Abstract
Increasing evidence suggests that mitoNEET, a target of the type II diabetes drug pioglitazone, is a key regulator of energy metabolism in mitochondria. MitoNEET is anchored to the mitochondrial outer membrane via its N-terminal α helix domain and hosts a redox-active [2Fe-2S] cluster in its C-terminal cytosolic region. The mechanism by which mitoNEET regulates energy metabolism in mitochondria, however, is not fully understood. Previous studies have shown that mitoNEET specifically interacts with the reduced flavin mononucleotide (FMNH2) and that FMNH2 can quickly reduce the mitoNEET [2Fe-2S] clusters. Here we report that the reduced mitoNEET [2Fe-2S] clusters can be readily oxidized by oxygen. In the presence of FMN, NADH, and flavin reductase, which reduces FMN to FMNH2 using NADH as the electron donor, mitoNEET mediates oxidation of NADH with a concomitant reduction of oxygen. Ubiquinone-2, an analog of ubiquinone-10, can also oxidize the reduced mitoNEET [2Fe-2S] clusters under anaerobic or aerobic conditions. Compared with oxygen, ubiquinone-2 is more efficient in oxidizing the mitoNEET [2Fe-2S] clusters, suggesting that ubiquinone could be an intrinsic electron acceptor of the reduced mitoNEET [2Fe-2S] clusters in mitochondria. Pioglitazone or its analog NL-1 appears to inhibit the electron transfer activity of mitoNEET by forming a unique complex with mitoNEET and FMNH2. The results suggest that mitoNEET is a redox enzyme that may promote oxidation of NADH to facilitate enhanced glycolysis in the cytosol and that pioglitazone may regulate energy metabolism in mitochondria by inhibiting the electron transfer activity of mitoNEET.
Keywords: electron paramagnetic resonance (EPR), electron transfer, FMN, iron-sulfur protein, type 2 diabetes
Introduction
Pioglitazone is a drug prescribed for the treatment of type II diabetes (1). In addition to acting on peroxisome proliferator-activated receptor γ (PPARγ), which regulates the expression of genes related to insulin sensitivity (2), pioglitazone also has a rapid pharmacological effect on energy metabolism in mitochondria (3). A search of pioglitazone-binding targets revealed a new protein, mitoNEET, that localizes in the mitochondrial outer membrane (4). In mice, deletion of mitoNEET results in a decrease in oxidative phosphorylation capacity in mitochondria by about 30% (5). On the other hand, overexpression of mitoNEET enhances lipid storage and decreases oxidative damage in adipocytes (6, 7) and inhibits ferroptosis in human hepatocellular carcinoma cells (8). In β cells, increased expression of mitoNEET leads to hyperglycemia and glucose intolerance (9). In spinal neurons, microRNA-127 has been shown to specifically target the expression of mitoNEET, and depletion of mitoNEET results in neuronal loss and apoptosis (10). In breast cancer cells, mitoNEET is highly expressed, and depletion of mitoNEET inhibits cancer cell proliferation (11, 12). It has thus been postulated that mitoNEET is a key regulator of energy metabolism in mitochondria (13) and an attractive chemotherapeutic target for treating mitochondrion-related human diseases (14–17).
Crystallographic studies have shown that mitoNEET is a homodimer, with each monomer hosting a [2Fe-2S] cluster via an unusual ligand arrangement of three cysteine residues and one histidine residue in its C-terminal cytosolic domain (18–21). The N terminus of mitoNEET has a transmembrane α helix that anchors the protein to the mitochondrial outer membrane (4). However, the mechanism by which mitoNEET regulates energy metabolism in mitochondria still remains elusive. One hypothesis states that mitoNEET may transfer its [2Fe-2S] clusters to apo-proteins such as apo-ferredoxin (22), iron regulatory protein-1 (IRP-1) (23), or anamorsin, a protein required for iron-sulfur cluster assembly in cytosol (24). Because mitochondria are the primary sites for iron-sulfur cluster biogenesis in cells (25), it is tempting to propose that mitoNEET is a carrier transporting the clusters assembled in mitochondria to target proteins in the cytosol (22–24). However, the observed cluster transfer occurs only when the mitoNEET [2Fe-2S] clusters are oxidized (22, 23), and the mitoNEET [2Fe-2S] clusters are mostly in a reduced state in cells under normal physiological conditions (26–29). Thus, the exact role of mitoNEET in iron-sulfur cluster biogenesis has yet to be further defined. An alternative hypothesis is that mitoNEET may regulate mitochondrial functions via protein-protein interactions. This hypothesis is based on the observations that mitoNEET interacts with mitochondrial proteins, including the Parkinson's disease-associated protein Parkin (9), mitochondrial outer membrane import complex protein 1 (MTX1) (30), glutathione reductase (28), and glutamate dehydrogenase 1 (31). Nevertheless, no direct evidence is available suggesting that the function of mitochondrial proteins are regulated by mitoNEET.
In previous studies, we have found that human mitoNEET has a specific interaction with the reduced flavin mononucleotide (FMNH2) and that FMNH2 can rapidly reduce the mitoNEET [2Fe-2S] clusters (29). Here we report that the reduced mitoNEET [2Fe-2S] clusters can be oxidized by oxygen or ubiquinone-2. Compared with oxygen, ubiquinone-2 appears to be more efficient in oxidizing the mitoNEET [2Fe-2S] clusters, indicating that ubiquinone may act as a native electron acceptor of the reduced mitoNEET [2Fe-2S] clusters in mitochondria. In the presence of flavin reductase, which reduces FMN to FMNH2 using NADH as the electron donor, mitoNEET mediates oxidation of NADH with a concomitant reduction of oxygen or ubiquinone-2. Furthermore, pioglitazone can effectively inhibit the electron transfer activity of mitoNEET by forming a unique complex with mitoNEET and FMNH2. The results led us to propose that mitoNEET is a redox enzyme that may promote oxidation of NADH to enhance glycolysis in the cytosol and that pioglitazone may regulate energy metabolism in mitochondria by inhibiting the electron transfer activity of mitoNEET.
Results
Reduction of the mitoNEET [2Fe-2S] clusters under anaerobic and aerobic conditions
Previous studies have shown that human mitoNEET has a specific interaction with FMNH2 and that FMNH2 can quickly reduce the mitoNEET [2Fe-2S] clusters (29). As oxygen would be present in mitochondria, we asked whether oxygen will have any effects on the FMNH2-mediated reduction of the mitoNEET [2Fe-2S] clusters.
