GlcNAc-modified NSL3 by OGT1 was visualized in the presence of donor UDP-N-acetylglucosamine (UDP-GlcNAc). Moreover, this O-GlcNAcylation on NSL3 was removed by adding the O-GlcNAc hydrolase OGA in the presence of either UDP-[3H]GlcNAc (Fig. 4D, upper, lanes 5 and 6) or cold UDP-GlcNAc (Fig. 4D, lower, lane 4). Thus, there is no doubt that OGT1 can O-GlcNAcylate NSL3 protein in vitro. WGA is a lectin that can bind O-GlcNAc residues, and it is often used to detect intracellular O-GlcNAcylated proteins (20). To further demonstrate the O-GlcNAc modification on NSL3 in cells, WGA lectin affinity purification of total cell lysates from 293T cells (Fig. 4E, upper panel) was carried out. As shown in Fig. 4E (lower panel), NSL3 was detected by immunoblot analysis of the precipitate (lanes 4 and 6). Specificity was shown by adding free GlcNAc during WGA lectin affinity purification (lane 3 and 5). Consistent with this, dose-dependent reduction of O-GlcNAc-modified endogenous NSL3 was confirmed by WGA affinity purification in FLAG-OGA-transfected cells (Fig. 4F). These results suggest the occurrence of O-GlcNAc modification on NSL3 in cells.
Figure 4.
NSL3 can be O-GlcNAcylated by OGT1 in vitro and in vivo. A, O-GlcNAcylation of stably expressed FLAG-NSL3 and its associated proteins. Anti-FLAG-agarose eluates from FLAG-NSL3-expressing HEK293FRT cells and parental 293FRT cells (as a negative control) were subjected to SDS-PAGE in a 4–20% gradient gel, and O-GlcNAc-modified proteins were visualized by Western blotting with anti-GlcNAc antibody. B, confirmation of O-GlcNAc transferase activity of the recombinant OGT1. Insect cell-expressed/purified full-length OGT1 was visualized by Coomassie Brilliant Blue staining (upper). An in vitro O-GlcNAc transferase assay was performed by mixing recombinant OGT1, UDP-[3H]GlcNAc, and Nup62. O-GlcNAc transferase activity was measured by autoradiography (lower). C, O-GlcNAcylation of recombinant NSL3 by OGT1. Insect cell-expressed/purified full-length NSL3 (FL-NSL3) (upper) was used in an O-GlcNAc transferase assay. Modified O-GlcNAcylation was measured by anti-GlcNAc antibody (lower). D, OGA antagonistic effect on OGT1-produced O-GlcNAcylation of NSL3. O-GlcNAc transferase assays were performed as indicated in the presence or absence of OGA. Modified O-GlcNAcylation was visualized by autoradiography (upper panel) or by Western blotting with anti-GlcNAc antibody (lower panel). E, WGA lectin affinity purification. Whole-cell lysates (WCL) from 293T cells (upper panel) were subjected to WGA affinity purification. Binding reactions were as indicated. The precipitates were then analyzed by Western blotting for endogenous NSL3. Free GlcNAc (20 mm) was added for control of specificity (lower panel). Lanes 3 and 4 and lanes 5 and 6 represent two biological experimental repeats. Input 1 and Input 2 indicate whole-cell lysates harvested at different times. F, WGA lectin affinity purification in FLAG-OGA transfected cells. Prepared whole-cell lysates of FLAG-OGA (5 or 10 μg/10-cm dish) transiently transfected (48 h) 293T cells were subjected to WGA affinity purification. Endogenous O-GlcNAcylated NSL3 was detected by Western blotting with NSL3 antibody. IB, immunoblot; Fl, FLAG.
O-GlcNAc transferase activity of OGT1 was tightly associated with its stabilization of NSL3
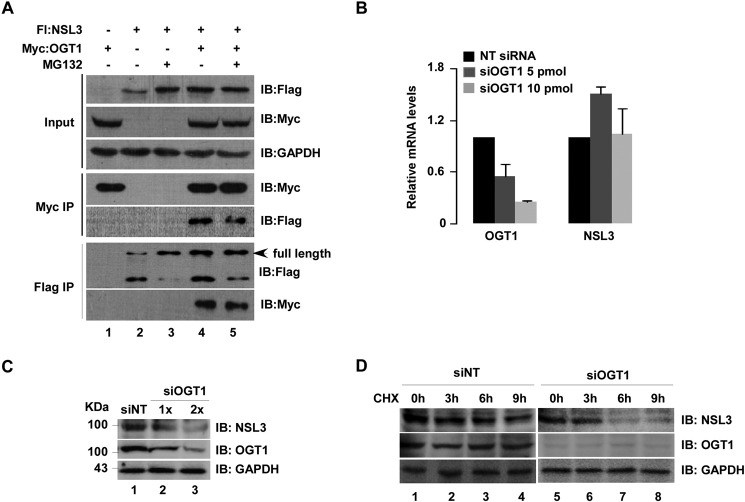
In fact, during the experiments, higher protein levels of NSL3, either full length or deletion mutants, were observed in Input (Fig. 5A) after co-transfection of OGT1 and NSL3, suggesting a role of OGT1 in stabilizing the NSL3 protein. To confirm this phenomenon, we designed and performed experiments as indicated in Fig. 5A. Compared with NSL3-only transfection (first row, lane 2), the protein levels of NSL3 in Input increased by treating cells with MG132 (first row, lane 3) or co-transfection with OGT1 (first row, lane 4). Similar results were seen after FLAG IP/anti-FLAG (full-length NSL3; lanes 3–5). To further verify this phenomenon, knockdown of OGT1 with siRNA was carried out. There was no obvious change in NSL3 mRNA level (Fig. 5B), but the NSL3 protein level decreased in a dose-dependent manner (Fig. 5C, lanes 2 and 3). This behavior was further confirmed by detecting NSL3 protein at different time points (0, 3, 6, and 9 h) after treating cells with the protein synthesis inhibitor cycloheximide (CHX; 3 μg/ml). As shown in Fig. 5D, knockdown of OGT1 with siRNA accelerated the degradation of NSL3 (lanes 5–8) compared with that in the non-targeting siRNA group (lanes 1–4), suggesting a role of OGT1 in regulating the stabilization of NSL3 in a post-translational manner.