To test this idea, human mitoNEET was preincubated with FMN and NADH under anaerobic or aerobic conditions. The reaction was initiated by adding a catalytic amount of Escherichia coli flavin reductase (32) to reduce FMN using NADH as the electron donor (29). Fig. 1A shows that, under anaerobic conditions, the oxidized mitoNEET [2Fe-2S] clusters (indicated by the absorption peaks at 455 nm and 540 nm (27)) were quickly reduced upon addition of flavin reductase, as reported previously (29). About 10 μm NADH was oxidized when 10 μm mitoNEET [2Fe-2S] clusters were reduced in the incubation solution under anaerobic conditions (Fig. 1C). Under aerobic conditions, the mitoNEET [2Fe-2S] clusters were also fully reduced after addition of flavin reductase (Fig. 1B). However, NADH was continuously oxidized even after the mitoNEET [2Fe-2S] clusters were fully reduced (Fig. 1C). The observed oxidation of NADH was mitoNEET-dependent, as only a very small amount of NADH was oxidized in the same incubation solution without mitoNEET under aerobic conditions (Fig. 1D). Thus, mitoNEET is able to promote oxidation of NADH in the presence of flavin reductase and FMN under aerobic conditions.
Figure 1.
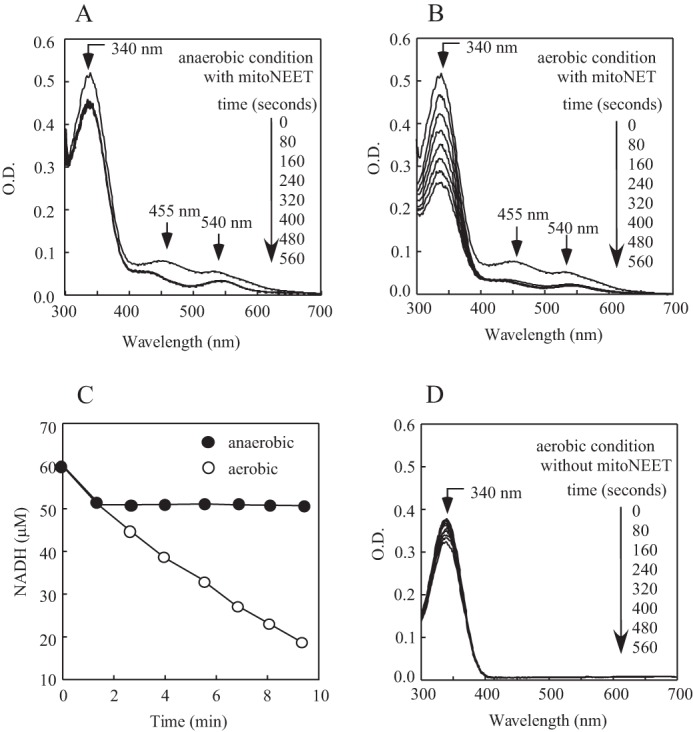
Reduction of the mitoNEET [2Fe-2S] clusters by FMNH2 under aerobic and anaerobic conditions. A and B, MitoNEET (containing 10 μm [2Fe-2S] clusters) was incubated with FMN (0.1 μm) and NADH (60 μm) under anaerobic (A) and aerobic (B) conditions. Fre (0.1 μm) was then added to the solutions to initiate the reaction. UV-visible absorption spectra were taken every 80 s after addition of Fre for 560 s. Absorption peaks at 455 nm and 540 nm represent the oxidized mitoNEET [2Fe-2S] clusters. O.D., optical density. C, time courses of NADH oxidation in the incubation solutions containing mitoNEET, Fre, and FMN under anaerobic (closed circles) or aerobic (open circles) conditions. The amount of NADH in the incubation solutions was measured from the absorption peak at 340 nm after subtracting the absorption of proteins in the incubation solutions. D, the same as in B, except no mitoNEET was included. The data are representative of three independent experiments.
The reduced mitoNEET [2Fe-2S] clusters are oxidized by oxygen under aerobic conditions
In the presence of flavin reductase, FMN and excess NADH, the mitoNEET [2Fe-2S] clusters were fully reduced under aerobic conditions (Fig. 1B). One explanation could be that the reduced mitoNEET [2Fe-2S] clusters are oxidized by oxygen and rapidly rereduced by FMNH2/NADH/flavin reductase under aerobic conditions.
To test this idea, mitoNEET was incubated with a limited amount of NADH and FMN under aerobic conditions. As shown in Fig. 2, the mitoNEET [2Fe-2S] clusters were fully reduced upon addition of a catalytic amount of flavin reductase. However, when NADH in the incubation solution was completely oxidized, the reduced mitoNEET [2Fe-2S] clusters were gradually reoxidized under aerobic conditions, demonstrating that the reduced mitoNEET [2Fe-2S] clusters are indeed oxidized by oxygen under aerobic conditions.
Figure 2.
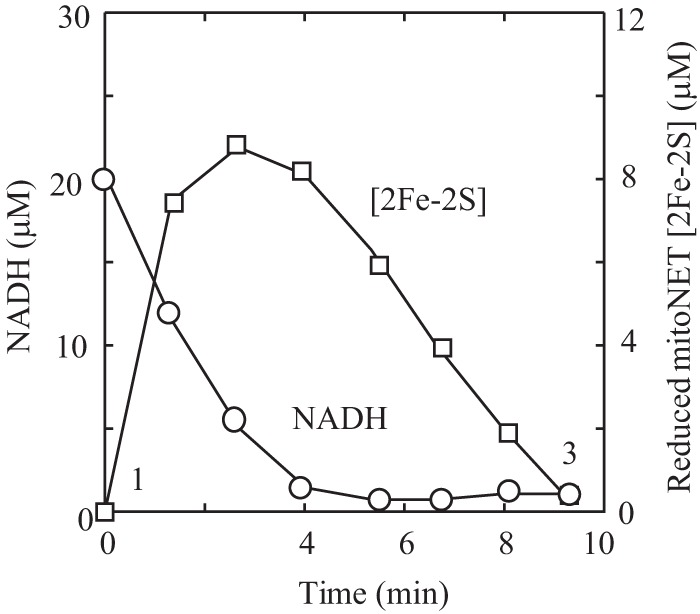
Oxidation kinetics of the reduced mitoNEET [2Fe-2S] clusters by oxygen. MitoNEET (containing 10 μm [2Fe-2S] clusters) was incubated with FMN (0.1 μm) and NADH (20 μm) under aerobic conditions. Fre (0.1 μm) was then added to the incubation solution to initiate the reaction. The amount of NADH in the incubation solution was measured from the absorption peak at 340 nm. The amount of the reduced mitoNEET [2Fe-2S] clusters was measured from the different absorption at absorption peaks of 455 nm and 420 nm. The amounts of NADH and the reduced mitoNEET [2Fe-2S] clusters in the incubation solution were plotted as a function of incubation time. The data are representative of three independent experiments.
The reduced mitoNEET [2Fe-2S] clusters can also be oxidized by ubiquinone-2 under anaerobic conditions
In mitochondria, NADH-ubiquinone oxidoreductase (complex I) oxidizes NADH and reduces ubiquinone via FMN and a chain of iron-sulfur clusters (33). Ubiquinone is a key electron transport component in the respiratory chain (34) and is present in both the mitochondrial inner and outer membranes (35). As mitoNEET is a mitochondrial outer membrane protein (4), and the [2Fe-2S] clusters in mitoNEET are positioned close to the membrane (18–21), we postulated that the reduced mitoNEET [2Fe-2S] clusters may also reduce ubiquinone in the mitochondrial outer membrane.
In the experiments, the mitoNEET [2Fe-2S] clusters were prereduced with flavin reductase, FMN, and an excess amount of NADH under anaerobic conditions. Ubiquinone-2, a ubiquinone-10 analog, was then injected into the incubation solution anaerobically using a gas-tight syringe. Fig. 3 shows that the reduced mitoNEET [2Fe-2S] clusters were immediately oxidized upon injection of ubiquinone-2 under anaerobic conditions. As a control, injection of degassed water had no effect on the reduced mitoNEET [2Fe-2S] clusters (data not shown). After further incubation, the ubiquinone-2-oxidized mitoNEET[2Fe-2S] clusters were fully rereduced in the incubation solution (Fig. 3A), suggesting that oxidation of the mitoNEET [2Fe-2S] clusters by ubiquinone-2 is reversible.
Figure 3.
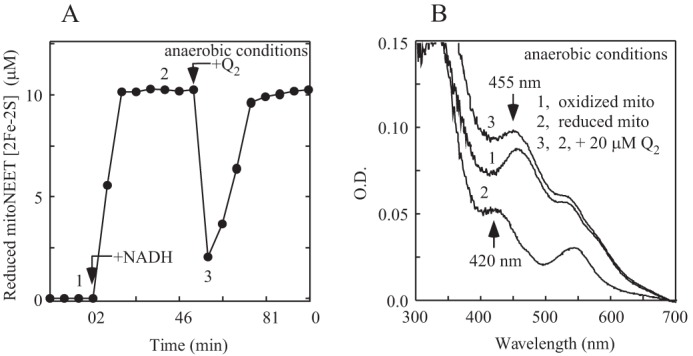
Oxidation of the reduced mitoNEET [2Fe-2S] clusters by ubiquinone-2 under anaerobic condition. A, oxidation of the reduced mitoNEET [2Fe-2S] clusters by ubiquinone-2 under anaerobic conditions. MitoNEET (containing 10 μm [2Fe-2S] clusters) was reduced with FMN (0.1 μm) and NADH (50 μm) and flavin reductase (0.1 μm) under anaerobic conditions. Ubiquinone-2 (Q2, 20 μm) was injected into the incubation solution anaerobically as indicated. The reduced mitoNEET [2Fe-2S] clusters were measured from the different absorption at absorption peaks of 455 nm and 420 nm and plotted as a function of the incubation time. B, UV-visible absorption spectra of mitoNEET. Spectra were taken before (spectrum 1) and after (spectrum 2) addition of NADH (50 μm). Spectrum 3, right after anaerobic addition of ubiquinone-2 (20 μm). The experiments were repeated three times. Similar results were obtained. O.D., optical density.
We also tested whether the reduced mitoNEET [2Fe-2S] clusters could be oxidized by other oxidants. In the experiments, the mitoNEET [2Fe-2S] clusters were prereduced with flavin reductase, FMN, and an excess amount of NADH under anaerobic conditions, followed by injection of oxidants anaerobically. We found that dichloroindophenol, K3Fe(CN)6, or menadione could all quickly oxidize the reduced mitoNEET [2Fe-2S] clusters (supplemental Fig. 1), suggesting that the reduced mitoNEET [2Fe-2S] clusters may also be oxidized by other oxidants in the cytosol. Oxidation of the mitoNEET [2Fe-2S] clusters by dichloroindophenol or K3Fe(CN)6 was very similar to that of ubiquinone-2. However, menadione appeared to be less active to oxidize the mitoNEET [2Fe-2S] clusters under the experimental conditions (supplemental Fig. 1). Because ubiquinone is a physiological oxidant and is in the vicinity of the mitoNEET [2Fe-2S] clusters in mitochondria, we propose that ubiquinone could be a native electron acceptor for the reduced mitoNEET [2Fe-2S] clusters in mitochondria.
Oxidation of the reduced mitoNEET [2Fe-2S] clusters by ubiquinone-2 under aerobic conditions
Compared with oxygen (Fig. 2), ubiquinone-2 is much more efficient in oxidizing the reduced mitoNEET [2Fe-2S] clusters (Fig. 3A). To further explore the reactivity of the reduced mitoNEET [2Fe-2S] clusters with oxygen and ubiquinone-2, we incubated mitoNEET with FMN, NADH, and ubiquinone-2 under aerobic conditions. Fig. 4A shows that NADH was continuously oxidized after flavin reductase was added to the incubation solution. The observed NADH oxidation was mitoNEET-dependent, as only a very small amount of NADH was oxidized in the incubation solution without mitoNEET under aerobic conditions (Fig. 4B). Importantly, despite oxidation of NADH in the incubation solution containing ubiquinone-2, the mitoNEET [2Fe-2S] clusters remained oxidized during the incubation process (Fig. 4A). This result indicates that the reduced mitoNEET [2Fe-2S] clusters might be rapidly oxidized by ubiquinone-2, which subsequently transfers electron to oxygen under aerobic conditions. Indeed, when the incubation solution in Fig. 4A was purged with pure argon gas to remove oxygen, the mitoNEET [2Fe-2S] clusters were rereduced upon addition of NADH (Fig. 4, C and D). Thus, the results further suggest that ubiquinone in mitochondria may act as an intrinsic electron acceptor for the mitoNEET [2Fe-2S] clusters under anaerobic or aerobic conditions.
Figure 4.