Figure 5.
Stabilization of NSL3 by OGT1 was observed in cells. A, stabilization of NSL3 by OGT1 in 293T cells. Cells were co-transfected with FLAG-NSL3 and Myc-OGT1 as indicated with or without MG132 (10 μm; 12 h). 48 h later, co-IP was performed with prepared whole-cell lysates. Bound proteins were confirmed by Western blotting with anti-FLAG or anti-Myc antibody. B and C, expression level of NSL3 in siOGT1-treated 293T cells. mRNA and protein levels of NSL3 were measured using quantitative real time PCR (B) and Western blotting (C), respectively, in OGT1 knockdown 293T cells. D, expression level of NSL3 in OGT1 knockdown cells treated with CHX. OGT1 knockdown 293T cells were treated with CHX (3 μg/ml) for 0, 3, 6, and 9 h. Proteins in whole-cell lysates were detected using specific antibodies. IB, immunoblot; Fl, FLAG; NT, non-targeting.
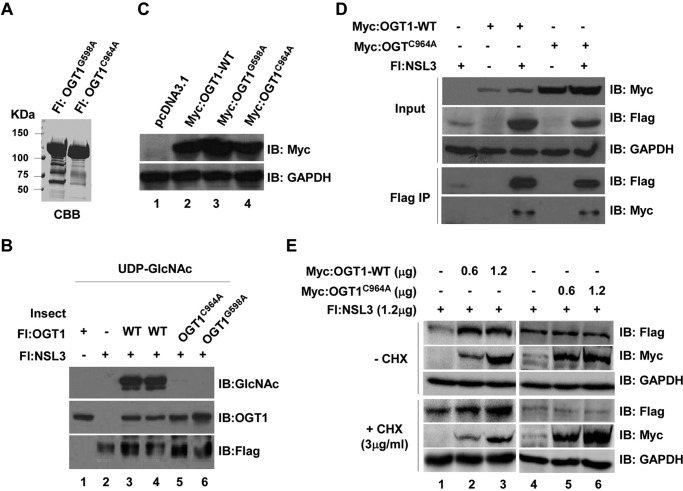
Because NSL3 can be O-GlcNAcylated by OGT1, we hypothesized that the stabilizing function of OGT1 on NSL3 might be associated with its O-GlcNAcylation of NSL3. To address this hypothesis, point mutants of OGT1 (OGT1G598A and OGT1C964A) were expressed and purified from insect cells (Fig. 6A, left). The enzyme activity of both OGT1G598A and OGT1C964A was then tested using in vitro O-GlcNAc transferase assays. Compared with wild-type OGT1 (OGT1-WT) (lanes 3 and 4), no signals were detected in point mutant of OGT1 (OGT1mt) lanes by Western blotting with anti-GlcNAc antibody, suggesting that both OGT1G598A and OGT1C964A mutants lost the enzyme activity (Fig. 6B, lanes 5 and 6). To further confirm whether the catalytically dead mutants abolish the interaction between OGT1 and NSL3, OGT1G598A and OGT1C964A mutants were subcloned into a mammalian expression vector. Protein expression was confirmed by Western blotting (Fig. 6C). Then Myc-tagged OGT1C964A was used in co-transfection/IP experiments. As shown in Fig. 6D, although OGT1C964A lost the enzyme activity, like OGT1-WT, OGT1C964A also interacted with NSL3 (lane 5, Flag IP/IB: Myc). However, the ability to stabilize NSL3 was obviously decreased because the expression level of OGT1C964A relative to OGT1-WT was significantly higher (Input/IB: Myc). To further investigate whether OGT1C964A affects the stability of NSL3 protein, increasing amounts of OGT1-WT or OGT1C964A were co-transfected with 1.2 μg of NSL3 with or without CHX treatment. Clearly, the NSL3 was stabilized by OGT1-WT (Fig. 6E, lanes 2 and 3) but not by OGT1C964A (Fig. 6E, lanes 5 and 6) in cells. These results strongly suggest that the mechanism of O-GlcNAcylation of OGT1 is involved in stabilizing NSL3.
Figure 6.
The O-GlcNAc transferase activity of OGT1 is tightly associated with its stabilization of NSL3. A, purified OGT1mt proteins. Insect cell-expressed and purified point-mutated FLAG-tagged OGT1 proteins (OGT1G598A and OGT1C964A) were visualized by Coomassie Brilliant Blue (CBB) staining. B, loss of O-GlcNAc transferase activity of OGT1mt. An O-GlcNAc transferase assay was performed using OGT1-WT or OGT1mt. Modified NSL3 was detected by Western blotting with anti-GlcNAc antibody. C, validation of OGT1-WT and OGT1mt mammalian expression plasmids. Myc-tagged OGT1-WT and OGT1mt plasmids were transiently transfected into 293T cells. Expressed proteins were checked by Western blotting with anti-Myc antibody. pcDNA3.1 vector is a negative control. D, binding activity of OGT1mt and NSL3. Cells were co-transfected with FLAG-NSL3 and Myc-OGT1-WT/OGTC964A as indicated. 48 h later, FLAG-IP was performed with prepared whole-cell lysates. Bound proteins were analyzed by Western blotting. E, stability of NSL3 protein in OGT1-WT- or OGT1mt-overexpressing 293T cells in the presence or absence of CHX. 293T cells were treated with the translation inhibitor CHX (3 μg/ml) 6 h before harvesting. Western blot analysis was performed after anti-FLAG affinity purification. IB, immunoblot; Fl, FLAG.
OGT1 regulates global histone H4 acetylation via stabilization of the NSL3
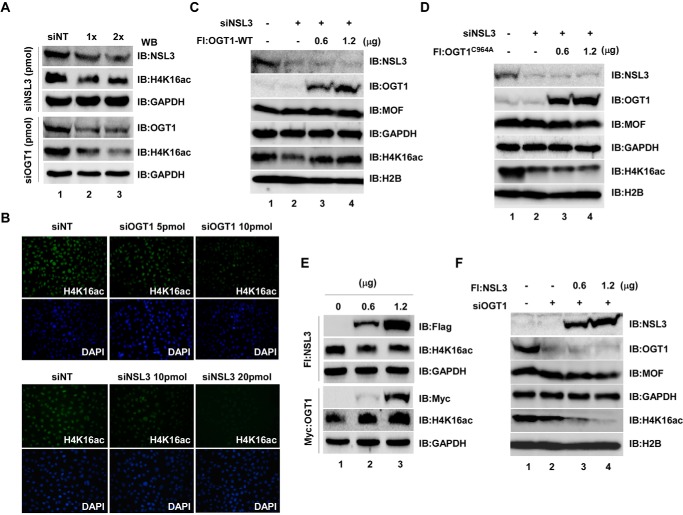
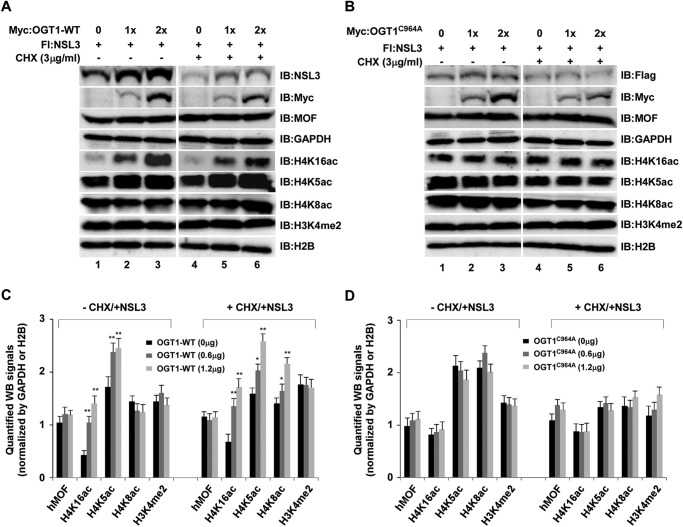
Because both OGT1 and NSL3 are subunits of the NSL complex, it is necessary to know whether OGT1 regulates the enzyme activity of the NSL complex through stabilization of NSL3. To gain a clear idea of the functions of OGT1 in the NSL complex, the coordination of OGT1 and NSL3 in global histone H4K16ac was investigated. At first, knockdown of either OGT1 or NSL3 with siRNA in 293T (Fig. 7A) or HeLa (Fig. 7B) cells significantly decreased global H4K16ac in a dose-dependent manner. However, overexpression of OGT1-WT (Fig. 7C), but not OGT1C964A (Fig. 7D), in NSL3 knockdown 293T cells restored the global H4K16ac to normal levels (Fig. 7, C, row 5, and D, row 5). Interestingly, although overexpression of OGT1 in 293T cells dramatically increased global H4K16ac (Fig. 7E, lower), reduction or lesser changes of global H4K16ac were observed in NSL3-overexpressing cells (Fig. 7E, upper), and this reduction was more obvious in OGT1 knockdown 293T cells (Fig. 7F, lanes 3 and 4, row 5), suggesting the importance of the O-GlcNAcylation of NSL3 in regulating global H4K16ac in cells. Although both hMOF-containing HAT MSL and NSL complexes can acetylate H4 Lys-16 (21, 22), the NSL complex can simultaneously acetylate histone H4 at Lys-5 and Lys-8 (15). Therefore, to further clarify the coordination of OGT1 and NSL3 in regulating the global H4K16ac, H4K5ac, and H4K8ac, co-transfection of NSL3 and OGT1-WT or OGT1C964A was carried out. As shown in Fig. 8, 1.2 μg of NSL3 was co-transfected with increasing amounts of OGT1-WT or OGT1C964A in the presence or absence of CHX (3 μg/ml). OGT1-WT significantly increased global H4K16ac and H4K5ac in cells with or without CHX. Although the changes of H4K8ac were not obvious in co-transfection of NSL3 and OGT1-WT in the absence of CHX, the global H4K8ac was dose-dependently raised in CHX-exposed cells (Fig. 8A, right panel). However, no obvious increase in NSL3 and no dose-dependent increase in H4K16ac, H4K5ac, or H4K8ac were observed in cells after co-transfection of NSL3 and OGT1C964A in the absence or presence of CHX (3 μg/ml) (Fig. 8B). Quantified protein levels of Fig. 8, A and B, are shown in Fig. 8, C and D, respectively.