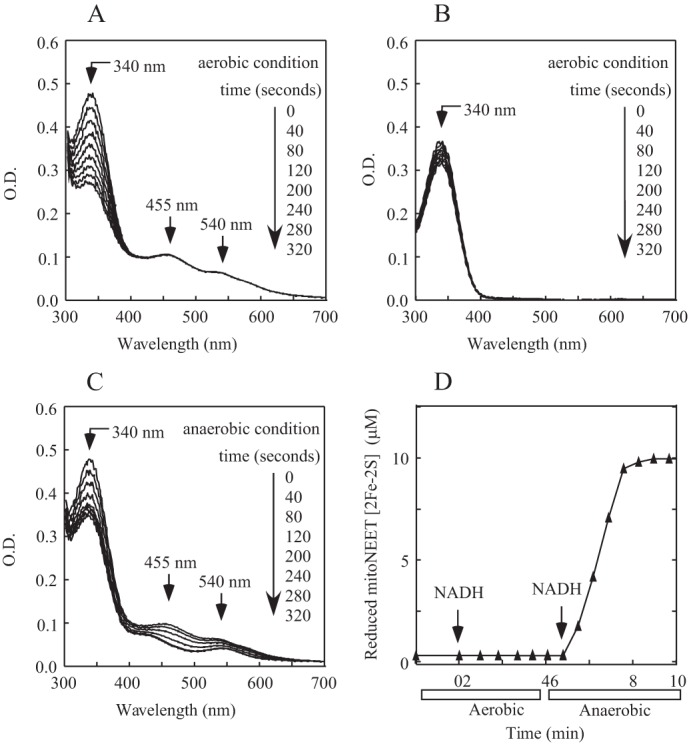
Oxidation of the reduced mitoNEET [2Fe-2S] clusters by ubiquinone-2 under aerobic condition. A, mitoNEET (containing 10 μm [2Fe-2S] clusters) was preincubated with FMN (0.1 μm), NADH (50 μm), and ubiquinone-2 (20 μm) under aerobic conditions. UV-visible absorption spectra were taken every 40 s after addition of Fre (0.1 μm) for 320 s. The amount of NADH in the incubation solution was measured from the absorption peak at 340 nm. O.D., optical density. B, the same as A, except mitoNEET was not included in the incubation solution. C, the incubation solution in A was purged with pure argon gas for 10 min, followed by injection of NADH (50 μm) anaerobically. UV-visible absorption spectra were taken every 40 s for 320 s. D, redox transition of the mitoNEET [2Fe-2S] clusters under aerobic and anaerobic conditions. The data are from A and C. The amount of the reduced mitoNEET [2Fe-2S] clusters was measured from the different absorption at peaks of 455 nm and 420 nm in each spectrum. The data are representative of three independent experiments.
A pioglitazone analog, NL-1, inhibits the electron transfer activity of mitoNEET
MitoNEET was identified as a target of the type II diabetes drug pioglitazone (1). However, pioglitazone has a very low solubility in water (<1 mg/ml), making it difficult to characterize its biochemical activity (28). A new pioglitazone analog, NL-1 or 5-(3,5-di-tert-butyl-4-hydroxybenzyl)-4-hydroxythiazol-2(5H)-one, was designed and synthesized by Geldenhuys et al. (36). NL-1 has been improved for its solubility in water and binding specificity to mitoNEET. It has similar pharmacological activities as pioglitazone to decrease the maximal respiration rate of mitochondria by 45%, reduce the production of reactive oxygen species (36), and improve the survival of cardiac stem cells during oxidative stress (37).
Here we explored the effect of NL-1 on the electron transfer activity of mitoNEET. In the experiments, mitoNEET was preincubated with increasing concentrations of NL-1 under aerobic conditions, followed by addition of flavin reductase and FMN. The reaction was initiated by adding NADH. The amounts of the reduced mitoNEET [2Fe-2S] clusters in the incubation solutions were monitored spectroscopically and plotted as a function of incubation time. Fig. 5 shows that, as the concentration of NL-1 increased, the reduction of the mitoNEET [2Fe-2S] clusters in the incubation solution gradually decreased. At 250 μm NL-1, the reduction of the mitoNEET [2Fe-2S] clusters (10 μm) in the incubation solution was inhibited by about 70%, indicating that NL-1 can effectively block the electron transfer activity of mitoNEET.
Figure 5.
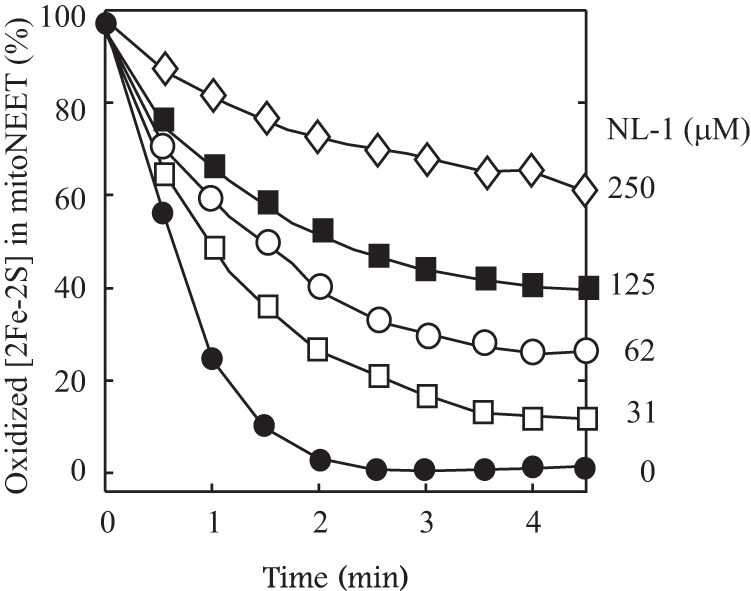
Inhibition of the electron transfer activity of mitoNEET by the pioglitazone analog NL-1. MitoNEET (containing 10 μm [2Fe-2S] clusters) was preincubated with increasing concentrations of NL-1 (from 0–250 μm) at room temperature for 30 min under aerobic conditions, followed by addition of FMN (0.1 μm) and flavin reductase (0.1 μm). NADH (50 μm) was then added to the incubation solutions to initiate the reaction. The amount of the reduced mitoNEET [2Fe-2S] clusters in the incubation solutions was measured from the different absorption at absorption peaks of 455 nm and 420 nm and plotted as function of the reaction time after injection of NADH. The data are representative of three independent experiments.