Figure 7.
O-GlcNAc modification of NSL3 by OGT1 is essential for acetylating the global H4 Lys-16 in cells. A and B, decreased global H4K16ac in both OGT1 and NSL3 knockdown 293T or HeLa cells. Cells were transfected with NSL3- or OGT1-specific siRNA SMARTpool. 48 h after transfection, Western blot (WB) analysis (A) and immunofluorescence staining (B) were carried out to detect the global histone H4K16ac status. C and D, restoration of the global histone H4K16ac in NSL3 knockdown 293T cells. Cells were transfected with siNSL3. 12 h later, the cells were continuously transfected with OGT1-WT (C) or OGT1mt (OGT1C964A) (D) plasmids. 48 h after transfection of plasmids, cells were harvested, and the prepared whole-cell lysates were subjected to Western blot analysis. E, effect of regulation of overexpression of OGT1 or NSL3 on global H4K16ac in cells. 293T cells were transfected with NSL3 or OGT1 plasmids. 48 h after transfection, Western blotting was performed to check the global H4K16ac level in whole-cell lysates. F, additive effect of the global H4K16ac by overexpression of NSL3 in OGT1 knockdown cells. siOGT1 knockdown 293T cells were transfected with 0, 0.6, and 1.2 μg of FLAG-NSL3 plasmids. IB, immunoblot; Fl, FLAG; NT, non-targeting.
Figure 8.
O-GlcNAcylation of NSL3 by OGT1 facilitates the global acetylation of histone H4 residues Lys-5, Lys-8, and Lys-16 in cells. A and B, coordination of OGT1-NSL3 in global histone H4 Lys-5/8/16 acetylation in 293T cells with or without CHX treatment. Cells were co-transfected with the indicated plasmids. 48 h later, Western blot analysis was performed using the indicated antibodies. GAPDH and H2B are internal controls. Cells were treated with CHX (3 μg/ml) 6 h before harvesting. Quantified Western blot (WB) signals of A and B was shown in C and D, respectively. IB, immunoblot; Fl, FLAG.
Discussion
In this study, using an in vitro O-GlcNAc transferase assay and WGA affinity purification combined with knockdown/overexpression approaches, we found that that OGT1 can regulate global histone H4 acetylation at Lys-5, Lys-8, and Lys-16 by stabilizing NSL3. In cells, ubiquitously expressed and conserved glycosyltransferase OGT1 not only O-GlcNAc-modifies the substrate proteins but also often forms complexes to participate in cell function (11–14). Although OGT1 is characterized as a subunit of the NSL complex (15), the precise functions of OGT1 assembled in complex remain unclear. Previous reports suggest that cleavage of the HCF1pro repeat (HCF1 is also a subunit of the NSL complex) is tightly associated with its N-terminal O-GlcNAc modification by OGT1 (8). However, whether some other subunits of the NSL complex can be modified by OGT1 has not been explored. Based on in vitro O-GlcNAc transferase assays, we verified that OGT1 O-GlcNAcylated the NSL3 subunit. In line with this, WGA lectin affinity purification further confirmed the occurrence of O-GlcNAc modification on NSL3 in cells. Functional domain studies have proved that OGT1 is composed of two important regions, an N-terminal 13.5 TPR superhelical structure and a C-terminal catalytic domain (23, 24). The TPR domain is often required for the protein substrate recognition (25). Consistent with this view, an interaction between N-terminal OGT1 and N-terminal NSL3, including the abhydrolase domain, was demonstrated, suggesting that NSL3 as a substrate protein might be recognized by OGT1.
In fact, numerous intracellular proteins are stabilized by OGT1 through O-GlcNAc modification. For instance, tumor suppressor gene p53 can be stabilized by OGT1-mediated O-GlcNAcylation by blocking its ubiquitin pathway (26, 27). Also, O-GlcNAcylation of MLL5 at Ser-435 and Thr-440 decreased its ubiquitin modification and thus maintained the intracellular protein levels (28). In our study, co-transfection of OGT1 with NSL3 in cells increased the NSL3 protein levels in the presence or absence of the proteasome inhibitor MG132, suggesting that mechanisms outside of the ubiquitin pathway might also be involved in the stabilization of NSL3. Moreover, in our knockdown experiments, an obvious reduction of NSL3 proteins was observed in OGT1 knockdown cells with or without CHX treatment, indicating that the stabilization of OGT1 on NSL3 might be associated with its enzyme activity. Consistent with this idea, the stabilization of NSL3 was lost by co-transfecting the catalytically dead mutant OGT1C964A with NSL3.
Recently, increasing evidence shows that OGT1-mediated O-GlcNAcylation is often implicated in chromatin-mediated processes in a coordinated manner (29). For instance, O-GlcNAcylation at histone H2A Thr-101 relaxes chromatin structure through destabilization of H2A-H2B dimers (30). In contrast, O-GlcNAcylation at histone H2B Ser-112 may preserve stable chromatin at the early stage of adipocyte differentiation, thus repressing gene transcription in cell fate determination (31). In addition, O-GlcNAcylation of H2B Ser-112 facilitates H2B Lys-120 ubiquitination, which further acts as a platform to recruit the SET1/COMPASS complex, thereby activating gene transcription by trimethylating histone H3 Lys-4 (11). Given that both OGT1 and NSL3 are components of the NSL complex (15), we speculate that they might play an important role in regulating the whole enzyme activity of the NSL complex. In line with this speculation, acetylation of histone H4 residues Lys-16, Lys-5, and Lys-8 was down-regulated by knockdown of OGT1 or NSL3. It has been reported that histone H4 acetylation is one of the most important mechanisms to regulate chromatin structure, and it functions in fundamental cellular processes such as DNA repair, cell autophagy, and tumorigenesis (2). The HAT NSL complex is mainly responsible for acetylating histone H4 at Lys-16, Lys-5, and Lys-8 in cells. Besides promoting histone H3 Lys-4 dimethylation by cooperating with MLL-SET complexes, the NSL complex can be recruited by c-Jun and regulates the c-Jun target genes (32). Conversely, OGT1 is an important and indispensable enzyme in the cell metabolic pathway. Thus, it is easy to imagine functional links between O-GlcNAcylation and fundamental biological processes in cells. This is why unbalanced O-GlcNAc modification on substrate proteins leads to various kinds of diseases such as diabetes, neurologic disorders, cardiovascular disease, and cancer (33). Taken together, it is not difficult to understand that interaction between OGT1 and NSL3 may play a critical role in regulation of the HAT activity of the NSL complex and other functions in cells. More recently, Akhtar and co-workers (34) found that NSL3 as an autonomous microtubule minus-end-binding protein localized to the mitotic spindle, demonstrating that NSL3 is essential for spindle assembly and chromosome segregation. In addition, NSL3 can localize in mitochondria and mediates the binding of MOF and mitochondrial DNA to regulate oxidative phosphorylation and related gene expression (35). However, how the interaction of OGT1 and NSL3 is involved in the above functions remains to be further investigated.
In summary, we provide a new collaborative mechanism that OGT1 impacts the enzyme activity of the NSL complex by regulating global histone H4 acetylation. However, there are still some issues to be discussed. For example, although overexpression of OGT1 increased global histone H4 acetylation, an unexpected decline in H4K16ac was observed in NSL3-overexpressing cells. Furthermore, additive reduction of global H4K16ac was detected by overexpression of NSL3 in OGT1 knockdown cells, suggesting that the enzyme activity of the NSL complex may be required for O-GlcNAc-modified NSL3 assembly in the complex. One possible reason for this is that there is insufficient endogenous OGT1 to O-GlcNAcylate transient overexpressed NSL3; therefore the ratio of non-O-GlcNAcylated NSL3 proteins is dramatically increased. This may explain why the global H4K16ac was further decreased after overexpression of NSL3 in OGT1 knockdown cells. A second possible reason is that reduction of O-GlcNAc-modified NSL3 resulted in instability of NSL3, thus affecting the formation of NSL complex, and third, there might be a regulatory pathway beyond NSL3-OGT1 co-regulation.
Materials and methods
Antibodies
Anti-FLAG (M2)- and anti-Myc-agarose (A7470), anti-FLAG M2 (F3165), anti-H4K16ac (H9164) polyclonal antibody, UDP-GlcNAc (U4375), and MG132 (Z-Leu-Leu-al) were from Sigma. Anti-OGT1 (H300, sc-32921) polyclonal antibody, anti-GlcNAc (IgM), anti-α-tubulin (sc-58666) monoclonal antibody, and anti-c-Myc (9E10) monoclonal antibody were obtained from Santa Cruz Biotechnology. Anti-H4K5ac (07-327), anti-H4K8ac (07-328), and anti-H3K4me2 (07-030) polyclonal antibodies were purchased from Merck Millipore (Germany). Anti-histone H4 (16047-1-AP) polyclonal antibody was from Proteintech Group (Wuhan, China). Anti-MSL2 (ab83911) was from Abcam (UK). Anti-MOF (BM1942) monoclonal antibody was obtained from Boster (China). Anti-NSL3, anti-MSL3L1, anti-H2B, and anti-GAPDH rabbit polyclonal antibodies were raised against bacterially expressed proteins (Jilin University). CHX was purchased from GenView.
Cell culture
HEK293FRT, HEK293T, or HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (KangYuan Biology, China) and 1% penicillin-streptomycin mixture (Thermo Fisher Scientific) at 37 °C in the presence of 5% CO2.