Pioglitazone/NL-1 and FMNH2 forms a unique complex with the reduced mitoNEET [2Fe-2S] clusters
To further assess the effects of pioglitazone and its analog NL-1 on the electron transfer activity of mitoNEET, mitoNEET was incubated with FMN, pioglitazone, NL-1, or combinations at room temperature for 1 h under aerobic conditions. The samples were then reduced with sodium dithionite for electron paramagnetic resonance (EPR)2 measurements of the reduced mitoNEET [2Fe-2S] clusters. Fig. 6A shows that incubation with FMNH2 results in a small EPR signal at g = 1.85 of the reduced mitoNEET [2Fe-2S] clusters (spectrum 2), as reported previously (29). However, unlike FMNH2, incubation with pioglitazone (4) (Fig. 6A, spectrum 3) or NL-1 (36) (Fig. 6A, spectrum 4) did not change the EPR spectrum of the reduced mitoNEET [2Fe-2S] clusters. Nevertheless, incubation of mitoNEET with FMNH2 and pioglitazone completely shifted the typical EPR spectrum at g = 1.94 of the reduced mitoNEET [2Fe-2S] clusters to a new EPR spectrum with g = 1.85 (Fig. 6A, spectrum 5). The same result was also observed when mitoNEET was incubated with FMNH2 and NL-1 (Fig. 6A, spectrum 6). While a small EPR signal at g = 1.86 of the reduced mitoNEET [2Fe-2S] clusters was noticed previously (38), the interpretation of the EPR signal at g = 1.85 remains elusive. Nevertheless, the dramatic change of the EPR spectrum strongly suggests that FMNH2 and pioglitazone/NL-1 may have a close interaction with the reduced [2Fe-2S] clusters in mitoNEET or that binding of FMNH2 and pioglitazone/NL-1 may result in conformational changes that alter the structural orientation of the cluster ligands in mitoNEET.
Figure 6.
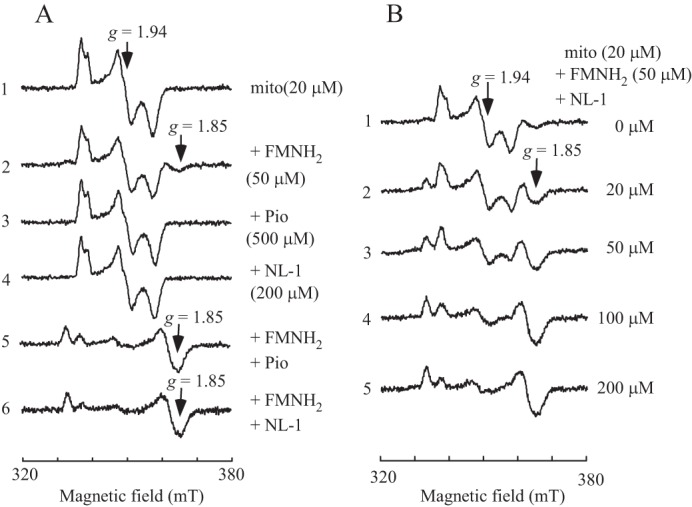
Synergistic effect of FMNH2 and pioglitazone/NL-1 on the reduced mitoNEET [2Fe-2S] clusters. A, EPR spectra of the reduced mitoNEET [2Fe-2S] clusters. MitoNEET (containing 20 μm [2Fe-2S] clusters, spectrum 1) was incubated with FMN (50 μm, spectrum 2), pioglitazone (Pio, 500 μm, spectrum 3), NL-1 (200 μm, spectrum 4), FMN + pioglitazone (spectrum 5), and FMN + NL-1 (spectrum 6) at room temperature for 60 min. After incubation, samples were reduced with freshly prepared sodium dithionite (4 mm), transferred to EPR tubes, and immediately frozen in liquid nitrogen for EPR measurements. mT, millitesla. B, titration of NL-1. MitoNEET (containing 20 μm [2Fe-2S] clusters) was incubated with FMN (50 μm) and increasing concentrations of NL-1 (0 to 200 μm) (spectra 1–5) at room temperature for 60 min. After incubation, samples were reduced with freshly prepared sodium dithionite (4 mm), transferred to EPR tubes, and immediately frozen in liquid nitrogen for EPR measurements. The data are representative of three independent experiments.
MitoNEET was further incubated with a fixed concentration of FMN and increasing concentrations of NL-1 at room temperature for 1 h under aerobic conditions, followed by reduction with sodium dithionite. Fig. 6B shows that, as the concentration of NL-1 increased, the EPR signal at g = 1.85 gradually increased. About 200 μm NL-1 was sufficient to change the EPR spectrum of g = 1.94 to that of g = 1.85 of the reduced mitoNEET [2Fe-2S] clusters. The results suggest that FMNH2 and pioglitazone/NL-1 have a synergistic effect on the reduced mitoNEET [2Fe-2S] clusters.
Discussion
MitoNEET is a founding member of a small family of proteins that contain a CDGSH motif (39, 40). Although it has been reported that mitoNEET is a key regulator of energy metabolism in mitochondria (13), the underlying mechanism has not been fully understood. Several research groups proposed that mitoNEET may transfer its [2Fe-2S] clusters for maturation of iron-sulfur proteins in the cytosol (22–24). Others suggested that mitoNEET may regulate mitochondrial functions via specific protein-protein interactions (9). Here we report that mitoNEET is a novel redox enzyme that transfers electron from FMNH2 to oxygen or ubiquinone in the mitochondrial outer membrane via its [2Fe-2S] clusters. In this process, FMNH2, which may be reduced by flavin reductase (41) and NADH in the cytosol, binds to mitoNEET and reduces the [2Fe-2S] clusters in mitoNEET (29). The reduced mitoNEET [2Fe-2S] clusters are readily oxidized by oxygen or ubiquinone (Fig. 7). Together with flavin reductase (41) and FMN, mitoNEET may effectively promote oxidation of NADH in the cytosol with a concomitant reduction of oxygen or ubiquinone in the mitochondrial outer membrane.
Figure 7.
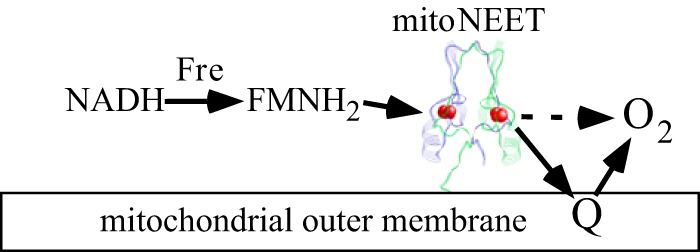
A proposed model for the electron transfer of mitoNEET in the mitochondria outer membrane. In the cytosol, FMNH2 is reduced by flavin reductase, using NADH as the electron donor. FMNH2 then reduces the mitoNEET [2Fe-2S] clusters, which, in turn, transfer electrons to oxygen or ubiquinone in the mitochondrial outer membrane. Together with flavin reductase and FMN, mitoNEET promotes NADH oxidation with a concomitant reduction of oxygen or ubiquinone in mitochondria.