Construction of plasmids
Full-length cDNAs encoding the human PHF20 (NM_016436), OGT1 (NM_181672), and NSL3 (FLJ10081; BC063792) proteins were subcloned with FLAG tag into pcDNA5/FRT and introduced into HEK293/FRT cells using the Invitrogen Flp-In system (15). In addition, cDNAs encoding full-length PHF20, full-length OGT1, point mutants OGT1G598A and OGT1C964A, deletion mutants OGT1-1 (aa 1–494) and OGT1-2 (aa 467–1046), full-length NSL3, and deletion mutants of NSL3 including NSL3-1 (aa 1–320), NSL3-2 (aa 330–548), NSL3-3 (aa 535–878), NSL3dc (aa 1–537), and NSL3dD (aa 1–878) were subcloned with FLAG or Myc tag into pcDNA3.1(−).
Co-transfection and co-IP
293T cells cultured in 10-cm tissue culture plates (5 × 106 cells) were transiently transfected with cDNAs encoding FLAG- or Myc-tagged full-length OGT1, NSL3, and PHF20; different mutants of OGT1; and different mutants of NSL3 using polyethyleneimine (PEI) (catalog number 23966, Polysciences). Total transfected plasmids in a 10-cm plate were 10 μg. 48 h after transfection, whole-cell lysates were prepared. FLAG-tagged or Myc-tagged proteins and their associated proteins were purified on anti-FLAG (M2)- or anti-Myc-agarose beads and then eluted with buffer containing 0.1 m KCl and 0.2 mg/ml FLAG or Myc peptide. Bound proteins were detected by Western blotting with anti-FLAG or anti-Myc antibodies.
Expression of recombinant proteins in Sf21 insect cells
N-terminal FLAG-tagged human OGT1-WT, point mutants OGT1C964A and OGT1G598A, and NSL3 were subcloned into pBacPAK8. Recombinant baculoviruses were generated with the BacPAK baculovirus expression system (Clontech). Sf21 cells were cultured at 27 °C in Sf-900 II serum-free medium with 5% fetal calf serum, 100 units/ml penicillin, and 100 g/ml streptomycin and infected with the recombinant baculoviruses. 60 h after infection, cells were lysed, and the FLAG-tagged OGT1 and NSL3 proteins were purified with anti-FLAG (M2)-agarose affinity chromatography.
RNAi treatment
HeLa or 293T cells were transfected with non-targeting siRNA (D-001206), OGT1 siRNA (M-019111), and NSL3 siRNA (M-016928) SMARTpool (Dharmacon). 48 h later, cells were harvested and lysed for immunoblotting, IP, and HAT experiments.
HAT assay
HAT assays were performed essentially as described (15).
O-GlcNAc transferase assay
Reaction mixtures (25 μl) containing 50 mm Tris-HCl (pH 7.4), 12.5 mm MgCl2, 1 mm dithiothreitol (DTT), 0.4 μCi of UDP-[3H]GlcNAc (0.1 μCi/μl; Amersham Biosciences) or 1 μg/μl cold UDP-GlcNAc, anti-FLAG-agarose eluates prepared from insect cell-expressed FLAG-OGT1 protein, and candidate substrates Nup62 and NSL3 were incubated at 37 °C for 90 min. The reaction was stopped by the addition of 4× SDS sample buffer and then subjected to SDS-PAGE. Proteins were analyzed by Western blotting with anti-GlcNAc antibody. Radiolabeled proteins were visualized by autoradiography.
WGA lectin affinity purification
Prepared whole-cell lysates of 293T cells were incubated with WGA-agarose (lectin from Triticum vulgaris (wheat)-agarose; Sigma-Aldrich, L1394) for 3 h at 4 °C. Proteins bound to WGA-agarose were collected and washed five times with wash buffer. For control of specificity, 20 mm GlcNAc was added in the binding reaction. Precipitated NSL3 protein was eluted by boiling in SDS sample buffer (36).
Reverse transcription-PCR (RT-PCR)
1 μg of isolated total RNA was used as a template to produce cDNA with a PrimeScript 1st Strand cDNA Synthesis kit (Takara). OGT1, NSL3, and GAPDH mRNAs were measured by quantitative real time PCR with an EcoTM Real-Time PCR System (Illumina, Gene Co. Ltd.). All PCRs were performed using the following protocol: initial denaturation step was at 95 °C for 30 s followed by 40 cycles of denaturation at 95 °C for 5 s and annealing at 60 °C for 30 s. Primer sets used in quantitative RT-PCR were as follows: OGT1, 5′-ACTGCAGGAAGCTCTGATGC-3′ (forward) and 5′-ATCCTTATGAATGGAAGCCAG-3′ (reverse), yielding a 195-bp product; NSL3, 5′-ATCCCTGTAGCCACCCATCT-3′ (forward) and 5′-CACAGTAAGCAGAGGAAACCC-3′ (reverse), yielding a 230-bp product; GAPDH, 5′-ATCACTGCCACCCAGAAGAC-3′ (forward) and 5′-ATGAGGTCCACCACCCTGTT-3′ (reverse), yielding a 460-bp product.
Immunofluorescence staining
HeLa cells, grown to ∼30% confluence in 24-well plates containing a coverslip (8D1007, Nest) on each well, were transfected with specific siRNAs using RNAiMAX (Invitrogen). 48 h later, cells were immunostained according to the method of Liu et al. (37).
Author contributions
D. W., L. Z., Y. C., and J. J. conceived and designed the experiments. D. W., L. Z., C. Y., J. D., Z. F., L. W., F. W., D. L., H. Z., and F. X. performed the experiments. J. J., D. W., and L. Z. analyzed the data. J. W. C. and R. C. C. contributed reagents, materials, and analysis tools. D. W., Y. C., and J. J. wrote the paper.
This work was supported by National Natural Science Foundation of China Grants 31571316 and 31371311; Project of Jilin Province Science and Technology Development Program Grant 20140414057GH; and Key Laboratory of Regenerative Biology, Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences Grants KLRB201303 and KLRB201504. The authors declare that they have no conflicts of interest with the contents of this article.
- O-GlcNAc
- O-linked N-acetylglucosamine
- NSL
- nonspecific lethal
- MOF
- males absent on the first
- OGT
- O-GlcNAc transferase
- WGA
- wheat germ agglutinin
- OGA
- O-GlcNAcase
- TET
- ten-eleven translocation
- MLL
- mixed lineage leukemia
- HAT
- histone acetyltransferase
- COMPASS
- complex of proteins associated with SET1
- hMOF
- human MOF
- HCF1
- host cell factor 1
- MSL
- male-specific lethal
- H4K16ac
- histone H4 Lys-16 acetylation
- H3K4me2
- histone H3 Lys-4 dimethylation
- MudPIT
- multidimensional protein identification technology
- IP
- immunoprecipitation
- TPR
- tetratricopeptide repeat
- CHX
- cycloheximide
- OGT1mt
- point mutant of OGT1
- Z
- carbobenzoxy
- FRT
- flippase recognition target
- aa
- amino acids.
References
- 1. Wells L., and Hart G. W. (2003) O-GlcNAc turns twenty: functional implications for post-translational modification of nuclear and cytosolic proteins with a sugar. FEBS Lett. 546, 154–158 [DOI] [PubMed] [Google Scholar]
- 2. Gao Y., Wells L., Comer F. I., Parker G. J., and Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 [DOI] [PubMed] [Google Scholar]
- 3. Chen Q., Chen Y., Bian C., Fujiki R., and Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Casey P. J. (1995) Protein lipidation in cell signaling. Science 268, 221–225 [DOI] [PubMed] [Google Scholar]
- 5. Bullen J. W., Balsbaugh J. L., Chanda D., Shabanowitz J., Hunt D. F., Neumann D., and Hart G. W. (2014) Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK). J. Biol. Chem. 289, 10592–10606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butkinaree C., Cheung W. D., Park S., Park K., Barber M., and Hart G. W. (2008) Characterization of β-N-acetylglucosaminidase cleavage by caspase-3 during apoptosis. J. Biol. Chem. 283, 23557–23566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sakabe K., Wang Z., and Hart G. W. (2010) β-N-Acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., and Herr W. (2011) O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 [DOI] [PubMed] [Google Scholar]
- 9. Fujiki R., Hashiba W., Sekine H., Yokoyama A., Chikanishi T., Ito S., Imai Y., Kim J., He H. H., Igarashi K., Kanno J., Ohtake F., Kitagawa H., Roeder R. G., Brown M., et al. (2011) GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakamura K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D. V., Shimada M., Tauchi H., Suzuki H., Tashiro S., Zou L., and Komatsu K. (2011) Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell 41, 515–528 [DOI] [PubMed] [Google Scholar]
- 11. Deplus R., Delatte B., Schwinn M. K., Defrance M., Méndez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., et al. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vella P., Scelfo A., Jammula S., Chiacchiera F., Williams K., Cuomo A., Roberto A., Christensen J., Bonaldi T., Helin K., and Pasini D. (2013) Tet proteins connect the O-linked N-acetylglucosamine transferase OGT to chromatin in embryonic stem cells. Mol. Cell 49, 645–656 [DOI] [PubMed] [Google Scholar]
- 13. Dehennaut V., Leprince D., and Lefebvre T. (2014) O-GlcNAcylation, an epigenetic mark, focus on the histone code, TET family proteins, and polycomb group proteins. Front. Endocrinol. 5, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ito R., Katsura S., Shimada H., Tsuchiya H., Hada M., Okumura T., Sugawara A., and Yokoyama A. (2014) TET3-OGT interaction increases the stability and the presence of OGT in chromatin. Genes Cells 19, 52–65 [DOI] [PubMed] [Google Scholar]
- 15. Cai Y., Jin J., Swanson S. K., Cole M. D., Choi S. H., Florens L., Washburn M. P., Conaway J. W., and Conaway R. C. (2010) Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J. Biol. Chem. 285, 4268–4272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rea S., Xouri G., and Akhtar A. (2007) Males absent on the first (MOF): from flies to humans. Oncogene 26, 5385–5394 [DOI] [PubMed] [Google Scholar]