The intracellular concentration of FMN in human cells is in the nanomolar range (42). Here we found that 100 nm FMN is sufficient to reduce 10 μm mitoNEET [2Fe-2S] clusters in the presence of flavin reductase and NADH in less than 2 min (Fig. 1), indicating that FMN may act as an electron shuttle, reducing the mitoNEET [2Fe-2S] clusters (29). In the cytosol, FMN is reduced to FMNH2 by flavin reductase (41), using NADH as the electron donor. Because glycolysis will produce NADH, which must be oxidized to sustain glycolysis activity (43), mitoNEET may enhance glycolysis by promoting oxidation of NADH in the cytosol (Fig. 7). In primary and metastatic cancer cells, glycolysis is highly up-regulated, resulting in increased glucose consumption (44). Interestingly, mitoNEET is also highly expressed in cancer cells, and overexpression of mitoNEET is essential for cancer cell proliferation (11, 12). In this context, we propose that overproduced mitoNEET may enhance glycolysis in the cytosol by promoting oxidation of NADH in cancer cells. Accordingly, when mitoNEET is deleted, oxidation of NADH in the cytosol could be diminished, leading to a decrease in glycolysis in the cytosol and oxidative phosphorylation in mitochondria. This notion is consistent with a previous report showing that deletion of mitoNEET decreases the oxidative phosphorylation capacity in mitochondria by about 30% (5) and inhibits cancer cell proliferation (11, 12). Nevertheless, additional experiments are required to elucidate the physiological link between the electron transfer activity of mitoNEET and glycolysis in cells.
Iron-sulfur clusters in proteins are often sensitive to oxygen and reactive oxygen species (45). However, the mitoNEET [2Fe-2S] clusters are highly resistant to oxygen and hydrogen peroxide (27). In fact, the reduced mitoNEET [2Fe-2S] clusters can be oxidized by oxygen without disruption of the cluster (Fig. 2). Importantly, compared with oxygen, ubiquinone-2 is more efficient in oxidizing the reduced mitoNEET [2Fe-2S] clusters (Figs. 3 and 4). Because mitoNEET is a mitochondrial outer membrane protein (4), and the [2Fe-2S] clusters in mitoNEET are positioned close to the membrane (18–21), we propose that ubiquinone could be an intrinsic electron acceptor for the reduced mitoNEET [2Fe-2S] clusters (Fig. 7). Upon the single-electron reduction by the reduced mitoNEET [2Fe-2S] cluster, ubiquinone is converted to semiquinone, which may be further reduced to ubihydroquinone, or transfer the electron to oxygen to produce superoxide or hydrogen peroxide (34). Although other oxidants in the cytosol may also oxidize the reduced mitoNEET [2Fe-2S] clusters in the cytosol, the proposed electron transfer path from NADH to ubiquinone catalyzed by the mitoNEET [2Fe-2S] clusters (Fig. 7) is reminiscent of the electron transfer path in complex I, in which electrons in NADH are transferred to ubiquinone via FMN and a chain of iron-sulfur clusters (33). The observed reduction of oxygen by the mitoNEET [2Fe-2S] clusters may reflect the possible electron transfer leak in mitoNEET, as reported in complex I (33). The final products of the electron transfer reaction catalyzed by mitoNEET in mitochondria remain to be determined.
Pioglitazone has two major targets in cells: peroxisome proliferator-activated receptor γ, which regulates the expression of genes involved in insulin sensitivity (2), and mitoNEET (4), which modulates energy metabolism in mitochondria (3). It was reported previously that binding of pioglitazone stabilizes the mitoNEET [2Fe-2S] clusters (21) and shifts the redox midpoint potential of the [2Fe-2S] clusters by about 100 mV (26). Here we found that pioglitazone and its analog NL-1 (36) can inhibit the electron transfer activity of mitoNEET (Fig. 5) by forming a unique complex with mitoNEET and FMNH2 that has an unusual EPR signal at g = 1.85 (Fig. 6). Based on molecular docking modeling (29, 36), FMN and pioglitazone/NL-1 have distinct binding sites but with significant overlap in mitoNEET, suggesting that pioglitazone/NL-1 may interfere with FMNH2 binding in mitoNEET and inhibit the electron transfer activity of mitoNEET in mitochondria. As mitoNEET is proposed as a chemotherapeutic target for treating type II diabetes (4), breast cancer (16, 17), and neurodegenerative diseases (10), a high-throughput screening approach combining the electron transfer activity assay and EPR measurements of mitoNEET may help identify new drugs that specifically target the mitoNEET [2Fe-2S] clusters in mitochondria.
Experimental procedures
Protein preparation
The human mitochondrial outer membrane protein mitoNEET33–108 (containing residues 33–108) was purified as described in Ref. 28. E. coli flavin reductase (Fre) was prepared using an E. coli strain hosting an expression plasmid encoding Fre from the ASKA (A Complete Set of E. coli K-12 ORF Archive) library (46). Both mitoNEET and Fre were overproduced in E. coli cells and purified using nickel-agarose columns, followed by passing through a High-Trap desalting column. The purity of purified proteins was greater than 95%, as judged by electrophoresis analysis on a 15% polyacrylamide gel containing SDS followed by staining with Coomassie Blue. The protein concentrations of mitoNEET and E. coli Fre were measured at 280 nm using the molar extinction coefficients of 8.6 and 26.4 mm−1cm−1, respectively. The molar extinction coefficients were calculated based on the protein primary sequence.
Redox state of the mitoNEET [2Fe-2S] clusters
Reduction and oxidation of the mitoNEET [2Fe-2S] clusters in incubation solutions were monitored using a Beckman DU640 UV-visible absorption spectrometer equipped with a temperature controller. Oxidized mitoNEET [2Fe-2S] clusters have two major absorption peaks at 455 nm and 540 nm, whereas reduced mitoNEET [2Fe-2S] clusters have absorption peaks at 420 nm and 540 nm (29). The redox state of the mitoNEET [2Fe-2S] clusters was further determined by EPR measurements. Oxidation of NADH in the incubation solution was monitored at 340 nm after subtracting the basal absorption of proteins in the incubation solution. Anaerobic conditions were achieved by purging the incubation solutions with pure argon gas for 15 min in a sealed vial.
EPR measurements
The X-band EPR spectra were recorded using a Bruker model ESR-300 spectrometer equipped with an Oxford Instruments 910 continuous flow cryostat. Routine EPR conditions were as follows: microwave frequency, 9.47 GHz; microwave power, 10.0 milliwatt; modulation frequency, 100 kHz; modulation amplitude, 1.2 millitesla; temperature, 30 K; receiver gain, 2 × 105.
Chemicals
NADH, NADPH, isopropyl β-d-1-thiogalactopyranoside, kanamycin, and ampicillin were purchased from Research Products International. FMN, FAD, ubiquinone-2, pioglitazone, NL-1, and other chemicals were purchased from Sigma. The molar extinction coefficients of 6.2 mm−1cm−1 at 340 nm, 12.5 mm−1cm−1 at 445 nm, and 11.3 mm−1cm−1 at 450 nm were used for measuring the concentration of NADH/NADPH, FMN, and FAD, respectively (43). The molar extinction coefficient of 14.9 mm−1cm−1 at 278 nm was used for ubiquinone-2.