- 17. Zhao X., Su J., Wang F., Liu D., Ding J., Yang Y., Conaway J. W., Conaway R. C., Cao L., Wu D., Wu M., Cai Y., and Jin J. (2013) Crosstalk between NSL histone acetyltransferase and MLL/SET complexes: NSL complex functions in promoting histone H3K4 di-methylation activity by MLL/SET complexes. PLoS Genet. 9, e1003940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mazars R., Gonzalez-de-Peredo A., Cayrol C., Lavigne A. C., Vogel J. L., Ortega N., Lacroix C., Gautier V., Huet G., Ray A., Monsarrat B., Kristie T. M., and Girard J. P. (2010) The THAP-Zinc finger protein THAP1 associates with coactivator HCF-1 and O-GlcNAc transferase. J. Biol. Chem. 285, 13364–13371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roquemore E. P., Chou T. Y., and Hart G. W. (1994) Detection of O-linked N-acetylglucosamine (O-GlcNAc) on cytoplasmic and nuclear proteins. Methods Enzymol. 230, 443–460 [DOI] [PubMed] [Google Scholar]
- 21. Smith E. R., Cayrou C., Huang R., Lane W. S., Côté J., and Lucchesi J. C. (2005) A human protein complex homologous to the Drosophila MSL complex is responsible for the majority of histone H4 acetylation at lysine 16. Mol. Cell. Biol. 25, 9175–9188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Taipale M., Rea S., Richter K., Vilar A., Lichter P., Imhof A., and Akhtar A. (2005) hMOF histone acetyltransferases is required for histone H4 lysine 16 acetylation in mammalian cells. Mol. Cell. Biol. 25, 6798–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jínek M., Rehwinkel J., Lazarus B. D., Izaurralde E., Hanover J. A., and Conti E. (2004) The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat. Struct. Mol. Biol. 11, 1001–1007 [DOI] [PubMed] [Google Scholar]
- 24. Kreppel L. K., and Hart G. W. (1999) Regulation of a cytosolic and nuclear O-GlcNAc transferase. Role of the tetratricopeptide repeats. J. Biol. Chem. 274, 32015–32022 [DOI] [PubMed] [Google Scholar]
- 25. Lubas W. A., and Hanover J. A. (2000) Functional expression of O-linked GlcNAc transferase. Domain structure and substrate specificity. J. Biol. Chem. 275, 10983–10988 [DOI] [PubMed] [Google Scholar]
- 26. Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., and Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 27. de Queiroz R. M., Madan R., Chien J., Dias W. B., and Slawson C. (2016) Changes in O-Linked N-acetylglucosamine (O-GlcNAc) homeostasis activate the p53 pathway in ovarian cancer cells. J. Biol. Chem. 291, 18897–18914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ding X., Jiang W., Zhou P., Liu L., Wan X., Yuan X., Wang X., Chen M., Chen J., Yang J., Kong C., Li B., Peng C., Wong C. C., Hou F., et al. (2015) Mixed lineage leukemia 5 (MLL5) protein stability Is cooperatively regulated by O-GlcNac transferase (OGT) and ubiquitin specific protease 7 (USP7). PLoS One 10, e0145023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hardivillé S., and Hart G. W. (2016) Nutrient regulation of gene expression by O-GlcNAcylation of chromatin. Curr. Opin. Chem. Biol. 33, 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lercher L., Raj R., Patel N. A., Price J., Mohammed S., Robinson C. V., Schofield C. J., and Davis B. G. (2015) Generation of a synthetic GlcNAcylated nucleosome reveals regulation of stability by H2A-Thr101 GlcNAcylation. Nat. Commun. 6, 7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rønningen T., Shah A., Oldenburg A. R., Vekterud K., Delbarre E., Moskaug J. Ø., and Collas P. (2015) Prepatterning of differentiation-driven nuclear lamin A/C-associated chromatin domains by GlcNAcylated histone H2B. Genome Res. 25, 1825–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Su J., Wang F., Cai Y., and Jin J. (2016) The functional analysis of histone acetyltransferase MOF in tumorigenesis. Int. J. Mol. Sci. 17, E99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Y., Long Y., Xing Z., and Zhang D. (2015) c-Jun recruits the NSL complex to regulate its target gene expression by modulating H4K16 acetylation and promoting the release of the repressive NuRD complex. Oncotarget 6, 14497–14506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Meunier S., Shvedunova M., Van Nguyen N., Avila L., Vernos I., and Akhtar A. (2015) An epigenetic regulator emerges as microtubule minus-end binding and stabilizing factor in mitosis. Nat. Commun. 6, 7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chatterjee A., Seyfferth J., Lucci J., Gilsbach R., Preissl S., Böttinger L., Mårtensson C. U., Panhale A., Stehle T., Kretz O., Sahyoun A. H., Avilov S., Eimer S., Hein L., Pfanner N., et al. (2016) MOF acetyl transferase regulates transcription and respiration in mitochondria. Cell 167, 722–738.e23 [DOI] [PubMed] [Google Scholar]
- 36. Hansen B. F., Glendorf T., Hegelund A. C., Lundby A., Lützen A., Slaaby R., and Stidsen C. E. (2012) Molecular characterisation of long-acting insulin analogues in comparison with human insulin, IGF-1 and insulin X10. PLoS One 7, e34274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu D., Wu D., Zhao L., Yang Y., Ding J., Dong L., Hu L., Wang F., Zhao X., Cai Y., and Jin J. (2015) Arsenic trioxide reduces global histone H4 acetylation at lysine 16 through direct binding to histone acetyltransferase hMOF in human cells. PLoS One 10, e0141014. [DOI] [PMC free article] [PubMed] [Google Scholar]