Author contributions
Y. W., A. P. L., and H. D. designed, performed, and analyzed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
This research was supported in part by National Institutes of Health Grant R15GM109399 and American Heart Association Grant 13GRNT16890014. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- EPR
- electron paramagnetic resonance
- Fre
- flavin reductase
- g
- the g-factor of an unpaired electron.
References
- 1. Hippisley-Cox J., and Coupland C. (2016) Diabetes treatments and risk of heart failure, cardiovascular disease, and all cause mortality: cohort study in primary care. BMJ 354, i3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wright M. B., Bortolini M., Tadayyon M., and Bopst M. (2014) Minireview: challenges and opportunities in development of PPAR agonists. Mol. Endocrinol. 28, 1756–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feinstein D. L., Spagnolo A., Akar C., Weinberg G., Murphy P., Gavrilyuk V., and Dello Russo C. (2005) Receptor-independent actions of PPAR thiazolidinedione agonists: is mitochondrial function the key? Biochem. Pharmacol. 70, 177–188 [DOI] [PubMed] [Google Scholar]
- 4. Colca J. R., McDonald W. G., Waldon D. J., Leone J. W., Lull J. M., Bannow C. A., Lund E. T., and Mathews W. R. (2004) Identification of a novel mitochondrial protein (“mitoNEET”) cross-linked specifically by a thiazolidinedione photoprobe. Am. J. Physiol. Endocrinol. Metab. 286, E252–E260 [DOI] [PubMed] [Google Scholar]
- 5. Wiley S. E., Murphy A. N., Ross S. A., van der Geer P., and Dixon J. E. (2007) MitoNEET is an iron-containing outer mitochondrial membrane protein that regulates oxidative capacity. Proc. Natl. Acad. Sci. U.S.A. 104, 5318–5323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kusminski C. M., Holland W. L., Sun K., Park J., Spurgin S. B., Lin Y., Askew G. R., Simcox J. A., McClain D. A., Li C., and Scherer P. E. (2012) MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat. Med. 18, 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kusminski C. M., Park J., and Scherer P. E. (2014) MitoNEET-mediated effects on browning of white adipose tissue. Nat. Commun. 5, 3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan H., Li X., Zhang X., Kang R., and Tang D. (2016) CISD1 inhibits ferroptosis by protection against mitochondrial lipid peroxidation. Biochem. Biophys. Res. Commun. 478, 838–844 [DOI] [PubMed] [Google Scholar]
- 9. Kusminski C. M., Chen S., Ye R., Sun K., Wang Q. A., Spurgin S. B., Sanders P. E., Brozinick J. T., Geldenhuys W. J., Li W. H., Unger R. H., and Scherer P. E. (2016) MitoNEET-Parkin effects in pancreatic α- and β-cells, cellular survival, and intrainsular cross talk. Diabetes 65, 1534–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He Q. Q., Xiong L. L., Liu F., He X., Feng G. Y., Shang F. F., Xia Q. J., Wang Y. C., Qiu D. L., Luo C. Z., Liu J., and Wang T. H. (2016) MicroRNA-127 targeting of mitoNEET inhibits neurite outgrowth, induces cell apoptosis and contributes to physiological dysfunction after spinal cord transection. Sci. Rep. 6, 35205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sohn Y. S., Tamir S., Song L., Michaeli D., Matouk I., Conlan A. R., Harir Y., Holt S. H., Shulaev V., Paddock M. L., Hochberg A., Cabanchick I. Z., Onuchic J. N., Jennings P. A., Nechushtai R., and Mittler R. (2013) NAF-1 and mitoNEET are central to human breast cancer proliferation by maintaining mitochondrial homeostasis and promoting tumor growth. Proc. Natl. Acad. Sci. U.S.A. 110, 14676–14681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Salem A. F., Whitaker-Menezes D., Howell A., Sotgia F., and Lisanti M. P. (2012) Mitochondrial biogenesis in epithelial cancer cells promotes breast cancer tumor growth and confers autophagy resistance. Cell Cycle 11, 4174–4180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tamir S., Paddock M. L., Darash-Yahana-Baram M., Holt S. H., Sohn Y. S., Agranat L., Michaeli D., Stofleth J. T., Lipper C. H., Morcos F., Cabantchik I. Z., Onuchic J. N., Jennings P. A., Mittler R., and Nechushtai R. (2015) Structure-function analysis of NEET proteins uncovers their role as key regulators of iron and ROS homeostasis in health and disease. Biochim. Biophys. Acta 1853, 1294–1315 [DOI] [PubMed] [Google Scholar]
- 14. Bieganski R. M., and Yarmush M. L. (2011) Novel ligands that target the mitochondrial membrane protein mitoNEET. J. Mol. Graph. Model. 29, 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi T., Yamamoto M., Amikura K., Kato K., Serizawa T., Serizawa K., Akazawa D., Aoki T., Kawai K., Ogasawara E., Hayashi J., Nakada K., and Kainoh M. (2015) A novel MitoNEET ligand, TT01001, improves diabetes and ameliorates mitochondrial function in db/db mice. J. Pharmacol. Exp. Ther. 352, 338–345 [DOI] [PubMed] [Google Scholar]
- 16. Bai F., Morcos F., Sohn Y. S., Darash-Yahana M., Rezende C. O., Lipper C. H., Paddock M. L., Song L., Luo Y., Holt S. H., Tamir S., Theodorakis E. A., Jennings P. A., Onuchic J. N., Mittler R., and Nechushtai R. (2015) The Fe-S cluster-containing NEET proteins mitoNEET and NAF-1 as chemotherapeutic targets in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 112, 3698–3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geldenhuys W. J., Yonutas H. M., Morris D. L., Sullivan P. G., Darvesh A. S., and Leeper T. C. (2016) Identification of small molecules that bind to the mitochondrial protein mitoNEET. Bioorg. Med. Chem. Lett. 26, 5350–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wiley S. E., Paddock M. L., Abresch E. C., Gross L., van der Geer P., Nechushtai R., Murphy A. N., Jennings P. A., and Dixon J. E. (2007) The outer mitochondrial membrane protein mitoNEET contains a novel redox-active 2Fe-2S cluster. J. Biol. Chem. 282, 23745–23749 [DOI] [PubMed] [Google Scholar]
- 19. Hou X., Liu R., Ross S., Smart E. J., Zhu H., and Gong W. (2007) Crystallographic studies of human MitoNEET. J. Biol. Chem. 282, 33242–33246 [DOI] [PubMed] [Google Scholar]
- 20. Lin J., Zhou T., Ye K., and Wang J. (2007) Crystal structure of human mitoNEET reveals distinct groups of iron-sulfur proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 14640–14645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paddock M. L., Wiley S. E., Axelrod H. L., Cohen A. E., Roy M., Abresch E. C., Capraro D., Murphy A. N., Nechushtai R., Dixon J. E., and Jennings P. A. (2007) MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. U.S.A. 104, 14342–14347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zuris J. A., Harir Y., Conlan A. R., Shvartsman M., Michaeli D., Tamir S., Paddock M. L., Onuchic J. N., Mittler R., Cabantchik Z. I., Jennings P. A., and Nechushtai R. (2011) Facile transfer of [2Fe-2S] clusters from the diabetes drug target mitoNEET to an apo-acceptor protein. Proc. Natl. Acad. Sci. U.S.A. 108, 13047–13052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Golinelli-Cohen M. P., Lescop E., Mons C., Gonçalves S., Clémancey M., Santolini J., Guittet E., Blondin G., Latour J. M., and Bouton C. (2016) Redox control of the human iron-sulfur repair protein MitoNEET activity via its iron-sulfur cluster. J. Biol. Chem. 291, 7583–7593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lipper C. H., Paddock M. L., Onuchic J. N., Mittler R., Nechushtai R., and Jennings P. A. (2015) Cancer-related NEET proteins transfer 2Fe-2S clusters to anamorsin, a protein required for cytosolic iron-sulfur cluster biogenesis. PLoS ONE 10, e0139699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paul V. D., and Lill R. (2015) Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 1853, 1528–1539 [DOI] [PubMed] [Google Scholar]
- 26. Bak D. W., Zuris J. A., Paddock M. L., Jennings P. A., and Elliott S. J. (2009) Redox characterization of the FeS protein MitoNEET and impact of thiazolidinedione drug binding. Biochemistry 48, 10193–10195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landry A. P., and Ding H. (2014) Redox control of human mitochondrial outer membrane protein MitoNEET [2Fe-2S] clusters by biological thiols and hydrogen peroxide. J. Biol. Chem. 289, 4307–4315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Landry A. P., Cheng Z., and Ding H. (2015) Reduction of mitochondrial protein mitoNEET [2Fe-2S] clusters by human glutathione reductase. Free Radic. Biol. Med. 81, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Landry A. P., Wang Y., Cheng Z., Crochet R. B., Lee Y. H., and Ding H. (2017) Flavin nucleotides act as electron shuttles mediating reduction of the [2Fe-2S] clusters in mitochondrial outer membrane protein mitoNEET. Free Radic. Biol. Med. 102, 240–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Havugimana P. C., Hart G. T., Nepusz T., Yang H., Turinsky A. L., Li Z., Wang P. I., Boutz D. R., Fong V., Phanse S., Babu M., Craig S. A., Hu P., Wan C., Vlasblom J., et al. (2012) A census of human soluble protein complexes. Cell 150, 1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roberts M. E., Crail J. P., Laffoon M. M., Fernandez W. G., Menze M. A., and Konkle M. E. (2013) Identification of disulfide bond formation between MitoNEET and glutamate dehydrogenase 1. Biochemistry 52, 8969–8971 [DOI] [PubMed] [Google Scholar]
- 32. Ingelman M., Ramaswamy S., Nivière V., Fontecave M., and Eklund H. (1999) Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry 38, 7040–7049 [DOI] [PubMed] [Google Scholar]
- 33. Baradaran R., Berrisford J. M., Minhas G. S., and Sazanov L. A. (2013) Crystal structure of the entire respiratory complex I. Nature 494, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Song Y., and Buettner G. R. (2010) Thermodynamic and kinetic considerations for the reaction of semiquinone radicals to form superoxide and hydrogen peroxide. Free Radic. Biol. Med. 49, 919–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lapointe J., Wang Y., Bigras E., and Hekimi S. (2012) The submitochondrial distribution of ubiquinone affects respiration in long-lived Mclk1+/− mice. J. Cell Biol. 199, 215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Geldenhuys W. J., Funk M. O., Barnes K. F., and Carroll R. T. (2010) Structure-based design of a thiazolidinedione which targets the mitochondrial protein mitoNEET. Bioorg. Med. Chem. Lett. 20, 819–823 [DOI] [PubMed] [Google Scholar]
- 37. Logan S. J., Yin L., Geldenhuys W. J., Enrick M. K., Stevanov K. M., Carroll R. T., Ohanyan V. A., Kolz C. L., and Chilian W. M. (2015) Novel thiazolidinedione mitoNEET ligand-1 acutely improves cardiac stem cell survival under oxidative stress. Basic Res. Cardiol. 110, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iwasaki T., Samoilova R. I., Kounosu A., Ohmori D., and Dikanov S. A. (2009) Continuous-wave and pulsed EPR characterization of the [2Fe-2S](Cys)3(His)1 cluster in rat MitoNEET. J. Am. Chem. Soc. 131, 13659–13667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lin J., Zhang L., Lai S., and Ye K. (2011) Structure and molecular evolution of CDGSH iron-sulfur domains. PLoS ONE 6, e24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Inupakutika M. A., Sengupta S., Nechushtai R., Jennings P. A., Onuchic J. N., Azad R. K., Padilla P., and Mittler R. (2017) Phylogenetic analysis of eukaryotic NEET proteins uncovers a link between a key gene duplication event and the evolution of vertebrates. Sci. Rep. 7, 42571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith L. J., Browne S., Mulholland A. J., and Mantle T. J. (2008) Computational and experimental studies on the catalytic mechanism of biliverdin-IXβ reductase. Biochem. J. 411, 475–484 [DOI] [PubMed] [Google Scholar]
- 42. Hühner J., Ingles-Prieto Á., Neusüß C., Lämmerhofer M., and Janovjak H. (2015) Quantification of riboflavin, flavin mononucleotide, and flavin adenine dinucleotide in mammalian model cells by CE with LED-induced fluorescence detection. Electrophoresis 36, 518–525 [DOI] [PubMed] [Google Scholar]
- 43. Heikal A. A. (2010) Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark. Med. 4, 241–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gatenby R. A., and Gillies R. J. (2004) Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 4, 891–899 [DOI] [PubMed] [Google Scholar]
- 45. Imlay J. A. (2006) Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 59, 1073–1082 [DOI] [PubMed] [Google Scholar]
- 46. Kitagawa M., Ara T., Arifuzzaman M., Ioka-Nakamichi T., Inamoto E., Toyonaga H., and Mori H. (2005) Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 12, 291–299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


